Identifying GPSM Family Members as Potential Biomarkers in Breast Cancer: A Comprehensive Bioinformatics Analysis
Abstract
:1. Introduction
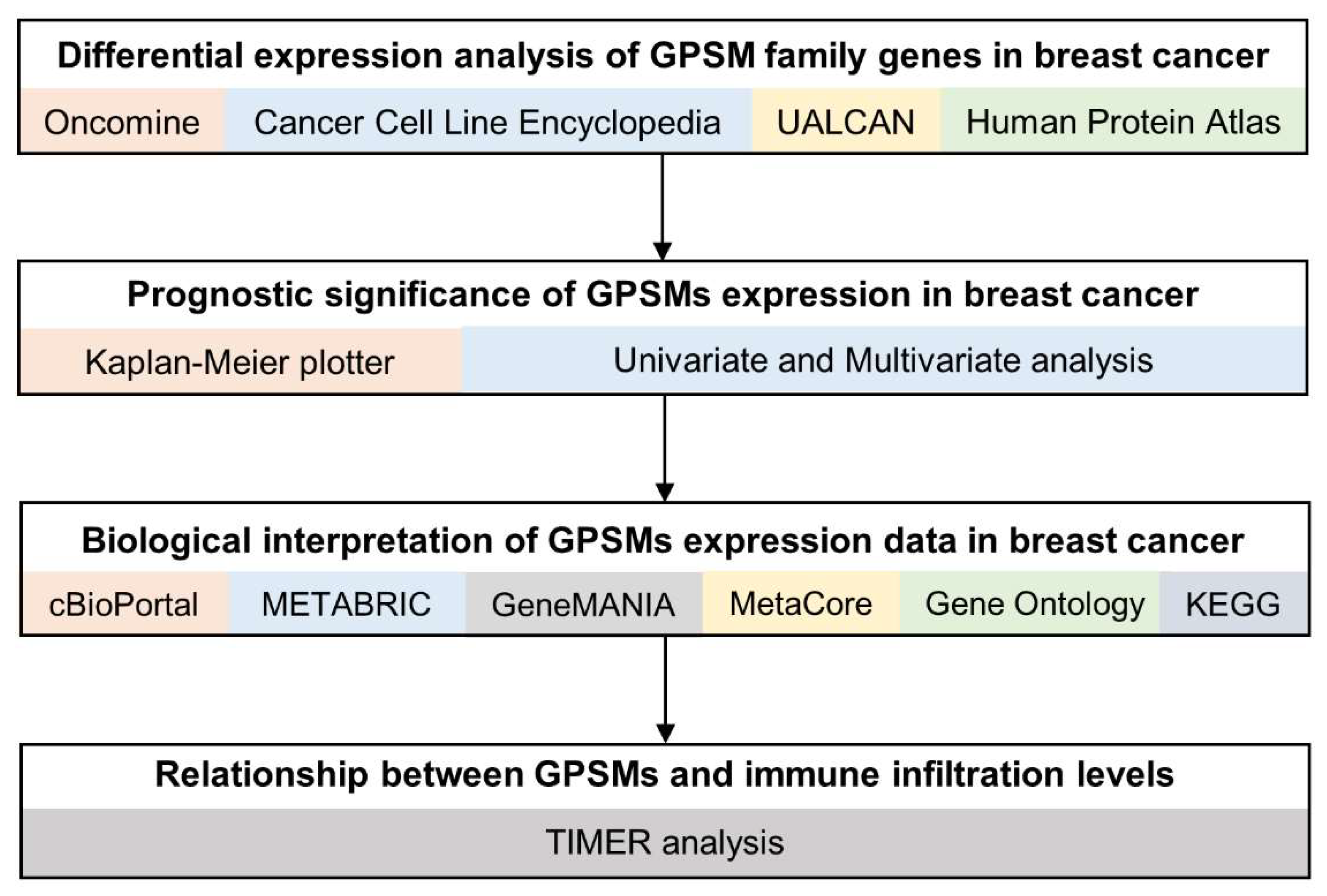
2. Materials and Methods
2.1. Oncomine
2.2. CCLE (Cancer Cell Line Encyclopedia)
2.3. UALCAN
2.4. Human Protein Atlas
2.5. Survival Analysis
2.6. Functional Enrichment Analysis
2.7. TIMER (Tumor IMmune Estimation Resource)
3. Results
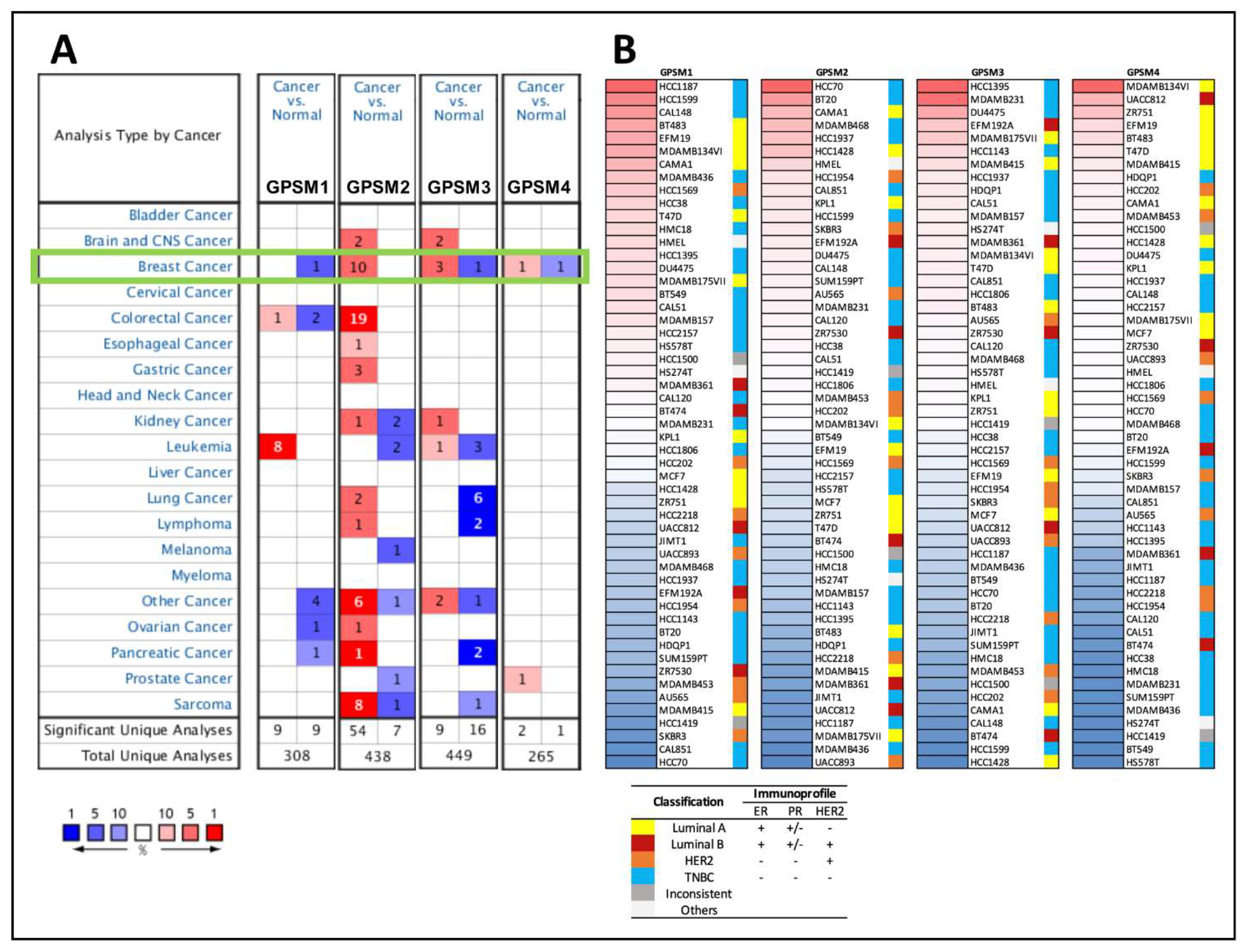
3.1. Differential Expression of GPSM Family Genes in Breast Cancer
3.2. GPSMs Expression in Subgroups of Individuals with Breast Cancer
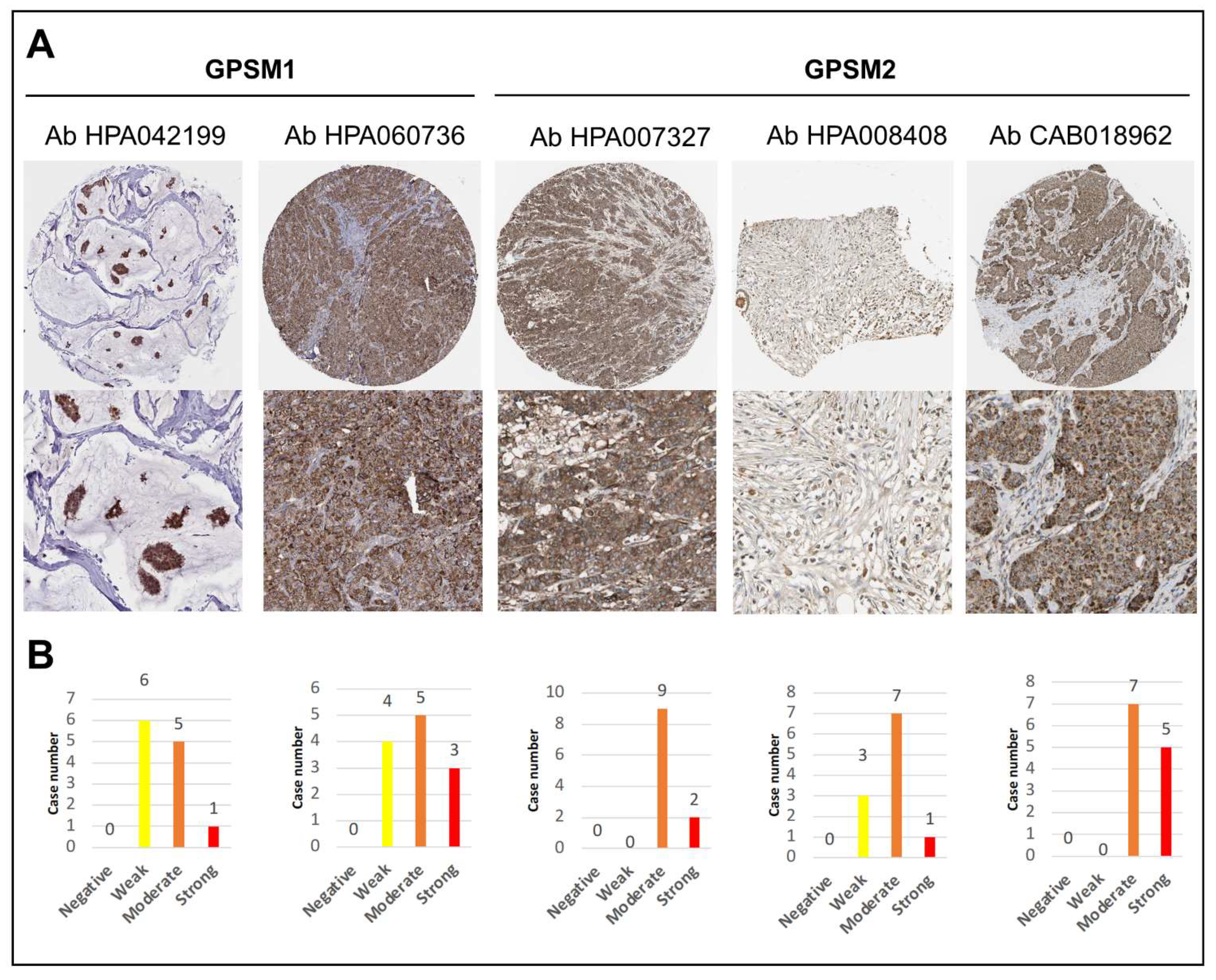
3.3. GPSMs Expression Profiles on mRNA and Protein Level
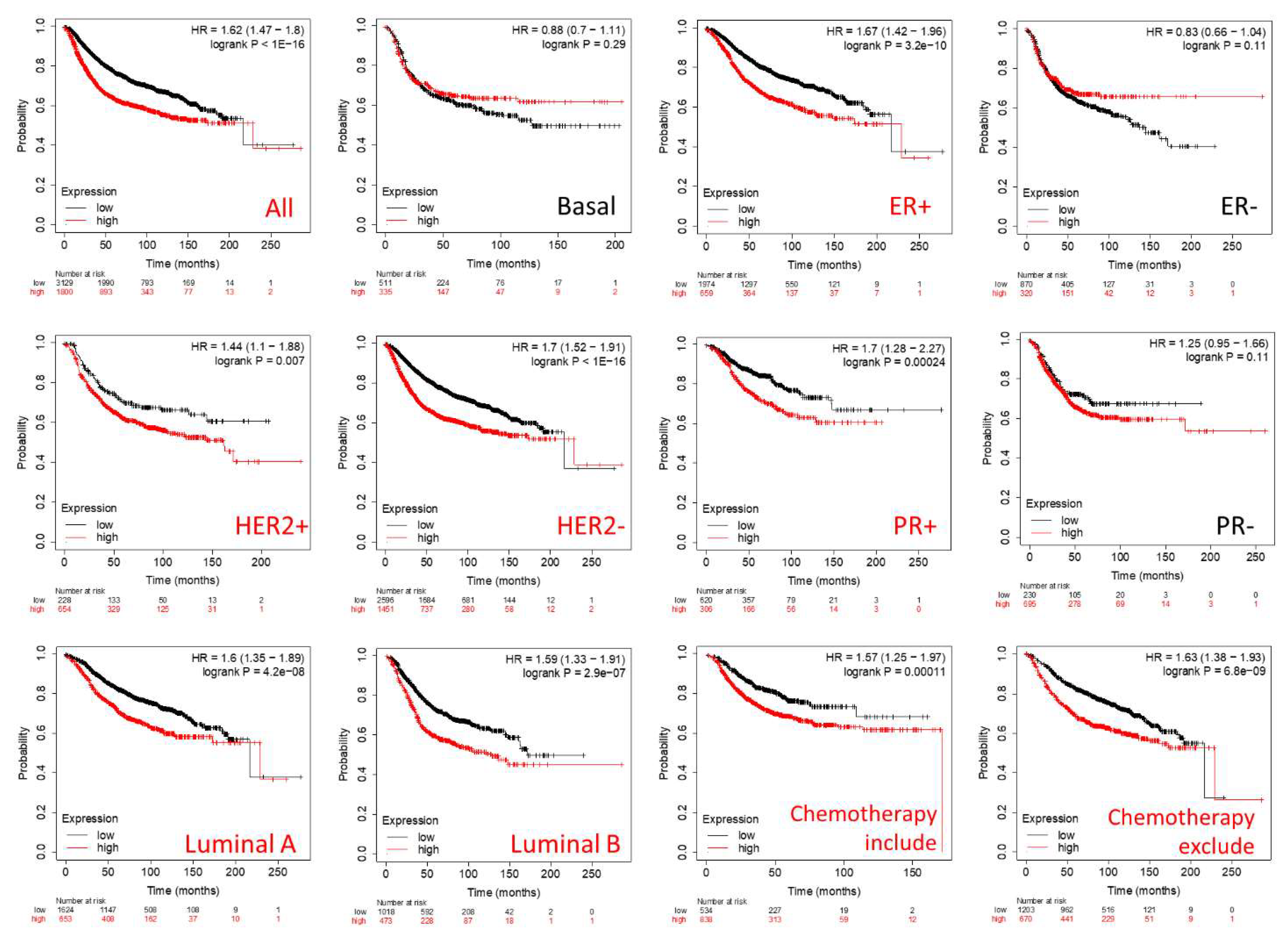
3.4. Prognostic Roles of GPSMs in Breast Cancer
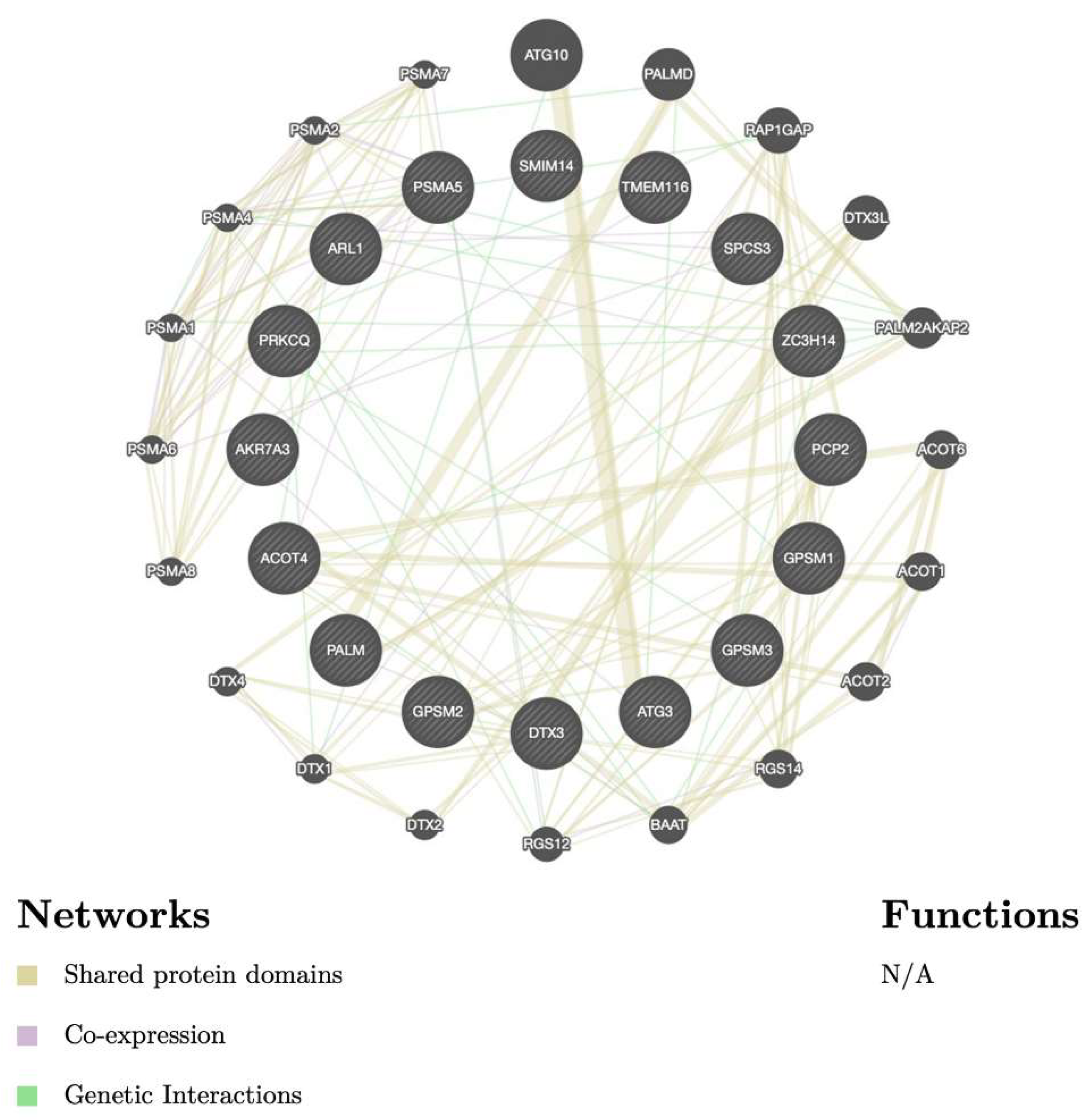
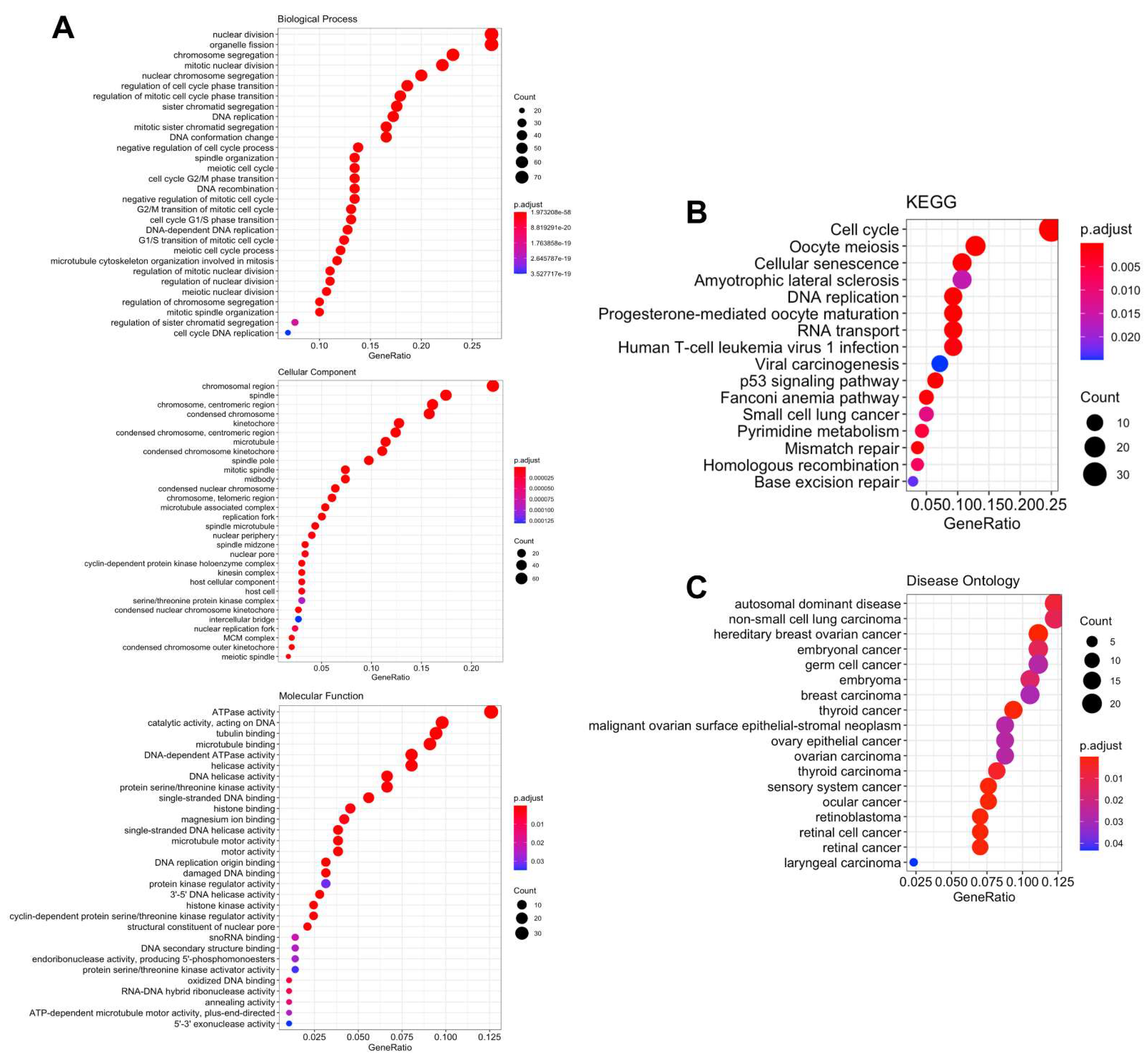
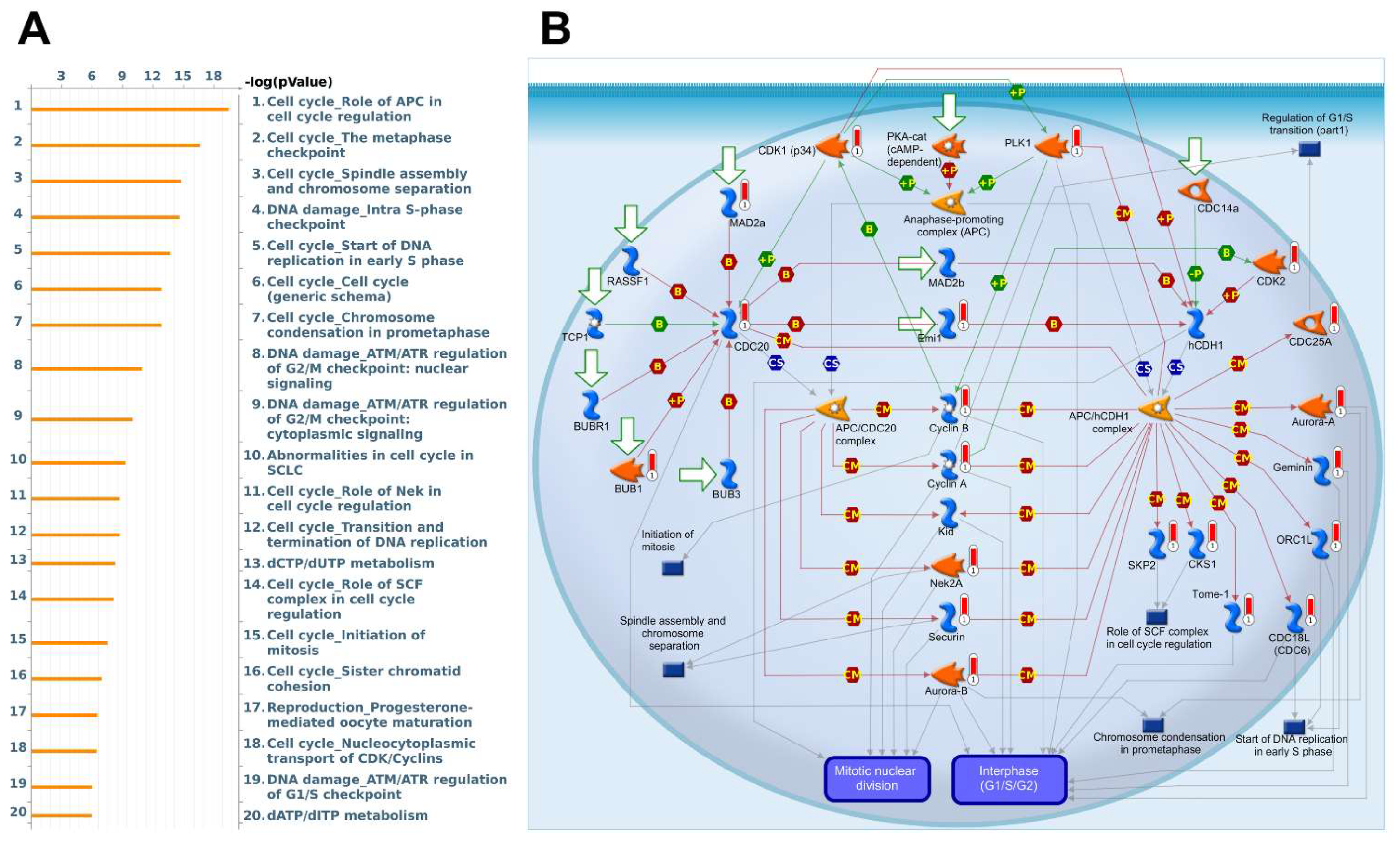
3.5. Biological Functions and Pathway Enrichment Analysis
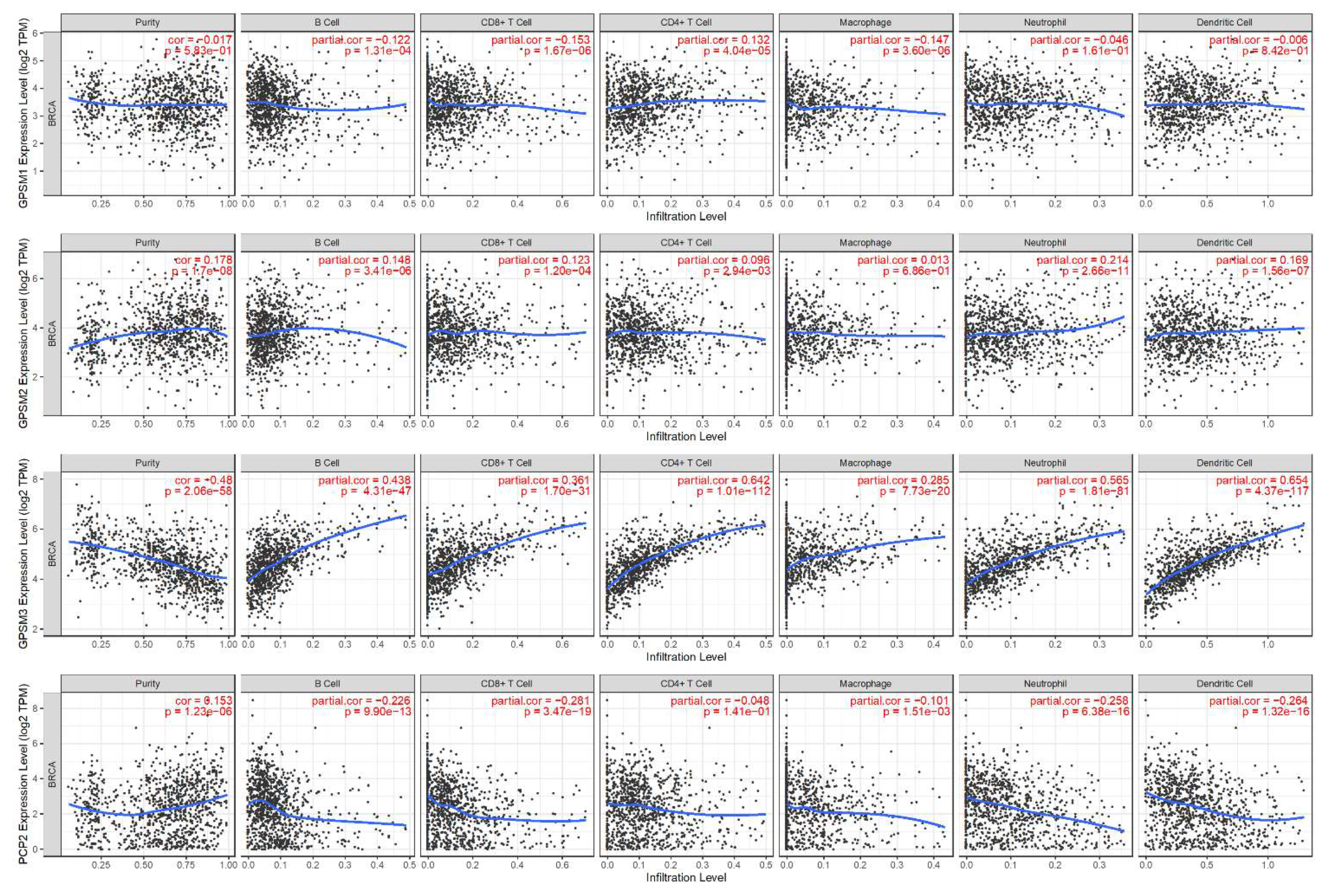
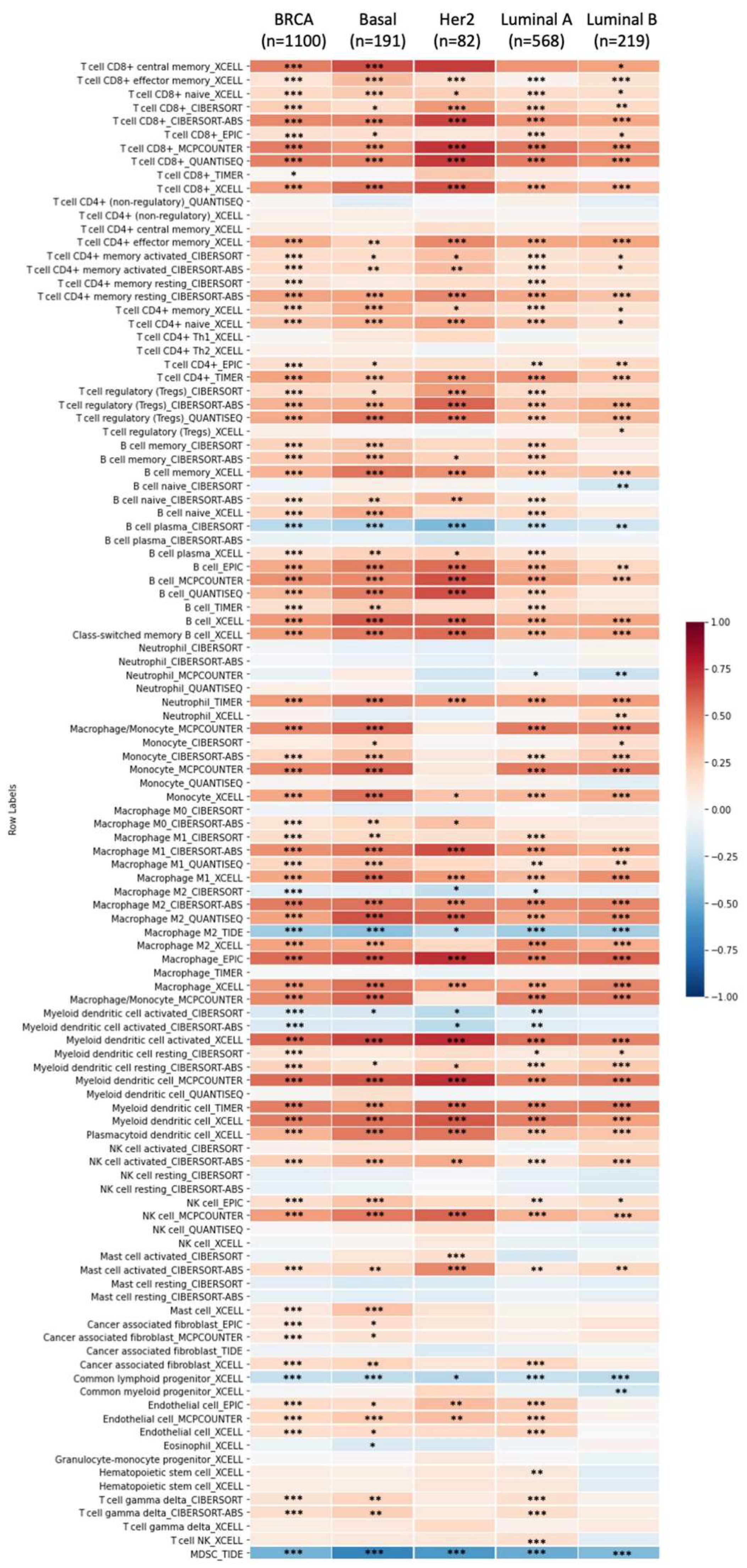
3.6. Correlation between GPSMs and Tumor Purity and Immune Infiltrate in Breast Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Primers 2019, 5, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Loibl, S.; Poortmans, P.; Morrow, M.; Denkert, C.; Curigliano, G. Breast cancer. Lancet 2021, 397, 1750–1769. [Google Scholar] [CrossRef]
- Sever, R.; Brugge, J.S. Signal Transduction in Cancer. Cold Spring Harb. Perspect. Med. 2015, 5, a006098. [Google Scholar] [CrossRef] [Green Version]
- Tuteja, N. Signaling through G protein coupled receptors. Plant Signal. Behav. 2009, 4, 942–947. [Google Scholar] [CrossRef] [PubMed]
- Kiyomitsu, T.; Cheeseman, I.M. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 2012, 14, 311–317. [Google Scholar] [CrossRef] [Green Version]
