Cannabis sativa: Interdisciplinary Strategies and Avenues for Medical and Commercial Progression Outside of CBD and THC
Abstract
1. Introduction
2. The Endocannabinoid System and Its Associated Molecular Targets
2.1. An Overview of the Endocannabinoid System
2.2. The Expanded Cannabinoid System and Its Less Characterised Receptors
| Receptor | Cannabinoid | Disease/Interaction | Study Type | Reference |
|---|---|---|---|---|
| CB1 | Anandamide | Appetite | Murine models | [173,174] |
| Met-F-AEA | Thyroid cancer | in vitro human | [175] | |
| THCB (PA) | Pain | Murine models | [176] | |
| THC (PA) | Epilepsy | Murine models | [177] | |
| Sleep | Various studies | [178] | ||
| THCP (Ag) | Pain, anxiety, hypothermia, catalepsy | Murine models | [22] | |
| THCV (^) | Pain, anxiety, hypothermia, catalepsy | Murine models | [179,180] | |
| Parkinson’s disease | Murine models | [181] | ||
| Obesity | Murine models | [182] | ||
| Epilepsy | in vitro murine | [183] | ||
| THC, WIN55,212-2, CP55, 940 | Emesis | Animal models | [184,185,186,187,188] | |
| WIN55,212-2 | Parkinson’s disease | Murine model | [189] | |
| Prostate cancer | in vitro human | [190] | ||
| WIN55,212-2, JWH-133 | Breast, lung cancer | in vitro human | [191,192] | |
| CB2 | CBC (Ag) | Inflammation | in vitro models | [193] |
| CBG (PA) | Inflammatory bowel disease | Murine models | [194] | |
| HU-308, AM630 | Parkinson’s disease | Murine models | [195,196] | |
| THCP (Ag) | Pain, anxiety, hypothermia, catalepsy | Murine models | [22] | |
| THCV (^) | Inflammation | Murine models | [180] | |
| CB2 | THCV (^) | Parkinson’s disease | Murine models | [181] |
| Pain, anxiety, hypothermia, catalepsy | Murine models | [179] | ||
| WIN55,212-2 | Prostate cancer | in vitro human | [190] | |
| WIN55,212-2, JWH-133 | Breast, lung cancer | in vitro human | [191,192] | |
| GPR55 | Abnormal CBD | Parkinson’s disease | Murine models | [103] |
| GPR55 | Abnormal CBD | Pain/arthritis | Murine models | [105] |
| CBD (An) | Gastrointestinal disorders | Canine, murine models | [93,94,95,96] | |
| CBDV (An) | Rett syndrome | Murine models | [197] | |
| LPI inhibitor | in vitro | [198] | ||
| THC, anandamide, JWH015 | Pain | in vitro HEK239 | [97] | |
| TRPV1 | CBDV (Ag) | Anti-seizure | in vitro HEK239 | [199] |
| CBG (Ag), CBGV, CBD (Ag), CBDV (Ag), THCV (Ag) | Receptor desensitisation | in vitro HEK239 | [200] | |
| TRPV2 | CBD (Ag), CBGV, CBG (Ag), THCV (Ag), CBDV (Ag), CBN (Ag) | Receptor desensitisation | in vitro HEK239 | [200] |
| TRPV3 | CBGV, CBGA (Ag) | Receptor desensitisation | in vitro HEK239 | [201] |
| TRPV4 | CBGV, CBGA, CBN, CBG | Receptor desensitisation | in vitro HEK239 | [201] |
| TRPM8 | CBG (An), CBC (An), CBD (An), CBDV (An), THC (An), THCA (An) | Colorectal cancer | in vitro model | [200,202,203] |
| TRPA1 | CBC (Ag), CBN (Ag), THC (Ag), THCV (Ag), THCA (Ag), CBDA, CBG (Ag) | Receptor desensitisation | in vitro HEK239 | [200,202] |
| CBDV (Ag) | Ulcerative colitis | in vitro human | [204] | |
| Muscular dystrophy | in vitro studies | [205] |
2.3. Examples of the Potential Medicinal Use of Cannabinoids
3. The Cannabinoid and Terpene Pathways of Cannabis
4. Minor Cannabinoids and Their Biological Interactions
5. Directions in Cannabis Development for Secondary Metabolite Production
5.1. Next-Generation Sequencing of the Cannabis Plant and Its Potential for Genetic Manipulation
5.2. Synthetic Production of Cannabinoids
5.3. Phenotypic Parameters Affecting Cannabis Yield and Potency
5.4. Papaver somniferum: Potential Parallels for Future Cannabis Research
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Clarke, R.C.; Merlin, M.D. Letter to the Editor: Small, Ernest. 2015. Evolution and Classification of Cannabis sativa (Marijuana, Hemp) in Relation to Human Utilization. Bot. Rev. 2015, 81, 295–305. [Google Scholar] [CrossRef]
- Schultes, R.E.; Klein, W.M.; Plowman, T.; Lockwood, T.E. Cannabis: An Example of Taxonomic Neglect. Bot. Museum Leafl. Harvard Univ. 1974, 23, 337–367. [Google Scholar]
- Small, E. Evolution and Classification of Cannabis sativa (Marijuana, Hemp) in Relation to Human Utilization. Bot. Rev. 2015, 81, 189–294. [Google Scholar] [CrossRef]
- Small, E.; Cronquist, A. A Practical and Natural Taxonomy for Cannabis. Taxon 1976, 25, 405–435. [Google Scholar] [CrossRef]
- Small, E.; Naraine, S.G.U. Size Matters: Evolution of Large Drug-Secreting Resin Glands in Elite Pharmaceutical Strains of Cannabis sativa (Marijuana). Genet. Resour. Crop Evol. 2016, 254, 349–359. [Google Scholar] [CrossRef]
- Salentijn, E.M.J.; Zhang, Q.; Amaducci, S.; Yang, M.; Trindade, L.M. New Developments in Fiber Hemp (Cannabis sativa L.) Breeding. Ind. Crops Prod. 2015, 68, 32–41. [Google Scholar] [CrossRef]
- Mead, A. The Legal Status of Cannabis (Marijuana) and Cannabidiol (CBD) under U.S. Law. Epilepsy Behav. 2017, 70, 288–291. [Google Scholar] [CrossRef]
- Amaducci, S.; Scordia, D.; Liu, F.H.; Zhang, Q.; Guo, H.; Testa, G.; Cosentino, S.L. Key Cultivation Techniques for Hemp in Europe and China. Ind. Crop. Prod. 2015, 68, 2–16. [Google Scholar] [CrossRef]
- Bielecka, M.; Kaminski, F.; Adams, I.; Poulson, H.; Sloan, R.; Li, Y.; Larson, T.R.; Winzer, T.; Graham, I.A. Targeted Mutation of Δ12 and Δ15 Desaturase Genes in Hemp Produce Major Alterations in Seed Fatty Acid Composition Including a High Oleic Hemp Oil. Plant Biotechnol. J. 2014, 12, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Callaway, J.C.; Tennilä, T.; Pate, D.W. Occurence of “Omega 3” Stearidonic Acid (Cis-6,9,12,15-Ocadecatetraenoic Acid) in Hemp (Cannabis sativa L.) Seed. J. Int. Hemp Assoc. 1996, 3, 61–63. [Google Scholar]
- Dimić, E.; Romanić, R.; Vujasinović, V. Essential Fatty Acids, Nutritive Value and Oxidative Stability of Cold Pressed Hempseed (Cannabis sativa L.) Oil from Different Varieties. Acta Aliment. 2009, 38, 229–236. [Google Scholar] [CrossRef]
- Deferne, J.; Pate, D.W. Hemp Seed Oil: A Source of Valuable Essential Fatty Acids. J. Int. Hemp Assoc. 1996, 3, 4–7. [Google Scholar]
- Erasmus, U. Fats That Heal, Fats That Kill: The Complete Guide to Fats, Oils, Cholesterol, and Human Health, 3rd ed.; Alive Books: Burnaby, BC, Canada, 1993. [Google Scholar]
- Stubbs, C.D.; Smith, A.D. The Modification of Mammalian Membrane Polyunsaturated Fatty Acid Composition in Relation to Membrane Fluidity and Function. Biochim. Biophys. Acta 1984, 779, 89–137. [Google Scholar] [CrossRef]
- Callaway, J.C. Hempseed as a Nutritional Resource: An Overview. Euphytica 2004, 140, 65–72. [Google Scholar] [CrossRef]
- Harbige, L.S.; Layward, L.; Morris-Downes, M.M.; Dumonde, D.C.; Amor, S. The Protective Effects of Omega-6 Fatty Acids in Experimental Autoimmune Encephalomyelitis (EAE) in Relation to Transforming Growth Factor-Beta 1 (TGF-Β1) up-Regulation and Increased Prostaglandin E2 (PGE2) Production. Clin. Exp. Immunol. 2000, 122, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Prociuk, M.A.; Edel, A.L.; Richard, M.N.; Gavel, N.T.; Ander, B.P.; Dupasquier, C.M.C.; Pierce, G.N. Cholesterol-Induced Stimulation of Platelet Aggregation Is Prevented by a Hempseed-Enriched Diet. Can. J. Physiol. Pharmacol. 2008, 86, 153–159. [Google Scholar] [CrossRef]
- Clarke, R.C.; Merlin, M.D. Cannabis Domestication, Breeding History, Present-Day Genetic Diversity, and Future Prospects. CRC Crit. Rev. Plant Sci. 2016, 35, 293–327. [Google Scholar] [CrossRef]
- Dayanandan, P.; Kaufman, P.B. Trichomes of Cannabis sativa L. (Cannabaceae). Am. J. Bot. 1976, 63, 578–591. [Google Scholar] [CrossRef]
- ElSohly, M.A.; Slade, D. Chemical Constituents of Marijuana: The Complex Mixture of Natural Cannabinoids. Life Sci. 2005, 78, 539–548. [Google Scholar] [CrossRef]
- Pertwee, R.G. (Ed.) Handbook of Cannabis; Oxford University Press: Oxford, UK, 2015. [Google Scholar] [CrossRef]
- Citti, C.; Linciano, P.; Russo, F.; Luongo, L.; Iannotta, M.; Maione, S.; Laganà, A.; Capriotti, A.L.; Forni, F.; Vandelli, M.A.; et al. A Novel Phytocannabinoid Isolated from Cannabis sativa L. with an in Vivo Cannabimimetic Activity Higher than Δ9-Tetrahydrocannabinol: Δ9-Tetrahydrocannabiphorol. Sci. Rep. 2019, 9, 20335. [Google Scholar] [CrossRef] [PubMed]
- Babaei, M.; Ajdanian, L. Screening of Different Iranian Ecotypes of Cannabis under Water Deficit Stress. Sci. Hortic. 2020, 260. [Google Scholar] [CrossRef]
- Linger, P.; Ostwald, A.; Haensler, J. Cannabis sativa L. Growing on Heavy Metal Contaminated Soil: Growth, Cadmium Uptake and Photosynthesis. Biol. Plant. 2005, 49, 567–576. [Google Scholar] [CrossRef]
- Bouquet, R.J. Cannabis. Bull. Narc. 1950, 2, 14–30. [Google Scholar]
- Amaducci, S.; Zatta, A.; Raffanini, M.; Venturi, G. Characterisation of Hemp (Cannabis sativa L.) Roots under Different Growing Conditions. Plant Soil 2008, 313. [Google Scholar] [CrossRef]
- Small, E.; Marcus, D.; Butler, G.; McElroy, A.R. Apparent Increase in Biomass and Seed Productivity in Hemp (Cannabis sativa) Resulting from Branch Proliferation Caused by the European Corn Borer (Ostrinia nubilalis). J. Ind. Hemp 2007, 12, 15–26. [Google Scholar] [CrossRef]
- Government of Canada. Department of Justice. Available online: https://www.justice.gc.ca/eng/cj-jp/cannabis/ (accessed on 15 January 2020).
