Expression of SARS-CoV-2 Entry Factors in Human Alveolar Type II Cells in Aging and Emphysema
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Primary ATII Cell Isolation
2.2. Western Blotting
2.3. RT-PCR
2.4. miRNA Analysis
2.5. miR142 Overexpression
2.6. Statistical Analysis
3. Results
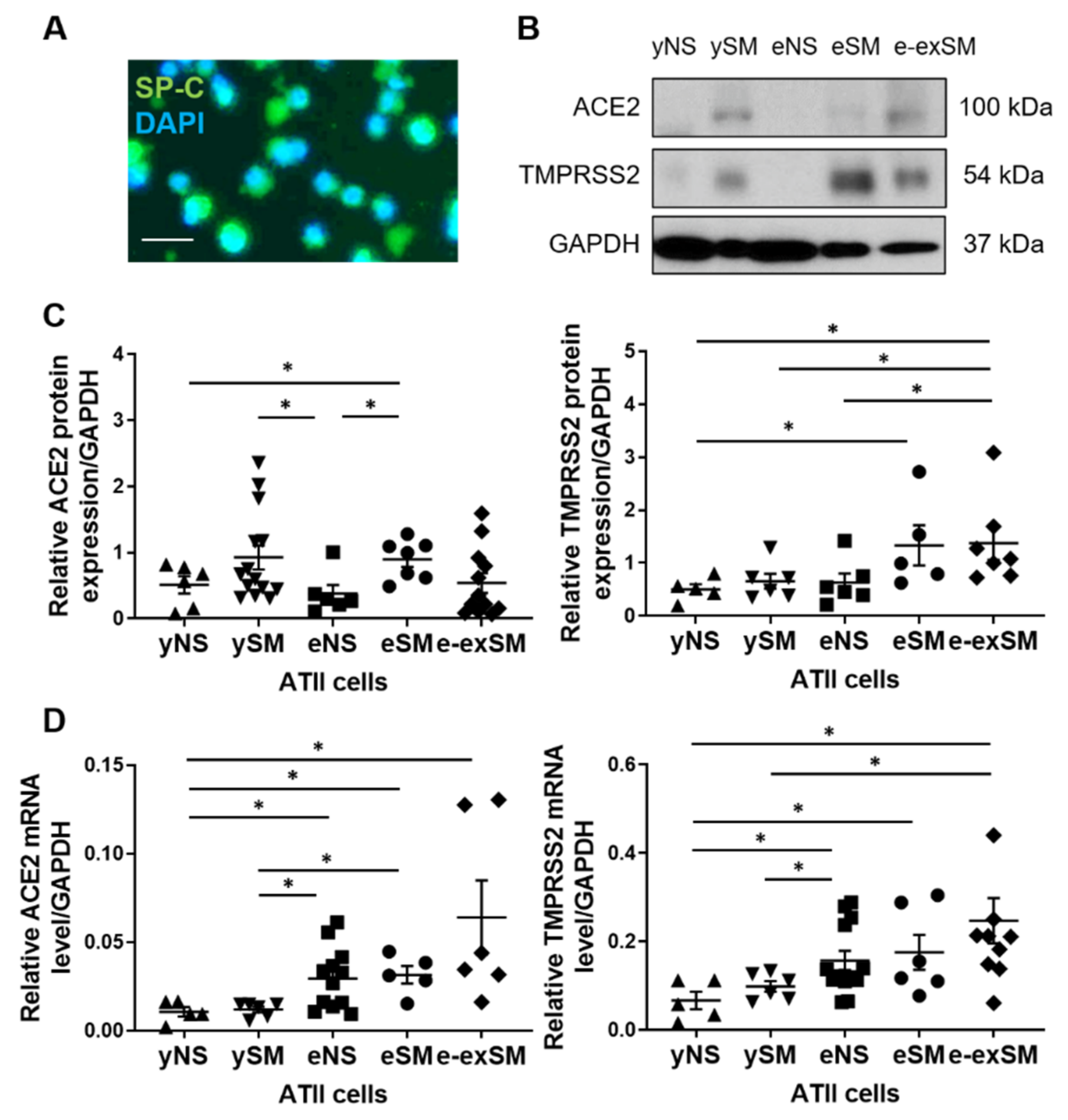
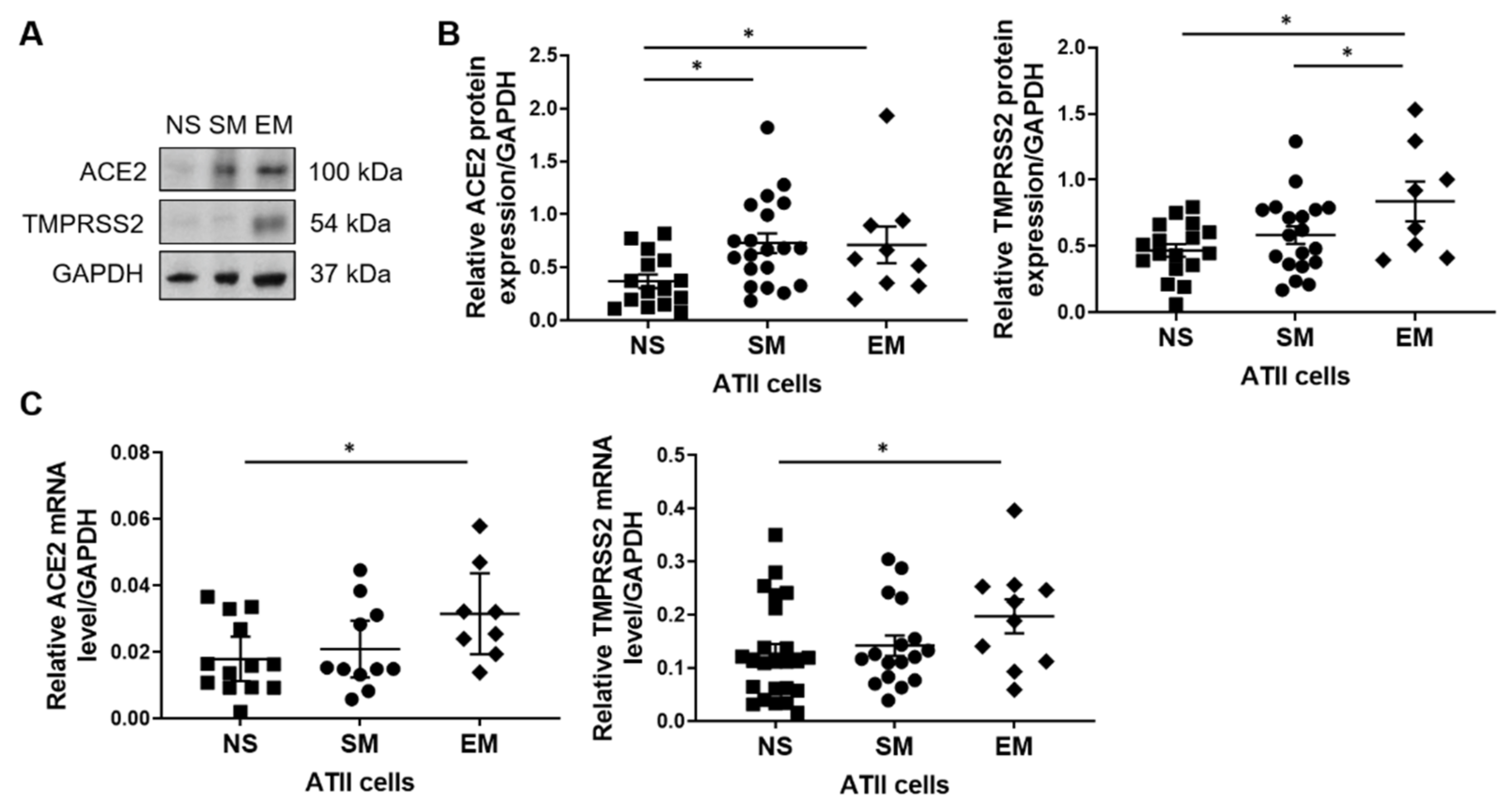
3.1. ACE2 and TMPRRS2 Levels in ATII Cells
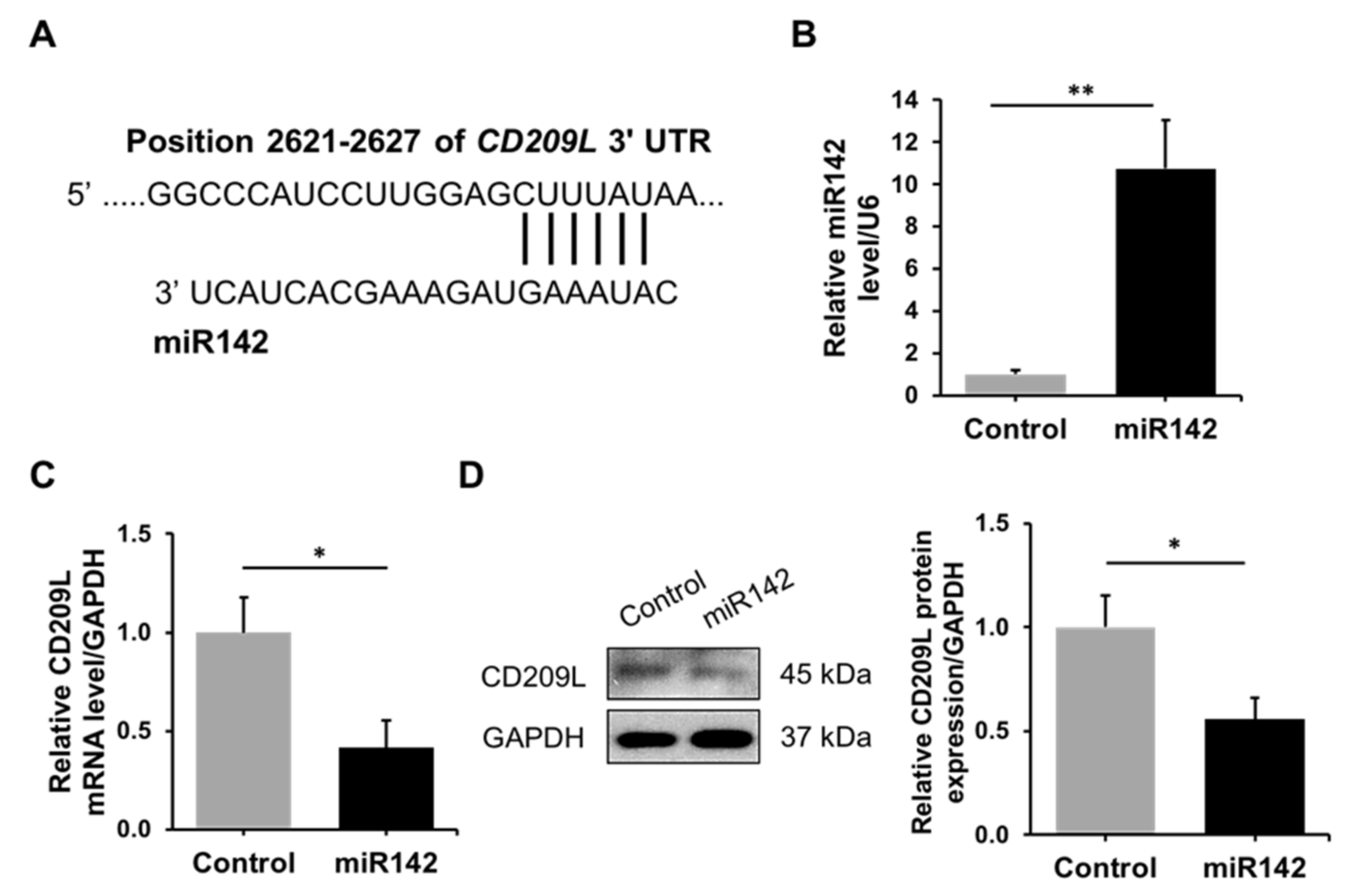
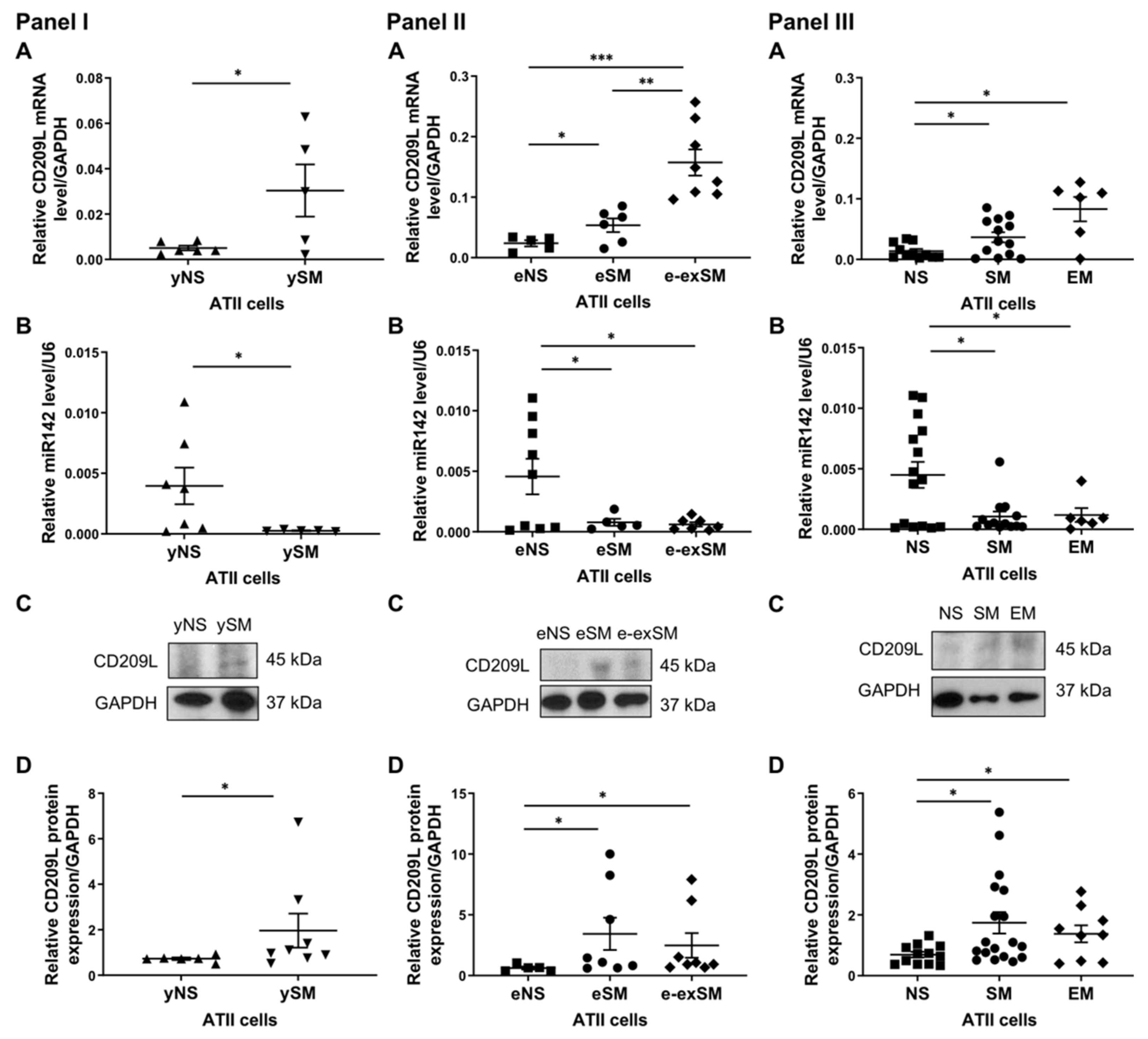
3.2. Regulation of CD209L by miR142 in ATII Cells
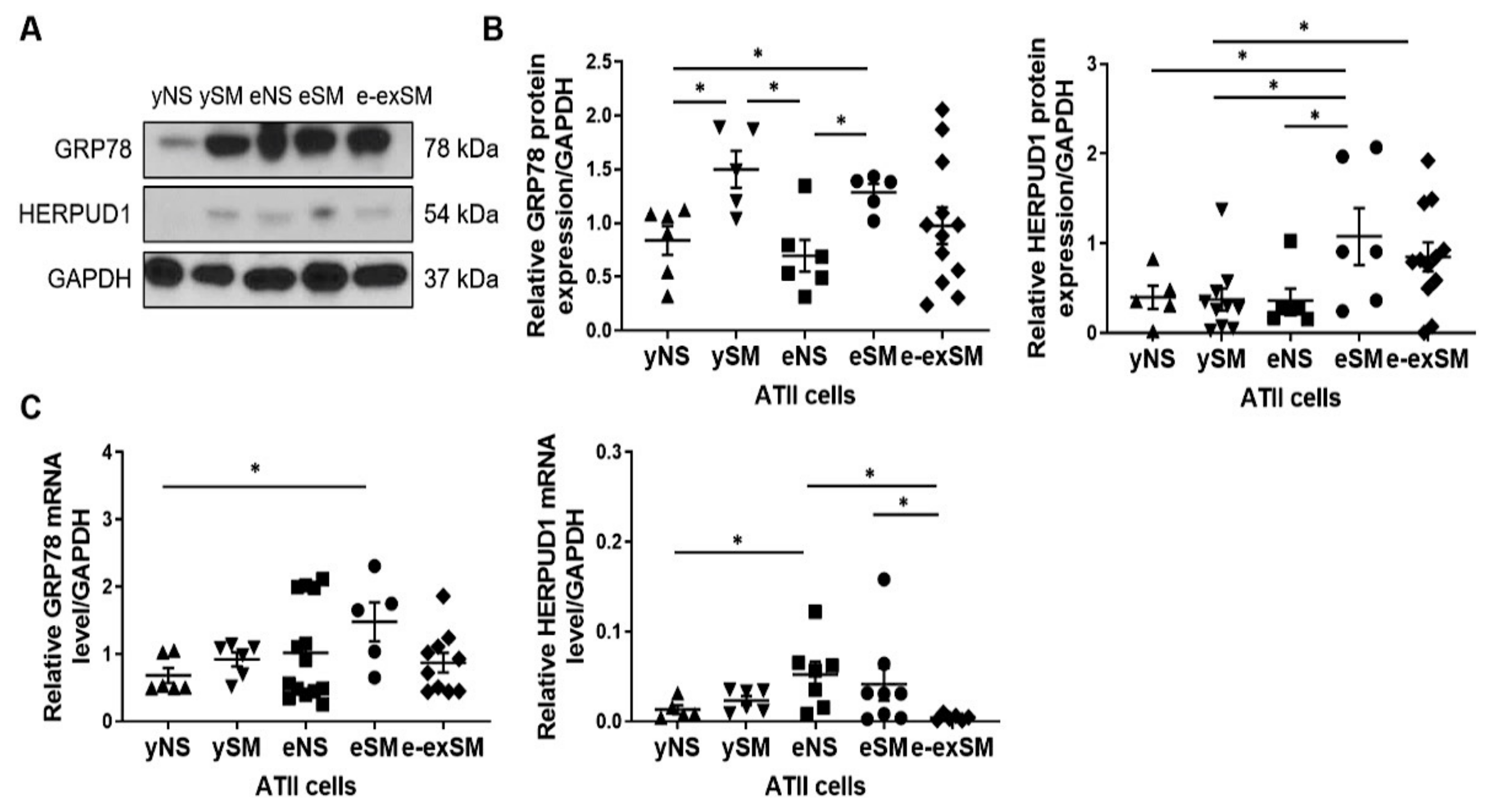
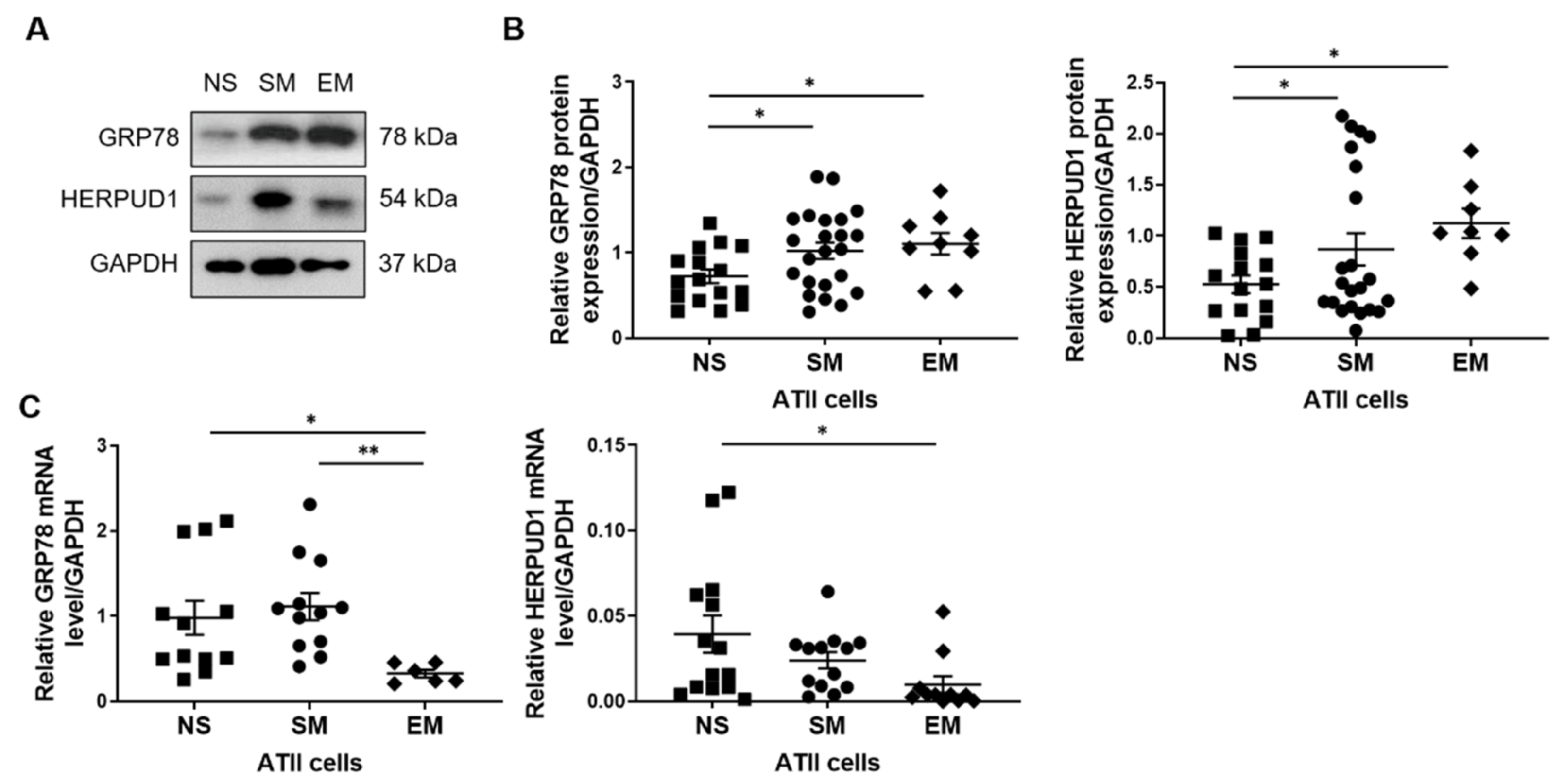
3.3. Increased GRP78 and HERPUD1 Protein Levels in ATII Cells in Smokers and Emphysema Patients
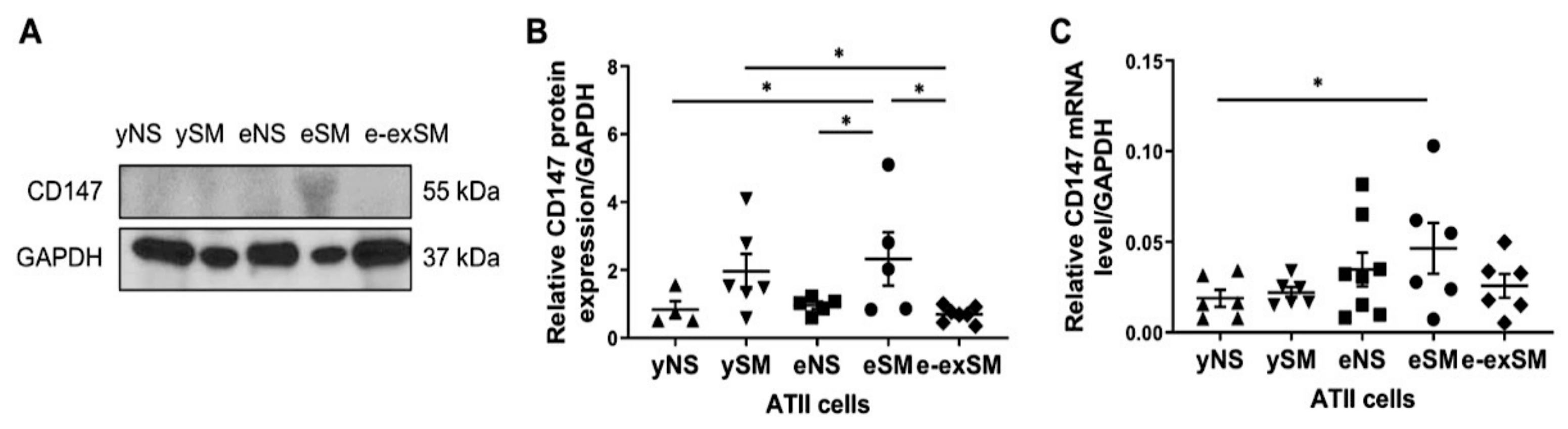
3.4. High CD147 Expression in ATII Cells in Elderly Smokers
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mason, R.J. Biology of alveolar type II cells. Respirology 2006, 11, S12–S15. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, W.; Yang, L.; You, R. Physiological and pathological regulation of ACE2, the SARS-CoV-2 receptor. Pharmacol. Res. 2020, 157, 104833. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in COVID-19. N. Engl. J. Med. 2020, 383, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Zamorano Cuervo, N.; Grandvaux, N. ACE2: Evidence of role as entry receptor for SARS-CoV-2 and implications in comorbidities. eLife 2020, 9, e61390. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Jeffers, S.A.; Tusell, S.M.; Gillim-Ross, L.; Hemmila, E.M.; Achenbach, J.E.; Babcock, G.J.; Thomas, W.D., Jr.; Thackray, L.B.; Young, M.D.; Mason, R.J.; et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 2004, 101, 15748–15753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal. Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Mi, L.; Xu, J.; Yu, J.; Wang, X.; Jiang, J.; Xing, J.; Shang, P.; Qian, A.; Li, Y.; et al. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2005, 191, 755–760. [Google Scholar] [CrossRef] [Green Version]
- Glingston, R.S.; Deb, R.; Kumar, S.; Nagotu, S. Organelle dynamics and viral infections: At cross roads. Microbes. Infect. 2019, 21, 20–32. [Google Scholar] [CrossRef]
- Chu, H.; Chan, C.M.; Zhang, X.; Wang, Y.; Yuan, S.; Zhou, J.; Au-Yeung, R.K.; Sze, K.H.; Yang, D.; Shuai, H.; et al. Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J. Biol. Chem. 2018, 293, 11709–11726. [Google Scholar] [CrossRef] [Green Version]
- COVID-19 Dashboard. Available online: https://coronavirus.jhu.edu/map.html (accessed on 21 June 2021).
- Wang, A.; Chiou, J.; Poirion, O.B.; Buchanan, J.; Valdez, M.J.; Verheyden, J.M.; Hou, X.; Kudtarkar, P.; Narendra, S.; Newsome, J.M.; et al. Single-cell multiomic profiling of human lungs reveals cell-type-specific and age-dynamic control of SARS-CoV2 host genes. eLife 2020, 9, e62522. [Google Scholar] [CrossRef]