- Fukukawa, C.; Ueda, K.; Nishidate, T.; Katagiri, T.; Nakamura, Y. Critical roles of LGN/GPSM2 phosphorylation by PBK/TOPK in cell division of breast cancer cells. Genes Chromosomes Cancer 2010, 49, 861–872. [Google Scholar] [CrossRef]
- Deng, M.; Zhang, Z.; Liu, B.; Hou, K.; Che, X.; Qu, X.; Liu, Y.; Hu, X.; Zhang, Y.; Lv, Q. Localization of GPSM2 in the Nucleus of Invasive Breast Cancer Cells Indicates a Poor Prognosis. Front. Oncol. 2020, 10, 227. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Li, Z.; Deng, M.; Liu, B.; Xin, X.; Zhao, Z.; Zhang, Y.; Lv, Q. Downregulation of GPSM2 is associated with primary resistance to paclitaxel in breast cancer. Oncol. Rep. 2020, 43, 965–974. [Google Scholar] [CrossRef]
- Deng, M.; Liu, B.; Zhang, Z.; Chen, Y.; Wang, Y.; Wang, X.; Lv, Q.; Yang, X.; Hou, K.; Che, X.; et al. Knockdown of G-protein-signaling modulator 2 promotes metastasis of non-small-cell lung cancer by inducing the expression of Snail. Cancer Sci. 2020, 111, 3210–3221. [Google Scholar] [CrossRef] [PubMed]
- Adekoya, T.O.; Smith, N.; Aladeniyi, T.; Blumer, J.B.; Chen, X.L.; Richardson, R.M. Activator of G protein signaling 3 modulates prostate tumor development and progression. Carcinogenesis 2019, 40, 1504–1513. [Google Scholar] [CrossRef] [PubMed]
- Dang, S.-C.; Qian, X.-B.; Jin, W.; Cui, L.; Chen, J.-X.; Gu, M. G-protein-signaling modulator 2 expression and role in a CD133+ pancreatic cancer stem cell subset. OncoTargets Ther. 2019, 12, 785–794. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Dong, X.; Xie, B.; Ding, N.; Chen, J.; Li, Y.; Zhang, Q.; Qu, H.; Fang, X. Databases and Web Tools for Cancer Genomics Study. Genom. Proteom. Bioinform. 2015, 13, 46–50. [Google Scholar] [CrossRef] [Green Version]
- Anuraga, G.; Tang, W.-C.; Phan, N.N.; Ta, H.D.; Liu, Y.-H.; Wu, Y.-F.; Lee, K.-H.; Wang, C.-Y. Comprehensive Analysis of Prognostic and Genetic Signatures for General Transcription Factor III (GTF3) in Clinical Colorectal Cancer Patients Using Bioinformatics Approaches. Curr. Issues Mol. Biol. 2021, 43, 2–20. [Google Scholar] [CrossRef]
