Tumor-Associated Macrophages—Implications for Molecular Oncology and Imaging
Abstract
1. Formation of the Tumor Microenvironment
2. Myeloid Cells: Monocytes and M-MDSCs
3. Recruitment and Polarization of M-MDSCs and Macrophages
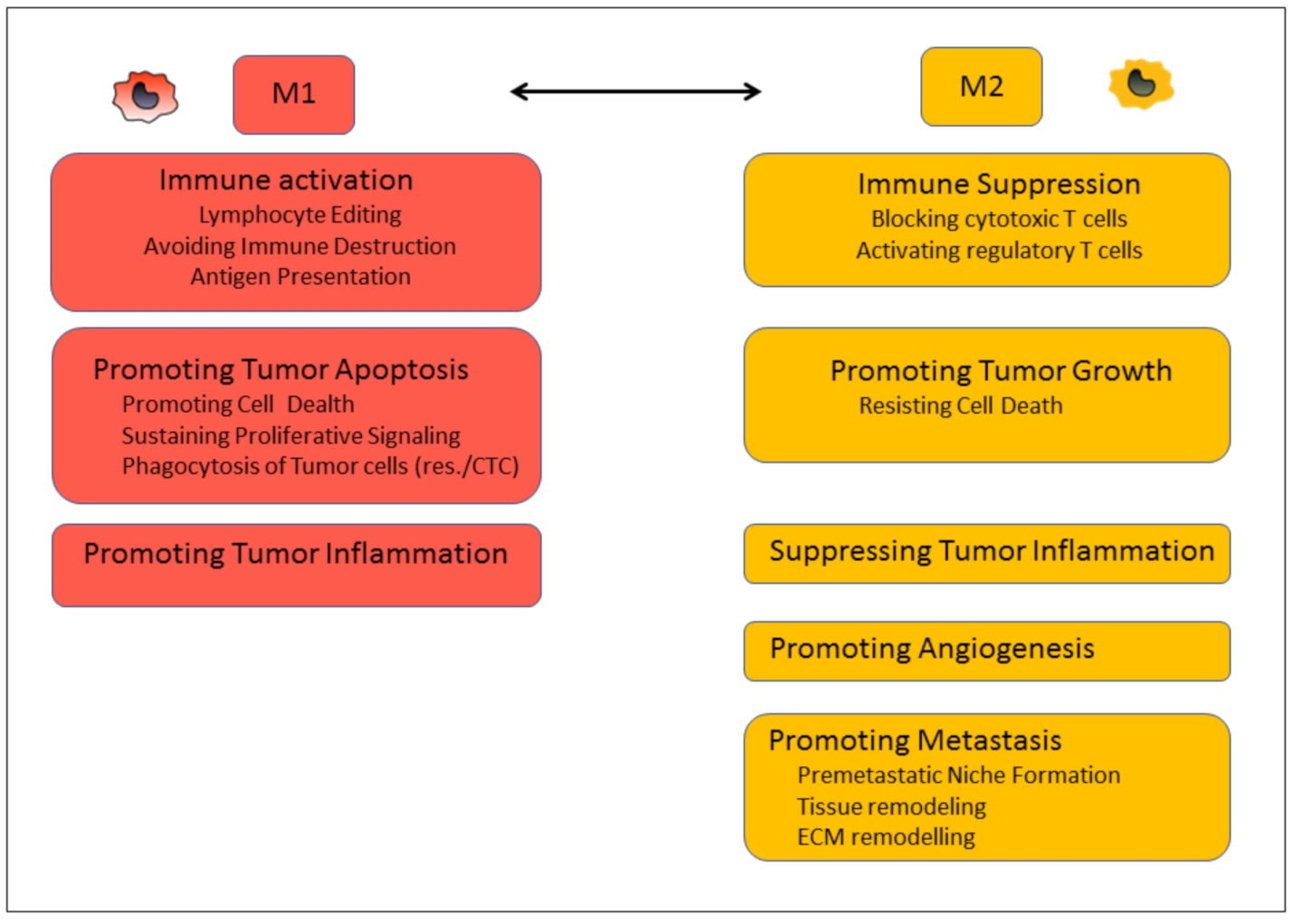
4. TAM Targeting
| M1 | M2 | |
|---|---|---|
| CD80, CD86 [128,129] | CD surface receptor | CD163, CD206, CD200R [128,130,131] |
| CXCL8, CXCL9, CXCL10, CCL2, CCL3, CCL5 [132] | Chemokines | CXCL12, CCL2,3,4,5,18,20 [133,134] |
| IL-1β, IL-2, IL-6, IL-12, IL-23, IFN-γ, TNF-α [135,136,137] | Cytokines | IL-4, IL-6, IL-10, IL-13, TGF-β, EGF [130,138,139,140,141,142,143,144,145,146] |
| MHC class II [147,148] | Biomarker | S100A8, S100A9, MMP2, MMP9, STAB1 [149,150,151,152,153] |
| Vasculature marker | VEGF [154,155] | |
| STAT1, IRF3, IRF5, HIF1α, AP1 [156,157,158,159] | Transcription Factors | STAT3, IRF4, FIZZ1, YM1 [160,161,162,163] |
| Checkpoint proteins | PD-L1 [164,165] | |
| iNOS, NO, ROS, IDO, PFKFB3, PKM2, ACOD1 [166,167,168,169,170,171] | Metabolites | ARG1, IDO, CARKL, GS [140,149,171,172,173,174,175] |
| miR-9, miR-18, miR-19a/b, miR-21, miR-26, miR-27a/b, miR-29b, miR-33, miR-125b, miR-127, miR-130a, miR-143, miR-145, miR-147, miR-155, miR-216a, miR-330 [176,177,178,179] | miRNA | miR-21, miR-23a/b, miR-24, miR-27a, miR-29, miR-34a, miR-124, miR-125a, miR-132, miR-146a, miR-155, miR-181, miR-188, miR-223, miR-511 [176,179,180,181,182] |
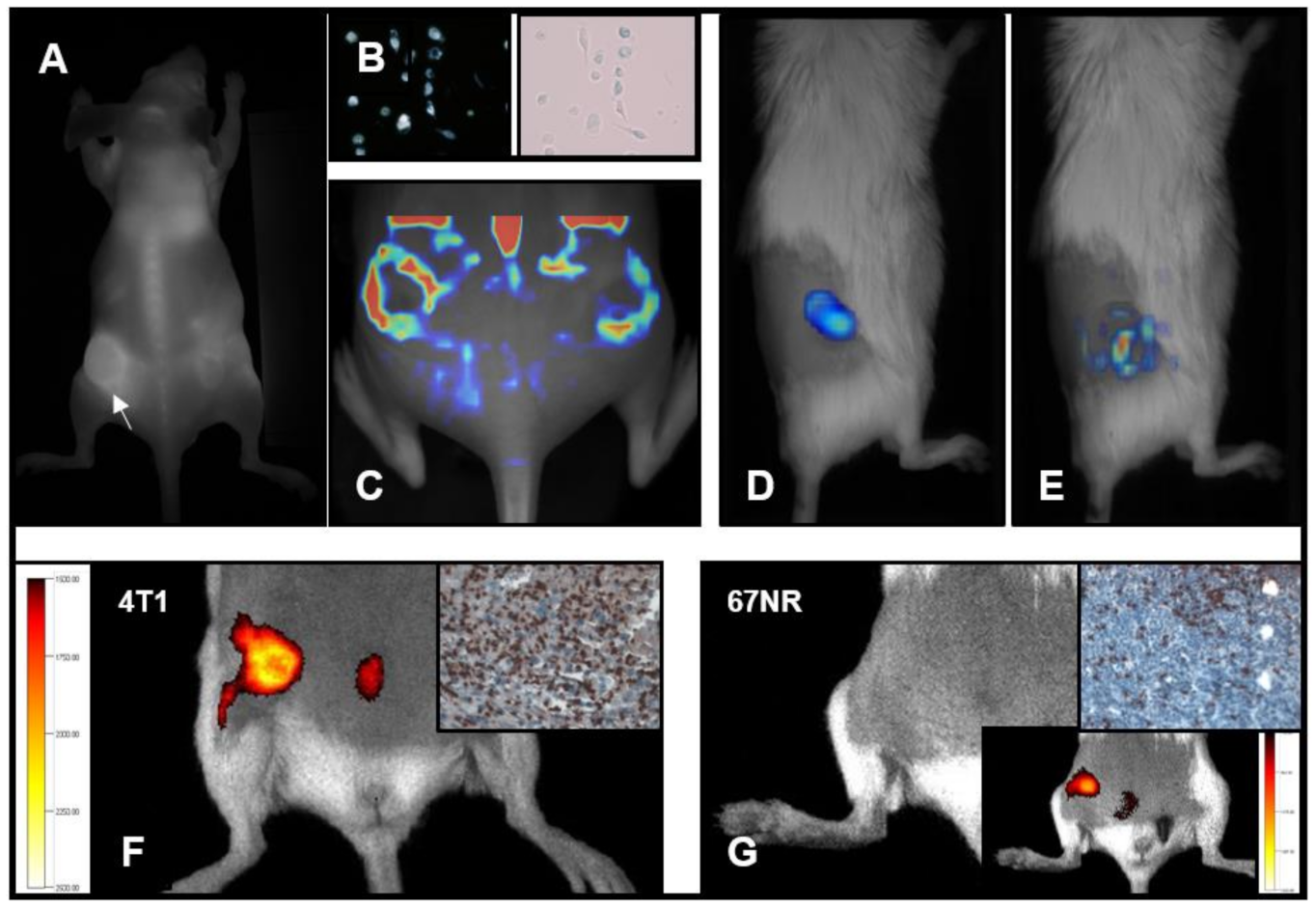
5. TAM Tracking and Imaging
6. Perspective
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stratton, M.R. Exploring the Genomes of Cancer Cells: Progress and Promise. Science 2011, 331, 1553–1558. [Google Scholar] [CrossRef] [PubMed]
- Marcus, A.; Gowen, B.G.; Thompson, T.W.; Iannello, A.; Ardolino, M.; Deng, W.; Wang, L.; Shifrin, N.; Raulet, D.H. Chapter Three Recognition of Tumors by the Innate Immune System and Natural Killer Cells. Adv. Immunol. 2014, 122, 91–128. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Zhang, H.; Zheng, J.; Zhang, L. Turning Cold into Hot: Firing up the Tumor Microenvironment. Trends Cancer 2020, 6, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Nagelkerke, A.; Bussink, J.; Rowan, A.E.; Span, P.N. The Mechanical Microenvironment in Cancer: How Physics Affects Tumours. Semin. Cancer Biol. 2015, 35, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Ferrao, P.T.; Behren, A.; Anderson, R.L.; Thompson, E.W. Editorial: Cellular and Phenotypic Plasticity in Cancer. Front. Oncol. 2015, 5, 171. [Google Scholar] [CrossRef]
- Rice, A.J.; Cortes, E.; Lachowski, D.; Cheung, B.C.H.; Karim, S.A.; Morton, J.P.; del Rio Hernández, A. Matrix Stiffness Induces Epithelial–Mesenchymal Transition and Promotes Chemoresistance in Pancreatic Cancer Cells. Oncogenesis 2017, 6, e352. [Google Scholar] [CrossRef]
- Choquet, D.; Felsenfeld, D.P.; Sheetz, M.P. Extracellular Matrix Rigidity Causes Strengthening of Integrin–Cytoskeleton Linkages. Cell 1997, 88, 39–48. [Google Scholar] [CrossRef]
- Labani-Motlagh, A.; Ashja-Mahdavi, M.; Loskog, A. The Tumor Microenvironment: A Milieu Hindering and Obstructing Antitumor Immune Responses. Front. Immunol. 2020, 11, 940. [Google Scholar] [CrossRef]
- Lau, A.N.; Heiden, M.G.V. Metabolism in the Tumor Microenvironment. Annu. Rev. Cancer Biol. 2019, 4, 1–24. [Google Scholar] [CrossRef]
- Vito, A.; El-Sayes, N.; Mossman, K. Hypoxia-Driven Immune Escape in the Tumor Microenvironment. Cells 2020, 9, 992. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Calar, K.; de la Puente, P. Mimicking Tumor Hypoxia and Tumor-Immune Interactions Employing Three-Dimensional in Vitro Models. J. Exp. Clin. Cancer Res. 2020, 39, 75. [Google Scholar] [CrossRef]
- Wang, W.; Wang, L.; Ruan, L.; Oh, J.; Dong, X.; Zhuge, Q.; Su, D.-M. Extracellular Vesicles Extracted from Young Donor Serum Attenuate Inflammaging via Partially Rejuvenating Aged T-cell Immunotolerance. FASEB J. 2018, 32, 5899–5912. [Google Scholar] [CrossRef]
- Li, I.; Nabet, B.Y. Exosomes in the Tumor Microenvironment as Mediators of Cancer Therapy Resistance. Mol. Cancer 2019, 18, 32. [Google Scholar] [CrossRef]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key Players in Cancer and Potential Therapeutic Strategy. Signal. Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef]
- Vilgelm, A.E.; Richmond, A. Chemokines Modulate Immune Surveillance in Tumorigenesis, Metastasis, and Response to Immunotherapy. Front. Immunol. 2019, 10, 333. [Google Scholar] [CrossRef]
- Atretkhany, K.-S.N.; Drutskaya, M.S.; Nedospasov, S.A.; Grivennikov, S.I.; Kuprash, D.V. Chemokines, Cytokines and Exosomes Help Tumors to Shape Inflammatory Microenvironment. Pharmacol. Therapeut. 2016, 168, 98–112. [Google Scholar] [CrossRef]
- Xiao, Z.; Dai, Z.; Locasale, J.W. Metabolic Landscape of the Tumor Microenvironment at Single Cell Resolution. Nat. Commun. 2019, 10, 3763. [Google Scholar] [CrossRef]
- Jin, M.-Z.; Jin, W.-L. The Updated Landscape of Tumor Microenvironment and Drug Repurposing. Signal. Transduct. Target. Ther. 2020, 5, 166. [Google Scholar] [CrossRef]
- Murciano-Goroff, Y.R.; Warner, A.B.; Wolchok, J.D. The Future of Cancer Immunotherapy: Microenvironment-Targeting Combinations. Cell Res. 2020, 30, 507–519. [Google Scholar] [CrossRef]
- Popel, A.S. Immunoactivating the Tumor Microenvironment Enhances Immunotherapy as Predicted by Integrative Computational Model. Proc. Natl. Acad. Sci. USA 2020, 117, 4447–4449. [Google Scholar] [CrossRef]
- Mpekris, F.; Voutouri, C.; Baish, J.W.; Duda, D.G.; Munn, L.L.; Stylianopoulos, T.; Jain, R.K. Combining Microenvironment Normalization Strategies to Improve Cancer Immunotherapy. Proc. Natl. Acad. Sci. USA 2020, 117, 3728–3737. [Google Scholar] [CrossRef]
- Zheng, J.; Gao, P. Toward Normalization of the Tumor Microenvironment for Cancer Therapy. Integr. Cancer Ther. 2019, 18, 1534735419862352. [Google Scholar] [CrossRef]
- Jain, R.K. Normalizing Tumor Microenvironment to Treat Cancer: Bench to Bedside to Biomarkers. J. Clin. Oncol. 2013, 31, 2205–2218. [Google Scholar] [CrossRef] [PubMed]
- Colotta, F.; Allavena, P.; Sica, A.; Garlanda, C.; Mantovani, A. Cancer-Related Inflammation, the Seventh Hallmark of Cancer: Links to Genetic Instability. Carcinogenesis 2009, 30, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Teng, M.W.L.; Galon, J.; Fridman, W.-H.; Smyth, M.J. From Mice to Humans: Developments in Cancer Immunoediting. J. Clin. Investig. 2015, 125, 3338–3346. [Google Scholar] [CrossRef] [PubMed]
- Vesely, M.D.; Kershaw, M.H.; Schreiber, R.D.; Smyth, M.J. Natural Innate and Adaptive Immunity to Cancer. Annu. Rev. Immunol. 2011, 29, 235–271. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the Immune System in Cancer: From Tumor Initiation to Metastatic Progression. Gene Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef] [PubMed]
- Ostroumov, D.; Fekete-Drimusz, N.; Saborowski, M.; Kühnel, F.; Woller, N. CD4 and CD8 T Lymphocyte Interplay in Controlling Tumor Growth. Cell. Mol. Life Sci. 2017, 75, 689–713. [Google Scholar] [CrossRef] [PubMed]
- Tomić, S.; Joksimović, B.; Bekić, M.; Vasiljević, M.; Milanović, M.; Čolić, M.; Vučević, D. Prostaglanin-E2 Potentiates the Suppressive Functions of Human Mononuclear Myeloid-Derived Suppressor Cells and Increases Their Capacity to Expand IL-10-Producing Regulatory T Cell Subsets. Front. Immunol. 2019, 10, 475. [Google Scholar] [CrossRef]
- Batlle, E.; Massagué, J. Transforming Growth Factor-β Signaling in Immunity and Cancer. Immunity 2019, 50, 924–940. [Google Scholar] [CrossRef]
- Tamura, R.; Tanaka, T.; Akasaki, Y.; Murayama, Y.; Yoshida, K.; Sasaki, H. The Role of Vascular Endothelial Growth Factor in the Hypoxic and Immunosuppressive Tumor Microenvironment: Perspectives for Therapeutic Implications. Med. Oncol. 2019, 37, 2. [Google Scholar] [CrossRef]