- Nikolai, L.A.; Meyer, C.G.; Kremsner, P.G.; Velavan, T.P. Asymptomatic SARS Coronavirus 2 infection: Invisible yet invincible. Int. J. Infect. Dis. 2020, 100, 112–116. [Google Scholar] [CrossRef]
- Liu, K.; Chen, Y.; Lin, R.; Han, K. Clinical features of COVID-19 in elderly patients: A comparison with young and middle-aged patients. J. Infect. 2020, 80, e14–e18. [Google Scholar] [CrossRef] [Green Version]
- Nikolich-Zugich, J.; Knox, K.S.; Rios, C.T.; Natt, B.; Bhattacharya, D.; Fain, M.J. SARS-CoV-2 and COVID-19 in older adults: What we may expect regarding pathogenesis, immune responses, and outcomes. Geroscience 2020, 42, 505–514. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; He, W.; Yu, X.; Hu, D.; Bao, M.; Liu, H.; Zhou, J.; Jiang, H. Coronavirus disease 2019 in elderly patients: Characteristics and prognostic factors based on 4-week follow-up. J. Infect. 2020, 80, 639–645. [Google Scholar] [CrossRef]
- Cai, G.; Bosse, Y.; Xiao, F.; Kheradmand, F.; Amos, C.I. Tobacco Smoking Increases the Lung Gene Expression of ACE2, the Receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020, 201, 1557–1559. [Google Scholar] [CrossRef]
- Zhang, H.; Rostami, M.R.; Leopold, P.L.; Mezey, J.G.; O’Beirne, S.L.; Strulovici-Barel, Y.; Crystal, R.G. Expression of the SARS-CoV-2 ACE2 Receptor in the Human Airway Epithelium. Am. J. Respir. Crit. Care Med. 2020, 202, 219–229. [Google Scholar] [CrossRef]
- Patanavanich, R.; Glantz, S.A. Smoking Is Associated with COVID-19 Progression: A Meta-analysis. Nicotine Tob. Res. 2020, 22, 1653–1656. [Google Scholar] [CrossRef]