- Khoa Ta, H.D.; Tang, W.-C.; Phan, N.N.; Anuraga, G.; Hou, S.-Y.; Chiao, C.-C.; Liu, Y.-H.; Wu, Y.-F.; Lee, K.-H.; Wang, C.-Y. Analysis of LAGEs Family Gene Signature and Prognostic Relevance in Breast Cancer. Diagnostics 2021, 11, 726. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Kalyana-Sundaram, S.; Mahavisno, V.; Varambally, R.; Yu, J.; Briggs, B.B.; Barrette, T.R.; Anstet, M.J.; Kincead-Beal, C.; Kulkarni, P.; et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007, 9, 166–180. [Google Scholar] [CrossRef] [Green Version]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Asplund, A.; Edqvist, P.-H.D.; Schwenk, J.M.; Pontén, F. Antibodies for profiling the human proteome-The Human Protein Atlas as a resource for cancer research. Proteomics 2012, 12, 2067–2077. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Á.; Munkácsy, G.; Győrffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep 2021, 11, 6047. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Yan, G.-R.; He, Q.-Y. DOSE: An R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 2015, 31, 608–609. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [Green Version]
- Mulcahy Levy, J.M.; Thorburn, A. Autophagy in cancer: Moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ. 2020, 27, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Valla, M.; Opdahl, S.; Ytterhus, B.; Bofin, A.M. DTX3 copy number increase in breast cancer: A study of associations to molecular subtype, proliferation and prognosis. Breast Cancer Res. Treat. 2021, 187, 57–67. [Google Scholar] [CrossRef]
- Albrecht, I.; Bieri, R.; Leu, A.; Granacher, P.; Hagmann, J.; Kilimann, M.W.; Christofori, G. Paralemmin-1 is expressed in lymphatic endothelial cells and modulates cell migration, cell maturation and tumor lymphangiogenesis. Angiogenesis 2013, 16, 795–807. [Google Scholar] [CrossRef] [Green Version]