- Cervantes-Villagrana, R.D.; Albores-García, D.; Cervantes-Villagrana, A.R.; García-Acevez, S.J. Tumor-Induced Neurogenesis and Immune Evasion as Targets of Innovative Anti-Cancer Therapies. Signal. Transduct. Target. Ther. 2020, 5, 99. [Google Scholar] [CrossRef]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the Cancer Microenvironment and Their Relevance in Cancer Immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef]
- Kitamura, T.; Qian, B.-Z.; Pollard, J.W. Immune Cell Promotion of Metastasis. Nat. Rev. Immunol. 2015, 15, 73–86. [Google Scholar] [CrossRef]
- Zemek, R.M.; Chin, W.L.; Nowak, A.K.; Millward, M.J.; Lake, R.A.; Lesterhuis, W.J. Sensitizing the Tumor Microenvironment to Immune Checkpoint Therapy. Front. Immunol. 2020, 11, 223. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, Z.; Xu, X.; Yu, Z.; Mi, J. The Influence of Microenvironment on Tumor Immunotherapy. FEBS J. 2019, 286, 4160–4175. [Google Scholar] [CrossRef]
- Guilliams, M.; Mildner, A.; Yona, S. Developmental and Functional Heterogeneity of Monocytes. Immunity 2018, 49, 595–613. [Google Scholar] [CrossRef] [PubMed]
- Trzebanski, S.; Jung, S. Plasticity of Monocyte Development and Monocyte Fates. Immunol. Lett. 2020, 227, 66–78. [Google Scholar] [CrossRef]
- Paul, W.E. Bridging Innate and Adaptive Immunity. Cell 2011, 147, 1212–1215. [Google Scholar] [CrossRef]
- Olingy, C.E.; Dinh, H.Q.; Hedrick, C.C. Monocyte Heterogeneity and Functions in Cancer. J. Leukoc. Biol. 2019, 106, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Canè, S.; Ugel, S.; Trovato, R.; Marigo, I.; Sanctis, F.D.; Sartoris, S.; Bronte, V. The Endless Saga of Monocyte Diversity. Front. Immunol. 2019, 10, 1786. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.A.; Zhang, Y.; Fullerton, J.N.; Boelen, L.; Rongvaux, A.; Maini, A.A.; Bigley, V.; Flavell, R.A.; Gilroy, D.W.; Asquith, B.; et al. The Fate and Lifespan of Human Monocyte Subsets in Steady State and Systemic InflammationHuman Monocyte Kinetics. J. Exp. Med. 2017, 214, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Kapellos, T.S.; Bonaguro, L.; Gemünd, I.; Reusch, N.; Saglam, A.; Hinkley, E.R.; Schultze, J.L. Human Monocyte Subsets and Phenotypes in Major Chronic Inflammatory Diseases. Front. Immunol. 2019, 10, 2035. [Google Scholar] [CrossRef]
- Kratofil, R.M.; Kubes, P.; Deniset, J.F. Monocyte Conversion during Inflammation and Injury. Arterioscler. Thromb. Vasc Biol. 2017, 37, 35–42. [Google Scholar] [CrossRef]
- Buscher, K.; Marcovecchio, P.; Hedrick, C.C.; Ley, K. Patrolling Mechanics of Non-Classical Monocytes in Vascular Inflammation. Front. Cardiovasc. Med. 2017, 4, 80. [Google Scholar] [CrossRef]
- Grage-Griebenow, E.; Flad, H.-D.; Ernst, M. Heterogeneity of Human Peripheral Blood Monocyte Subsets. J. Leukoc. Biol. 2001, 69, 11–20. [Google Scholar] [CrossRef]
- Merah-Mourah, F.; Cohen, S.O.; Charron, D.; Mooney, N.; Haziot, A. Identification of Novel Human Monocyte Subsets and Evidence for Phenotypic Groups Defined by Interindividual Variations of Expression of Adhesion Molecules. Sci. Rep. 2020, 10, 4397. [Google Scholar] [CrossRef]
- Sanford, D.E.; Belt, B.A.; Panni, R.Z.; Mayer, A.; Deshpande, A.D.; Carpenter, D.; Mitchem, J.B.; Plambeck-Suess, S.M.; Worley, L.A.; Goetz, B.D.; et al. Inflammatory Monocyte Mobilization Decreases Patient Survival in Pancreatic Cancer: A Role for Targeting the CCL2/CCR2 Axis. Clin. Cancer Res. 2013, 19, 3404–3415. [Google Scholar] [CrossRef]
- Auffray, C.; Fogg, D.; Garfa, M.; Elain, G.; Join-Lambert, O.; Kayal, S.; Sarnacki, S.; Cumano, A.; Lauvau, G.; Geissmann, F. Monitoring of Blood Vessels and Tissues by a Population of Monocytes with Patrolling Behavior. Science 2007, 317, 666–670. [Google Scholar] [CrossRef]
- Bianchini, M.; Duchêne, J.; Santovito, D.; Schloss, M.J.; Evrard, M.; Winkels, H.; Aslani, M.; Mohanta, S.K.; Horckmans, M.; Blanchet, X.; et al. PD-L1 Expression on Nonclassical Monocytes Reveals Their Origin and Immunoregulatory Function. Sci. Immunol. 2019, 4, eaar3054. [Google Scholar] [CrossRef]
- Satoh, T.; Nakagawa, K.; Sugihara, F.; Kuwahara, R.; Ashihara, M.; Yamane, F.; Minowa, Y.; Fukushima, K.; Ebina, I.; Yoshioka, Y.; et al. Identification of an Atypical Monocyte and Committed Progenitor Involved in Fibrosis. Nature 2017, 541, 96–101. [Google Scholar] [CrossRef]
- Gabrilovich, D.I. Myeloid-Derived Suppressor Cells. Cancer Immunol. Res. 2017, 5, 3–8. [Google Scholar] [CrossRef]
- Furusawa, J.; Mizoguchi, I.; Chiba, Y.; Hisada, M.; Kobayashi, F.; Yoshida, H.; Nakae, S.; Tsuchida, A.; Matsumoto, T.; Ema, H.; et al. Promotion of Expansion and Differentiation of Hematopoietic Stem Cells by Interleukin-27 into Myeloid Progenitors to Control Infection in Emergency Myelopoiesis. PLoS Pathog. 2016, 12, e1005507. [Google Scholar] [CrossRef]
- Mandruzzato, S.; Brandau, S.; Britten, C.M.; Bronte, V.; Damuzzo, V.; Gouttefangeas, C.; Maurer, D.; Ottensmeier, C.; van der Burg, S.H.; Welters, M.J.P.; et al. Toward Harmonized Phenotyping of Human Myeloid-Derived Suppressor Cells by Flow Cytometry: Results from an Interim Study. Cancer Immunol. Immunother. 2016, 65, 161–169. [Google Scholar] [CrossRef]
- Haile, L.A.; Greten, T.F.; Korangy, F. Immune Suppression: The Hallmark of Myeloid Derived Suppressor Cells. Immunol. Investig. 2012, 41, 581–594. [Google Scholar] [CrossRef]
- Yang, Y.; Li, C.; Liu, T.; Dai, X.; Bazhin, A.V. Myeloid-Derived Suppressor Cells in Tumors: From Mechanisms to Antigen Specificity and Microenvironmental Regulation. Front. Immunol. 2020, 11, 1371. [Google Scholar] [CrossRef]
- Bruse, N.; Leijte, G.P.; Pickkers, P.; Kox, M. New Frontiers in Precision Medicine for Sepsis-Induced Immunoparalysis. Expert Rev. Clin. Immunol. 2018, 15, 251–263. [Google Scholar] [CrossRef]
- Schrijver, I.T.; Théroude, C.; Roger, T. Myeloid-Derived Suppressor Cells in Sepsis. Front. Immunol. 2019, 10, 327. [Google Scholar] [CrossRef]
- Davidov, V.; Jensen, G.; Mai, S.; Chen, S.-H.; Pan, P.-Y. Analyzing One Cell at a TIME: Analysis of Myeloid Cell Contributions in the Tumor Immune Microenvironment. Front. Immunol. 2020, 11, 1842. [Google Scholar] [CrossRef]
- Sica, A.; Bronte, V. Altered Macrophage Differentiation and Immune Dysfunction in Tumor Development. J. Clin. Investig. 2007, 117, 1155–1166. [Google Scholar] [CrossRef]
- Lee, W.-C.; Hsu, P.-Y.; Hsu, H.-Y. Stem Cell Factor Produced by Tumor Cells Expands Myeloid-Derived Suppressor Cells in Mice. Sci. Rep. 2020, 10, 11257. [Google Scholar] [CrossRef]
- Horikawa, N.; Abiko, K.; Matsumura, N.; Hamanishi, J.; Baba, T.; Yamaguchi, K.; Yoshioka, Y.; Koshiyama, M.; Konishi, I. Expression of Vascular Endothelial Growth Factor in Ovarian Cancer Inhibits Tumor Immunity through the Accumulation of Myeloid-Derived Suppressor Cells. Clin. Cancer Res. 2017, 23, 587–599. [Google Scholar] [CrossRef]
- Johnson, B.S.; Mueller, L.; Si, J.; Collins, S.J. The Cytokines IL-3 and GM-CSF Regulate the Transcriptional Activity of Retinoic Acid Receptors in Different in Vitro Models of Myeloid Differentiation. Blood 2002, 99, 746–753. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, J.; Zhang, W.; Zhang, R.; Ye, Y.; Liu, P.; Yu, W.; Wei, F.; Ren, X.; Yu, J. Interleukin-6 Trans-Signaling Pathway Promotes Immunosuppressive Myeloid-Derived Suppressor Cells via Suppression of Suppressor of Cytokine Signaling 3 in Breast Cancer. Front. Immunol. 2017, 8, 1840. [Google Scholar] [CrossRef]
- Tobin, R.P.; Jordan, K.R.; Kapoor, P.; Spongberg, E.; Davis, D.; Vorwald, V.M.; Couts, K.L.; Gao, D.; Smith, D.E.; Borgers, J.S.W.; et al. IL-6 and IL-8 Are Linked With Myeloid-Derived Suppressor Cell Accumulation and Correlate With Poor Clinical Outcomes in Melanoma Patients. Front. Oncol. 2019, 9, 1223. [Google Scholar] [CrossRef]
- Zhao, F.; Hoechst, B.; Duffy, A.; Gamrekelashvili, J.; Fioravanti, S.; Manns, M.P.; Greten, T.F.; Korangy, F. S100A9 a New Marker for Monocytic Human Myeloid-derived Suppressor Cells. Immunology 2012, 136, 176–183. [Google Scholar] [CrossRef]