- Barnes, P.J. Chronic obstructive pulmonary disease. N. Engl. J. Med. 2000, 343, 269–280. [Google Scholar] [CrossRef] [Green Version]
- Caliskan, T.; Saylan, B. Smoking and comorbidities are associated with COVID-19 severity and mortality in 565 patients treated in Turkey: A retrospective observational study. Rev. Assoc. Med. Bras. 2020, 66, 1679–1684. [Google Scholar] [CrossRef]
- Kosmider, B.; Mason, R.J.; Bahmed, K. Isolation and Characterization of Human Alveolar Type II Cells. Methods Mol. Biol. 2018, 1809, 83–90. [Google Scholar]
- Kosmider, B.; Lin, C.R.; Karim, L.; Tomar, D.; Vlasenko, L.; Marchetti, N.; Bolla, S.; Madesh, M.; Criner, G.J.; Bahmed, K. Mitochondrial dysfunction in human primary alveolar type II cells in emphysema. EBioMedicine 2019, 46, 305–316. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.R.; Bahmed, K.; Criner, G.J.; Marchetti, N.; Tuder, R.M.; Kelsen, S.; Bolla, S.; Mandapati, C.; Kosmider, B. S100A8 Protects Human Primary Alveolar Type II Cells against Injury and Emphysema. Am. J. Respir. Cell Mol. Biol. 2019, 60, 299–307. [Google Scholar] [CrossRef]
- PrimerBank. Available online: https://pga.mgh.harvard.edu/primerbank/ (accessed on 10 May 2020).
- Search for Predicted microRNA Targets in Mammals. Available online: http://www.targetscan.org/vert_72/ (accessed on 5 May 2020).