- Ney, L.J.; Matthews, A.; Bruno, R.; Felmingham, K.L. Cannabinoid Interventions for PTSD: Where to Next? Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2019, 93, 124–140. [Google Scholar] [CrossRef]
- Lattanzi, S.; Brigo, F.; Trinka, E.; Zaccara, G.; Cagnetti, C.; Del Giovane, C.; Silvestrini, M. Efficacy and Safety of Cannabidiol in Epilepsy: A Systematic Review and Meta-Analysis. Drugs 2018, 78, 1791–1804. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Hill, M.N.; Cheer, J.F.; Wotjak, C.T.; Holmes, A. The Endocannabinoid System as a Target for Novel Anxiolytic Drugs. Neurosci. Biobehav. Rev. 2017, 76, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Whiting, P.F.; Wolff, R.F.; Deshpande, S.; Di Nisio, M.; Duffy, S.; Hernandez, A.V.; Keurentjes, J.C.; Lang, S.; Misso, K.; Ryder, S.; et al. Cannabinoids for Medical Use: A Systematic Review and Meta-Analysis. JAMA J. Am. Med. Assoc. 2015, 313, 2456–2473. [Google Scholar] [CrossRef] [PubMed]
- Wade, D.T.; Collin, C.; Stott, C.; Duncombe, P. Meta-Analysis of the Efficacy and Safety of Sativex (Nabiximols), on Spasticity in People with Multiple Sclerosis. Mult. Scler. 2010, 16, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Stevens, A.J.; Higgins, M.D. A Systematic Review of the Analgesic Efficacy of Cannabinoid Medications in the Management of Acute Pain. Acta Anaesthesiol. Scand. 2017, 61, 268–280. [Google Scholar] [CrossRef]
- Morales, P.; Jagerovic, N. Novel Approaches and Current Challenges with Targeting the Endocannabinoid System. Expert Opin. Drug Discov. 2020, 15, 917–930. [Google Scholar] [CrossRef]
- Morales, P.; Hurst, D.P.; Reggio, P.H. Molecular Targets of the Phytocannabinoids: A Complex Picture. Prog. Chem. Org. Nat. Prod. 2017, 103, 103–131. [Google Scholar] [CrossRef]
- Sampson, P.B. Phytocannabinoid Pharmacology: Medicinal Properties of Cannabis sativa Constituents Aside from the “Big Two”. J. Nat. Prod. 2020, 84, 142–160. [Google Scholar] [CrossRef]
- Gaoni, Y.; Mechoulam, R. Isolation, Structure, and Partial Synthesis of an Active Constituent of Hashish. J. Am. Chem. Soc. 1964, 86, 1646–1647. [Google Scholar] [CrossRef]
- Mechoulam, R.; Gaoni, Y. A Total Synthesis of Dl-Δ1-Tetrahydrocannabinol, the Active Constituent of Hashish. J. Am. Chem. Soc. 1965, 87, 3273–3275. [Google Scholar] [CrossRef] [PubMed]
- Devane, W.A.; Dysarz, F.A.; Johnson, M.R.; Melvin, L.S.; Howlett, A.C. Determination and Characterization of a Cannabinoid Receptor in Rat Brain. Mol. Pharmacol. 1988, 34, 605–613. [Google Scholar]
- Munro, S.; Thomas, K.L.; Abu-Shaar, M. Molecular Characterization of a Peripheral Receptor for Cannabinoids. Nature 1993, 365, 61–65. [Google Scholar] [CrossRef]
- Herkenham, M.; Lynn, A.B.; Little, M.D.; Johnson, M.R.; Melvin, L.S.; De Costa, B.R.; Rice, K.C. Cannabinoid Receptor Localization in Brain. Proc. Natl. Acad. Sci. USA 1990, 87, 1932–1936. [Google Scholar] [CrossRef] [PubMed]
- Devane, W.A.; Hanuš, L.; Breuer, A.; Pertwee, R.G.; Stevenson, L.A.; Griffin, G.; Gibson, D.; Mandelbaum, A.; Etinger, A.; Mechoulam, R. Isolation and Structure of a Brain Constituent That Binds to the Cannabinoid Receptor. Science 1992, 258, 1946–1949. [Google Scholar] [CrossRef]
- Mechoulam, R.; Ben-Shabat, S.; Hanus, L.; Ligumsky, M.; Kaminski, N.E.; Schatz, A.R.; Gopher, A.; Almog, S.; Martin, B.R.; Compton, D.R.; et al. Identification of an Endogenous 2-Monoglyceride, Present in Canine Gut, That Binds to Cannabinoid Receptors. Biochem. Pharmacol. 1995, 50, 83–90. [Google Scholar] [CrossRef]
- Sugiura, T.; Kondo, S.; Sukagawa, A.; Nakane, S.; Shinoda, A.; Itoh, K.; Yamashita, A.; Waku, K. 2-Arachidonoylglycerol: A Possible Endogenous Cannabinoid Receptor Ligand in Brain. Biochem. Biophys. Res. Commun. 1995, 215, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, L.; Harvey-White, J.; Osei-Hyiaman, D.; Razdan, R.; Gong, Q.; Chan, A.C.; Zhou, Z.; Huang, B.X.; Kim, H.Y.; et al. A Biosynthetic Pathway for Anandamide. Proc. Natl. Acad. Sci. USA 2006, 103, 13345–13350. [Google Scholar] [CrossRef]
- Mechoulam, R.; Parker, L.A. The Endocannabinoid System and the Brain. Annu. Rev. Psychol. 2013, 64, 21–47. [Google Scholar] [CrossRef]
- Di Marzo, V. The Endocannabinoid System: Its General Strategy of Action, Tools for Its Pharmacological Manipulation and Potential Therapeutic Exploitation. Pharmacol. Res. 2009, 60, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Kano, M.; Ohno-Shosaku, T.; Hashimotodani, Y.; Uchigashima, M.; Watanabe, M. Endocannabinoid-Mediated Control of Synaptic Transmission. Physiol. Rev. 2009, 89, 309–380. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.K.P. A Perspective Review on Fatty Acid Amide Hydrolase (FAAH) Inhibitors as Potential Therapeutic Agents. Eur. J. Med. Chem. 2020, 188, 111953. [Google Scholar] [CrossRef] [PubMed]
- Ligresti, A.; Cascio, M.G.; Pryce, G.; Kulasegram, S.; Beletskaya, I.; De Petrocellis, L.; Saha, B.; Mahadevan, A.; Visintin, C.; Wiley, J.L.; et al. New Potent and Selective Inhibitors of Anandamide Reuptake with Antispastic Activity in a Mouse Model of Multiple Sclerosis. Br. J. Pharmacol. 2006, 147, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Glass, M.; Dragunow, M.; Faull, R.L.M. Cannabinoid Receptors in the Human Brain: A Detailed Anatomical and Quantitative Autoradiographic Study in the Fetal, Neonatal and Adult Human Brain. Neuroscience 1997, 77, 299–318. [Google Scholar] [CrossRef]
- Hua, T.; Vemuri, K.; Pu, M.; Qu, L.; Han, G.W.; Wu, Y.; Zhao, S.; Shui, W.; Li, S.; Korde, A.; et al. Crystal Structure of the Human Cannabinoid Receptor CB1. Cell 2016, 167, 750–762. [Google Scholar] [CrossRef]
- Li, X.; Hua, T.; Vemuri, K.; Ho, J.H.; Wu, Y.; Wu, L.; Popov, P.; Benchama, O.; Zvonok, N.; Locke, K.; et al. Crystal Structure of the Human Cannabinoid Receptor CB2. Cell 2019, 176, 459–467. [Google Scholar] [CrossRef]
- Katona, I.; Sperlágh, B.; Sík, A.; Käfalvi, A.; Vizi, E.S.; Mackie, K.; Freund, T.F. Presynaptically Located CB1 Cannabinoid Receptors Regulate GABA Release from Axon Terminals of Specific Hippocampal Interneurons. J. Neurosci. 1999, 19, 4544–4558. [Google Scholar] [CrossRef]
- Derkinderen, P.; Ledent, C.; Parmentier, M.; Girault, J.A. Cannabinoids Activate P38 Mitogen-Activated Protein Kinases through CB1 Receptors in Hippocampus. J. Neurochem. 2001, 77, 957–960. [Google Scholar] [CrossRef]
- Herkenham, M.; Lynn, A.B.; de Costa, B.R.; Richfield, E.K. Neuronal Localization of Cannabinoid Receptors in the Basal Ganglia of the Rat. Brain Res. 1991, 547, 267–274. [Google Scholar] [CrossRef]
- Mátyás, F.; Yanovsky, Y.; Mackie, K.; Kelsch, W.; Misgeld, U.; Freund, T.F. Subcellular Localization of Type 1 Cannabinoid Receptors in the Rat Basal Ganglia. Neuroscience 2006, 137, 337–361. [Google Scholar] [CrossRef]
- Bidaut-Russell, M.; Devane, W.A.; Howlett, A.C. Cannabinoid Receptors and Modulation of Cyclic AMP Accumulation in the Rat Brain. J. Neurochem. 1990, 55, 21–26. [Google Scholar] [CrossRef]
- Kawamura, Y.; Fukaya, M.; Maejima, T.; Yoshida, T.; Miura, E.; Watanabe, M.; Ohno-Shosaku, T.; Kano, M. The CB1 Cannabinoid Receptor Is the Major Cannabinoid Receptor at Excitatory Presynaptic Sites in the Hippocampus and Cerebellum. J. Neurosci. 2006, 26, 2991–3001. [Google Scholar] [CrossRef]
- Kreitzer, A.C.; Regehr, W.G. Cerebellar Depolarization-Induced Suppression of Inhibition Is Mediated by Endogenous Cannabinoids. J. Neurosci. 2001, 21, RC174. [Google Scholar] [CrossRef] [PubMed]
- Lévénès, C.; Daniel, H.; Soubrié, P.; Crépel, F. Cannabinoids Decrease Excitatory Synaptic Transmission and Impair Long-Term Depression in Rat Cerebellar Purkinje Cells. J. Physiol. 1998, 510, 867–879. [Google Scholar] [CrossRef]
- Ohno-Shosaku, T.; Maejima, T.; Kano, M. Endogenous Cannabinoids Mediate Retrograde Signals from Depolarized Postsynaptic Neurons to Presynaptic Terminals. Neuron 2001, 29, 729–738. [Google Scholar] [CrossRef]
- Ney, L.J.; Akhurst, J.; Bruno, R.; Laing, P.A.F.; Matthews, A.; Felmingham, K.L. Dopamine, Endocannabinoids and Their Interaction in Fear Extinction and Negative Affect in PTSD. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2021, 105, 110118. [Google Scholar] [CrossRef] [PubMed]
- Balsevich, G.; Petrie, G.N.; Hill, M.N. Endocannabinoids: Effectors of Glucocorticoid Signaling. Front. Neuroendocrinol. 2017, 47, 86–108. [Google Scholar] [CrossRef] [PubMed]
- González-Mariscal, I.; Krzysik-Walker, S.M.; Doyle, M.E.; Liu, Q.R.; Cimbro, R.; Santa-Cruz Calvo, S.; Ghosh, S.; Cieala, A.; Moaddel, R.; Carlson, O.D.; et al. Human CB1 Receptor Isoforms, Present in Hepatocytes and β-Cells, Are Involved in Regulating Metabolism. Sci. Rep. 2016, 6, 33302. [Google Scholar] [CrossRef]
- Nogueiras, R.; Veyrat-Durebex, C.; Suchanek, P.M.; Klein, M.; Tschöp, J.; Caldwell, C.; Woods, S.C.; Wittmann, G.; Watanabe, M.; Liposits, Z.; et al. Peripheral, but Not Central, CB1 Antagonism Provides Food Intake-Independent Metabolic Benefits in Diet-Induced Obese Rats. Diabetes 2008, 57, 2977–2991. [Google Scholar] [CrossRef]
- Ashton, J.C.; Friberg, D.; Darlington, C.L.; Smith, P.F. Expression of the Cannabinoid CB2 Receptor in the Rat Cerebellum: An Immunohistochemical Study. Neurosci. Lett. 2006, 396, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Núñez, E.; Benito, C.; Pazos, M.R.; Barbachano, A.; Fajardo, O.; González, S.; Tolón, R.M.; Romero, J. Cannabinoid CB2 Receptors Are Expressed by Perivascular Microglial Cells in the Human Brain: An Immunohistochemical Study. Synapse 2004, 53, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Ross, R.A.; Coutts, A.A.; McFarlane, S.M.; Anavi-Goffer, S.; Irving, A.J.; Pertwee, R.G.; MacEwan, D.J.; Scott, R.H. Actions of Cannabinoid Receptor Ligands on Rat Cultured Sensory Neurones: Implications for Antinociception. Neuropharmacology 2001, 40, 221–232. [Google Scholar] [CrossRef]
- Van Sickle, M.D.; Duncan, M.; Kingsley, P.J.; Mouihate, A.; Urbani, P.; Mackie, K.; Stella, N.; Makriyannis, A.; Piomelli, D.; Davison, J.S.; et al. Identification and Functional Characterization of Brainstem Cannabinoid CB2 Receptors. Science 2005, 310, 329–332. [Google Scholar] [CrossRef]
- Wotherspoon, G.; Fox, A.; McIntyre, P.; Colley, S.; Bevan, S.; Winter, J. Peripheral Nerve Injury Induces Cannabinoid Receptor 2 Protein Expression in Rat Sensory Neurons. Neuroscience 2005, 135, 235–245. [Google Scholar] [CrossRef]
- Galiègue, S.; Mary, S.; Marchand, J.; Dussossoy, D.; Carrière, D.; Carayon, P.; Bouaboula, M.; Shire, D.; LE Fur, G.; Casellas, P. Expression of Central and Peripheral Cannabinoid Receptors in Human Immune Tissues and Leukocyte Subpopulations. Eur. J. Biochem. 1995, 232, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Lynn, A.B.; Herkenham, M. Localization of Cannabinoid Receptors and Nonsaturable High-Density Cannabinoid Binding Sites in Peripheral Tissues of the Rat: Implications for Receptor-Mediated Immune Modulation by Cannabinoids. J. Pharmacol. Exp. Ther. 1994, 268, 1612–1623. [Google Scholar]
- Facci, L.; Dal Toso, R.; Romanello, S.; Buriani, A.; Skaper, S.D.; Leon, A. Mast Cells Express a Peripheral Cannabinoid Receptor with Differential Sensitivity to Anandamide and Palmitoylethanolamide. Proc. Natl. Acad. Sci. USA 1995, 92, 3376–3380. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.M.W.; Marczylo, T.H.; El-Talatini, M.; Finney, M.; Nallendran, V.; Taylor, A.H.; Konje, J.C. Ultra Performance Liquid Chromatography Tandem Mass Spectrometry Method for the Measurement of Anandamide in Human Plasma. Anal. Biochem. 2008, 380, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, F.; Di Lallo, V.D.; Belluomo, I.; De Iasio, R.; Baccini, M.; Casadio, E.; Gasparini, D.I.; Colavita, M.; Gambineri, A.; Grossi, G.; et al. Estimation of Reference Intervals of Five Endocannabinoids and Endocannabinoid Related Compounds in Human Plasma by Two Dimensional-LC/MS/MS. J. Lipid Res. 2012, 53, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Krumbholz, A.; Anielski, P.; Reisch, N.; Schelling, G.; Thieme, D. Diagnostic Value of Concentration Profiles of Glucocorticosteroids and Endocannabinoids in Hair. Ther. Drug Monit. 2013, 35, 600–607. [Google Scholar] [CrossRef]
- Mwanza, C.; Chen, Z.; Zhang, Q.; Chen, S.; Wang, W.; Deng, H. Simultaneous HPLC-APCI-MS/MS Quantification of Endogenous Cannabinoids and Glucocorticoids in Hair. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2016, 1028, 1–10. [Google Scholar] [CrossRef]
- Voegel, C.D.; Baumgartner, M.R.; Kraemer, T.; Wüst, S.; Binz, T.M. Simultaneous Quantification of Steroid Hormones and Endocannabinoids (ECs) in Human Hair Using an Automated Supported Liquid Extraction (SLE) and LC-MS/MS—Insights into EC Baseline Values and Correlation to Steroid Concentrations. Talanta 2021, 222, 121499. [Google Scholar] [CrossRef]
- Ney, L.J.; Felmingham, K.L.; Bruno, R.; Matthews, A.; Nichols, D.S. Simultaneous Quantification of Endocannabinoids, Oleoylethanolamide and Steroid Hormones in Human Plasma and Saliva. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2020, 1152, 122252. [Google Scholar] [CrossRef]