- Lim, S.Y.; Yuzhalin, A.E.; Gordon-Weeks, A.N.; Muschel, R.J. Tumor-Infiltrating Monocytes/Macrophages Promote Tumor Invasion and Migration by Upregulating S100A8 and S100A9 Expression in Cancer Cells. Oncogene 2016, 35, 5735–5745. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-Derived Suppressor Cells as Regulators of the Immune System. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Umemura, N.; Saio, M.; Suwa, T.; Kitoh, Y.; Bai, J.; Nonaka, K.; Ouyang, G.; Okada, M.; Balazs, M.; Adany, R.; et al. Tumor-infiltrating Myeloid-derived Suppressor Cells Are Pleiotropic-inflamed Monocytes/Macrophages That Bear M1- and M2-type Characteristics. J. Leukoc. Biol. 2008, 83, 1136–1144. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Pan, P.-Y.; Eisenstein, S.; Divino, C.M.; Lowell, C.A.; Takai, T.; Chen, S.-H. Paired Immunoglobin-like Receptor-B Regulates the Suppressive Function and Fate of Myeloid-Derived Suppressor Cells. Immunity 2011, 34, 385–395. [Google Scholar] [CrossRef]
- He, W.; Liang, P.; Guo, G.; Huang, Z.; Niu, Y.; Dong, L.; Wang, C.; Zhang, J. Re-Polarizing Myeloid-Derived Suppressor Cells (MDSCs) with Cationic Polymers for Cancer Immunotherapy. Sci. Rep. 2016, 6, 24506. [Google Scholar] [CrossRef]
- Ochando, J.C.; Chen, S.H. Myeloid-Derived Suppressor Cells in Transplantation and Cancer. Immunol. Res. 2012, 54, 275–285. [Google Scholar] [CrossRef][Green Version]
- Yang, W.-C.; Ma, G.; Chen, S.-H.; Pan, P.-Y. Polarization and Reprogramming of Myeloid-Derived Suppressor Cells. J. Mol. Cell Biol. 2013, 5, 207–209. [Google Scholar] [CrossRef]
- Elliott, L.A.; Doherty, G.A.; Sheahan, K.; Ryan, E.J. Human Tumor-Infiltrating Myeloid Cells: Phenotypic and Functional Diversity. Front. Immunol. 2017, 8, 86. [Google Scholar] [CrossRef]
- Laviron, M.; Boissonnas, A. Ontogeny of Tumor-Associated Macrophages. Front. Immunol. 2019, 10, 1799. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, Z.; Wu, L.; Zhang, M.; Li, W.; Ding, J.; Zhu, J.; Wei, H.; Zhao, K. Circulating and Tumor-Infiltrating Myeloid-Derived Suppressor Cells in Patients with Colorectal Carcinoma. PLoS ONE 2013, 8, e57114. [Google Scholar] [CrossRef]
- Mahmoud, S.M.A.; Lee, A.H.S.; Paish, E.C.; Macmillan, R.D.; Ellis, I.O.; Green, A.R. Tumour-Infiltrating Macrophages and Clinical Outcome in Breast Cancer. J. Clin. Pathol. 2012, 65, 159. [Google Scholar] [CrossRef]
- Fogg, D.K.; Sibon, C.; Miled, C.; Jung, S.; Aucouturier, P.; Littman, D.R.; Cumano, A.; Geissmann, F. A Clonogenic Bone Marrow Progenitor Specific for Macrophages and Dendritic Cells. Science 2006, 311, 83–87. [Google Scholar] [CrossRef]
- Hettinger, J.; Richards, D.M.; Hansson, J.; Barra, M.M.; Joschko, A.-C.; Krijgsveld, J.; Feuerer, M. Origin of Monocytes and Macrophages in a Committed Progenitor. Nat. Immunol. 2013, 14, 821–830. [Google Scholar] [CrossRef]
- Ginhoux, F.; Guilliams, M. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity 2016, 44, 439–449. [Google Scholar] [CrossRef]
- Theret, M.; Mounier, R.; Rossi, F. The Origins and Non-Canonical Functions of Macrophages in Development and Regeneration. Development 2019, 146, dev156000. [Google Scholar] [CrossRef]
- Linehan, E.; Fitzgerald, D. Ageing and the Immune System: Focus on Macrophages. Eur. J. Microbiol. Immunol. 2015, 5, 14–24. [Google Scholar] [CrossRef]
- Ajami, B.; Bennett, J.L.; Krieger, C.; Tetzlaff, W.; Rossi, F.M.V. Local Self-Renewal Can Sustain CNS Microglia Maintenance and Function throughout Adult Life. Nat. Neurosci. 2007, 10, 1538–1543. [Google Scholar] [CrossRef]
- Epelman, S.; Lavine, K.J.; Beaudin, A.E.; Sojka, D.K.; Carrero, J.A.; Calderon, B.; Brija, T.; Gautier, E.L.; Ivanov, S.; Satpathy, A.T.; et al. Embryonic and Adult-Derived Resident Cardiac Macrophages Are Maintained through Distinct Mechanisms at Steady State and during Inflammation. Immunity 2014, 40, 91–104. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, Z.; Xiong, S.; Sun, F.; Qin, G.; Hu, G.; Wang, J.; Zhao, L.; Liang, Y.-X.; Wu, T.; et al. Repopulated Microglia Are Solely Derived from the Proliferation of Residual Microglia after Acute Depletion. Nat. Neurosci. 2018, 21, 530–540. [Google Scholar] [CrossRef]
- Guillot, A.; Tacke, F. Liver Macrophages: Old Dogmas and New Insights. Hepatol. Commun. 2019, 3, 730–743. [Google Scholar] [CrossRef] [PubMed]
- MacParland, S.A.; Liu, J.C.; Ma, X.-Z.; Innes, B.T.; Bartczak, A.M.; Gage, B.K.; Manuel, J.; Khuu, N.; Echeverri, J.; Linares, I.; et al. Single Cell RNA Sequencing of Human Liver Reveals Distinct Intrahepatic Macrophage Populations. Nat. Commun. 2018, 9, 4383. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Li, N.; Tang, H.; Lou, Z.; Chong, X.; Zhang, C.; Su, J.; Dong, X. Kupffer Cell-Derived TNF-α Promotes Hepatocytes to Produce CXCL1 and Mobilize Neutrophils in Response to Necrotic Cells. Cell Death Dis. 2018, 9, 323. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Liu, F.; Qin, Z.; Zhang, J.; Chen, J.; Ding, W.-X.; Feng, D.; Ji, Y.; Qin, X. Kupffer Cells Promote T-Cell Hepatitis by Producing CXCL10 and Limiting Liver Sinusoidal Endothelial Cell Permeability. Theranostics 2020, 10, 7163–7177. [Google Scholar] [CrossRef]
- Blériot, C.; Dupuis, T.; Jouvion, G.; Eberl, G.; Disson, O.; Lecuit, M. Liver-Resident Macrophage Necroptosis Orchestrates Type 1 Microbicidal Inflammation and Type-2-Mediated Tissue Repair during Bacterial Infection. Immunity 2015, 42, 145–158. [Google Scholar] [CrossRef]
- Mass, E.; Ballesteros, I.; Farlik, M.; Halbritter, F.; Günther, P.; Crozet, L.; Jacome-Galarza, C.E.; Händler, K.; Klughammer, J.; Kobayashi, Y.; et al. Specification of Tissue-Resident Macrophages during Organogenesis. Science 2016, 353, aaf4238. [Google Scholar] [CrossRef]
- Wang, J.; Kubes, P. A Reservoir of Mature Cavity Macrophages That Can Rapidly Invade Visceral Organs to Affect Tissue Repair. Cell 2016, 165, 668–678. [Google Scholar] [CrossRef]
- Swirski, F.K.; Nahrendorf, M.; Etzrodt, M.; Wildgruber, M.; Cortez-Retamozo, V.; Panizzi, P.; Figueiredo, J.-L.; Kohler, R.H.; Chudnovskiy, A.; Waterman, P.; et al. Identification of Splenic Reservoir Monocytes and Their Deployment to Inflammatory Sites. Science 2009, 325, 612–616. [Google Scholar] [CrossRef]
- Aoyama, T.; Kuwahara-Arai, K.; Uchiyama, A.; Kon, K.; Okubo, H.; Yamashina, S.; Ikejima, K.; Kokubu, S.; Miyazaki, A.; Watanabe, S. Spleen-Derived Lipocalin-2 in the Portal Vein Regulates Kupffer Cells Activation and Attenuates the Development of Liver Fibrosis in Mice. Lab. Investig. 2017, 97, 890–902. [Google Scholar] [CrossRef]
- Li, L.; Wei, W.; Li, Z.; Chen, H.; Li, Y.; Jiang, W.; Chen, W.; Kong, G.; Yang, J.; Li, Z. The Spleen Promotes the Secretion of CCL2 and Supports an M1 Dominant Phenotype in Hepatic Macrophages During Liver Fibrosis. Cell Physiol. Biochem. 2018, 51, 557–574. [Google Scholar] [CrossRef]
- Cortez-Retamozo, V.; Etzrodt, M.; Newton, A.; Ryan, R.; Pucci, F.; Sio, S.W.; Kuswanto, W.; Rauch, P.J.; Chudnovskiy, A.; Iwamoto, Y.; et al. Angiotensin II Drives the Production of Tumor-Promoting Macrophages. Immunity 2013, 38, 296–308. [Google Scholar] [CrossRef]
- Wu, C.; Hua, Q.; Zheng, L. Generation of Myeloid Cells in Cancer: The Spleen Matters. Front. Immunol. 2020, 11, 1126. [Google Scholar] [CrossRef]
- Long, X.; Wang, J.; Zhao, J.; Liang, H.; Zhu, P.; Cheng, Q.; Chen, Q.; Wu, Y.; Zhang, Z.; Zhang, B.; et al. Splenectomy Suppresses Growth and Metastasis of Hepatocellular Carcinoma through Decreasing Myeloid-Derived Suppressor Cells in Vivo. J. Huazhong Univ. Sci. Technol. Med Sci. 2016, 36, 667–676. [Google Scholar] [CrossRef]
- Sugase, T.; Lam, B.Q.; Danielson, M.; Terai, M.; Aplin, A.E.; Gutkind, J.S.; Sato, T. Development and Optimization of Orthotopic Liver Metastasis Xenograft Mouse Models in Uveal Melanoma. J. Transl. Med. 2020, 18, 208. [Google Scholar] [CrossRef]
- Bercovici, N.; Guérin, M.V.; Trautmann, A.; Donnadieu, E. The Remarkable Plasticity of Macrophages: A Chance to Fight Cancer. Front. Immunol. 2019, 10, 1563. [Google Scholar] [CrossRef]