- Zhao, Y.; Ridge, K.; Zhao, J. Acute Lung Injury, Repair, and Remodeling: Pulmonary Endothelial and Epithelial Biology. Mediat. Inflamm. 2017, 2017, 9081521. [Google Scholar] [CrossRef] [Green Version]
- Cattaruzza, M.S.; Gorini, G.; Bosetti, C.; Boffi, R.; Lugo, A.; Veronese, C.; Carreras, G.; Santucci, C.; Stival, C.; Pacifici, R.; et al. COVID-19 and the role of smoking: The protocol of the multicentric prospective study COSMO-IT (COVID19 and SMOking in ITaly). Acta Biomed. 2020, 91, e2020062. [Google Scholar]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Moller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Eapen, M.S.; Lu, W.; Hackett, T.L.; Singhera, G.K.; Thompson, I.E.; McAlinden, K.D.; Hardikar, A.; Weber, H.C.; Haug, G.; Wark, P.A.B.; et al. Dysregulation of endocytic machinery and ACE2 in small airways of smokers and COPD patients can augment their susceptibility to SARS-CoV-2 (COVID-19) infections. Am. J. Physiol. Lung Cell Mol. Physiol. 2020, 320, L158–L163. [Google Scholar] [CrossRef]
- Mossel, E.C.; Wang, J.; Jeffers, S.; Edeen, K.E.; Wang, S.; Cosgrove, G.P.; Funk, C.J.; Manzer, R.; Miura, T.A.; Pearson, L.D.; et al. SARS-CoV replicates in primary human alveolar type II cell cultures but not in type I-like cells. Virology 2008, 372, 127–135. [Google Scholar] [CrossRef] [Green Version]
- Hou, Y.J.; Okuda, K.; Edwards, C.E.; Martinez, D.R.; Asakura, T.; Dinnon, K.H., III; Kato, T.; Lee, R.E.; Yount, B.L.; Mascenik, T.M.; et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell 2020, 182, 429–446.e14. [Google Scholar] [CrossRef]