- Azimi, F.; Scolyer, R.A.; Rumcheva, P.; Moncrieff, M.; Murali, R.; McCarthy, S.W.; Saw, R.P.; Thompson, J.F. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J. Clin. Oncol. 2012, 30, 2678–2683. [Google Scholar] [CrossRef] [PubMed]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. (Lausanne) 2017, 4, 227. [Google Scholar] [CrossRef] [Green Version]
- Novel Biomarkers in the Continuum of Breast Cancer; Stearns, V. (Ed.) Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar]
- He, Y.; Deng, F.; Yang, S.; Wang, D.; Chen, X.; Zhong, S.; Zhao, J.; Tang, J. Exosomal microRNA: A novel biomarker for breast cancer. Biomark. Med. 2018, 12, 177–188. [Google Scholar] [CrossRef]
- Shao, Y.; Yu, Y.; He, Y.; Chen, Q.; Liu, H. Serum ATX as a novel biomarker for breast cancer. Medicine 2019, 98, e14973. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, H.; Li, J.; Zhang, J.; Wu, J.; Li, Y.; Piao, Y.; Pan, L.; Xiang, R.; Yue, S. FZR1 as a novel biomarker for breast cancer neoadjuvant chemotherapy prediction. Cell Death Dis. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Ortega, M.A.; Fraile-Martínez, O.; Asúnsolo, Á.; Buján, J.; García-Honduvilla, N.; Coca, S. Signal Transduction Pathways in Breast Cancer: The Important Role of PI3K/Akt/mTOR. J. Oncol. 2020, 2020, 9258396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pires, B.R.B.; Amorim, Í.S.S.D.; Souza, L.D.E.; Rodrigues, J.A.; Mencalha, A.L. Targeting Cellular Signaling Pathways in Breast Cancer Stem Cells and its Implication for Cancer Treatment. Anticancer Res. 2016, 36, 5681–5691. [Google Scholar] [CrossRef] [Green Version]