- Ney, L.J.; Stone, C.; Nichols, D.; Felmingham, K.L.; Bruno, R.; Matthews, A. Endocannabinoid Reactivity to Acute Stress: Investigation of the Relationship between Salivary and Plasma Levels. Biol. Psychol. 2021, 159, 108022. [Google Scholar] [CrossRef]
- Matias, I.; Gatta-Cherifi, B.; Tabarin, A.; Clark, S.; Leste-Lasserre, T.; Marsicano, G.; Piazza, P.V.; Cota, D. Endocannabinoids Measurement in Human Saliva as Potential Biomarker of Obesity. PLoS ONE 2012, 7, e42399. [Google Scholar] [CrossRef] [PubMed]
- Schuel, H.; Burkman, L.J.; Lippes, J.; Crickard, K.; Forester, E.; Piomelli, D.; Giuffrida, A. N-Acylethanolamines in Human Reproductive Fluids. Chem. Phys. Lipids 2002, 121, 211–227. [Google Scholar] [CrossRef]
- Lam, M.P.Y.; Siu, S.O.; Lau, E.; Mao, X.; Sun, H.Z.; Chiu, P.C.N.; Yeung, W.S.B.; Cox, D.M.; Chu, I.K. Online Coupling of Reverse-Phase and Hydrophilic Interaction Liquid Chromatography for Protein and Glycoprotein Characterization. Anal. Bioanal. Chem. 2010, 398, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.E.M.; Rapino, C.; Di Tommaso, M.; Pucci, M.; Battista, N.; Paro, R.; Simon, L.; Lutton, D.; Maccarrone, M. Differences in the Endocannabinoid System of Sperm from Fertile and Infertile Men. PLoS ONE 2012, 7, e47704. [Google Scholar] [CrossRef]
- Malan, T.P.; Ibrahim, M.M.; Deng, H.; Liu, Q.; Mata, H.P.; Vanderah, T.; Porreca, F.; Makriyannis, A. CB2 Cannabinoid Receptor-Mediated Peripheral Antinociception. Pain 2001, 93, 239–245. [Google Scholar] [CrossRef]
- Jaggar, S.I.; Hasnie, F.S.; Sellaturay, S.; Rice, A.S.C. The Anti-Hyperalgesic Actions of the Cannabinoid Anandamide and the Putative CB2 Receptor Agonist Palmitoylethanolamide in Visceral and Somatic Inflammatory Pain. Pain 1998, 76, 189–199. [Google Scholar] [CrossRef]
- Katona, I.; Freund, T.F. Multiple Functions of Endocannabinoid Signaling in the Brain. Annu. Rev. Neurosci. 2012, 35, 529–558. [Google Scholar] [CrossRef] [PubMed]
- Ligresti, A.; De Petrocellis, L.; Di Marzo, V. From Phytocannabinoids to Cannabinoid Receptors and Endocannabinoids: Pleiotropic Physiological and Pathological Roles through Complex Pharmacology. Physiol. Rev. 2016, 96, 1593–1659. [Google Scholar] [CrossRef]
- Ryberg, E.; Larsson, N.; Sjögren, S.; Hjorth, S.; Hermansson, N.O.; Leonova, J.; Elebring, T.; Nilsson, K.; Drmota, T.; Greasley, P.J. The Orphan Receptor GPR55 Is a Novel Cannabinoid Receptor. Br. J. Pharmacol. 2007, 152, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Fichna, J.; Schicho, R.; Saur, D.; Bashashati, M.; MacKie, K.; Li, Y.; Zimmer, A.; Göke, B.; Sharkey, K.A.; et al. A Role for O-1602 and G Protein-Coupled Receptor GPR55 in the Control of Colonic Motility in Mice. Neuropharmacology 2013, 71, 255–263. [Google Scholar] [CrossRef]
- Ross, G.R.; Lichtman, A.; Dewey, W.L.; Akbarali, H.I. Evidence for the Putative Cannabinoid Receptor (GPR55)-Mediated Inhibitory Effects on Intestinal Contractility in Mice. Pharmacology 2012, 90, 55–65. [Google Scholar] [CrossRef]
- Lin, X.H.; Yuece, B.; Li, Y.Y.; Feng, Y.J.; Feng, J.Y.; Yu, L.Y.; Li, K.; Li, Y.N.; Storr, M. A Novel CB Receptor GPR55 and Its Ligands Are Involved in Regulation of Gut Movement in Rodents. Neurogastroenterol. Motil. 2011, 23, 862-e342. [Google Scholar] [CrossRef] [PubMed]
- Galiazzo, G.; Giancola, F.; Stanzani, A.; Fracassi, F.; Bernardini, C.; Forni, M.; Pietra, M.; Chiocchetti, R. Localization of Cannabinoid Receptors CB1, CB2, GPR55, and PPARα in the Canine Gastrointestinal Tract. Histochem. Cell Biol. 2018, 150, 187–205. [Google Scholar] [CrossRef] [PubMed]
- Oka, S.; Nakajima, K.; Yamashita, A.; Kishimoto, S.; Sugiura, T. Identification of GPR55 as a Lysophosphatidylinositol Receptor. Biochem. Biophys. Res. Commun. 2007, 362, 928–934. [Google Scholar] [CrossRef] [PubMed]
- Lauckner, J.E.; Jensen, J.B.; Chen, H.Y.; Lu, H.C.; Hille, B.; Mackie, K. GPR55 Is a Cannabinoid Receptor That Increases Intracellular Calcium and Inhibits M Current. Proc. Natl. Acad. Sci. USA 2008, 105, 2699–2704. [Google Scholar] [CrossRef] [PubMed]
- Waldeck-Welermair, M.; Zoratti, C.; Osibow, K.; Balenga, N.; Goessnitzer, E.; Waldhoer, M.; Malli, R.; Graier, W.F. Integrin Clustering Enables Anandamide-Induced Ca2+ Signaling in Endothelial Cells via GPR55 by Protection against CB1-Receptor-Triggered Repression. J. Cell Sci. 2008, 121, 1704–1717. [Google Scholar] [CrossRef]
- Liu, Z.; Lee, J.; Krummey, S.; Lu, W.; Cai, H.; Lenardo, M.J. The Kinase LRRK2 Is a Regulator of the Transcription Factor NFAT That Modulates the Severity of Inflammatory Bowel Disease. Nat. Immunol. 2011, 12, 1063–1070. [Google Scholar] [CrossRef]
- Henstridge, C.M.; Balenga, N.A.; Schröder, R.; Kargl, J.K.; Platzer, W.; Martini, L.; Arthur, S.; Penman, J.; Whistler, J.L.; Kostenis, E.; et al. GPR55 Ligands Promote Receptor Coupling to Multiple Signalling Pathways. Br. J. Pharmacol. 2010, 160, 604–614. [Google Scholar] [CrossRef]
- Badrichani, A.Z.; Stroka, D.M.; Bilbao, G.; Curiel, D.T.; Bach, F.H.; Ferran, C. Bcl-2 and Bcl-X(L) Serve an Anti-Inflammatory Function in Endothelial Cells through Inhibition of NF-ΚB. J. Clin. Investig. 1999, 103, 543–553. [Google Scholar] [CrossRef]
- Wu, C.S.; Chen, H.; Sun, H.; Zhu, J.; Jew, C.P.; Wager-Miller, J.; Straiker, A.; Spencer, C.; Bradshaw, H.; Mackie, K.; et al. GPR55, a G-Protein Coupled Receptor for Lysophosphatidylinositol, Plays a Role in Motor Coordination. PLoS ONE 2013, 8, e60314. [Google Scholar] [CrossRef]
- Celorrio, M.; Rojo-Bustamante, E.; Fernández-Suárez, D.; Sáez, E.; Estella-Hermoso de Mendoza, A.; Müller, C.E.; Ramírez, M.J.; Oyarzábal, J.; Franco, R.; Aymerich, M.S. GPR55: A Therapeutic Target for Parkinson’s Disease? Neuropharmacology 2017, 125, 319–332. [Google Scholar] [CrossRef]
- Sawzdargo, M.; Nguyen, T.; Lee, D.K.; Lynch, K.R.; Cheng, R.; Heng, H.H.Q.; George, S.R.; O’Dowd, B.F. Identification and Cloning of Three Novel Human G Protein-Coupled Receptor Genes GPR52, ΨGPR53 and GPR55: GPR55 Is Extensively Expressed in Human Brain. Mol. Brain Res. 1999, 64, 193–198. [Google Scholar] [CrossRef]
- Schuelert, N.; McDougall, J.J. The Abnormal Cannabidiol Analogue O-1602 Reduces Nociception in a Rat Model of Acute Arthritis via the Putative Cannabinoid Receptor GPR55. Neurosci. Lett. 2011, 500, 72–76. [Google Scholar] [CrossRef]
- Deliu, E.; Sperow, M.; Console-Bram, L.; Carter, R.L.; Tilley, D.G.; Kalamarides, D.J.; Kirby, L.G.; Brailoiu, G.C.; Brailoiu, E.; Benamar, K.; et al. The Lysophosphatidylinositol Receptor GPR55 Modulates Pain Perception in the Periaqueductal Gray. Mol. Pharmacol. 2015, 88, 265–272. [Google Scholar] [CrossRef]
- Carey, L.M.; Gutierrez, T.; Deng, L.; Lee, W.H.; Mackie, K.; Hohmann, A.G. Inflammatory and Neuropathic Nociception Is Preserved in GPR55 Knockout Mice. Sci. Rep. 2017, 7, 944. [Google Scholar] [CrossRef]
- Gantz, I.; Muraoka, A.; Yang, Y.K.; Samuelson, L.C.; Zimmerman, E.M.; Cook, H.; Yamada, T. Cloning and Chromosomal Localization of a Gene (GPR18) Encoding a Novel Seven Transmembrane Receptor Highly Expressed in Spleen and Testis. Genomics 1997, 42, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Kohno, M.; Hasegawa, H.; Inoue, A.; Muraoka, M.; Miyazaki, T.; Oka, K.; Yasukawa, M. Identification of N-Arachidonylglycine as the Endogenous Ligand for Orphan G-Protein-Coupled Receptor GPR18. Biochem. Biophys. Res. Commun. 2006, 347, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Fukuoka, T.; Obata, K.; Yamanaka, H.; Dai, Y.; Tokunaga, A.; Noguchi, K. Distinct Expression of TRPM8, TRPA1, and TRPV1 MRNAs in Rat Primary Afferent Neurons with Aδ/C-Fibers and Colocalization with Trk Receptors. J. Comp. Neurol. 2005, 493, 596–606. [Google Scholar] [CrossRef]
- Cavanaugh, D.J.; Chesler, A.T.; Jackson, A.C.; Sigal, Y.M.; Yamanaka, H.; Grant, R.; O’Donnell, D.; Nicoll, R.A.; Shah, N.M.; Julius, D.; et al. Trpv1 Reporter Mice Reveal Highly Restricted Brain Distribution and Functional Expression in Arteriolar Smooth Muscle Cells. J. Neurosci. 2011, 31, 5067–5077. [Google Scholar] [CrossRef] [PubMed]
- Avelino, A.; Cruz, F. TRPV1 (Vanilloid Receptor) in the Urinary Tract: Expression, Function and Clinical Applications. Naunyn. Schmiedebergs. Arch. Pharmacol. 2006, 373, 287–299. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, H.; Wang, Z.; Mergler, S.; Wolosin, J.M.; Reinach, P.S. Functional TRPV1 Expression in Human Corneal Fibroblasts. Exp. Eye Res. 2013, 107, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Tóth, A.; Boczán, J.; Kedei, N.; Lizanecz, E.; Bagi, Z.; Papp, Z.; Édes, I.; Csiba, L.; Blumberg, P.M. Expression and Distribution of Vanilloid Receptor 1 (TRPV1) in the Adult Rat Brain. Mol. Brain Res. 2005, 135, 162–168. [Google Scholar] [CrossRef]
- Caterina, M.J.; Rosen, T.A.; Tominaga, M.; Brake, A.J.; Julius, D. A Capsaicin-Receptor Homologue with a High Threshold for Noxious Heat. Nature 1999, 398, 436–441. [Google Scholar] [CrossRef]
- Frederick, J.; Buck, M.E.; Matson, D.J.; Cortright, D.N. Increased TRPA1, TRPM8, and TRPV2 Expression in Dorsal Root Ganglia by Nerve Injury. Biochem. Biophys. Res. Commun. 2007, 358, 1058–1064. [Google Scholar] [CrossRef] [PubMed]
- Shimosato, G.; Amaya, F.; Ueda, M.; Tanaka, Y.; Decosterd, I.; Tanaka, M. Peripheral Inflammation Induces Up-Regulation of TRPV2 Expression in Rat DRG. Pain 2005, 119, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Saunders, C.I.; Kunde, D.A.; Crawford, A.; Geraghty, D.P. Expression of Transient Receptor Potential Vanilloid 1 (TRPV1) and 2 (TRPV2) in Human Peripheral Blood. Mol. Immunol. 2007, 44, 1429–1435. [Google Scholar] [CrossRef]
- Santoni, G.; Amantini, C.; Maggi, F.; Marinelli, O.; Santoni, M.; Nabissi, M.; Morelli, M.B. The TRPV2 Cation Channels: From Urothelial Cancer Invasiveness to Glioblastoma Multiforme Interactome Signature. Lab. Investig. 2020, 100, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Link, T.M.; Park, U.; Vonakis, B.M.; Raben, D.M.; Soloski, M.J.; Caterina, M.J. TRPV2 Has a Pivotal Role in Macrophage Particle Binding and Phagocytosis. Nat. Immunol. 2010, 11, 232–241. [Google Scholar] [CrossRef]
- Zhang, D.; Spielmann, A.; Wang, L.; Ding, G.; Huang, F.; Gu, Q.; Schwarz, W. Mast-Cell Degranulation Induced by Physical Stimuli Involves the Activation of Transient-Receptor-Potential Channel TRPV2. Physiol. Res. 2012, 61, 113–124. [Google Scholar] [CrossRef]
- Iwata, Y.; Ohtake, H.; Suzuki, O.; Matsuda, J.; Komamura, K.; Wakabayashi, S. Blockade of Sarcolemmal TRPV2 Accumulation Inhibits Progression of Dilated Cardiomyopathy. Cardiovasc. Res. 2013, 99, 760–768. [Google Scholar] [CrossRef] [PubMed]
- Lorin, C.; Vögeli, I.; Niggli, E. Dystrophic Cardiomyopathy: Role of TRPV2 Channels in Stretch-Induced Cell Damage. Cardiovasc. Res. 2015, 106, 153–162. [Google Scholar] [CrossRef]
- Iwata, Y.; Katanosaka, Y.; Arai, Y.; Shigekawa, M.; Wakabayashi, S. Dominant-Negative Inhibition of Ca2+ Influx via TRPV2 Ameliorates Muscular Dystrophy in Animal Models. Hum. Mol. Genet. 2009, 18, 84–834. [Google Scholar] [CrossRef]
- Iwata, Y.; Wakabayashi, S.; Ito, S.; Kitakaze, M. Production of TRPV2-Targeting Functional Antibody Ameliorating Dilated Cardiomyopathy and Muscular Dystrophy in Animal Models. Lab. Investig. 2020, 100, 324–337. [Google Scholar] [CrossRef]
- Hisanaga, E.; Nagasawa, M.; Ueki, K.; Kulkarni, R.N.; Mori, M.; Kojima, I. Regulation of Calcium-Permeable TRPV2 Channel by Insulin in Pancreatic β-Cells. Diabetes 2009, 58, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, M.; Zhang, Y.Q.; Mashima, H.; Li, L.; Shibata, H.; Kojima, I. Translocation of a Calcium-Permeable Cation Channel Induced by Insulin-like Growth Factor-I. Nat. Cell Biol. 1999, 1, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Aoyagi, K.; Ohara-Imaizumi, M.; Nishiwaki, C.; Nakamichi, Y.; Nagamatsu, S. Insulin/Phosphoinositide 3-Kinase Pathway Accelerates the Glucose-Induced First-Phase Insulin Secretion through TrpV2 Recruitment in Pancreatic β-Cells. Biochem. J. 2010, 432, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Caterina, M.J.; Schumacher, M.A.; Tominaga, M.; Rosen, T.A.; Levine, J.D.; Julius, D. The Capsaicin Receptor: A Heat-Activated Ion Channel in the Pain Pathway. Nature 1997, 389, 816–824. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.D.; Gunthorpe, M.J.; Kelsell, R.E.; Hayes, P.D.; Reilly, P.; Facer, P.; Wright, J.E.; Jerman, J.C.; Walhin, J.P.; Ooi, L.; et al. TRPV3 Is a Temperature-Sensitive Vanilloid Receptor-like Protein. Nature 2002, 418, 186–190. [Google Scholar] [CrossRef]
- Güler, A.D.; Lee, H.; Iida, T.; Shimizu, I.; Tominaga, M.; Caterina, M. Heat-Evoked Activation of the Ion Channel, TRPV4. J. Neurosci. 2002, 22, 6408–6414. [Google Scholar] [CrossRef]
- Xu, H.; Ramsey, I.S.; Kotecha, S.A.; Moran, M.M.; Chong, J.A.; Lawson, D.; Ge, P.; Lilly, J.; Silos-Santiago, I.; Xie, Y.; et al. TRPV3 Is a Calcium-Permeable Temperature-Sensitive Cation Channel. Nature 2002, 418, 181–186. [Google Scholar] [CrossRef]