- Bradley, B.T.; Maioli, H.; Johnston, R.; Chaudhry, I.; Fink, S.L.; Xu, H.; Najafian, B.; Deutsch, G.; Lacy, J.M.; Williams, T.; et al. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: A case series. Lancet 2020, 396, 320–332. [Google Scholar] [CrossRef]
- Imai, Y.; Kuba, K.; Rao, S.; Huan, Y.; Guo, F.; Guan, B.; Yang, P.; Sarao, R.; Wada, T.; Leong-Poi, H.; et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 2005, 436, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Sarzani, R.; Giulietti, F.; Di Pentima, C.; Giordano, P.; Spannella, F. Disequilibrium between the classic renin-angiotensin system and its opposing arm in SARS-CoV-2-related lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 2020, 319, L325–L336. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Petitjean, S.J.L.; Koehler, M.; Zhang, Q.; Dumitru, A.C.; Chen, W.; Derclaye, S.; Vincent, S.P.; Soumillion, P.; Alsteens, D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020, 11, 4541. [Google Scholar] [CrossRef]
- Lukassen, S.; Chua, R.L.; Trefzer, T.; Kahn, N.C.; Schneider, M.A.; Muley, T.; Winter, H.; Meister, M.; Veith, C.; Boots, A.W.; et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020, 39, e105114. [Google Scholar] [CrossRef]
- Iwata-Yoshikawa, N.; Okamura, T.; Shimizu, Y.; Hasegawa, H.; Takeda, M.; Nagata, N. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J. Virol. 2019, 93, e01815-18. [Google Scholar] [CrossRef] [Green Version]