- Lappano, R.; Jacquot, Y.; Maggiolini, M. GPCR Modulation in Breast Cancer. Int. J. Mol. Sci. 2018, 19, 3840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usman, S.; Khawer, M.; Rafique, S.; Naz, Z.; Saleem, K. The current status of anti-GPCR drugs against different cancers. J. Pharm. Anal. 2020, 10, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Ting, S.; Lee, N.; Li, M.; Russell, S.; Godde, N.; Humbert, P.; Matsuzaki, F.; Deneault, E.; Sauvageau, G. Perturbation of Gpsm2/Lgn Enhances Haematopoietic Stem Cell Function. Blood 2013, 122, 1176. [Google Scholar] [CrossRef]
- Yang, C.H.; Snyder, M. The nuclear-mitotic apparatus protein is important in the establishment and maintenance of the bipolar mitotic spindle apparatus. Mol. Biol. Cell 1992, 3, 1259–1267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Q.; Taylor, L.; Compton, D.A.; Macara, I.G. LGN blocks the ability of NuMA to bind and stabilize microtubules. A mechanism for mitotic spindle assembly regulation. Curr. Biol. 2002, 12, 1928–1933. [Google Scholar] [CrossRef] [Green Version]
- Mochizuki, N.; Cho, G.; Wen, B.; Insel, P.A. Identification and cDNA cloning of a novel human mosaic protein, LGN, based on interaction with G alpha i2. Gene 1996, 181, 39–43. [Google Scholar] [CrossRef]
- He, X.-Q.; Zhang, Y.-F.; Yu, J.-J.; Gan, Y.-Y.; Han, N.-N.; Zhang, M.-X.; Ge, W.; Deng, J.-J.; Zheng, Y.-F.; Xu, X.-M. High expression of G-protein signaling modulator 2 in hepatocellular carcinoma facilitates tumor growth and metastasis by activating the PI3K/AKT signaling pathway. Tumour Biol. 2017, 39, 1010428317695971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, M.; Liu, B.; Zhang, Z.; Chen, Y.; Wang, Y.; Wang, X.; Lv, Q.; Yang, X.; Hou, K.; Che, X.; et al. Loss of G-protein-signaling modulator 2 accelerates proliferation of lung adenocarcinoma via EGFR signaling pathway. Int. J. Biochem. Cell Biol. 2020, 122, 105716. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Lian, M.; Shen, Q.; Feng, G.; Huang, D.; Lu, X.; Gu, Z.; Li, L.; Zhang, J.; Huang, S.; et al. AGS3 is involved in TNF-α medicated osteogenic differentiation of human dental pulp stem cells. Differentiation 2015, 89, 128–136. [Google Scholar] [CrossRef]