- Ginhoux, F.; Schultze, J.L.; Murray, P.J.; Ochando, J.; Biswas, S.K. New Insights into the Multidimensional Concept of Macrophage Ontogeny, Activation and Function. Nat. Immunol. 2016, 17, 34–40. [Google Scholar] [CrossRef]
- Cassetta, L.; Pollard, J.W. Repolarizing Macrophages Improves Breast Cancer Therapy. Cell Res. 2017, 27, 963–964. [Google Scholar] [CrossRef]
- Vitale, I.; Manic, G.; Coussens, L.M.; Kroemer, G.; Galluzzi, L. Macrophages and Metabolism in the Tumor Microenvironment. Cell Metab. 2019, 30, 36–50. [Google Scholar] [CrossRef]
- Müller, S.; Kohanbash, G.; Liu, S.J.; Alvarado, B.; Carrera, D.; Bhaduri, A.; Watchmaker, P.B.; Yagnik, G.; Lullo, E.D.; Malatesta, M.; et al. Single-Cell Profiling of Human Gliomas Reveals Macrophage Ontogeny as a Basis for Regional Differences in Macrophage Activation in the Tumor Microenvironment. Genome Biol. 2017, 18, 234. [Google Scholar] [CrossRef]
- Arora, S.; Dev, K.; Agarwal, B.; Das, P.; Syed, M.A. Macrophages: Their Role, Activation and Polarization in Pulmonary Diseases. Immunobiology 2018, 223, 383–396. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, S.; Wu, H.; Rong, X.; Guo, J. M2b Macrophage Polarization and Its Roles in Diseases. J. Leukoc. Biol. 2019, 106, 345–358. [Google Scholar] [CrossRef]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The Chemokine System in Diverse Forms of Macrophage Activation and Polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef]
- Mosser, D.M.; Edwards, J.P. Exploring the Full Spectrum of Macrophage Activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef]
- Rőszer, T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediat. Inflamm. 2015, 2015, 1–16. [Google Scholar] [CrossRef]
- Chen, C.; Liu, J.; Luo, Y. MicroRNAs in Tumor Immunity: Functional Regulation in Tumor-Associated Macrophages. J. Zhejiang Univ. Sci. B 2020, 21, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Szebeni, G.J.; Vizler, C.; Kitajka, K.; Puskas, L.G. Inflammation and Cancer: Extra- and Intracellular Determinants of Tumor-Associated Macrophages as Tumor Promoters. Mediat. Inflamm. 2017, 2017, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Curtale, G.; Rubino, M.; Locati, M. MicroRNAs as Molecular Switches in Macrophage Activation. Front. Immunol. 2019, 10, 799. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Yin, Y.; Li, N.; Zhu, D.; Zhang, J.; Zhang, C.-Y.; Zen, K. Re-Polarization of Tumor-Associated Macrophages to pro-Inflammatory M1 Macrophages by MicroRNA. J. Mol. Cell Biol. 2012, 4, 341–343. [Google Scholar] [CrossRef]
- Ying, H.; Kang, Y.; Zhang, H.; Zhao, D.; Xia, J.; Lu, Z.; Wang, H.; Xu, F.; Shi, L. MiR-127 Modulates Macrophage Polarization and Promotes Lung Inflammation and Injury by Activating the JNK Pathway. J. Immunol. 2015, 194, 1239–1251. [Google Scholar] [CrossRef]
- Chaudhuri, A.A.; So, A.Y.-L.; Sinha, N.; Gibson, W.S.J.; Taganov, K.D.; O’Connell, R.M.; Baltimore, D. MicroRNA-125b Potentiates Macrophage Activation. J. Immunol. 2011, 187, 5062–5068. [Google Scholar] [CrossRef]
- Wang, P.; Wang, H.; Huang, Q.; Peng, C.; Yao, L.; Chen, H.; Qiu, Z.; Wu, Y.; Wang, L.; Chen, W. Exosomes from M1-Polarized Macrophages Enhance Paclitaxel Antitumor Activity by Activating Macrophages-Mediated Inflammation. Theranostics 2019, 9, 1714–1727. [Google Scholar] [CrossRef]
- Qian, B.-Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 Recruits Inflammatory Monocytes to Facilitate Breast-Tumour Metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef]
- Pathria, P.; Louis, T.L.; Varner, J.A. Targeting Tumor-Associated Macrophages in Cancer. Trends Immunol. 2019, 40, 310–327. [Google Scholar] [CrossRef]
- Scala, S. Molecular Pathways: Targeting the CXCR4–CXCL12 Axis—Untapped Potential in the Tumor Microenvironment. Clin. Cancer Res. 2015, 21, 4278–4285. [Google Scholar] [CrossRef]
- Rogers, T.L.; Holen, I. Tumour Macrophages as Potential Targets of Bisphosphonates. J. Transl. Med. 2011, 9, 177. [Google Scholar] [CrossRef]
- Gül, N.; Babes, L.; Siegmund, K.; Korthouwer, R.; Bögels, M.; Braster, R.; Vidarsson, G.; ten Hagen, T.L.M.; Kubes, P.; van Egmond, M. Macrophages Eliminate Circulating Tumor Cells after Monoclonal Antibody Therapy. J. Clin. Investig. 2014, 124, 812–823. [Google Scholar] [CrossRef]
- Guerriero, J.L. Macrophages: The Road Less Traveled, Changing Anticancer Therapy. Trends Mol. Med. 2018, 24, 472–489. [Google Scholar] [CrossRef]
- Mantovani, A.; Marchesi, F.; Malesci, A.; Laghi, L.; Allavena, P. Tumour-Associated Macrophages as Treatment Targets in Oncology. Nat. Rev. Clin. Oncol. 2017, 14, 399–416. [Google Scholar] [CrossRef]
- Hu, G.; Guo, M.; Xu, J.; Wu, F.; Fan, J.; Huang, Q.; Yang, G.; Lv, Z.; Wang, X.; Jin, Y. Nanoparticles Targeting Macrophages as Potential Clinical Therapeutic Agents Against Cancer and Inflammation. Front. Immunol. 2019, 10, 1998. [Google Scholar] [CrossRef]
- Wang, H.-F.; Liu, Y.; Yang, G.; Zhao, C.-X. Macrophage-Mediated Cancer Drug Delivery. Mater. Today Sustain. 2020, 100055. [Google Scholar] [CrossRef]
- Xia, Y.; Rao, L.; Yao, H.; Wang, Z.; Ning, P.; Chen, X. Engineering Macrophages for Cancer Immunotherapy and Drug Delivery. Adv. Mater. 2020, 32, 2002054. [Google Scholar] [CrossRef]
- Pettersen, J.S.; Fuentes-Duculan, J.; Suárez-Fariñas, M.; Pierson, K.C.; Pitts-Kiefer, A.; Fan, L.; Belkin, D.A.; Wang, C.Q.F.; Bhuvanendran, S.; Johnson-Huang, L.M.; et al. Tumor-Associated Macrophages in the Cutaneous SCC Microenvironment Are Heterogeneously Activated. J. Investig. Dermatol. 2011, 131, 1322–1330. [Google Scholar] [CrossRef]
- Bertani, F.R.; Mozetic, P.; Fioramonti, M.; Iuliani, M.; Ribelli, G.; Pantano, F.; Santini, D.; Tonini, G.; Trombetta, M.; Businaro, L.; et al. Classification of M1/M2-Polarized Human Macrophages by Label-Free Hyperspectral Reflectance Confocal Microscopy and Multivariate Analysis. Sci. Rep. 2017, 7, 8965. [Google Scholar] [CrossRef]
- Buechler, C.; Ritter, M.; Orsó, E.; Langmann, T.; Klucken, J.; Schmitz, G. Regulation of Scavenger Receptor CD163 Expression in Human Monocytes and Macrophages by Pro- and Antiinflammatory Stimuli. J. Leukoc. Biol. 2000, 67, 97–103. [Google Scholar] [CrossRef]
- Koning, N.; van Eijk, M.; Pouwels, W.; Brouwer, M.S.M.; Voehringer, D.; Huitinga, I.; Hoek, R.M.; Raes, G.; Hamann, J. Expression of the Inhibitory CD200 Receptor Is Associated with Alternative Macrophage Activation. J. Innate Immun. 2010, 2, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Xuan, W.; Qu, Q.; Zheng, B.; Xiong, S.; Fan, G. The Chemotaxis of M1 and M2 Macrophages Is Regulated by Different Chemokines. J. Leukoc. Biol. 2015, 97, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Ruytinx, P.; Proost, P.; Damme, J.V.; Struyf, S. Chemokine-Induced Macrophage Polarization in Inflammatory Conditions. Front. Immunol. 2018, 9, 1930. [Google Scholar] [CrossRef] [PubMed]
- Conti, I.; Rollins, B.J. CCL2 (Monocyte Chemoattractant Protein-1) and Cancer. Semin. Cancer Biol. 2004, 14, 149–154. [Google Scholar] [CrossRef]
- Biswas, S.K.; Mantovani, A. Macrophage Plasticity and Interaction with Lymphocyte Subsets: Cancer as a Paradigm. Nat. Immunol. 2010, 11, 889–896. [Google Scholar] [CrossRef]
- Scheller, J.; Garbers, C.; Rose-John, S. Interleukin-6: From Basic Biology to Selective Blockade of pro-Inflammatory Activities. Semin. Immunol. 2014, 26, 2–12. [Google Scholar] [CrossRef]
- Martinez, F.O.; Gordon, S. The M1 and M2 Paradigm of Macrophage Activation: Time for Reassessment. F1000prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef]
- Högger, P.; Dreier, J.; Droste, A.; Buck, F.; Sorg, C. Identification of the Integral Membrane Protein RM3/1 on Human Monocytes as a Glucocorticoid-Inducible Member of the Scavenger Receptor Cysteine-Rich Family (CD163). J. Immunol. 1998, 161, 1883–1890. [Google Scholar]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage Activation and Polarization: Nomenclature and Experimental Guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef]