- Moqrich, A.; Hwang, S.W.; Earley, T.J.; Petrus, M.J.; Murray, A.N.; Spencer, K.S.R.; Andahazy, M.; Story, G.M.; Patapoutian, A. Impaired Thermosensation in Mice Lacking TRPV3, a Heat and Camphor Sensor in the Skin. Science 2005, 307, 1468–1472. [Google Scholar] [CrossRef] [PubMed]
- Mandadi, S.; Sokabe, T.; Shibasaki, K.; Katanosaka, K.; Mizuno, A.; Moqrich, A.; Patapoutian, A.; Fukumi-Tominaga, T.; Mizumura, K.; Tominaga, M. TRPV3 in Keratinocytes Transmits Temperature Information to Sensory Neurons via ATP. Pflugers Arch. Eur. J. Physiol. 2009, 458, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.K.; Lee, H.; Mizuno, A.; Suzuki, M.; Caterina, M.J. TRPV3 and TRPV4 Mediate Warmth-Evoked Currents in Primary Mouse Keratinocytes. J. Biol. Chem. 2004, 279, 21569–21575. [Google Scholar] [CrossRef]
- Todaka, H.; Taniguchi, J.; Satoh, J.I.; Mizuno, A.; Suzuki, M. Warm Temperature-Sensitive Transient Receptor Potential Vanilloid 4 (TRPV4) Plays an Essential Role in Thermal Hyperalgesia. J. Biol. Chem. 2004, 279, 35133–35138. [Google Scholar] [CrossRef]
- Watanabe, H.; Vriens, J.; Suh, S.H.; Benham, C.D.; Droogmans, G.; Nilius, B. Heat-Evoked Activation of TRPV4 Channels in a HEK293 Cell Expression System and in Native Mouse Aorta Endothelial Cells. J. Biol. Chem. 2002, 277, 47044–47051. [Google Scholar] [CrossRef] [PubMed]
- Peier, A.M.; Reeve, A.J.; Andersson, D.A.; Moqrich, A.; Earley, T.J.; Hergarden, A.C.; Story, G.M.; Colley, S.; Hogenesch, J.B.; McIntyre, P.; et al. A Heat-Sensitive TRP Channel Expressed in Keratinocytes. Science 2002, 296, 2046–2049. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Delling, M.; Jun, J.C.; Clapham, D.E. Oregano, Thyme and Clove-Derived Flavors and Skin Sensitizers Activate Specific TRP Channels. Nat. Neurosci. 2006, 9, 628–635. [Google Scholar] [CrossRef]
- Bang, S.; Yoo, S.; Yang, T.J.; Cho, H.; Hwang, S.W. Isopentenyl Pyrophosphate Is a Novel Antinociceptive Substance That Inhibits TRPV3 and TRPA1 Ion Channels. Pain 2011, 152, 1156–1164. [Google Scholar] [CrossRef]
- Liu, X.; Bandyopadhyay, B.; Nakamoto, T.; Singh, B.; Liedtke, W.; Melvin, J.E.; Ambudkar, I. A Role for AQP5 in Activation of TRPV4 by Hypotonicity: Concerted Involvement of AQP5 and TRPV4 in Regulation of Cell Volume Recovery. J. Biol. Chem. 2006, 281, 15485–15495. [Google Scholar] [CrossRef]
- Becker, D.; Blase, C.; Bereiter-Hahn, J.; Jendrach, M. TRPV4 Exhibits a Functional Role in Cell-Volume Regulation. J. Cell Sci. 2005. [Google Scholar] [CrossRef]
- Vriens, J.; Watanabe, H.; Janssens, A.; Droogmans, G.; Voets, T.; Nilius, B. Cell Swelling, Heat, and Chemical Agonists Use Distinct Pathways for the Activation of the Cation Channel TRPV4. Proc. Natl. Acad. Sci. USA 2004, 101, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Liedtke, W.; Choe, Y.; Martí-Renom, M.A.; Bell, A.M.; Denis, C.S.; Šali, A.; Hudspeth, A.J.; Friedman, J.M.; Heller, S. Vanilloid Receptor-Related Osmotically Activated Channel (VR-OAC), a Candidate Vertebrate Osmoreceptor. Cell 2000, 103, 525–535. [Google Scholar] [CrossRef]
- Shibasaki, K.; Tominaga, M.; Ishizaki, Y. Hippocampal Neuronal Maturation Triggers Post-Synaptic Clustering of Brain Temperature-Sensor TRPV4. Biochem. Biophys. Res. Commun. 2015, 458, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Papadopoulos, P.; Hamel, E. Endothelial TRPV4 Channels Mediate Dilation of Cerebral Arteries: Impairment and Recovery in Cerebrovascular Pathologies Related to Alzheimer’s Disease. Br. J. Pharmacol. 2013, 170, 661–670. [Google Scholar] [CrossRef]
- Wissenbach, U.; Bödding, M.; Freichel, M.; Flockerzi, V. Trp12, a Novel Trp Related Protein from Kidney. FEBS Lett. 2000, 485, 127–134. [Google Scholar] [CrossRef]
- Liedtke, W.; Friedman, J.M. Abnormal Osmotic Regulation in Trpv4-/- Mice. Proc. Natl. Acad. Sci. USA 2003, 100, 13698–13703. [Google Scholar] [CrossRef] [PubMed]
- Heckel, E.; Boselli, F.; Roth, S.; Krudewig, A.; Belting, H.G.; Charvin, G.; Vermot, J. Oscillatory Flow Modulates Mechanosensitive Klf2a Expression through Trpv4 and Trpp2 during Heart Valve Development. Curr. Biol. 2015, 25, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Davis, J.B.; Smart, D.; Jerman, J.C.; Smith, G.D.; Hayes, P.; Vriens, J.; Cairns, W.; Wissenbach, U.; Prenen, J.; et al. Activation of TRPV4 Channels (HVRL-2/MTRP12) by Phorbol Derivatives. J. Biol. Chem. 2002, 277, 13569–13577. [Google Scholar] [CrossRef]
- Watanabe, H.; Vriens, J.; Prenen, J.; Droogmans, G.; Voets, T.; Nillus, B. Anandamide and Arachidonic Acid Use Epoxyeicosatrienoic Acids to Activate TRPV4 Channels. Nature 2003, 424, 434–438. [Google Scholar] [CrossRef] [PubMed]
- McKemy, D.D.; Neuhausser, W.M.; Julius, D. Identification of a Cold Receptor Reveals a General Role for TRP Channels in Thermosensation. Nature 2002, 416, 52–58. [Google Scholar] [CrossRef]
- Peier, A.M.; Moqrich, A.; Hergarden, A.C.; Reeve, A.J.; Andersson, D.A.; Story, G.M.; Earley, T.J.; Dragoni, I.; McIntyre, P.; Bevan, S.; et al. A TRP Channel That Senses Cold Stimuli and Menthol. Cell 2002, 108, 705–715. [Google Scholar] [CrossRef]
- Del Camino, D.; Murphy, S.; Heiry, M.; Barrett, L.B.; Earley, T.J.; Cook, C.A.; Petrus, M.J.; Zhao, M.; D’Amours, M.; Deering, N.; et al. TRPA1 Contributes to Cold Hypersensitivity. J. Neurosci. 2010, 30, 15165–15174. [Google Scholar] [CrossRef] [PubMed]
- Story, G.M.; Peier, A.M.; Reeve, A.J.; Eid, S.R.; Mosbacher, J.; Hricik, T.R.; Earley, T.J.; Hergarden, A.C.; Andersson, D.A.; Hwang, S.W.; et al. ANKTM1, a TRP-like Channel Expressed in Nociceptive Neurons, Is Activated by Cold Temperatures. Cell 2003, 112, 819–829. [Google Scholar] [CrossRef]
- Bandell, M.; Story, G.M.; Hwang, S.W.; Viswanath, V.; Eid, S.R.; Petrus, M.J.; Earley, T.J.; Patapoutian, A. Noxious Cold Ion Channel TRPA1 Is Activated by Pungent Compounds and Bradykinin. Neuron 2004, 41, 849–857. [Google Scholar] [CrossRef]
- Karashima, Y.; Talavera, K.; Everaerts, W.; Janssens, A.; Kwan, K.Y.; Vennekens, R.; Nilius, B.; Voets, T. TRPA1 Acts as a Cold Sensor in Vitro and in Vivo. Proc. Natl. Acad. Sci. USA 2009, 106, 1273–1278. [Google Scholar] [CrossRef]
- McNamara, C.R.; Mandel-Brehm, J.; Bautista, D.M.; Siemens, J.; Deranian, K.L.; Zhao, M.; Hayward, N.J.; Chong, J.A.; Julius, D.; Moran, M.M.; et al. TRPA1 Mediates Formalin-Induced Pain. Proc. Natl. Acad. Sci. USA 2007, 104, 13525–13530. [Google Scholar] [CrossRef] [PubMed]
- Jordt, S.E.; Bautista, D.M.; Chuang, H.H.; McKemy, D.D.; Zygmunt, P.M.; Högestätt, E.D.; Meng, I.D.; Julius, D. Mustard Oils and Cannabinoids Excite Sensory Nerve Fibres through the TRP Channel ANKTM1. Nature 2004, 427, 260–265. [Google Scholar] [CrossRef]
- Dai, Y.; Wang, S.; Tominaga, M.; Yamamoto, S.; Fukuoka, T.; Higashi, T.; Kobayashi, K.; Obata, K.; Yamanaka, H.; Noguchi, K. Sensitization of TRPA1 by PAR2 Contributes to the Sensation of Inflammatory Pain. J. Clin. Investig. 2007, 117, 1979–1987. [Google Scholar] [CrossRef] [PubMed]
- Nagata, K.; Duggan, A.; Kumar, G.; García-Añoveros, J. Nociceptor and Hair Cell Transducer Properties of TRPA1, a Channel for Pain and Hearing. J. Neurosci. 2005, 25, 4052–4061. [Google Scholar] [CrossRef]
- Bautista, D.M.; Jordt, S.E.; Nikai, T.; Tsuruda, P.R.; Read, A.J.; Poblete, J.; Yamoah, E.N.; Basbaum, A.I.; Julius, D. TRPA1 Mediates the Inflammatory Actions of Environmental Irritants and Proalgesic Agents. Cell 2006, 124, 1269–1282. [Google Scholar] [CrossRef]
- Kremeyer, B.; Lopera, F.; Cox, J.J.; Momin, A.; Rugiero, F.; Marsh, S.; Woods, C.G.; Jones, N.G.; Paterson, K.J.; Fricker, F.R.; et al. A Gain-of-Function Mutation in TRPA1 Causes Familial Episodic Pain Syndrome. Neuron 2010, 66, 671–680. [Google Scholar] [CrossRef]
- Xiong, W.; Cui, T.; Cheng, K.; Yang, F.; Chen, S.R.; Willenbring, D.; Guan, Y.; Pan, H.L.; Ren, K.; Xu, Y.; et al. Cannabinoids Suppress Inflammatory and Neuropathic Pain by Targeting Α3 Glycine Receptors. J. Exp. Med. 2012, 209, 1121–1134. [Google Scholar] [CrossRef]
- Hejazi, N.; Zhou, C.; Oz, M.; Sun, H.; Jiang, H.Y.; Zhang, L. Δ9-Tetrahydrocannabinol and Endogenous Cannabinoid Anandamide Directly Potentiate the Function of Glycine Receptors. Mol. Pharmacol. 2006, 69, 991–997. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Cheng, K.; Cui, T.; Godlewski, G.; Rice, K.C.; Xu, Y.; Zhang, L. Cannabinoid Potentiation of Glycine Receptors Contributes to Cannabis-Induced Analgesia. Nat. Chem. Biol. 2011, 7, 296–303. [Google Scholar] [CrossRef]
- O’Sullivan, S.E. An Update on PPAR Activation by Cannabinoids. Br. J. Pharmacol. 2016, 173, 1899–1910. [Google Scholar] [CrossRef]
- Russo, E.B.; Burnett, A.; Hall, B.; Parker, K.K. Agonistic Properties of Cannabidiol at 5-HT1a Receptors. Neurochem. Res. 2005, 30, 1037–1043. [Google Scholar] [CrossRef] [PubMed]
- Franklin, J.M.; Carrasco, G.A. Cannabinoid-Induced Enhanced Interaction and Protein Levels of Serotonin 5-HT2A and Dopamine D2 Receptors in Rat Prefrontal Cortex. J. Psychopharmacol. 2012, 26, 1333–1347. [Google Scholar] [CrossRef]
- Franklin, J.M.; Carrasco, G.A. Cannabinoid Receptor Agonists Upregulate and Enhance Serotonin 2A (5-HT2A) Receptor Activity via ERK1/2 Signaling. Synapse 2013, 67, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Hill, M.N.; Campolongo, P.; Yehuda, R.; Patel, S. Integrating Endocannabinoid Signaling and Cannabinoids into the Biology and Treatment of Posttraumatic Stress Disorder. Neuropsychopharmacology 2018, 43, 80–102. [Google Scholar] [CrossRef]
- Blessing, E.M.; Steenkamp, M.M.; Manzanares, J.; Marmar, C.R. Cannabidiol as a Potential Treatment for Anxiety Disorders. Neurotherapeutics 2015, 12, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Harrold, J.A.; Elliott, J.C.; King, P.J.; Widdowson, P.S.; Williams, G. Down-Regulation of Cannabinoid-1 (CB-1) Receptors in Specific Extrahypothalamic Regions of Rats with Dietary Obesity: A Role for Endogenous Cannabinoids in Driving Appetite for Palatable Food? Brain Res. 2002, 952, 232–238. [Google Scholar] [CrossRef]
- Jamshidi, N.; Taylor, D.A. Anandamide Administration into the Ventromedial Hypothalamus Stimulates Appetite in Rats. Br. J. Pharmacol. 2001, 134, 1151–1154. [Google Scholar] [CrossRef]
- Portella, G.; Laezza, C.; Laccetti, P.; De Petrocellis, L.; Di Marzo, V.; Bifulco, M. Inhibitory Effects of Cannabinoid CB1 Receptor Stimulation on Tumor Growth and Metastatic Spreading: Actions on Signals Involved in Angiogenesis and Metastasis. FASEB J. 2003, 17, 1771–1773. [Google Scholar] [CrossRef]
- Linciano, P.; Citti, C.; Luongo, L.; Belardo, C.; Maione, S.; Vandelli, M.A.; Forni, F.; Gigli, G.; Laganà, A.; Montone, C.M.; et al. Isolation of a High-Affinity Cannabinoid for the Human CB1 Receptor from a Medicinal Cannabis sativa Variety: Δ9-Tetrahydrocannabutol, the Butyl Homologue of Δ9-Tetrahydrocannabinol. J. Nat. Prod. 2020, 83, 88–98. [Google Scholar] [CrossRef]
- Wallace, M.J.; Blair, R.E.; Falenski, K.W.; Martin, B.R.; DeLorenzo, R.J. The Endogenous Cannabinoid System Regulates Seizure Frequency and Duration in a Model of Temporal Lobe Epilepsy. J. Pharmacol. Exp. Ther. 2003, 307, 129–137. [Google Scholar] [CrossRef]
- Murillo-Rodríguez, E. The Role of the CB1 Receptor in the Regulation of Sleep. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2008, 32, 1420–1427. [Google Scholar] [CrossRef]
- Zagzoog, A.; Mohamed, K.A.; Kim, H.J.J.; Kim, E.D.; Frank, C.S.; Black, T.; Jadhav, P.D.; Holbrook, L.A.; Laprairie, R.B. In Vitro and in Vivo Pharmacological Activity of Minor Cannabinoids Isolated from Cannabis sativa. Sci. Rep. 2020, 10, 20405. [Google Scholar] [CrossRef]
- Bolognini, D.; Costa, B.; Maione, S.; Comelli, F.; Marini, P.; Di Marzo, V.; Parolaro, D.; Ross, R.A.; Gauson, L.A.; Cascio, M.G.; et al. The Plant Cannabinoid Δ 9-Tetrahydrocannabivarin Can Decrease Signs of Inflammation and Inflammatory Pain in Mice. Br. J. Pharmacol. 2010, 160, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Espadas, I.; Keifman, E.; Palomo-Garo, C.; Burgaz, S.; García, C.; Fernández-Ruiz, J.; Moratalla, R. Beneficial Effects of the Phytocannabinoid Δ9-THCV in L-DOPA-Induced Dyskinesia in Parkinson’s Disease. Neurobiol. Dis. 2020, 141, 104892. [Google Scholar] [CrossRef] [PubMed]
- Wargent, E.T.; Zaibi, M.S.; Silvestri, C.; Hislop, D.C.; Stocker, C.J.; Stott, C.G.; Guy, G.W.; Duncan, M.; Di Marzo, V.; Cawthorne, M.A. The Cannabinoid Δ9-Tetrahydrocannabivarin (THCV) Ameliorates Insulin Sensitivity in Two Mouse Models of Obesity. Nutr. Diabetes 2013, 3, e68. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.J.; Weston, S.E.; Jones, N.A.; Smith, I.; Bevan, S.A.; Williamson, E.M.; Stephens, G.J.; Williams, C.M.; Whalley, B.J. 9-Tetrahydrocannabivarin Suppresses in Vitro Epileptiform and in Vivo Seizure Activity in Adult Rats. Epilepsia 2010, 51, 1522–1532. [Google Scholar] [CrossRef]