- Marzi, A.; Gramberg, T.; Simmons, G.; Moller, P.; Rennekamp, A.J.; Krumbiegel, M.; Geier, M.; Eisemann, J.; Turza, N.; Saunier, B.; et al. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004, 78, 12090–12095. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, A.; Czinn, S.J.; Reiter, R.J.; Blanchard, T.G. Crosstalk between endoplasmic reticulum stress and anti-viral activities: A novel therapeutic target for COVID-19. Life Sci. 2020, 255, 117842. [Google Scholar] [CrossRef]
- Aksoy, M.O.; Kim, V.; Cornwell, W.D.; Rogers, T.J.; Kosmider, B.; Bahmed, K.; Barrero, C.; Merali, S.; Shetty, N.; Kelsen, S.G. Secretion of the endoplasmic reticulum stress protein, GRP78, into the BALF is increased in cigarette smokers. Respir. Res. 2017, 18, 78. [Google Scholar] [CrossRef] [Green Version]
- Torrealba, N.; Navarro-Marquez, M.; Garrido, V.; Pedrozo, Z.; Romero, D.; Eura, Y.; Villalobos, E.; Roa, J.C.; Chiong, M.; Kokame, K.; et al. Herpud1 negatively regulates pathological cardiac hypertrophy by inducing IP3 receptor degradation. Sci. Rep. 2017, 7, 13402. [Google Scholar] [CrossRef]
| ACE2 | F | CGAAGCCGAAGACCTGTTCTA |
| R | GGGCAAGTGTGGACTGTTCC | |
| CD147 | F | GAAGTCGTCAGAACACATCAACG |
| R | TTCCGGCGCTTCTCGTAGA | |
| CD209L | F | TCAAGCAGTATTGGAACAGAGGA |
| R | CAGGAGGCTGCGGACTTTTT | |
| GRP78 | F | GAAAGAAGGTTACCCATGCAGT |
| R | CAGGCCATAAGCAATAGCAGC | |
| HERPUD1 | F | ATGGAGTCCGAGACCGAAC |
| R | TTGGTGATCCAACAACAGCTT | |