- Faulknor, R.A.; Olekson, M.A.; Ekwueme, E.C.; Krzyszczyk, P.; Freeman, J.W.; Berthiaume, F. Hypoxia Impairs Mesenchymal Stromal Cell-Induced Macrophage M1 to M2 Transition. Technology 2017, 05, 81–86. [Google Scholar] [CrossRef]
- Zhao, G.; Liu, L.; Peek, R.M.; Hao, X.; Polk, D.B.; Li, H.; Yan, F. Activation of Epidermal Growth Factor Receptor in Macrophages Mediates Feedback Inhibition of M2 Polarization and Gastrointestinal Tumor Cell Growth. J. Biol. Chem. 2016, 291, 20462–20472. [Google Scholar] [CrossRef]
- Zhu, Y.; Yang, J.; Xu, D.; Gao, X.-M.; Zhang, Z.; Hsu, J.L.; Li, C.-W.; Lim, S.-O.; Sheng, Y.-Y.; Zhang, Y.; et al. Disruption of Tumour-Associated Macrophage Trafficking by the Osteopontin-Induced Colony-Stimulating Factor-1 Signalling Sensitises Hepatocellular Carcinoma to Anti-PD-L1 Blockade. Gut 2019, 68, 1653. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, H.; Wang, X.; Jiang, G.; Liu, H.; Zhang, G.; Wang, H.; Fang, R.; Bu, X.; Cai, S.; et al. TGF-β Induces M2-like Macrophage Polarization via SNAIL-Mediated Suppression of a pro-Inflammatory Phenotype. Oncotarget 2015, 7, 52294–52306. [Google Scholar] [CrossRef]
- Chen, L.; Wang, S.; Wang, Y.; Zhang, W.; Ma, K.; Hu, C.; Zhu, H.; Liang, S.; Liu, M.; Xu, N. IL-6 Influences the Polarization of Macrophages and the Formation and Growth of Colorectal Tumor. Oncotarget 2018, 9, 17443–17454. [Google Scholar] [CrossRef]
- Chuang, Y.; Hung, M.E.; Cangelose, B.K.; Leonard, J.N. Regulation of the IL-10-Driven Macrophage Phenotype under Incoherent Stimuli. Innate Immun. 2016, 22, 647–657. [Google Scholar] [CrossRef]
- Benner, B.; Scarberry, L.; Suarez-Kelly, L.P.; Duggan, M.C.; Campbell, A.R.; Smith, E.; Lapurga, G.; Jiang, K.; Butchar, J.P.; Tridandapani, S.; et al. Generation of Monocyte-Derived Tumor-Associated Macrophages Using Tumor-Conditioned Media Provides a Novel Method to Study Tumor-Associated Macrophages in Vitro. J. Immunother. Cancer 2019, 7, 140. [Google Scholar] [CrossRef]
- Lee, J.; Tam, H.; Adler, L.; Ilstad-Minnihan, A.; Macaubas, C.; Mellins, E.D. The MHC Class II Antigen Presentation Pathway in Human Monocytes Differs by Subset and Is Regulated by Cytokines. PLoS ONE 2017, 12, e0183594. [Google Scholar] [CrossRef]
- Wang, B.; Li, Q.; Qin, L.; Zhao, S.; Wang, J.; Chen, X. Transition of Tumor-Associated Macrophages from MHC Class IIhi to MHC Class IIlow Mediates Tumor Progression in Mice. BMC Immunol. 2011, 12, 43. [Google Scholar] [CrossRef]
- Wynn1, T.; Barron1, L.; Bataller, R.; Brenner, D.; Iredale, J.; Friedman, S.; Friedman, S.; Wallace, K.; Burt, A.; Wright, M.; et al. Macrophages: Master Regulators of Inflammation and Fibrosis. Semin. Liver Dis. 2010, 30, 245. [Google Scholar] [CrossRef]
- Kwak, T.; Wang, F.; Deng, H.; Condamine, T.; Kumar, V.; Perego, M.; Kossenkov, A.; Montaner, L.J.; Xu, X.; Xu, W.; et al. Distinct Populations of Immune-Suppressive Macrophages Differentiate from Monocytic Myeloid-Derived Suppressor Cells in Cancer. Cell Rep. 2020, 33, 108571. [Google Scholar] [CrossRef]
- Schelbergen, R.F.; Blom, A.B.; de Munter, W.; Vogl, T.; Roth, J.; van den Berg, W.B.; van Lent, P.L. Alarmins S100A8 and S100A9 Stimulate Production of Pro-Inflammatory Cytokines in M2 Macrophages without Changing Their M2 Membrane Phenotype. Ann. Rheum. Dis. 2012, 71, A76. [Google Scholar] [CrossRef]
- Riabov, V.; Yin, S.; Song, B.; Avdic, A.; Schledzewski, K.; Ovsiy, I.; Gratchev, A.; Verdiell, M.L.; Sticht, C.; Schmuttermaier, C.; et al. Stabilin-1 Is Expressed in Human Breast Cancer and Supports Tumor Growth in Mammary Adenocarcinoma Mouse Model. Oncotarget 2015, 7, 31097–31110. [Google Scholar] [CrossRef] [PubMed]
- Hollmén, M.; Figueiredo, C.R.; Jalkanen, S. New Tools to Prevent Cancer Growth and Spread: A ‘Clever’ Approach. Br. J. Cancer 2020, 123, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Wahyuningtyas, R.; Aui, S.; Chang, K. Autocrine VEGF Signalling on M2 Macrophages Regulates PD-L1 Expression for Immunomodulation of T Cells. J. Cell Mol. Med. 2019, 23, 1257–1267. [Google Scholar] [CrossRef]
- Wu, W.-K.; Llewellyn, O.P.C.; Bates, D.O.; Nicholson, L.B.; Dick, A.D. IL-10 Regulation of Macrophage VEGF Production Is Dependent on Macrophage Polarisation and Hypoxia. Immunobiology 2010, 215, 796–803. [Google Scholar] [CrossRef]
- Takeda, N.; O’Dea, E.L.; Doedens, A.; Kim, J.; Weidemann, A.; Stockmann, C.; Asagiri, M.; Simon, M.C.; Hoffmann, A.; Johnson, R.S. Differential Activation and Antagonistic Function of HIF-α Isoforms in Macrophages Are Essential for NO Homeostasis. Gene Dev. 2010, 24, 491–501. [Google Scholar] [CrossRef]
- Huangfu, N.; Zheng, W.; Xu, Z.; Wang, S.; Wang, Y.; Cheng, J.; Li, Z.; Cheng, K.; Zhang, S.; Chen, X.; et al. RBM4 Regulates M1 Macrophages Polarization through Targeting STAT1-Mediated Glycolysis. Int. Immunopharmacol. 2020, 83, 106432. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Myasoedova, V.A.; Revin, V.V.; Orekhov, A.N.; Bobryshev, Y.V. The Impact of Interferon-Regulatory Factors to Macrophage Differentiation and Polarization into M1 and M2. Immunobiology 2018, 223, 101–111. [Google Scholar] [CrossRef]
- Srivastava, M.; Saqib, U.; Naim, A.; Roy, A.; Liu, D.; Bhatnagar, D.; Ravinder, R.; Baig, M.S. The TLR4–NOS1–AP1 Signaling Axis Regulates Macrophage Polarization. Inflamm. Res. 2017, 66, 323–334. [Google Scholar] [CrossRef]
- Raes, G.; Baetselier, P.D.; Noël, W.; Beschin, A.; Brombacher, F.; Gh, G.H. Differential Expression of FIZZ1 and Ym1 in Alternatively versus Classically Activated Macrophages. J. Leukoc. Biol. 2002, 71, 597–602. [Google Scholar] [CrossRef]
- Welch, J.S.; Escoubet-Lozach, L.; Sykes, D.B.; Liddiard, K.; Greaves, D.R.; Glass, C.K. TH2 Cytokines and Allergic Challenge Induce Ym1 Expression in Macrophages by a STAT6-Dependent Mechanism. J. Biol. Chem. 2002, 277, 42821–42829. [Google Scholar] [CrossRef]
- Satoh, T.; Takeuchi, O.; Vandenbon, A.; Yasuda, K.; Tanaka, Y.; Kumagai, Y.; Miyake, T.; Matsushita, K.; Okazaki, T.; Saitoh, T.; et al. The Jmjd3-Irf4 Axis Regulates M2 Macrophage Polarization and Host Responses against Helminth Infection. Nat. Immunol. 2010, 11, 936–944. [Google Scholar] [CrossRef]
- Yin, Z.; Ma, T.; Lin, Y.; Lu, X.; Zhang, C.; Chen, S.; Jian, Z. IL-6/STAT3 Pathway Intermediates M1/M2 Macrophage Polarization during the Development of Hepatocellular Carcinoma. J. Cell Biochem. 2018, 119, 9419–9432. [Google Scholar] [CrossRef]
- Shima, T.; Shimoda, M.; Shigenobu, T.; Ohtsuka, T.; Nishimura, T.; Emoto, K.; Hayashi, Y.; Iwasaki, T.; Abe, T.; Asamura, H.; et al. Infiltration of Tumor-associated Macrophages Is Involved in Tumor Programmed Death-ligand 1 Expression in Early Lung Adenocarcinoma. Cancer Sci. 2020, 111, 727–738. [Google Scholar] [CrossRef]
- Xiong, H.; Mittman, S.; Rodriguez, R.; Moskalenko, M.; Pacheco-Sanchez, P.; Yang, Y.; Nickles, D.; Cubas, R. Anti-PD-L1 Treatment Results in Functional Remodeling of the Macrophage Compartment. Cancer Res. 2019, 79, 1493–1506. [Google Scholar] [CrossRef]
- Viola, A.; Munari, F.; Sánchez-Rodríguez, R.; Scolaro, T.; Castegna, A. The Metabolic Signature of Macrophage Responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef]
- Palsson-McDermott, E.M.; Curtis, A.M.; Goel, G.; Lauterbach, M.A.R.; Sheedy, F.J.; Gleeson, L.E.; van den Bosch, M.W.M.; Quinn, S.R.; Domingo-Fernandez, R.; Johnston, D.G.W.; et al. Pyruvate Kinase M2 Regulates Hif-1α Activity and IL-1β Induction and Is a Critical Determinant of the Warburg Effect in LPS-Activated Macrophages. Cell Metab. 2015, 21, 65–80. [Google Scholar] [CrossRef]
- Mazurek, S.; Boschek, C.B.; Hugo, F.; Eigenbrodt, E. Pyruvate Kinase Type M2 and Its Role in Tumor Growth and Spreading. Semin. Cancer Biol. 2005, 15, 300–308. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, P.; Wang, C.; Han, C.; Meng, J.; Liu, X.; Xu, S.; Li, N.; Wang, Q.; Shi, X.; et al. Immune Responsive Gene 1 (IRG1) Promotes Endotoxin Tolerance by Increasing A20 Expression in Macrophages through Reactive Oxygen Species. J. Biol. Chem. 2013, 288, 16225–16234. [Google Scholar] [CrossRef]