- Abrahamov, A.; Abrahamov, A.; Mechoulam, R. An Efficient New Cannabinoid Antiemetic in Pediatric Oncology. Life Sci. 1995, 56, 2097–2102. [Google Scholar] [CrossRef]
- Darmani, N.A. Δ9-Tetrahydrocannabinol and Synthetic Cannabinoids Prevent Emesis Produced by the Cannabinoid CB1 Receptor Antagonist/Inverse Agonist SR 141716A. Neuropsychopharmacology 2001, 24, 198–203. [Google Scholar] [CrossRef]
- Darmani, N.A. The Cannabinoid CB1 Receptor Antagonist SR 141716A Reverses the Antiemetic and Motor Depressant Actions of WIN 55, 212–2. Eur. J. Pharmacol. 2001, 430, 49–58. [Google Scholar] [CrossRef]
- Darmani, N.A.; Sim-Selley, L.J.; Martin, B.R.; Janoyan, J.J.; Crim, J.L.; Parekh, B.; Breivogel, C.S. Antiemetic and Motor-Depressive Actions of CP55,940: Cannabinoid CB1 Receptor Characterization, Distribution, and G-Protein Activation. Eur. J. Pharmacol. 2003, 459, 83–95. [Google Scholar] [CrossRef]
- Darmani, N.A.; Janoyan, J.J.; Crim, J.; Ramirez, J. Receptor Mechanism and Antiemetic Activity of Structurally-Diverse Cannabinoids against Radiation-Induced Emesis in the Least Shrew. Eur. J. Pharmacol. 2007, 563, 187–196. [Google Scholar] [CrossRef]
- Lastres-Becker, I.; Cebeira, M.; De Ceballos, M.L.; Zeng, B.Y.; Jenner, P.; Ramos, J.A.; Fernández-Ruiz, J.J. Increased Cannabinoid CB1 Receptor Binding and Activation of GTP-Binding Proteins in the Basal Ganglia of Patients with Parkinson’s Syndrome and of MPTP-Treated Marmosets. Eur. J. Neurosci. 2001, 14, 1827–1832. [Google Scholar] [CrossRef] [PubMed]
- Sarfaraz, S.; Afaq, F.; Adhami, V.M.; Mukhtar, H. Cannabinoid Receptor as a Novel Target for the Treatment of Prostate Cancer. Cancer Res. 2005, 65, 1635–1641. [Google Scholar] [CrossRef]
- Qamri, Z.; Preet, A.; Nasser, M.W.; Bass, C.E.; Leone, G.; Barsky, S.H.; Ganju, R.K. Synthetic Cannabinoid Receptor Agonists Inhibit Tumor Growth and Metastasis of Breast Cancer. Mol. Cancer Ther. 2009, 8, 3117–3129. [Google Scholar] [CrossRef]
- Preet, A.; Qamri, Z.; Nasser, M.W.; Prasad, A.; Shilo, K.; Zou, X.; Groopman, J.E.; Ganju, R.K. Cannabinoid Receptors, CB1 and CB2, as Novel Targets for Inhibition of Non-Small Cell Lung Cancer Growth and Metastasis. Cancer Prev. Res. 2011, 4, 65–75. [Google Scholar] [CrossRef]
- Izzo, A.A.; Capasso, R.; Aviello, G.; Borrelli, F.; Romano, B.; Piscitelli, F.; Gallo, L.; Capasso, F.; Orlando, P.; Di Marzo, V. Inhibitory Effect of Cannabichromene, a Major Non-Psychotropic Cannabinoid Extracted from Cannabis sativa, on Inflammation-Induced Hypermotility in Mice. Br. J. Pharmacol. 2012, 166, 1444–1460. [Google Scholar] [CrossRef]
- Borrelli, F.; Fasolino, I.; Romano, B.; Capasso, R.; Maiello, F.; Coppola, D.; Orlando, P.; Battista, G.; Pagano, E.; Di Marzo, V.; et al. Beneficial Effect of the Non-Psychotropic Plant Cannabinoid Cannabigerol on Experimental Inflammatory Bowel Disease. Biochem. Pharmacol. 2013, 85, 1306–1316. [Google Scholar] [CrossRef]
- Gómez-Gálvez, Y.; Palomo-Garo, C.; Fernández-Ruiz, J.; García, C. Potential of the Cannabinoid CB2 Receptor as a Pharmacological Target against Inflammation in Parkinson’s Disease. Prog. Neuropsychopharmacol. Biol. Psychiatry 2016, 64, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Javed, H.; Azimullah, S.; Haque, M.E.; Ojha, S.K. Cannabinoid Type 2 (CB2) Receptors Activation Protects against Oxidative Stress and Neuroinflammation Associated Dopaminergic Neurodegeneration in Rotenone Model of Parkinson’s Disease. Front. Neurosci. 2016, 10, 321. [Google Scholar] [CrossRef]
- Vigli, D.; Cosentino, L.; Raggi, C.; Laviola, G.; Woolley-Roberts, M.; De Filippis, B. Chronic Treatment with the Phytocannabinoid Cannabidivarin (CBDV) Rescues Behavioural Alterations and Brain Atrophy in a Mouse Model of Rett Syndrome. Neuropharmacology 2018, 140, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Anavi-Goffer, S.; Baillie, G.; Irving, A.J.; Gertsch, J.; Greig, I.R.; Pertwee, R.G.; Ross, R.A. Modulation of L-α-Lysophosphatidylinositol/GPR55 Mitogen-Activated Protein Kinase (MAPK) Signaling by Cannabinoids. J. Biol. Chem. 2012, 287, 91–104. [Google Scholar] [CrossRef]
- Iannotti, F.A.; Hill, C.L.; Leo, A.; Alhusaini, A.; Soubrane, C.; Mazzarella, E.; Russo, E.; Whalley, B.J.; Di Marzo, V.; Stephens, G.J. Nonpsychotropic Plant Cannabinoids, Cannabidivarin (CBDV) and Cannabidiol (CBD), Activate and Desensitize Transient Receptor Potential Vanilloid 1 (TRPV1) Channels in Vitro: Potential for the Treatment of Neuronal Hyperexcitability. ACS Chem. Neurosci. 2014, 5, 1131–1141. [Google Scholar] [CrossRef]
- De Petrocellis, L.; Ligresti, A.; Moriello, A.S.; Allarà, M.; Bisogno, T.; Petrosino, S.; Stott, C.G.; Di Marzo, V. Effects of Cannabinoids and Cannabinoid-Enriched Cannabis Extracts on TRP Channels and Endocannabinoid Metabolic Enzymes. Br. J. Pharmacol. 2011, 163, 1479–1494. [Google Scholar] [CrossRef]
- De Petrocellis, L.; Orlando, P.; Moriello, A.S.; Aviello, G.; Stott, C.; Izzo, A.A.; di Marzo, V. Cannabinoid Actions at TRPV Channels: Effects on TRPV3 and TRPV4 and Their Potential Relevance to Gastrointestinal Inflammation. Acta Physiol. 2012, 204, 255–266. [Google Scholar] [CrossRef]
- De Petrocellis, L.; Vellani, V.; Schiano-Moriello, A.; Marini, P.; Magherini, P.C.; Orlando, P.; Di Marzo, V. Plant-Derived Cannabinoids Modulate the Activity of Transient Receptor Potential Channels of Ankyrin Type-1 and Melastatin Type-8. J. Pharmacol. Exp. Ther. 2008, 325, 1007–1015. [Google Scholar] [CrossRef]
- Borrelli, F.; Pagano, E.; Romano, B.; Panzera, S.; Maiello, F.; Coppola, D.; De Petrocellis, L.; Buono, L.; Orlando, P.; Izzo, A.A. Colon Carcinogenesis Is Inhibited by the TRPM8 Antagonist Cannabigerol, a Cannabis-Derived Non-Psychotropic Cannabinoid. Carcinogenesis 2014, 35, 2787–2797. [Google Scholar] [CrossRef] [PubMed]
- Pagano, E.; Romano, B.; Iannotti, F.A.; Parisi, O.A.; D’Armiento, M.; Pignatiello, S.; Coretti, L.; Lucafò, M.; Venneri, T.; Stocco, G.; et al. The Non-Euphoric Phytocannabinoid Cannabidivarin Counteracts Intestinal Inflammation in Mice and Cytokine Expression in Biopsies from UC Pediatric Patients. Pharmacol. Res. 2019, 149, 104464. [Google Scholar] [CrossRef]
- Iannotti, F.A.; Pagano, E.; Moriello, A.S.; Alvino, F.G.; Sorrentino, N.C.; D’Orsi, L.; Gazzerro, E.; Capasso, R.; De Leonibus, E.; De Petrocellis, L.; et al. Effects of Non-Euphoric Plant Cannabinoids on Muscle Quality and Performance of Dystrophic Mdx Mice. Br. J. Pharmacol. 2019, 176, 1568–1584. [Google Scholar] [CrossRef]
- García-Arencibia, M.; González, S.; de Lago, E.; Ramos, J.A.; Mechoulam, R.; Fernández-Ruiz, J. Evaluation of the Neuroprotective Effect of Cannabinoids in a Rat Model of Parkinson’s Disease: Importance of Antioxidant and Cannabinoid Receptor-Independent Properties. Brain Res. 2007, 1134, 162–170. [Google Scholar] [CrossRef]
- Di Marzo, V. Enhanced Levels of Endogenous Cannabinoids in the Globus Pallidus Are Associated with a Reduction in Movement in an Animal Model of Parkinson’s Disease. FASEB J. 2000, 14, 1432–1438. [Google Scholar] [CrossRef] [PubMed]
- Fox, S.H.; Henry, B.; Hill, M.; Crossman, A.; Brotchie, J. Stimulation of Cannabinoid Receptors Reduces Levodopa-Induced Dyskinesia in the MPTP-Lesioned Nonhuman Primate Model of Parkinson’s Disease. Mov. Disord. 2002, 17, 1180–1187. [Google Scholar] [CrossRef]
- Morgese, M.G.; Cassano, T.; Cuomo, V.; Giuffrida, A. Anti-Dyskinetic Effects of Cannabinoids in a Rat Model of Parkinson’s Disease: Role of CB1 and TRPV1 Receptors. Exp. Neurol. 2007, 208, 110–119. [Google Scholar] [CrossRef]
- Sañudo-Peña, M.C.; Patrick, S.L.; Khen, S.; Patrick, R.L.; Tsou, K.; Walker, J.M. Cannabinoid Effects in Basal Ganglia in a Rat Model of Parkinson’s Disease. Neurosci. Lett. 1998, 248, 171–174. [Google Scholar] [CrossRef]
- Donadelli, M.; Dando, I.; Zaniboni, T.; Costanzo, C.; Dalla Pozza, E.; Scupoli, M.T.; Scarpa, A.; Zappavigna, S.; Marra, M.; Abbruzzese, A.; et al. Gemcitabine/Cannabinoid Combination Triggers Autophagy in Pancreatic Cancer Cells through a ROS-Mediated Mechanism. Cell Death Dis. 2011, 2, E152. [Google Scholar] [CrossRef]
- Afrin, F.; Chi, M.; Eamens, A.L.; Duchatel, R.J.; Douglas, A.M.; Schneider, J.; Gedye, C.; Woldu, A.S.; Dun, M.D. Can Hemp Help? Low-THC Cannabis and Non-THC Cannabinoids for the Treatment of Cancer. Cancers 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- De Petrocellis, L.; Melck, D.; Palmisano, A.; Bisogno, T.; Laezza, C.; Bifulco, M.; Di Marzo, V. The Endogenous Cannabinoid Anandamide Inhibits Human Breast Cancer Cell Proliferation. Proc. Natl. Acad. Sci. USA 1998, 95, 8375–8380. [Google Scholar] [CrossRef]
- Cianchi, F.; Papucci, L.; Schiavone, N.; Lulli, M.; Magnelli, L.; Vinci, M.C.; Messerini, L.; Manera, C.; Ronconi, E.; Romagnani, P.; et al. Cannabinoid Receptor Activation Induces Apoptosis through Tumor Necrosis Factor α-Mediated Ceramide de Novo Synthesis in Colon Cancer Cells. Clin. Cancer Res. 2008, 14, 7691–7700. [Google Scholar] [CrossRef]
- Blázquez, C.; Casanova, M.L.; Planas, A.; Del Pulgar, T.G.; Villanueva, C.; Fernández-Aceñero, M.J.; Aragonés, J.; Huffman, J.W.; Jorcano, J.L.; Guzmán, M. Inhibition of Tumor Angiogenesis by Cannabinoids. FASEB J. 2003, 17, 529–531. [Google Scholar] [CrossRef] [PubMed]
- Cunha, J.M.; Carlini, E.A.; Pereira, A.E.; Ramos, O.L.; Pimentel, C.; Gagliardi, R.; Sanvito, W.L.; Lander, N.; Mechoulam, R. Chronic Administration of Cannabidiol to Healthy Volunteers and Epileptic Patients. Pharmacology 1980, 21, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.C.; Tsien, R.W.; Whalley, B.J.; Devinsky, O. Cannabinoids and Epilepsy. Neurotherapeutics 2015, 12, 747–768. [Google Scholar] [CrossRef] [PubMed]
- Stockings, E.; Zagic, D.; Campbell, G.; Weier, M.; Hall, W.D.; Nielsen, S.; Herkes, G.K.; Farrell, M.; Degenhardt, L. Evidence for Cannabis and Cannabinoids for Epilepsy: A Systematic Review of Controlled and Observational Evidence. J. Neurol. Neurosurg. Psychiatry 2018, 89, 741–753. [Google Scholar] [CrossRef]
- Karanian, D.A.; Karim, S.L.; Wood, J.A.T.; Williams, J.S.; Lin, S.; Makriyannis, A.; Bahr, B.A. Endocannabinoid Enhancement Protects against Kainic Acid-Induced Seizures and Associated Brain Damage. J. Pharmacol. Exp. Ther. 2007, 322, 1059–1066. [Google Scholar] [CrossRef]
- Naidoo, V.; Karanian, D.A.; Vadivel, S.K.; Locklear, J.R.; Wood, J.A.T.; Nasr, M.; Quizon, P.M.P.; Graves, E.E.; Shukla, V.; Makriyannis, A.; et al. Equipotent Inhibition of Fatty Acid Amide Hydrolase and Monoacylglycerol Lipase—Dual Targets of the Endocannabinoid System to Protect against Seizure Pathology. Neurotherapeutics 2012, 9, 801–813. [Google Scholar] [CrossRef]
- Devinsky, O.; Patel, A.D.; Cross, J.H.; Villanueva, V.; Wirrell, E.C.; Privitera, M.; Greenwood, S.M.; Roberts, C.; Checketts, D.; VanLandingham, K.E.; et al. Effect of Cannabidiol on Drop Seizures in the Lennox–Gastaut Syndrome. N. Engl. J. Med. 2018, 378, 1888–1897. [Google Scholar] [CrossRef]
- Devinsky, O.; Cross, J.H.; Laux, L.; Marsh, E.; Miller, I.; Nabbout, R.; Scheffer, I.E.; Thiele, E.A.; Wright, S. Trial of Cannabidiol for Drug-Resistant Seizures in the Dravet Syndrome. N. Engl. J. Med. 2017, 376, 2011–2020. [Google Scholar] [CrossRef] [PubMed]
- Press, C.A.; Knupp, K.G.; Chapman, K.E. Parental Reporting of Response to Oral Cannabis Extracts for Treatment of Refractory Epilepsy. Epilepsy Behav. 2015, 45, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Guggenhuber, S.; Monory, K.; Lutz, B.; Klugmann, M. AAV Vector-Mediated Overexpression of CB1 Cannabinoid Receptor in Pyramidal Neurons of the Hippocampus Protects against Seizure-Induced Excitoxicity. PLoS ONE 2010, 5, e15707. [Google Scholar] [CrossRef] [PubMed]
- Riva, N.; Mora, G.; Sorarù, G.; Lunetta, C.; Ferraro, O.E.; Falzone, Y.; Leocani, L.; Fazio, R.; Comola, M.; Comi, G.; et al. Safety and Efficacy of Nabiximols on Spasticity Symptoms in Patients with Motor Neuron Disease (CANALS): A Multicentre, Double-Blind, Randomised, Placebo-Controlled, Phase 2 Trial. Lancet Neurol. 2019, 18, 155–164. [Google Scholar] [CrossRef]
- Beal, J.E.; Olson, R.; Laubenstein, L.; Morales, J.O.; Bellman, P.; Yangco, B.; Lefkowitz, L.; Plasse, T.F.; Shepard, K.V. Dronabinol as a Treatment for Anorexia Associated with Weight Loss in Patients with AIDS. J. Pain Symptom Manag. 1995, 10, 89–97. [Google Scholar] [CrossRef]
- Foltin, R.W.; Fischman, M.W.; Byrne, M.F. Effects of Smoked Marijuana on Food Intake and Body Weight of Humans Living in a Residential Laboratory. Appetite 1988, 11, 1–14. [Google Scholar] [CrossRef]
- Mattes, R.D.; Engelman, K.; Shaw, L.M.; Elsohly, M.A. Cannabinoids and Appetite Stimulation. Pharmacol. Biochem. Behav. 1994, 49, 187–195. [Google Scholar] [CrossRef]
- Williams, C.M.; Rogers, P.J.; Kirkham, T.C. Hyperphagia in Pre-Fed Rats Following Oral Δ9-THC. Physiol. Behav. 1998, 65, 343–346. [Google Scholar] [CrossRef]
- Feinberg, I.; Jones, R.; Walker, J.M.; Cavness, C.; March, J. Effects of High Dosage Delta-9-Tetrahydrocannabinol on Sleep Patterns in Man. Clin. Pharmacol. Ther. 1975, 14, 458–466. [Google Scholar] [CrossRef]