- Blumer, J.B.; Cismowski, M.J.; Sato, M.; Lanier, S.M. AGS proteins: Receptor-independent activators of G-protein signaling. Trends Pharmacol. Sci. 2005, 26, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Nadella, R.; Blumer, J.B.; Jia, G.; Kwon, M.; Akbulut, T.; Qian, F.; Sedlic, F.; Wakatsuki, T.; Sweeney, W.E.; Wilson, P.D.; et al. Activator of G protein signaling 3 promotes epithelial cell proliferation in PKD. J. Am. Soc. Nephrol. 2010, 21, 1275–1280. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.; Fu, H.; Wang, Y.; Liu, Q.; Wang, X. Depletion of GPSM1 enhances ovarian granulosa cell apoptosis via cAMP-PKA-CREB pathway in vitro. J. Ovarian Res. 2020, 13, 136. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Tan, A.L.M.; Parra, E.J.; Cruz, M.; Sim, X.; Teo, Y.-Y.; Long, J.; Alsafar, H.; Petretto, E.; Tai, E.S.; et al. Genome-wide meta-analysis associates GPSM1 with type 2 diabetes, a plausible gene involved in skeletal muscle function. J. Hum. Genet. 2020, 65, 411–420. [Google Scholar] [CrossRef]
- Bourne, P.E. Is “bioinformatics” dead? PLoS Biol. 2021, 19, e3001165. [Google Scholar] [CrossRef]
- Giguère, P.M.; Billard, M.J.; Laroche, G.; Buckley, B.K.; Timoshchenko, R.G.; McGinnis, M.W.; Esserman, D.; Foreman, O.; Liu, P.; Siderovski, D.P.; et al. G-protein signaling modulator-3, a gene linked to autoimmune diseases, regulates monocyte function and its deficiency protects from inflammatory arthritis. Mol. Immunol. 2013, 54, 193–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robichaux, W.G.; Branham-O’Connor, M.; Hwang, I.-Y.; Vural, A.; Kehrl, J.H.; Blumer, J.B. Regulation of Chemokine Signal Integration by Activator of G-Protein Signaling 4 (AGS4). J. Pharmacol. Exp. Ther. 2017, 360, 424–433. [Google Scholar] [CrossRef] [Green Version]