- Van den Bossche, J.; Baardman, J.; Otto, N.A.; van der Velden, S.; Neele, A.E.; van den Berg, S.M.; Luque-Martin, R.; Chen, H.-J.; Boshuizen, M.C.S.; Ahmed, M.; et al. Mitochondrial Dysfunction Prevents Repolarization of Inflammatory Macrophages. Cell Rep. 2016, 17, 684–696. [Google Scholar] [CrossRef]
- Wang, X.-F.; Wang, H.-S.; Wang, H.; Zhang, F.; Wang, K.-F.; Guo, Q.; Zhang, G.; Cai, S.-H.; Du, J. The Role of Indoleamine 2,3-Dioxygenase (IDO) in Immune Tolerance: Focus on Macrophage Polarization of THP-1 Cells. Cell Immunol. 2014, 289, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Cannon, M.J.; Ghosh, D.; Gujja, S. Signaling Circuits and Regulation of Immune Suppression by Ovarian Tumor-Associated Macrophages. Vaccines 2015, 3, 448–466. [Google Scholar] [CrossRef] [PubMed]
- Haschemi, A.; Kosma, P.; Gille, L.; Evans, C.R.; Burant, C.F.; Starkl, P.; Knapp, B.; Haas, R.; Schmid, J.A.; Jandl, C.; et al. The Sedoheptulose Kinase CARKL Directs Macrophage Polarization through Control of Glucose Metabolism. Cell Metab. 2012, 15, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, E.M.; Menga, A.; Martín-Pérez, R.; Quinto, A.; Riera-Domingo, C.; Tullio, G.D.; Hooper, D.C.; Lamers, W.H.; Ghesquière, B.; McVicar, D.W.; et al. Pharmacologic or Genetic Targeting of Glutamine Synthetase Skews Macrophages toward an M1-like Phenotype and Inhibits Tumor Metastasis. Cell Rep. 2017, 20, 1654–1666. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage Polarization: Tumor-Associated Macrophages as a Paradigm for Polarized M2 Mononuclear Phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef]
- Essandoh, K.; Li, Y.; Huo, J.; Fan, G.-C. MiRNA-Mediated Macrophage Polarization and Its Potential Role in the Regulation of Inflammatory Response. Shock 2016, 46, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Abraham, E. MicroRNAs in Immune Response and Macrophage Polarization. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 170–177. [Google Scholar] [CrossRef]
- Yang, S.; Li, J.; Chen, Y.; Zhang, S.; Feng, C.; Hou, Z.; Cai, J.; Wang, Y.; Hui, R.; Lv, B.; et al. MicroRNA-216a Promotes M1 Macrophages Polarization and Atherosclerosis Progression by Activating Telomerase via the Smad3/NF-ΚB Pathway. Biochim. Biophysica Acta BBA Mol. Basis Dis. 2019, 1865, 1772–1781. [Google Scholar] [CrossRef]
- ZHANG, Y.; ZHANG, M.; ZHONG, M.; SUO, Q.; LV, K. Expression Profiles of MiRNAs in Polarized Macrophages. Int. J. Mol. Med. 2013, 31, 797–802. [Google Scholar] [CrossRef]
- Huang, C.; Liu, X.; Qun, Z.; Xie, J.; Ma, T.; Meng, X.; Li, J. MiR-146a Modulates Macrophage Polarization by Inhibiting Notch1 Pathway in RAW264.7 Macrophages. Int. Immunopharmacol. 2016, 32, 46–54. [Google Scholar] [CrossRef]
- Dang, C.P.; Leelahavanichkul, A. Over-Expression of MiR-223 Induces M2 Macrophage through Glycolysis Alteration and Attenuates LPS-Induced Sepsis Mouse Model, the Cell-Based Therapy in Sepsis. PLoS ONE 2020, 15, e0236038. [Google Scholar] [CrossRef]
- Karo-Atar, D.; Itan, M.; Pasmanik-Chor, M.; Munitz, A. MicroRNA Profiling Reveals Opposing Expression Patterns for MiR-511 in Alternatively and Classically Activated Macrophages. J. Asthma 2014, 52, 545–553. [Google Scholar] [CrossRef]
- Nywening, T.M.; Wang-Gillam, A.; Sanford, D.E.; Belt, B.A.; Panni, R.Z.; Cusworth, B.M.; Toriola, A.T.; Nieman, R.K.; Worley, L.A.; Yano, M.; et al. Targeting Tumour-Associated Macrophages with CCR2 Inhibition in Combination with FOLFIRINOX in Patients with Borderline Resectable and Locally Advanced Pancreatic Cancer: A Single-Centre, Open-Label, Dose-Finding, Non-Randomised, Phase 1b Trial. Lancet Oncol. 2016, 17, 651–662. [Google Scholar] [CrossRef]
- Tacke, F. Cenicriviroc for the Treatment of Non-Alcoholic Steatohepatitis and Liver Fibrosis. Expert Opin. Investig. Drugs 2018, 27, 301–311. [Google Scholar] [CrossRef]
- Yumimoto, K.; Sugiyama, S.; Mimori, K.; Nakayama, K.I. Potentials of C-C Motif Chemokine 2–C-C Chemokine Receptor Type 2 Blockers Including Propagermanium as Anticancer Agents. Cancer Sci. 2019, 110, 2090–2099. [Google Scholar] [CrossRef]
- D’Incalci, M.; Badri, N.; Galmarini, C.M.; Allavena, P. Trabectedin, a Drug Acting on Both Cancer Cells and the Tumour Microenvironment. Br. J. Cancer 2014, 111, 646–650. [Google Scholar] [CrossRef]
- Germano, G.; Frapolli, R.; Belgiovine, C.; Anselmo, A.; Pesce, S.; Liguori, M.; Erba, E.; Uboldi, S.; Zucchetti, M.; Pasqualini, F.; et al. Role of Macrophage Targeting in the Antitumor Activity of Trabectedin. Cancer Cell 2013, 23, 249–262. [Google Scholar] [CrossRef]
- Guerriero, J.L.; Sotayo, A.; Ponichtera, H.E.; Castrillon, J.A.; Pourzia, A.L.; Schad, S.; Johnson, S.F.; Carrasco, R.D.; Lazo, S.; Bronson, R.T.; et al. Class IIa HDAC Inhibition Reduces Breast Tumours and Metastases through Anti-Tumour Macrophages. Nature 2017, 543, 428–432. [Google Scholar] [CrossRef]
- Mannaerts, I.; Eysackers, N.; Onyema, O.O.; Beneden, K.V.; Valente, S.; Mai, A.; Odenthal, M.; van Grunsven, L.A. Class II HDAC Inhibition Hampers Hepatic Stellate Cell Activation by Induction of MicroRNA. PLoS ONE 2013, 8, e55786. [Google Scholar] [CrossRef]
- Zeng, Y.; Li, B.; Liang, Y.; Reeves, P.M.; Qu, X.; Ran, C.; Liu, Q.; Callahan, M.V.; Sluder, A.E.; Gelfand, J.A.; et al. Dual Blockade of CXCL12-CXCR4 and PD-1–PD-L1 Pathways Prolongs Survival of Ovarian Tumor–Bearing Mice by Prevention of Immunosuppression in the Tumor Microenvironment. FASEB J. 2019, 33, 6596–6608. [Google Scholar] [CrossRef]
- Zhang, W.; Zhu, X.-D.; Sun, H.-C.; Xiong, Y.-Q.; Zhuang, P.-Y.; Xu, H.-X.; Kong, L.-Q.; Wang, L.; Wu, W.-Z.; Tang, Z.-Y. Depletion of Tumor-Associated Macrophages Enhances the Effect of Sorafenib in Metastatic Liver Cancer Models by Antimetastatic and Antiangiogenic Effects. Clin. Cancer Res. 2010, 16, 3420–3430. [Google Scholar] [CrossRef]
- Gladue, R.P.; Paradis, T.; Cole, S.H.; Donovan, C.; Nelson, R.; Alpert, R.; Gardner, J.; Natoli, E.; Elliott, E.; Shepard, R.; et al. The CD40 Agonist Antibody CP-870,893 Enhances Dendritic Cell and B-Cell Activity and Promotes Anti-Tumor Efficacy in SCID-Hu Mice. Cancer Immunol. Immunother. 2011, 60, 1009. [Google Scholar] [CrossRef]
- Piechutta, M.; Berghoff, A.S. New Emerging Targets in Cancer Immunotherapy: The Role of Cluster of Differentiation 40 (CD40/TNFR5). ESMO Open 2019, 4, e000510. [Google Scholar] [CrossRef]
- Ries, C.H.; Cannarile, M.A.; Hoves, S.; Benz, J.; Wartha, K.; Runza, V.; Rey-Giraud, F.; Pradel, L.P.; Feuerhake, F.; Klaman, I.; et al. Targeting Tumor-Associated Macrophages with Anti-CSF-1R Antibody Reveals a Strategy for Cancer Therapy. Cancer Cell 2014, 25, 846–859. [Google Scholar] [CrossRef]
- Papadopoulos, K.P.; Gluck, L.; Martin, L.P.; Olszanski, A.J.; Tolcher, A.W.; Ngarmchamnanrith, G.; Rasmussen, E.; Amore, B.M.; Nagorsen, D.; Hill, J.S.; et al. First-in-Human Study of AMG 820, a Monoclonal Anti-Colony-Stimulating Factor 1 Receptor Antibody, in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2017, 23, 5703–5710. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, M.M.; Messer, K.S.; Ralainirina, N.; Li, H.; Leem, C.J.; Gorjestani, S.; Woo, G.; Nguyen, A.V.; Figueiredo, C.C.; Foubert, P.; et al. PI3Kγ Is a Molecular Switch That Controls Immune Suppression. Nature 2016, 539, 437–442. [Google Scholar] [CrossRef]
- Willingham, S.B.; Volkmer, J.-P.; Gentles, A.J.; Sahoo, D.; Dalerba, P.; Mitra, S.S.; Wang, J.; Contreras-Trujillo, H.; Martin, R.; Cohen, J.D.; et al. The CD47-Signal Regulatory Protein Alpha (SIRPa) Interaction Is a Therapeutic Target for Human Solid Tumors. Proc. Natl. Acad. Sci. USA 2012, 109, 6662–6667. [Google Scholar] [CrossRef] [PubMed]
- Alvey, C.M.; Spinler, K.R.; Irianto, J.; Pfeifer, C.R.; Hayes, B.; Xia, Y.; Cho, S.; Dingal, P.C.P.D.; Hsu, J.; Smith, L.; et al. SIRPA-Inhibited, Marrow-Derived Macrophages Engorge, Accumulate, and Differentiate in Antibody-Targeted Regression of Solid Tumors. Curr. Biol. 2017, 27, 2065–2077. [Google Scholar] [CrossRef] [PubMed]