- Freemon, F.R. The Effect of Chronically Administered Delta-9-Tetrahydrocannabinol upon the Polygraphically Monitored Sleep of Normal Volunteers. Drug Alcohol Depend. 1982, 10, 345–353. [Google Scholar] [CrossRef]
- Pivik, R.T.; Zarcone, V.; Dement, W.C.; Hollister, L.E. Delta-9-Tetrahydrocannabinol and Synhexl: Effects on Human Sleep Patterns. Clin. Pharmacol. Ther. 1972, 13, 426–435. [Google Scholar] [CrossRef]
- Agarwal, N.; Pacher, P.; Tegeder, I.; Amaya, F.; Constantin, C.E.; Brenner, G.J.; Rubino, T.; Michalski, C.W.; Marsicano, G.; Monory, K.; et al. Cannabinoids Mediate Analgesia Largely via Peripheral Type 1 Cannabinoid Receptors in Nociceptors. Nat. Neurosci. 2007, 10, 870–879. [Google Scholar] [CrossRef] [PubMed]
- Meng, I.D.; Manning, B.H.; Martin, W.J.; Fields, H.L. An Analgesia Circuit Activated by Cannabinoids. Nature 1998, 395, 381–383. [Google Scholar] [CrossRef] [PubMed]
- Ottani, A.; Leone, S.; Sandrini, M.; Ferrari, A.; Bertolini, A. The Analgesic Activity of Paracetamol Is Prevented by the Blockade of Cannabinoid CB1 Receptors. Eur. J. Pharmacol. 2006, 531, 280–281. [Google Scholar] [CrossRef]
- Walker, J.M.; Hohmann, A.G.; Martin, W.J.; Strangman, N.M.; Huang, S.M.; Tsou, K. The Neurobiology of Cannabinoid Analgesia. Life Sci. 1999, 65, 665–673. [Google Scholar] [CrossRef]
- Leweke, F.M.; Piomelli, D.; Pahlisch, F.; Muhl, D.; Gerth, C.W.; Hoyer, C.; Klosterkötter, J.; Hellmich, M.; Koethe, D. Cannabidiol Enhances Anandamide Signaling and Alleviates Psychotic Symptoms of Schizophrenia. Transl. Psychiatry 2012, 2, e94. [Google Scholar] [CrossRef]
- Lutz, B.; Marsicano, G.; Maldonado, R.; Hillard, C.J. The Endocannabinoid System in Guarding against Fear, Anxiety and Stress. Nat. Rev. Neurosci. 2015, 16, 705–718. [Google Scholar] [CrossRef] [PubMed]
- Navarro, M.; Hernández, E.; Muñoz, R.M.; Del Arco, I.; Villanúa, M.A.; Carrera, M.R.A.; Rodríguez De Fonseca, F. Acute Administration of the CB1 Cannabinoid Receptor Antagonist SR 141716A Induces Anxiety-like Responses in the Rat. Neuroreport 1997, 8, 491–496. [Google Scholar] [CrossRef]
- Moreira, F.A.; Aguiar, D.C.; Guimarães, F.S. Anxiolytic-like Effect of Cannabinoids Injected into the Rat Dorsolateral Periaqueductal Gray. Neuropharmacology 2007, 52, 958–965. [Google Scholar] [CrossRef]
- Rey, A.A.; Purrio, M.; Viveros, M.P.; Lutz, B. Biphasic Effects of Cannabinoids in Anxiety Responses: CB1 and GABA B Receptors in the Balance of Gabaergic and Glutamatergic Neurotransmission. Neuropsychopharmacology 2012, 37, 2624–2634. [Google Scholar] [CrossRef]
- Ney, L.J.; Matthews, A.; Bruno, R.; Felmingham, K.L. Modulation of the Endocannabinoid System by Sex Hormones: Implications for Posttraumatic Stress Disorder. Neurosci. Biobehav. Rev. 2018, 94, 302–320. [Google Scholar] [CrossRef]
- Carter, G.T.; Flanagan, A.M.; Earleywine, M.; Abrams, D.I.; Aggarwal, S.K.; Grinspoon, L. Cannabis in Palliative Medicine: Improving Care and Reducing Opioid-Related Morbidity. Am. J. Hosp. Palliat. Med. 2011, 28, 297–303. [Google Scholar] [CrossRef]
- Bar-Sela, G.; Vorobeichik, M.; Drawsheh, S.; Omer, A.; Goldberg, V.; Muller, E. The Medical Necessity for Medicinal Cannabis: Prospective, Observational Study Evaluating the Treatment in Cancer Patients on Supportive or Palliative Care. Evid.-Based Complement. Altern. Med. 2013, 2013, 510392. [Google Scholar] [CrossRef] [PubMed]
- Motwani, M.P.; Bennett, F.; Norris, P.C.; Maini, A.A.; George, M.J.; Newson, J.; Henderson, A.; Hobbs, A.J.; Tepper, M.; White, B.; et al. Potent Anti-Inflammatory and Pro-Resolving Effects of Anabasum in a Human Model of Self-Resolving Acute Inflammation. Clin. Pharmacol. Ther. 2018, 104, 675–686. [Google Scholar] [CrossRef]
- Lucas, P. Rationale for Cannabis-Based Interventions in the Opioid Overdose Crisis. Harm Reduct. J. 2017, 14, 58. [Google Scholar] [CrossRef]
- Boehnke, K.F.; Litinas, E.; Clauw, D.J. Medical Cannabis Use Is Associated with Decreased Opiate Medication Use in a Retrospective Cross-Sectional Survey of Patients with Chronic Pain. J. Pain 2016, 17, 739–744. [Google Scholar] [CrossRef]
- Groce, E. The Health Effects of Cannabis and Cannabinoids: The Current State of Evidence and Recommendations for Research; National Academies Press: Washington, DC, USA, 2017. [Google Scholar] [CrossRef]
- Dos Santos, R.G.; Guimarães, F.S.; Crippa, J.A.S.; Hallak, J.E.C.; Rossi, G.N.; Rocha, J.M.; Zuardi, A.W. Serious Adverse Effects of Cannabidiol (CBD): A Review of Randomized Controlled Trials. Expert Opin. Drug Metab. Toxicol. 2020, 16, 517–526. [Google Scholar] [CrossRef] [PubMed]
- White, C.M. A Review of Human Studies Assessing Cannabidiol’s (CBD) Therapeutic Actions and Potential. J. Clin. Pharmacol. 2019, 59, 923–934. [Google Scholar] [CrossRef] [PubMed]
- Pauli, C.S.; Conroy, M.; Vanden Heuvel, B.D.; Park, S.H. Cannabidiol Drugs Clinical Trial Outcomes and Adverse Effects. Front. Pharmacol. 2020, 11, 63. [Google Scholar] [CrossRef]
- Kuhathasan, N.; Dufort, A.; MacKillop, J.; Gottschalk, R.; Minuzzi, L.; Frey, B.N. The Use of Cannabinoids for Sleep: A Critical Review on Clinical Trials. Exp. Clin. Psychopharmacol. 2019, 27, 383–401. [Google Scholar] [CrossRef] [PubMed]
- Black, N.; Stockings, E.; Campbell, G.; Tran, L.T.; Zagic, D.; Hall, W.D.; Farrell, M.; Degenhardt, L. Cannabinoids for the Treatment of Mental Disorders and Symptoms of Mental Disorders: A Systematic Review and Meta-Analysis. Lancet Psychiatry 2019, 6, 995–1010. [Google Scholar] [CrossRef]
- Marks, M.D.; Tian, L.; Wenger, J.P.; Omburo, S.N.; Soto-Fuentes, W.; He, J.; Gang, D.R.; Weiblen, G.D.; Dixon, R.A. Identification of Candidate Genes Affecting Δ9-Tetrahydrocannabinol Biosynthesis in Cannabis sativa. J. Exp. Bot. 2009, 60, 3715–3726. [Google Scholar] [CrossRef]
- Stout, J.M.; Boubakir, Z.; Ambrose, S.J.; Purves, R.W.; Page, J.E. The Hexanoyl-CoA Precursor for Cannabinoid Biosynthesis Is Formed by an Acyl-Activating Enzyme in Cannabis sativa Trichomes. Plant J. 2012, 71, 353–365. [Google Scholar] [CrossRef]
- Kim, E.-S.; Mahlberg, P.G. Secretory Cavity Development in Glandular Trichomes of Cannabis sativa L. (Cannabaceae). Am. J. Bot. 1991, 78, 220–229. [Google Scholar] [CrossRef]
- Kim, E.S.; Mahlberg, P.G. Secretory Vesicle Formation in the Secretory Cavity of Glandular Trichomes of Cannabis sativa L. (Cannabaceae). Mol. Cells 2003, 15, 387–395. [Google Scholar] [CrossRef]
- Mahlberg, P.G.; Eun, S.K. Accumulation of Cannabinoids in Glandular Trichomes of Cannabis (Cannabaceae). J. Ind. Hemp 2004, 9, 15–36. [Google Scholar] [CrossRef]
- Mahlberg, P.G.; Kim, E.-S. Cuticle Development on Glandular Trichomes of Cannabis sativa (Cannabaceae). Am. J. Bot. 1991, 78, 1113–1122. [Google Scholar] [CrossRef]
- Arigoni, D.; Sagner, S.; Latzel, C.; Eisenreich, W.; Bacher, A.; Zenk, M.H. Terpenoid Biosynthesis from 1-Deoxy-D-Xylulose in Higher Plants by Intramolecular Skeletal Rearrangement. Proc. Natl. Acad. Sci. USA 1997, 94, 10600–10605. [Google Scholar] [CrossRef] [PubMed]
- Fellermeier, M.; Eisenreich, W.; Bacher, A.; Zenk, M.H. Biosynthesis of Cannabinoids: Incorporation Experiments with 13C-Labeled Glucoses. Eur. J. Biochem. 2001, 268, 1596–1604. [Google Scholar] [CrossRef]
- Schwender, J.; Zeidler, J.; Gröner, R.; Müller, C.; Focke, M.; Braun, S.; Lichtenthaler, F.W.; Lichtenthaler, H.K. Incorporation of 1-Deoxy-D-Xylulose into Isoprene and Phytol by Higher Plants and Algae. FEBS Lett. 1997. [Google Scholar] [CrossRef]
- Botella-Pavía, P.; Besumbes, Ó.; Phillips, M.A.; Carretero-Paulet, L.; Boronat, A.; Rodríguez-Concepción, M. Regulation of Carotenoid Biosynthesis in Plants: Evidence for a Key Role of Hydroxymethylbutenyl Diphosphate Reductase in Controlling the Supply of Plastidial Isoprenoid Precursors. Plant J. 2004, 40, 188–199. [Google Scholar] [CrossRef]
- Phillips, M.A.; León, P.; Boronat, A.; Rodríguez-Concepción, M. The Plastidial MEP Pathway: Unified Nomenclature and Resources. Trends Plant Sci. 2008, 13, 619–623. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.H.; Chang, C.Y.; Hsu, S.J.; Chen, J.J. Chloroplast Localization of Methylerythritol 4-Phosphate Pathway Enzymes and Regulation of Mitochondrial Genes in IspD and IspE Albino Mutants in Arabidopsis. Plant Mol. Biol. 2008, 66, 663–673. [Google Scholar] [CrossRef]
- Bick, J.A.; Lange, B.M. Metabolic Cross Talk between Cytosolic and Plastidial Pathways of Isoprenoid Biosynthesis: Unidirectional Transport of Intermediates across the Chloroplast Envelope Membrane. Arch. Biochem. Biophys. 2003, 415, 146–154. [Google Scholar] [CrossRef]
- Buhaescu, I.; Izzedine, H. Mevalonate Pathway: A Review of Clinical and Therapeutical Implications. Clin. Biochem. 2007, 40, 575–584. [Google Scholar] [CrossRef]
- Goldstein, J.L.; Brown, M.S. Regulation of the Mevalonate Pathway. Nature 1990, 343, 425–430. [Google Scholar] [CrossRef]
- Miziorko, H.M. Enzymes of the Mevalonate Pathway of Isoprenoid Biosynthesis. Arch. Biochem. Biophys. 2011, 505, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Guirimand, G.; Simkin, A.J.; Papon, N.; Besseau, S.; Burlat, V.; St-Pierre, B.; Giglioli-Guivarc’h, N.; Clastre, M.; Courdavault, V. Cycloheximide as a Tool to Investigate Protein Import in Peroxisomes: A Case Study of the Subcellular Localization of Isoprenoid Biosynthetic Enzymes. J. Plant Physiol. 2012, 169, 825–829. [Google Scholar] [CrossRef]
- Simkin, A.J.; Guirimand, G.; Papon, N.; Courdavault, V.; Thabet, I.; Ginis, O.; Bouzid, S.; Giglioli-Guivarc’h, N.; Clastre, M. Peroxisomal Localisation of the Final Steps of the Mevalonic Acid Pathway in Planta. Planta 2011, 234, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Vranová, E.; Coman, D.; Gruissem, W. Network Analysis of the MVA and MEP Pathways for Isoprenoid Synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef]
- Thabet, I.; Guirimand, G.; Courdavault, V.; Papon, N.; Godet, S.; Dutilleul, C.; Bouzid, S.; Giglioli-Guivarc’h, N.; Clastre, M.; Simkin, A.J. The Subcellular Localization of Periwinkle Farnesyl Diphosphate Synthase Provides Insight into the Role of Peroxisome in Isoprenoid Biosynthesis. J. Plant Physiol. 2011, 168, 2110–2116. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.; Hahn, F.M.; Poulter, C.D.; Leustek, T. Analysis of the Isopentenyl Disphosphate Isomerase Gene from Arabidopsis thaliana. Plant Mol. Biol. 1998, 36, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Okada, K.; Kasahara, H.; Yamaguchi, S.; Kawaide, H.; Kamiya, Y.; Nojiri, H.; Yamane, H. Genetic Evidence for the Role of Isopentenyl Diphosphate Isomerases in the Mevalonate Pathway and Plant Development in Arabidopsis. Plant Cell Physiol. 2008, 49, 604–616. [Google Scholar] [CrossRef]
- Sapir-Mir, M.; Mett, A.; Belausov, E.; Tal-Meshulam, S.; Frydman, A.; Gidoni, D.; Eya, Y. Peroxisomal Localization of Arabidopsis Isopentenyl Diphosphate Isomerases Suggests That Part of the Plant Isoprenoid Mevalonic Acid Pathway Is Compartmentalized to Peroxisomes. Plant Physiol. 2008, 148, 1219–1228. [Google Scholar] [CrossRef]
- Burke, C.C.; Wildung, M.R.; Croteau, R. Geranyl Diphosphate Synthase: Cloning, Expression, and Characterization of This Prenyltransferase as a Heterodimer. Proc. Natl. Acad. Sci. USA 1999, 96, 13062–13067. [Google Scholar] [CrossRef]
- Bouvier, F.; Suire, C.; D’Harlingue, A.; Backhaus, R.A.; Camara, B. Molecular Cloning of Geranyl Diphosphate Synthase and Compartmentation of Monoterpene Synthesis in Plant Cells. Plant J. 2000, 24, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Ogura, K.; Koyama, T. Enzymatic Aspects of Isoprenoid Chain Elongation. Chem. Rev. 1998, 98, 1263–1276. [Google Scholar] [CrossRef] [PubMed]
- Ohnuma, S.I.; Hirooka, K.; Tsuruoka, N.; Yano, M.; Ohto, C.; Nakane, H.; Nishino, T. A Pathway Where Polyprenyl Diphosphate Elongates in Prenyltransferase: Insight into a Common Mechanism of Chain Length Determination of Prenyltransferases. J. Biol. Chem. 1998, 273, 26705–26713. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Ohnuma, S.I. Chain-Length Determination Mechanism of Isoprenyl Diphosphate Synthases and Implications for Molecular Evolution. Trends Biochem. Sci. 1999, 24, 445–451. [Google Scholar] [CrossRef]
- Booth, J.K.; Bohlmann, J. Terpenes in Cannabis sativa—From Plant Genome to Humans. Plant Sci. 2019, 284, 67–72. [Google Scholar] [CrossRef]
- Oldfield, E.; Lin, F.Y. Terpene Biosynthesis: Modularity Rules. Angew. Chem. Int. Ed. Engl. 2012, 51, 1124–1137. [Google Scholar] [CrossRef]
- Gagne, S.J.; Stout, J.M.; Liu, E.; Boubakir, Z.; Clark, S.M.; Page, J.E. Identification of Olivetolic Acid Cyclase from Cannabis sativa Reveals a Unique Catalytic Route to Plant Polyketides. Proc. Natl. Acad. Sci. USA 2012, 109, 12811–12816. [Google Scholar] [CrossRef]
- Taura, F.; Tanaka, S.; Taguchi, C.; Fukamizu, T.; Tanaka, H.; Shoyama, Y.; Morimoto, S. Characterization of Olivetol Synthase, a Polyketide Synthase Putatively Involved in Cannabinoid Biosynthetic Pathway. FEBS Lett. 2009, 583, 2061–2066. [Google Scholar] [CrossRef]