- Allen, M.D.; Jones, L.J. The role of inflammation in progression of breast cancer: Friend or foe? (Review). Int. J. Oncol. 2015, 47, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Research, C.f.D.E.a. FDA Grants Accelerated Approval to Pembrolizumab for Locally Recurrent Unresectable or Metastatic Triple Negative Breast Cancer; FDA: Silver Spring, MD, USA, 2021.
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef]
- Burugu, S.; Asleh-Aburaya, K.; Nielsen, T.O. Immune infiltrates in the breast cancer microenvironment: Detection, characterization and clinical implication. Breast Cancer 2017, 24, 3–15. [Google Scholar] [CrossRef]
- Guan, J.; Luo, Y.; Denker, B.M. Purkinje cell protein-2 (Pcp2) stimulates differentiation in PC12 cells by Gbetagamma-mediated activation of Ras and p38 MAPK. Biochem. J. 2005, 392, 389–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes-Gibby, C.C.; Wang, J.; Yeung, S.-C.J.; Chaftari, P.; Yu, R.K.; Hanna, E.Y.; Shete, S. Genome-wide association study identifies genes associated with neuropathy in patients with head and neck cancer. Sci. Rep. 2018, 8, 8789. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Approved Symbol | HGNC ID | Gene ID | Aliases | Location on Chromosome |
|---|---|---|---|---|
| GPSM1 | 17858 | 26086 | AGS3 DKFZP727I051 | 9q34.3 |
| GPSM2 | 29501 | 29899 | LGN Pins | 1p13.3 |
| GPSM3 | 13945 | 63940 | NG1 G18 G18.1a G18.1b G18.2 AGS4 | 6p21.32 |
| GPSM4 | 30209 | 126006 | PCP2 MGC41903 | 19p13.2 |
| Number of Patients | Univariate | Multivariate | |||
|---|---|---|---|---|---|
| Variables | HR (95% CI) | p Value | HR (95% CI) | p Value | |
| Age (y) | |||||
| <60 | 366 | reference | reference | ||
| ≥60 | 287 | 2.032 (1.277–3.235) | 0.003 * | 2.361 (1.436–3.881) | 0.0004 * |
| Gender | |||||