- Viitala, M.K.; Virtakoivu, R.; Tadayon, S.; Rannikko, J.; Jalkanen, S.; Hollmén, M. Immunotherapeutic Blockade of Macrophage Clever-1 Reactivates the CD8+ T Cell Response Against Immunosuppressive Tumors. Clin. Cancer Res. 2019, 25, 3289–3303. [Google Scholar] [CrossRef]
- Gordon, S.R.; Maute, R.L.; Dulken, B.W.; Hutter, G.; George, B.M.; McCracken, M.N.; Gupta, R.; Tsai, J.M.; Sinha, R.; Corey, D.; et al. PD-1 Expression by Tumour-Associated Macrophages Inhibits Phagocytosis and Tumour Immunity. Nature 2017, 545, 495–499. [Google Scholar] [CrossRef]
- Feng, Y.; Mu, R.; Wang, Z.; Xing, P.; Zhang, J.; Dong, L.; Wang, C. A Toll-like Receptor Agonist Mimicking Microbial Signal to Generate Tumor-Suppressive Macrophages. Nat. Commun. 2019, 10, 2272. [Google Scholar] [CrossRef]
- Anand, S.; Majeti, B.K.; Acevedo, L.M.; Murphy, E.A.; Mukthavaram, R.; Scheppke, L.; Huang, M.; Shields, D.J.; Lindquist, J.N.; Lapinski, P.E.; et al. MicroRNA-132–Mediated Loss of P120RasGAP Activates the Endothelium to Facilitate Pathological Angiogenesis. Nat. Med. 2010, 16, 909–914. [Google Scholar] [CrossRef]
- Baker, D.W.; Zhou, J.; Tsai, Y.-T.; Patty, K.M.; Weng, H.; Tang, E.N.; Nair, A.; Hu, W.-J.; Tang, L. Development of Optical Probes for in Vivo Imaging of Polarized Macrophages during Foreign Body Reactions. Acta Biomater. 2014, 10, 2945–2955. [Google Scholar] [CrossRef]
- Jager, N.A.; Westra, J.; Golestani, R.; van Dam, G.M.; Low, P.S.; Tio, R.A.; Slart, R.H.J.A.; Boersma, H.H.; Bijl, M.; Zeebregts, C.J. Folate Receptor-β Imaging Using 99mTc-Folate to Explore Distribution of Polarized Macrophage Populations in Human Atherosclerotic Plaque. J. Nucl. Med. 2014, 55, 1945–1951. [Google Scholar] [CrossRef]
- Zhou, J.; Tsai, Y.-T.; Weng, H.; Baker, D.W.; Tang, L. Real Time Monitoring of Biomaterial-Mediated Inflammatory Responses via Macrophage-Targeting NIR Nanoprobes. Biomaterials 2011, 32, 9383–9390. [Google Scholar] [CrossRef]
- Sun, J.Y.; Shen, J.; Thibodeaux, J.; Huang, G.; Wang, Y.; Gao, J.; Low, P.S.; Dimitrov, D.S.; Sumer, B.D. In Vivo Optical Imaging of Folate Receptor-β in Head and Neck Squamous Cell Carcinoma. Laryngoscope 2014, 124, E312–E319. [Google Scholar] [CrossRef]
- Sun, X.; Guo, L.; Shang, M.; Shi, D.; Liang, P.; Jing, X.; Meng, D.; Liu, X.; Zhou, X.; Zhao, Y.; et al. Ultrasound Mediated Destruction of LMW-HA-Loaded and Folate-Conjugated Nanobubble for TAM Targeting and Reeducation. Int. J. Nanomed. 2020, 15, 1967–1981. [Google Scholar] [CrossRef]
- Movahedi, K.; Schoonooghe, S.; Laoui, D.; Houbracken, I.; Waelput, W.; Breckpot, K.; Bouwens, L.; Lahoutte, T.; Baetselier, P.D.; Raes, G.; et al. Nanobody-Based Targeting of the Macrophage Mannose Receptor for Effective In Vivo Imaging of Tumor-Associated Macrophages. Cancer Res. 2012, 72, 4165–4177. [Google Scholar] [CrossRef]
- Bathula, N.V.; Bommadevara, H.; Hayes, J.M. Nanobodies: The Future of Antibody-Based Immune Therapeutics. Cancer Biother. Radiopharm 2020. [Google Scholar] [CrossRef]
- Keliher, E.J.; Yoo, J.; Nahrendorf, M.; Lewis, J.S.; Marinelli, B.; Newton, A.; Pittet, M.J.; Weissleder, R. 89 Zr-Labeled Dextran Nanoparticles Allow in Vivo Macrophage Imaging. Bioconjug. Chem. 2011, 22, 2383–2389. [Google Scholar] [CrossRef]
- Berr, L.W.; Mayo, M.W.; Yoo, A.D.; Williams, M.B.; Berr, S.S. PET Imaging of Tumor Associated Macrophages Using Mannose Coated 64Cu Liposomes. Biomaterials 2012, 33, 7785–7793. [Google Scholar] [CrossRef]
- Rodell, C.B.; Koch, P.D.; Weissleder, R. Screening for New Macrophage Therapeutics. Theranostics 2019, 9, 7714–7729. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Chandra, R.; Cuccarese, M.F.; Pfirschke, C.; Engblom, C.; Stapleton, S.; Adhikary, U.; Kohler, R.H.; Mohan, J.F.; Pittet, M.J.; et al. Radiation Therapy Primes Tumors for Nanotherapeutic Delivery via Macrophage-Mediated Vascular Bursts. Sci. Transl. Med. 2017, 9, eaal0225. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.S.C.; Garlin, M.A.; Weissleder, R.; Miller, M.A. Improving Nanotherapy Delivery and Action through Image-Guided Systems Pharmacology. Theranostics 2020, 10, 968–997. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Medina, C.; Tang, J.; Abdel-Atti, D.; Hogstad, B.; Merad, M.; Fisher, E.A.; Fayad, Z.A.; Lewis, J.S.; Mulder, W.J.M.; Reiner, T. PET Imaging of Tumor-Associated Macrophages with 89Zr-Labeled High-Density Lipoprotein Nanoparticles. J. Nucl. Med. 2015, 56, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Helfen, A.; Roth, J.; Ng, T.; Eisenblaetter, M. In Vivo Imaging of Pro- and Antitumoral Cellular Components of the Tumor Microenvironment. J. Nucl. Med. 2018, 59, 183–188. [Google Scholar] [CrossRef]
- Helfen, A.; Schnepel, A.; Rieß, J.; Stölting, M.; Gerwing, M.; Masthoff, M.; Vogl, T.; Roth, J.; Höltke, C.; Wildgruber, M.; et al. S100A9-Imaging Enables Estimation of Early Therapy-Mediated Changes in the Inflammatory Tumor Microenvironment. Biomedicines 2021, 9, 29. [Google Scholar] [CrossRef]
- Becker, A.; Hokamp, N.G.; Zenker, S.; Flores-Borja, F.; Barzcyk, K.; Varga, G.; Roth, J.; Geyer, C.; Heindel, W.; Bremer, C.; et al. Optical In Vivo Imaging of the Alarmin S100A9 in Tumor Lesions Allows for Estimation of the Individual Malignant Potential by Evaluation of Tumor–Host Cell Interaction. J. Nucl. Med. 2015, 56, 450–456. [Google Scholar] [CrossRef]
- Eisenblaetter, M.; Flores-Borja, F.; Lee, J.J.; Wefers, C.; Smith, H.; Hueting, R.; Cooper, M.S.; Blower, P.J.; Patel, D.; Rodriguez-Justo, M.; et al. Visualization of Tumor-Immune Interaction-Target-Specific Imaging of S100A8/A9 Reveals Pre-Metastatic Niche Establishment. Theranostics 2017, 7, 2392–2401. [Google Scholar] [CrossRef]
- Eisenblätter, M.; Ehrchen, J.; Varga, G.; Sunderkötter, C.; Heindel, W.; Roth, J.; Bremer, C.; Wall, A. In Vivo Optical Imaging of Cellular Inflammatory Response in Granuloma Formation Using Fluorescence-Labeled Macrophages. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2009, 50, 1676–1682. [Google Scholar] [CrossRef][Green Version]
- Wildgruber, M.; Lee, H.; Chudnovskiy, A.; Yoon, T.-J.; Etzrodt, M.; Pittet, M.J.; Nahrendorf, M.; Croce, K.; Libby, P.; Weissleder, R.; et al. Monocyte Subset Dynamics in Human Atherosclerosis Can Be Profiled with Magnetic Nano-Sensors. PLoS ONE 2009, 4, e5663. [Google Scholar] [CrossRef] [PubMed]
- Settles, M.; Etzrodt, M.; Kosanke, K.; Schiemann, M.; Zimmermann, A.; Meier, R.; Braren, R.; Huber, A.; Rummeny, E.J.; Weissleder, R.; et al. Different Capacity of Monocyte Subsets to Phagocytose Iron-Oxide Nanoparticles. PLoS ONE 2011, 6, e25197. [Google Scholar] [CrossRef]
- Shih, Y.I.; Hsu, Y.; Duong, T.Q.; Lin, S.; Chow, K.N.; Chang, C. Longitudinal Study of Tumor-associated Macrophages during Tumor Expansion Using MRI. NMR Biomed. 2011, 24, 1353–1360. [Google Scholar] [CrossRef]
- Leimgruber, A.; Berger, C.; Cortez-Retamozo, V.; Etzrodt, M.; Newton, A.P.; Waterman, P.; Figueiredo, J.L.; Kohler, R.H.; Elpek, N.; Mempel, T.R.; et al. Behavior of Endogenous Tumor-Associated Macrophages Assessed In Vivo Using a Functionalized Nanoparticle. Neoplasia 2009, 11, 459–468. [Google Scholar] [CrossRef]
- Mukherjee, S.; Sonanini, D.; Maurer, A.; Daldrup-Link, H.E. The Yin and Yang of Imaging Tumor Associated Macrophages with PET and MRI. Theranostics 2019, 9, 7730–7748. [Google Scholar] [CrossRef] [PubMed]
- Aghighi, M.; Theruvath, A.J.; Pareek, A.; Pisani, L.; Alford, R.; Muehe, A.M.; Sethi, T.K.; Holdsworth, S.J.; Hazard, F.K.; Gratzinger, D.; et al. Magnetic Resonance Imaging of Tumor Associated Macrophages: Clinical Translation. Clin. Cancer Res. 2018, 24. [Google Scholar] [CrossRef]
- Zanganeh, S.; Hutter, G.; Spitler, R.; Lenkov, O.; Mahmoudi, M.; Shaw, A.; Pajarinen, J.S.; Nejadnik, H.; Goodman, S.; Moseley, M.; et al. Iron Oxide Nanoparticles Inhibit Tumour Growth by Inducing Pro-Inflammatory Macrophage Polarization in Tumour Tissues. Nat. Nanotechnol. 2016, 11, 986–994. [Google Scholar] [CrossRef]