- Fellermeier, M.; Zenk, M.H. Prenylation of Olivetolate by a Hemp Transferase Yields Cannabigerolic Acid, the Precursor of Tetrahydrocannabinol. FEBS Lett. 1998, 427, 283–285. [Google Scholar] [CrossRef]
- Luo, X.; Reiter, M.A.; D’Espaux, L.; Wong, J.; Denby, C.M.; Lechner, A.; Zhang, Y.; Grzybowski, A.T.; Harth, S.; Lin, W.; et al. Complete Biosynthesis of Cannabinoids and Their Unnatural Analogues in Yeast. Nature 2019, 567, 123–126. [Google Scholar] [CrossRef]
- Valliere, M.A.; Korman, T.P.; Woodall, N.B.; Khitrov, G.A.; Taylor, R.E.; Baker, D.; Bowie, J.U. A Cell-Free Platform for the Prenylation of Natural Products and Application to Cannabinoid Production. Nat. Commun. 2019, 10, 565. [Google Scholar] [CrossRef]
- Taura, F.; Morimoto, S.; Shoyama, Y. Purification and Characterization of Cannabidiolic-Acid Synthase from Cannabis sativa L. Biochemical Analysis of a Novel Enzyme That Catalyzes the Oxidocyclization of Cannabigerolic Acid to Cannabidiolic Acid. J. Biol. Chem. 1996, 271, 17411–17416. [Google Scholar] [CrossRef]
- Morimoto, S.; Komatsu, K.; Taura, F.; Shoyama, Y. Purification and Characterization of Cannabichromenic Acid Synthase from Cannabis sativa. Phytochemistry 1998, 49, 1525–1529. [Google Scholar] [CrossRef]
- Shoyama, Y.; Tamada, T.; Kurihara, K.; Takeuchi, A.; Taura, F.; Arai, S.; Blaber, M.; Shoyama, Y.; Morimoto, S.; Kuroki, R. Structure and Function of Δ1-Tetrahydrocannabinolic Acid (THCA) Synthase, the Enzyme Controlling the Psychoactivity of Cannabis sativa. J. Mol. Biol. 2012, 423, 96–105. [Google Scholar] [CrossRef]
- Taura, F.; Morimoto, S.; Shoyama, Y.; Mechoulam, R. First Direct Evidence for the Mechanism of Δ1-Tetrahydrocannabinolie Acid Biosynthesis. J. Am. Chem. Soc. 1995, 117, 9766–9767. [Google Scholar] [CrossRef]
- Taura, F.; Dono, E.; Sirikantaramas, S.; Yoshimura, K.; Shoyama, Y.; Morimoto, S. Production of Δ1-Tetrahydrocannabinolic Acid by the Biosynthetic Enzyme Secreted from Transgenic Pichia Pastoris. Biochem. Biophys. Res. Commun. 2007, 361, 675–680. [Google Scholar] [CrossRef] [PubMed]
- De Zeeuw, R.A.; Wijsbeek, J.; Brejmer, D.D.; Vree, T.B.; Van Ginneken, C.A.M.; Van Rossum, J.M. Cannabinoids with a Propyl Side Chain in Cannabis: Occurrence and Chromatographic Behavior. Science 1972, 175, 778–779. [Google Scholar] [CrossRef] [PubMed]
- De Meijer, E.P.M.; Bagatta, M.; Carboni, A.; Crucitti, P.; Moliterni, V.M.C.; Ranalli, P.; Mandolino, G. The Inheritance of Chemical Phenotype in Cannabis sativa L. Genetics 2003, 163, 335–346. [Google Scholar]
- Shoyama, Y.; Hirano, H.; Nishioka, I. Biosynthesis of Propyl Cannabinoid Acid and Its Biosynthetic Relationship with Pentyl and Methyl Cannabinoid Acids. Phytochemistry 1984, 23, 1909–1984. [Google Scholar] [CrossRef]
- Kanter, S.L.; Musumeci, M.R.; Hollister, L.E. Quantitative Determination of Δ9-Tetrahydrocannabinol and Δ9-Tetrahydrocannabinolic Acid in Marihuana by High-Pressure Liquid Chromatography. J. Chromatogr. A 1979, 171, 504–508. [Google Scholar] [CrossRef]
- Perrotin-Brunel, H.; Buijs, W.; Van Spronsen, J.; Roosmalen, M.J.E.V.; Peters, C.J.; Verpoorte, R.; Witkamp, G.J. Decarboxylation of Δ9-Tetrahydrocannabinol: Kinetics and Molecular Modeling. J. Mol. Struct. 2011, 987, 67–73. [Google Scholar] [CrossRef]
- Shoyama, Y.; Yagi, M.; Nishioka, I.; Yamauchi, T. Biosynthesis of Cannabinoid Acids. Phytochemistry 1975, 14, 2189–2192. [Google Scholar] [CrossRef]
- Veress, T.; Szanto, J.I.; Leisztner, L. Determination of Cannabinoid Acids by High-Performance Liquid Chromatography of Their Neutral Derivatives Formed by Thermal Decarboxylation. I. Study of the Decarboxylation Process in Open Reactors. J. Chromatogr. A 1990. [Google Scholar] [CrossRef]
- Hanuš, L.O.; Meyer, S.M.; Muñoz, E.; Taglialatela-Scafati, O.; Appendino, G. Phytocannabinoids: A Unified Critical Inventory. Nat. Prod. Rep. 2016, 33, 1357–1392. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.A.; Ross, S.A.; Slade, D.; Radwan, M.M.; Khan, I.A.; ElSohly, M.A. Structure Determination and Absolute Configuration of Cannabichromanone Derivatives from High Potency Cannabis sativa. Tetrahedron Lett. 2008, 49, 6050–6053. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.A.; Ross, S.A.; Slade, D.; Radwan, M.M.; Zulfiqar, F.; ElSohly, M.A. Cannabinoid Ester Constituents from High-Potency Cannabis sativa. J. Nat. Prod. 2008, 71, 536–542. [Google Scholar] [CrossRef]
- Radwan, M.M.; Ross, S.A.; Slade, D.; Ahmed, S.A.; Zulfiqar, F.; Elsohly, M.A. Isolation and Characterization of New Cannabis Constituents from a High Potency Variety. Planta Med. 2008, 74, 267–272. [Google Scholar] [CrossRef]
- Pagani, A.; Scala, F.; Chianese, G.; Grassi, G.; Appendino, G.; Taglialatela-Scafati, O. Cannabioxepane, a Novel Tetracyclic Cannabinoid from Hemp, Cannabis sativa L. Tetrahedron 2011, 67, 3369–3373. [Google Scholar] [CrossRef]
- Zulfiqar, F.; Ross, S.A.; Slade, D.; Ahmed, S.A.; Radwan, M.M.; Ali, Z.; Khan, I.A.; Elsohly, M.A. Cannabisol, a Novel Δ 9-THC Dimer Possessing a Unique Methylene Bridge, Isolated from Cannabis sativa. Tetrahedron Lett. 2012, 53, 3560–3562. [Google Scholar] [CrossRef] [PubMed]
- Pollastro, F.; Taglialatela-Scafati, O.; Allarà, M.; Muñoz, E.; Di Marzo, V.; De Petrocellis, L.; Appendino, G. Bioactive Prenylogous Cannabinoid from Fiber Hemp (Cannabis sativa). J. Nat. Prod. 2011, 74, 2019–2022. [Google Scholar] [CrossRef]
- Taglialatela-Scafati, O.; Pagani, A.; Scala, F.; De Petrocellis, L.; Di Marzo, V.; Grassi, G.; Appendino, G. Cannabimovone, a Cannabinoid with a Rearranged Terpenoid Skeleton from Hemp (Eur. J. Org. Chem. 11/2010). Eur. J. Org. Chem. 2010, 2010, 2023. [Google Scholar] [CrossRef]
- Thomas, A.; Stevenson, L.A.; Wease, K.N.; Price, M.R.; Baillie, G.; Ross, R.A.; Pertwee, R.G. Evidence That the Plant Cannabinoid Δ 9-Tetrahydrocannabivarin Is a Cannabinoid CB 1 and CB 2 Receptor Antagonist. Br. J. Pharmacol. 2005, 146, 917–926. [Google Scholar] [CrossRef] [PubMed]
- Pertwee, R.G.; Thomas, A.; Stevenson, L.A.; Ross, R.A.; Varvel, S.A.; Lichtman, A.H.; Martin, B.R.; Razdan, R.K. The Psychoactive Plant Cannabinoid, Δ 9-Tetrahydrocannabinol, Is Antagonized by Δ 8- and Δ 9-Tetrahydrocannabivarin in Mice in Vivo. Br. J. Pharmacol. 2007, 150, 586–594. [Google Scholar] [CrossRef]
- García, C.; Palomo-Garo, C.; García-Arencibia, M.; Ramos, J.A.; Pertwee, R.G.; Fernández-Ruiz, J. Symptom-Relieving and Neuroprotective Effects of the Phytocannabinoid Δ 9-THCV in Animal Models of Parkinson’s Disease. Br. J. Pharmacol. 2011, 163, 1495–1506. [Google Scholar] [CrossRef]
- Ma, Y.L.; Weston, S.E.; Whalley, B.J.; Stephens, G.J. The Phytocannabinoid Δ 9-Tetrahydrocannabivarin Modulates Inhibitory Neurotransmission in the Cerebellum. Br. J. Pharmacol. 2008, 154, 204–215. [Google Scholar] [CrossRef]
- Dennis, I.; Whalley, B.J.; Stephens, G.J. Effects of Δ 9-Tetrahydrocannabivarin on [35S]GTPγS Binding in Mouse Brain Cerebellum and Piriform Cortex Membranes. Br. J. Pharmacol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Cascio, M.G.; Zamberletti, E.; Marini, P.; Parolaro, D.; Pertwee, R.G. The Phytocannabinoid, Δ9-Tetrahydrocannabivarin, Can Act through 5-HT1A Receptors to Produce Antipsychotic Effects. Br. J. Pharmacol. 2015, 172, 1305–1318. [Google Scholar] [CrossRef]
- O’Sullivan, S.E.; Bennett, A.J.; Kendall, D.A.; Randall, M.D. Cannabinoids and Peroxisome Proliferator-Activated Receptor γ (PPARg). In Proceedings of the 16th Annual Symposium on the Cannabinoids, Tihany, Hungary, 24–28 June 2006; Volume 59. [Google Scholar]
- Englund, A.; Atakan, Z.; Kralj, A.; Tunstall, N.; Murray, R.; Morrison, P. The Effect of Five Day Dosing with THCV on THC-Induced Cognitive, Psychological and Physiological Effects in Healthy Male Human Volunteers: A Placebo-Controlled, Double-Blind, Crossover Pilot Trial. J. Psychopharmacol. 2016, 30, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Tudge, L.; Williams, C.; Cowen, P.J.; McCabe, C. Neural Effects of Cannabinoid CB1 Neutral Antagonist Tetrahydrocannabivarin on Food Reward and Aversion in Healthy Volunteers. Int. J. Neuropsychopharmacol. 2015, 18, Pyu094. [Google Scholar] [CrossRef]
- Rzepa, E.; Tudge, L.; McCabe, C. The CB1 Neutral Antagonist Tetrahydrocannabivarin Reduces Default Mode Network and Increases Executive Control Network Resting State Functional Connectivity in Healthy Volunteers. Int. J. Neuropsychopharmacol. 2016, 19, Pyv092. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moldzio, R.; Pacher, T.; Krewenka, C.; Kranner, B.; Novak, J.; Duvigneau, J.C.; Rausch, W.D. Effects of Cannabinoids Δ(9)-Tetrahydrocannabinol, Δ(9)-Tetrahydrocannabinolic Acid and Cannabidiol in MPP+ Affected Murine Mesencephalic Cultures. Phytomedicine 2012, 19, 819–824. [Google Scholar] [CrossRef]
- Verhoeckx, K.C.M.; Korthout, H.A.A.J.; Van Meeteren-Kreikamp, A.P.; Ehlert, K.A.; Wang, M.; Van Der Greef, J.; Rodenburg, R.J.T.; Witkamp, R.F. Unheated Cannabis sativa Extracts and Its Major Compound THC-Acid Have Potential Immuno-Modulating Properties Not Mediated by CB1 and CB2 Receptor Coupled Pathways. Int. Immunopharmacol. 2006, 6, 656–665. [Google Scholar] [CrossRef]
- Hollister, L.E.; Gillespie, H.K. Delta-8- and Delta-9-Tetrahydrocannabinol Comparison in Man by Oral and Intravenous Administration. Clin. Pharmacol. Ther. 1973, 14, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Rock, E.M.; Limebeer, C.L.; Navaratnam, R.; Sticht, M.A.; Bonner, N.; Engeland, K.; Downey, R.; Morris, H.; Jackson, M.; Parker, L.A. A Comparison of Cannabidiolic Acid with Other Treatments for Anticipatory Nausea Using a Rat Model of Contextually Elicited Conditioned Gaping. Psychopharmacology 2014, 231, 3207–3215. [Google Scholar] [CrossRef] [PubMed]
- Nadal, X.; del Río, C.; Casano, S.; Palomares, B.; Ferreiro-Vera, C.; Navarrete, C.; Sánchez-Carnerero, C.; Cantarero, I.; Bellido, M.L.; Meyer, S.; et al. Tetrahydrocannabinolic Acid Is a Potent PPARγ Agonist with Neuroprotective Activity. Br. J. Pharmacol. 2017, 174, 4263–4276. [Google Scholar] [CrossRef] [PubMed]
- Morano, A.; Cifelli, P.; Nencini, P.; Antonilli, L.; Fattouch, J.; Ruffolo, G.; Roseti, C.; Aronica, E.; Limatola, C.; Di Bonaventura, C.; et al. Cannabis in Epilepsy: From Clinical Practice to Basic Research Focusing on the Possible Role of Cannabidivarin. Epilepsia Open 2016, 1, 145–151. [Google Scholar] [CrossRef]
- Hill, T.D.M.; Cascio, M.G.; Romano, B.; Duncan, M.; Pertwee, R.G.; Williams, C.M.; Whalley, B.J.; Hill, A.J. Cannabidivarin-Rich Cannabis Extracts Are Anticonvulsant in Mouse and Rat via a CB1 Receptor-Independent Mechanism. Br. J. Pharmacol. 2013, 170, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.J.; Mercier, M.S.; Hill, T.D.M.; Glyn, S.E.; Jones, N.A.; Yamasaki, Y.; Futamura, T.; Duncan, M.; Stott, C.G.; Stephens, G.J.; et al. Cannabidivarin Is Anticonvulsant in Mouse and Rat. Br. J. Pharmacol. 2012, 167, 1629–1642. [Google Scholar] [CrossRef] [PubMed]
- Amada, N.; Yamasaki, Y.; Williams, C.M.; Whalley, B.J. Cannabidivarin (CBDV) Suppresses Pentylenetetrazole (PTZ)-Induced Increases in Epilepsy-Related Gene Expression. PeerJ 2013, 1, E214. [Google Scholar] [CrossRef] [PubMed]
- Huizenga, M.N.; Sepulveda-Rodriguez, A.; Forcelli, P.A. Preclinical Safety and Efficacy of Cannabidivarin for Early Life Seizures. Neuropharmacology 2019, 148, 189–198. [Google Scholar] [CrossRef]
- Qin, N.; Neeper, M.P.; Liu, Y.; Hutchinson, T.L.; Lubin, M.L.; Flores, C.M. TRPV2 Is Activated by Cannabidiol and Mediates CGRP Release in Cultured Rat Dorsal Root Ganglion Neurons. J. Neurosci. 2008, 28, 6231–6238. [Google Scholar] [CrossRef] [PubMed]
- Pretzsch, C.M.; Voinescu, B.; Lythgoe, D.; Horder, J.; Mendez, M.A.; Wichers, R.; Ajram, L.; Ivin, G.; Heasman, M.; Edden, R.A.E.; et al. Effects of Cannabidivarin (CBDV) on Brain Excitation and Inhibition Systems in Adults with and without Autism Spectrum Disorder (ASD): A Single Dose Trial during Magnetic Resonance Spectroscopy. Transl. Psychiatry 2019, 9, 313. [Google Scholar] [CrossRef]
- Eibach, L.; Scheffel, S.; Cardebring, M.; Lettau, M.; Özgür Celik, M.; Morguet, A.; Roehle, R.; Stein, C. Cannabidivarin for HIV-Associated Neuropathic Pain: A Randomized, Blinded, Controlled Clinical Trial. Clin. Pharmacol. Ther. 2020, 1–8. [Google Scholar] [CrossRef]
- Russo, C.; Ferk, F.; Mišík, M.; Ropek, N.; Nersesyan, A.; Mejri, D.; Holzmann, K.; Lavorgna, M.; Isidori, M.; Knasmüller, S. Low Doses of Widely Consumed Cannabinoids (Cannabidiol and Cannabidivarin) Cause DNA Damage and Chromosomal Aberrations in Human-Derived Cells. Arch. Toxicol. 2019, 93, 179–188. [Google Scholar] [CrossRef]
- Cascio, M.G.; Gauson, L.A.; Stevenson, L.A.; Ross, R.A.; Pertwee, R.G. Evidence That the Plant Cannabinoid Cannabigerol Is a Highly Potent α 2-Adrenoceptor Agonist and Moderately Potent 5HT 1A Receptor Antagonist. Br. J. Pharmacol. 2010, 159, 129–141. [Google Scholar] [CrossRef]