| TMPRSS2 | F | CAAGTGCTCCAACTCTGGGAT |
| R | AACACACCGATTCTCGTCCTC | |
| GAPDH | F | GGAGCGAGATCCCTCCAAAAT |
| R | GGCTGTTGTCATACTTCTCATGG | |
| miR142 | F | CGCGCATAAAGTAGAAAGCACT |
| Common Reverse | R | CGAGGAAGAAGACGGAAGAAT |
| U6 | F | GCTTCGGCAGCACATATACTAAAAT |
| R | CGCTTCACGAATTTGCGTGTCAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-R.; Bahmed, K.; Simborio, H.; Hayek, H.; Bolla, S.; Marchetti, N.; Criner, G.J.; Kosmider, B. Expression of SARS-CoV-2 Entry Factors in Human Alveolar Type II Cells in Aging and Emphysema. Biomedicines 2021, 9, 779. https://doi.org/10.3390/biomedicines9070779
Lin C-R, Bahmed K, Simborio H, Hayek H, Bolla S, Marchetti N, Criner GJ, Kosmider B. Expression of SARS-CoV-2 Entry Factors in Human Alveolar Type II Cells in Aging and Emphysema. Biomedicines. 2021; 9(7):779. https://doi.org/10.3390/biomedicines9070779
Chicago/Turabian StyleLin, Chih-Ru, Karim Bahmed, Hannah Simborio, Hassan Hayek, Sudhir Bolla, Nathaniel Marchetti, Gerard J. Criner, and Beata Kosmider. 2021. "Expression of SARS-CoV-2 Entry Factors in Human Alveolar Type II Cells in Aging and Emphysema" Biomedicines 9, no. 7: 779. https://doi.org/10.3390/biomedicines9070779
APA StyleLin, C.-R., Bahmed, K., Simborio, H., Hayek, H., Bolla, S., Marchetti, N., Criner, G. J., & Kosmider, B. (2021). Expression of SARS-CoV-2 Entry Factors in Human Alveolar Type II Cells in Aging and Emphysema. Biomedicines, 9(7), 779. https://doi.org/10.3390/biomedicines9070779