- Masthoff, M.; Buchholz, R.; Beuker, A.; Wachsmuth, L.; Kraupner, A.; Albers, F.; Freppon, F.; Helfen, A.; Gerwing, M.; Höltke, C.; et al. Introducing Specificity to Iron Oxide Nanoparticle Imaging by Combining 57 Fe-Based MRI and Mass Spectrometry. Nano Lett. 2019, 19, 7908–7917. [Google Scholar] [CrossRef]
- Niehoff , A.; Wachsmuth, L.; Schmid, F.; Sperling, M.; Faber, C.; Karst, U. Quantification of Manganese Enhanced Magnetic Resonance Imaging Based on Spatially Resolved Elemental Mass Spectrometry. ChemistrySelect 2016, 1, 264–266. [Google Scholar] [CrossRef]
- Masthoff, M.; Gran, S.; Zhang, X.; Wachsmuth, L.; Bietenbeck, M.; Helfen, A.; Heindel, W.; Sorokin, L.; Roth, J.; Eisenblätter, M.; et al. Temporal Window for Detection of Inflammatory Disease Using Dynamic Cell Tracking with Time-Lapse MRI. Sci. Rep. 2018, 8, 9563. [Google Scholar] [CrossRef]
- Melancon, M.P.; Lu, W.; Huang, Q.; Thapa, P.; Zhou, D.; Ng, C.; Li, C. Targeted Imaging of Tumor-Associated M2 Macrophages Using a Macromolecular Contrast Agent PG-Gd-NIR. Biomaterials 2010, 31, 6567–6573. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-Y.; Li, R.; Ng, T.S.C.; Courties, G.; Rodell, C.B.; Prytyskach, M.; Kohler, R.H.; Pittet, M.J.; Nahrendorf, M.; Weissleder, R.; et al. Quantitative Imaging of Tumor-Associated Macrophages and Their Response to Therapy Using 64 Cu-Labeled Macrin. ACS Nano 2018, 12, 12015–12029. [Google Scholar] [CrossRef] [PubMed]
- Martelli, C.; Dico, A.L.; Diceglie, C.; Lucignani, G.; Ottobrini, L. Optical Imaging Probes in Oncology. Oncotarget 2016, 7, 48753–48787. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Sarkar, S.; Yong, V.W.; Dunn, J.F. In Vivo MR Imaging of Tumor-Associated Macrophages: The Next Frontier in Cancer Imaging. Magn. Reson. Insights 2018, 11, 1178623X18771974. [Google Scholar] [CrossRef]
- Terry, S.Y.A.; Boerman, O.C.; Gerrits, D.; Franssen, G.M.; Metselaar, J.M.; Lehmann, S.; Oyen, W.J.G.; Gerdes, C.A.; Abiraj, K. 111In-Anti-F4/80-A3-1 Antibody: A Novel Tracer to Image Macrophages. Eur. J. Nucl. Med. Mol. I 2015, 42, 1430–1438. [Google Scholar] [CrossRef][Green Version]
- Rashidian, M.; Keliher, E.J.; Bilate, A.M.; Duarte, J.N.; Wojtkiewicz, G.R.; Jacobsen, J.T.; Cragnolini, J.; Swee, L.K.; Victora, G.D.; Weissleder, R.; et al. Noninvasive Imaging of Immune Responses. Proc. Natl. Acad. Sci. USA 2015, 112, 6146–6151. [Google Scholar] [CrossRef]
- Azad, A.K.; Rajaram, M.V.S.; Metz, W.L.; Cope, F.O.; Blue, M.S.; Vera, D.R.; Schlesinger, L.S. γ-Tilmanocept, a New Radiopharmaceutical Tracer for Cancer Sentinel Lymph Nodes, Binds to the Mannose Receptor (CD206). J. Immunol. 2015, 195, 2019–2029. [Google Scholar] [CrossRef]
- Blykers, A.; Schoonooghe, S.; Xavier, C.; D’hoe, K.; Laoui, D.; D’Huyvetter, M.; Vaneycken, I.; Cleeren, F.; Bormans, G.; Heemskerk, J.; et al. PET Imaging of Macrophage Mannose Receptor–Expressing Macrophages in Tumor Stroma Using 18F-Radiolabeled Camelid Single-Domain Antibody Fragments. J. Nucl. Med. 2015, 56, 1265–1271. [Google Scholar] [CrossRef]
- Eichendorff, S.; Svendsen, P.; Bender, D.; Keiding, S.; Christensen, E.I.; Deleuran, B.; Moestrup, S.K. Biodistribution and PET Imaging of a Novel [68Ga]-Anti-CD163-Antibody Conjugate in Rats with Collagen-Induced Arthritis and in Controls. Mol. Imaging Biol. 2015, 17, 87–93. [Google Scholar] [CrossRef]
- Unterrainer, M.; Mahler, C.; Vomacka, L.; Lindner, S.; Havla, J.; Brendel, M.; Böning, G.; Ertl-Wagner, B.; Kümpfel, T.; Milenkovic, V.M.; et al. TSPO PET with [18F]GE-180 Sensitively Detects Focal Neuroinflammation in Patients with Relapsing–Remitting Multiple Sclerosis. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 1423–1431. [Google Scholar] [CrossRef]
- Vicidomini, C.; Panico, M.; Greco, A.; Gargiulo, S.; Coda, A.R.D.; Zannetti, A.; Gramanzini, M.; Roviello, G.N.; Quarantelli, M.; Alfano, B.; et al. In Vivo Imaging and Characterization of [18F]DPA-714, a Potential New TSPO Ligand, in Mouse Brain and Peripheral Tissues Using Small-Animal PET. Nucl. Med. Biol. 2015, 42, 309–316. [Google Scholar] [CrossRef]
- Varnäs, K.; Cselényi, Z.; Jucaite, A.; Halldin, C.; Svenningsson, P.; Farde, L.; Varrone, A. PET Imaging of [11C]PBR28 in Parkinson’s Disease Patients Does Not Indicate Increased Binding to TSPO despite Reduced Dopamine Transporter Binding. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 367–375. [Google Scholar] [CrossRef]
- Betzel, T.; Müller, C.; Groehn, V.; Müller, A.; Reber, J.; Fischer, C.R.; Krämer, S.D.; Schibli, R.; Ametamey, S.M. Radiosynthesis and Preclinical Evaluation of 3′-Aza-2′-[ 18 F]Fluorofolic Acid: A Novel PET Radiotracer for Folate Receptor Targeting. Bioconjug. Chem. 2013, 24, 205–214. [Google Scholar] [CrossRef]
- He, H.; Chiu, A.C.; Kanada, M.; Schaar, B.T.; Krishnan, V.; Contag, C.H.; Dorigo, O. Imaging of Tumor-Associated Macrophages in a Transgenic Mouse Model of Orthotopic Ovarian Cancer. Mol. Imaging Biol. 2017, 19, 694–702. [Google Scholar] [CrossRef]
- Choi, Y.J.; Oh, S.-G.; Singh, T.D.; Ha, J.-H.; Kim, D.W.; Lee, S.W.; Jeong, S.Y.; Ahn, B.-C.; Lee, J.; Jeon, Y.H. Visualization of the Biological Behavior of Tumor-Associated Macrophages in Living Mice with Colon Cancer Using Multimodal Optical Reporter Gene Imaging. Neoplasia 2016, 18, 133–141. [Google Scholar] [CrossRef]
- Aalipour, A.; Chuang, H.-Y.; Murty, S.; D’Souza, A.L.; Park, S.; Gulati, G.S.; Patel, C.B.; Beinat, C.; Simonetta, F.; Martinić, I.; et al. Engineered Immune Cells as Highly Sensitive Cancer Diagnostics. Nat. Biotechnol. 2019, 37, 531–539. [Google Scholar] [CrossRef]

| TAM Recruitment | TAM Depletion | TAM Reprogramming |
|---|---|---|
| CCCR2-CCL2 inhibition [183,184,185] | Trabectedin [186,187] | Class IIa HDAC inhibitors [188,189] |
| CXCR4-CXCL12 inhibition [190] | Biphosphonates [121,191] | CD40 agonists [192,193] |
| Anti-CSF-1R [194,195] | PI3Kγ inhibitors [196] | |
| SIRPα inhibitors [197,198] | ||
| STAB1 inhibitors [199] | ||
| Checkpoint inhibitors [200] | ||
| TLRs agonists [201] | ||
| siRNA/miRNA [202] |
| Target Process/Molecule | Modality | Tracer |
|---|---|---|
| Phagocytosis | MRI, PET, optic, hybrid | Iron oxide nanoparticles [231], 64Cu-labeled polyglucose nanoparticle (macrin) [232], Cy5.5-VEGF [233], perfluorocarbon (PFC) [234] |
| Endocytosis | PET | 89Zr-PL-HDL, 89Zr-AI-HDL [215] |
| F4/80 | Flow cytometry, fluorescence microscopy | 111In-αF4/80-A3-1 mAb [235] |
| CD11b | PET | 18F-VHHDC13 [236] |
| MHC-II | PET | 18F-VHH7 [236] |
| CD206 | PET | γ-Tilmanocept [237], 18F-FB-anti-MMR 3.49 sdAb [238] |
| CD163 | PET | 68Ga-αCD163-mAb [239] |
| TSPO | PET | 18F-GE-180 [240], 18F-DPA-714 [241], 11C-PBR28 [242] |
| FR-ß | SPECT, PET | 3′-Aza-2′[18F]fluorofolic acid [243] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kimm, M.A.; Klenk, C.; Alunni-Fabbroni, M.; Kästle, S.; Stechele, M.; Ricke, J.; Eisenblätter, M.; Wildgruber, M. Tumor-Associated Macrophages—Implications for Molecular Oncology and Imaging. Biomedicines 2021, 9, 374. https://doi.org/10.3390/biomedicines9040374
Kimm MA, Klenk C, Alunni-Fabbroni M, Kästle S, Stechele M, Ricke J, Eisenblätter M, Wildgruber M. Tumor-Associated Macrophages—Implications for Molecular Oncology and Imaging. Biomedicines. 2021; 9(4):374. https://doi.org/10.3390/biomedicines9040374
Chicago/Turabian StyleKimm, Melanie A., Christopher Klenk, Marianna Alunni-Fabbroni, Sophia Kästle, Matthias Stechele, Jens Ricke, Michel Eisenblätter, and Moritz Wildgruber. 2021. "Tumor-Associated Macrophages—Implications for Molecular Oncology and Imaging" Biomedicines 9, no. 4: 374. https://doi.org/10.3390/biomedicines9040374
APA StyleKimm, M. A., Klenk, C., Alunni-Fabbroni, M., Kästle, S., Stechele, M., Ricke, J., Eisenblätter, M., & Wildgruber, M. (2021). Tumor-Associated Macrophages—Implications for Molecular Oncology and Imaging. Biomedicines, 9(4), 374. https://doi.org/10.3390/biomedicines9040374