| Male | 10 | reference | |||
| Female | 643 | 0.887 (0.122–6.417) | 0.906 | ||
| Tumor stage | |||||
| Stage I/II | 471 | reference | reference | ||
| Stage III/IV | 176 | 2.687 (1.657–4.356) | 6.15 × 10−05 * | 2.517 (1.516–4.18) | 0.0002 * |
| M stage | |||||
| M0 | 530 | reference | reference | ||
| M1 | 9 | 3.965 (1.828–8.59) | 0.0004 * | 2.28 (1.014–5.128) | 0.04 * |
| MX | 114 | 1.268 (0.622–2.584) | 0.513 | 0.726 (0.306–1.723) | 0.49 |
| T stage | |||||
| T1/T2 | 532 | reference | |||
| T3/T4 | 118 | 1.55 (0.920–2.609) | 0.099 | ||
| GPSM2 expression | |||||
| Low | 336 | reference | |||
| High | 317 | 0.888 (0.56–1.41) | 0.617 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dang, H.-H.; Ta, H.D.K.; Nguyen, T.T.T.; Anuraga, G.; Wang, C.-Y.; Lee, K.-H.; Le, N.Q.K. Identifying GPSM Family Members as Potential Biomarkers in Breast Cancer: A Comprehensive Bioinformatics Analysis. Biomedicines 2021, 9, 1144. https://doi.org/10.3390/biomedicines9091144
Dang H-H, Ta HDK, Nguyen TTT, Anuraga G, Wang C-Y, Lee K-H, Le NQK. Identifying GPSM Family Members as Potential Biomarkers in Breast Cancer: A Comprehensive Bioinformatics Analysis. Biomedicines. 2021; 9(9):1144. https://doi.org/10.3390/biomedicines9091144
Chicago/Turabian StyleDang, Huy-Hoang, Hoang Dang Khoa Ta, Truc T. T. Nguyen, Gangga Anuraga, Chih-Yang Wang, Kuen-Haur Lee, and Nguyen Quoc Khanh Le. 2021. "Identifying GPSM Family Members as Potential Biomarkers in Breast Cancer: A Comprehensive Bioinformatics Analysis" Biomedicines 9, no. 9: 1144. https://doi.org/10.3390/biomedicines9091144
APA StyleDang, H.-H., Ta, H. D. K., Nguyen, T. T. T., Anuraga, G., Wang, C.-Y., Lee, K.-H., & Le, N. Q. K. (2021). Identifying GPSM Family Members as Potential Biomarkers in Breast Cancer: A Comprehensive Bioinformatics Analysis. Biomedicines, 9(9), 1144. https://doi.org/10.3390/biomedicines9091144