- Valdeolivas, S.; Navarrete, C.; Cantarero, I.; Bellido, M.L.; Muñoz, E.; Sagredo, O. Neuroprotective Properties of Cannabigerol in Huntington’s Disease: Studies in R6/2 Mice and 3-Nitropropionate-Lesioned Mice. Neurotherapeutics 2015, 12, 185–199. [Google Scholar] [CrossRef]
- Gugliandolo, A.; Pollastro, F.; Grassi, G.; Bramanti, P.; Mazzon, E. In Vitro Model of Neuroinflammation: Efficacy of Cannabigerol, a Non-Psychoactive Cannabinoid. Int. J. Mol. Sci. 2018, 19, 1992. [Google Scholar] [CrossRef] [PubMed]
- Kathmann, M.; Flau, K.; Redmer, A.; Tränkle, C.; Schlicker, E. Cannabidiol Is an Allosteric Modulator at Mu- and Delta-Opioid Receptors. Naunyn. Schmiedebergs. Arch. Pharmacol. 2006, 372, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Ligresti, A.; Moriello, A.S.; Starowicz, K.; Matias, I.; Pisanti, S.; De Petrocellis, L.; Laezza, C.; Portella, G.; Bifulco, M.; Di Marzo, V. Antitumor Activity of Plant Cannabinoids with Emphasis on the Effect of Cannabidiol on Human Breast Carcinoma. J. Pharmacol. Exp. Ther. 2006, 318, 1375–1387. [Google Scholar] [CrossRef]
- Booker, L.; Naidu, P.S.; Razdan, R.K.; Mahadevan, A.; Lichtman, A.H. Evaluation of Prevalent Phytocannabinoids in the Acetic Acid Model of Visceral Nociception. Drug Alcohol Depend. 2009, 105, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Esposito, G.; Scuderi, C.; Valenza, M.; Togna, G.I.; Latina, V.; de Filippis, D.; Cipriano, M.; Carratù, M.R.; Iuvone, T.; Steardo, L. Cannabidiol Reduces Aβ-Induced Neuroinflammation and Promotes Hippocampal Neurogenesis through PPARγ Involvement. PLoS ONE 2011, 6, e28668. [Google Scholar] [CrossRef]
- Rosenthaler, S.; Pöhn, B.; Kolmanz, C.; Nguyen Huu, C.; Krewenka, C.; Huber, A.; Kranner, B.; Rausch, W.D.; Moldzio, R. Differences in Receptor Binding Affinity of Several Phytocannabinoids Do Not Explain Their Effects on Neural Cell Cultures. Neurotoxicol. Teratol. 2014, 46, 49–56. [Google Scholar] [CrossRef]
- Turner, C.E.; Elsohly, M.A. Biological Activity of Cannabichromene, Its Homologs and Isomers. J. Clin. Pharmacol. 1981, 21, 283S–291S. [Google Scholar] [CrossRef] [PubMed]
- DeLong, G.T.; Wolf, C.E.; Poklis, A.; Lichtman, A.H. Pharmacological Evaluation of the Natural Constituent of Cannabis sativa, Cannabichromene and Its Modulation by Δ9-Tetrahydrocannabinol. Drug Alcohol Depend. 2010, 112, 126–133. [Google Scholar] [CrossRef]
- Davis, W.M.; Hatoum, N.S. Neurobehavioral Actions of Cannabichromene and Interactions with Δ9-Tetrahydrocannabinol. Gen. Pharmacol. 1983, 14, 247–252. [Google Scholar] [CrossRef]
- Udoh, M.; Santiago, M.; Devenish, S.; McGregor, I.S.; Connor, M. Cannabichromene Is a Cannabinoid CB2 Receptor Agonist. Br. J. Pharmacol. 2019, 176, 4537–4547. [Google Scholar] [CrossRef]
- Romano, B.; Borrelli, F.; Fasolino, I.; Capasso, R.; Piscitelli, F.; Cascio, M.G.; Pertwee, R.G.; Coppola, D.; Vassallo, L.; Orlando, P.; et al. The Cannabinoid TRPA1 Agonist Cannabichromene Inhibits Nitric Oxide Production in Macrophages and Ameliorates Murine Colitis. Br. J. Pharmacol. 2013, 169, 213–229. [Google Scholar] [CrossRef] [PubMed]
- Shinjyo, N.; Di Marzo, V. The Effect of Cannabichromene on Adult Neural Stem/Progenitor Cells. Neurochem. Int. 2013, 63, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Mahadevan, A.; Siegel, C.; Martin, B.R.; Abood, M.E.; Beletskaya, I.; Razdan, R.K. Novel Cannabinol Probes for CB1 and CB2 Cannabinoid Receptors. J. Med. Chem. 2000, 43, 3778–3785. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, I.; Watanabe, K.; Kuzuoka, K.; Narimatsu, S.; Yoshimura, H. The Pharmacological Activity of Cannabinol and Its Major Metabolite, 11-Hydroxycannabinol. Chem. Pharm. Bull. 1987, 35, 2144–2147. [Google Scholar] [CrossRef]
- Watanabe, K.; Yamaori, S.; Funahashi, T.; Kimura, T.; Yamamoto, I. Cytochrome P450 Enzymes Involved in the Metabolism of Tetrahydrocannabinols and Cannabinol by Human Hepatic Microsomes. Life Sci. 2007, 80, 1415–1419. [Google Scholar] [CrossRef]
- Yamaori, S.; Kushihara, M.; Yamamoto, I.; Watanabe, K. Characterization of Major Phytocannabinoids, Cannabidiol and Cannabinol, as Isoform-Selective and Potent Inhibitors of Human CYP1 Enzymes. Biochem. Pharmacol. 2010, 79, 1691–1698. [Google Scholar] [CrossRef]
- Aiken, C.T.; Tobin, A.J.; Schweitzer, E.S. A Cell-Based Screen for Drugs to Treat Huntington’s Disease. Neurobiol. Dis. 2004, 16, 546–555. [Google Scholar] [CrossRef]
- Glass, M.; Faull, R.L.M.; Dragunow, M. Loss of Cannabinoid Receptors in the Substantia Nigra in Huntington’s Disease. Neuroscience 1993, 56, 523–527. [Google Scholar] [CrossRef]
- Blázquez, C.; Chiarlone, A.; Sagredo, O.; Aguado, T.; Pazos, M.R.; Resel, E.; Palazuelos, J.; Julien, B.; Salazar, M.; Börner, C.; et al. Loss of Striatal Type 1 Cannabinoid Receptors Is a Key Pathogenic Factor in Huntington’s Disease. Brain 2011, 134, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Weydt, P.; Hong, S.; Witting, A.; Möller, T.; Stella, N.; Kliot, M. Cannabinol Delays Symptom Onset in SOD1 (G93A) Transgenic Mice without Affecting Survival. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 2005, 6, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.; Cairns, B.E. Cannabidiol, Cannabinol and Their Combinations Act as Peripheral Analgesics in a Rat Model of Myofascial Pain. Arch. Oral Biol. 2019, 104, 33–39. [Google Scholar] [CrossRef]
- Baroi, S.; Saha, A.; Bachar, R.; Bachar, S.C. Cannabinoid as Potential Aromatase Inhibitor through Molecular Modeling and Screening for Anti-Cancer Activity. Dhaka Univ. J. Pharm. Sci. 2020, 19, 47–58. [Google Scholar] [CrossRef]
- Furqan, T.; Batool, S.; Habib, R.; Shah, M.; Kalasz, H.; Darvas, F.; Kuca, K.; Nepovimova, E.; Batool, S.; Nurulain, S.M. Cannabis Constituents and Acetylcholinesterase Interaction: Molecular Docking, in Vitro Studies and Association with CNR1 RS806368 and ACHE RS17228602. Biomolecules 2020, 10, 758. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Akiyama, Y.; Fukui, K.; Kamada, H.; Satoh, S. Characterization; Genome Sizes and Morphology of Sex Chromosomes in Hemp (Cannabis sativa L.). Cytologia 1998, 635, 459–464. [Google Scholar] [CrossRef]
- Sakamoto, K.; Shimomura, K.; Komeda, Y.; Kamada, H.; Satoh, S. A Male-Associated DNA Sequence in a Dioecious Plant, Cannabis sativa L. Plant Cell Physiol. 1995, 36, 1549–1554. [Google Scholar] [CrossRef] [PubMed]
- Alghanim, H.J.; Almirall, J.R. Development of Microsatellite Markers in Cannabis sativa for DNA Typing and Genetic Relatedness Analyses. Anal. Bioanal. Chem. 2003, 376, 1225–1233. [Google Scholar] [CrossRef]
- Gilmore, S.; Peakall, R.; Robertson, J. Short Tandem Repeat (STR) DNA Markers Are Hypervariable and Informative in Cannabis sativa: Implications for Forensic Investigations. Forensic Sci. Int. 2003, 131, 65–74. [Google Scholar] [CrossRef]
- Hsieh, H.M.; Hou, R.J.; Tsai, L.C.; Wei, C.S.; Liu, S.W.; Huang, L.H.; Kuo, Y.C.; Linacre, A.; Lee, J.C.I. A Highly Polymorphic STR Locus in Cannabis sativa. Forensic Sci. Int. 2003, 131, 53–58. [Google Scholar] [CrossRef]
- Van Bakel, H.; Stout, J.M.; Cote, A.G.; Tallon, C.M.; Sharpe, A.G.; Hughes, T.R.; Page, J.E. The Draft Genome and Transcriptome of Cannabis sativa. Genome Biol. 2011, 12, R102. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Wang, B.; Xie, S.; Xu, X.; Zhang, J.; Pei, L.; Yu, Y.; Yang, W.; Zhang, Y. A High-Quality Reference Genome of Wild Cannabis sativa. Hortic. Res. 2020, 7, 73. [Google Scholar] [CrossRef]
- Laverty, K.U.; Stout, J.M.; Sullivan, M.J.; Shah, H.; Gill, N.; Holbrook, L.; Deikus, G.; Sebra, R.; Hughes, T.R.; Page, J.E.; et al. A Physical and Genetic Map of Cannabis sativa Identifies Extensive Rearrangements at the THC/CBD Acid Synthase Loci. Genome Res. 2019, 29, 146–156. [Google Scholar] [CrossRef]
- Kojoma, M.; Seki, H.; Yoshida, S.; Muranaka, T. DNA Polymorphisms in the Tetrahydrocannabinolic Acid (THCA) Synthase Gene in “Drug-Type” and “Fiber-Type” Cannabis sativa L. Forensic Sci. Int. 2006, 159, 132–140. [Google Scholar] [CrossRef] [PubMed]
- McKernan, K.; Helbert, Y.; Tadigotla, V.; McLaughlin, S.; Spangler, J.; Zhang, L.; Smith, D. Single Molecule Sequencing of THCA Synthase Reveals Copy Number Variation in Modern Drug-Type Cannabis sativa L. bioRxiv 2015, 28654. [Google Scholar] [CrossRef]
- Onofri, C.; De Meijer, E.P.M.; Mandolino, G. Sequence Heterogeneity of Cannabidiolic- and Tetrahydrocannabinolic Acid-Synthase in Cannabis sativa L. and Its Relationship with Chemical Phenotype. Phytochemistry 2015, 116, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Weiblen, G.D.; Wenger, J.P.; Craft, K.J.; ElSohly, M.A.; Mehmedic, Z.; Treiber, E.L.; Marks, M.D. Gene Duplication and Divergence Affecting Drug Content in Cannabis sativa. New Phytol. 2015, 208, 1241–1250. [Google Scholar] [CrossRef] [PubMed]
- Welling, M.T.; Liu, L.; Shapter, T.; Raymond, C.A.; King, G.J. Characterisation of Cannabinoid Composition in a Diverse Cannabis sativa L. Germplasm Collection. Euphytica 2016, 208, 463–475. [Google Scholar] [CrossRef]
- Lynch, R.C.; Vergara, D.; Tittes, S.; White, K.; Schwartz, C.J.; Gibbs, M.J.; Ruthenburg, T.C.; DeCesare, K.; Land, D.P.; Kane, N.C. Genomic and Chemical Diversity in Cannabis. CRC. Crit. Rev. Plant Sci. 2016, 35, 349–363. [Google Scholar] [CrossRef]
- Gao, C.; Xin, P.; Cheng, C.; Tang, Q.; Chen, P.; Wang, C.; Zang, G.; Zhao, L. Diversity Analysis in Cannabis sativa based on Large-Scale Development of Expressed Sequence Tag-Derived Simple Sequence Repeat Markers. PLoS ONE 2014, 9, e110638. [Google Scholar] [CrossRef]
- Zhang, L.G.; Chang, Y.; Zhang, X.F.; Guan, F.Z.; Yuan, H.M.; Yu, Y.; Zhao, L.J. Analysis of the Genetic Diversity of Chinese Native Cannabis sativa Cultivars by Using ISSR and Chromosome Markers. Genet. Mol. Res. 2014, 13, 10490–10500. [Google Scholar] [CrossRef]
- White, K.H.; Vergara, D.; Keepers, K.G.; Kane, N.C. The Complete Mitochondrial Genome for Cannabis sativa. Mitochondrial DNA Part B Resour. 2016, 1, 715–716. [Google Scholar] [CrossRef]
- Oh, H.; Seo, B.; Lee, S.; Ahn, D.H.; Jo, E.; Park, J.K.; Min, G.S. Two Complete Chloroplast Genome Sequences of Cannabis sativa Varieties. Mitochondrial DNA 2015, 27, 2835–2837. [Google Scholar] [CrossRef]
- Booth, J.K.; Page, J.E.; Bohlmann, J. Terpene Synthases from Cannabis sativa. PLoS ONE 2017, 12, e0173911. [Google Scholar] [CrossRef]
- Zager, J.J.; Lange, I.; Srividya, N.; Smith, A.; Markus Lange, B. Gene Networks Underlying Cannabinoid and Terpenoid Accumulation in Cannabis. Plant Physiol. 2019, 180, 1877–1897. [Google Scholar] [CrossRef] [PubMed]
- Amano, E.; Smith, H.H. Mutations Induced by Ethyl Methanesulfonate in Maize. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1965, 2, 344–351. [Google Scholar] [CrossRef]
- Froese-Gertzen, E.E.; Konzak, C.F.; Nilan, R.A.; Heiner, R.E. The Effect of Ethyl Methanesulfonate on the Growth Response, Chromosome Structure and Mutation Rate in Barley. Radiat. Bot. 1964, 4, 61–69. [Google Scholar] [CrossRef]
- Jander, G.; Baerson, S.R.; Hudak, J.A.; Gonzalez, K.A.; Gruys, K.J.; Last, R.L. Ethylmethanesulfonate Saturation Mutagenesis in Arabidopsis to Determine Frequency of Herbicide Resistance. Plant Physiol. 2003, 131, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Luan, Y.S.; Zhang, J.; Gao, X.R.; An, L.J. Mutation Induced by Ethylmethanesulphonate (EMS), in Vitro Screening for Salt Tolerance and Plant Regeneration of Sweet Potato (Ipomoea batatas L.). Plant Cell. Tissue Organ Cult. 2007, 88, 77–81. [Google Scholar] [CrossRef]
- Stavreva, D.A.; Ptáček, O.; Plewa, M.J.; Gichner, T. Single Cell Gel Electrophoresis Analysis of Genomic Damage Induced by Ethyl Methanesulfonate in Cultured Tobacco Cells. Mutat Res. 1998, 422, 323–330. [Google Scholar] [CrossRef]
- Watanabe, S.; Mizoguchi, T.; Aoki, K.; Kubo, Y.; Mori, H.; Imanishi, S.; Yamazaki, Y.; Shibata, D.; Ezura, H. Ethylmethanesulfonate (EMS) Mutagenesis of Solanum Lycopersicum Cv. Micro-Tom for Large-Scale Mutant Screens. Plant Biotechnol. 2007, 24, 33–38. [Google Scholar] [CrossRef]
- Koornneeff, M.; Dellaert, L.W.M.; van der Veen, J.H. EMS- and Relation-Induced Mutation Frequencies at Individual Loci in Arabidopsis thaliana (L.) Heynh. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1982, 93, 109–123. [Google Scholar] [CrossRef]
- Li, X.; Song, Y.; Century, K.; Straight, S.; Ronald, P.; Dong, X.; Lassner, M.; Zhang, Y. A Fast Neutron Deletion Mutagenesis-Based Reverse Genetics System for Plants. Plant J. 2001, 27, 235–242. [Google Scholar] [CrossRef]
- Shirley, B.W.; Hanley, S.; Goodman, H.M. Effects of Ionizing Radiation on a Plant Genome: Analysis of Two Arabidopsis Transparent Testa Mutations. Plant Cell 1992, 4, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Bevan, M.; Bancroft, I.; Bent, E.; Love, K.; Goodman, H.; Dean, C.; Bergkamp, R.; Dirkse, W.; Van Staveren, M.; Stiekema, W.; et al. Analysis of 1.9 Mb of Contiguous Sequence from Chromosome 4 of Arabidopsis thaliana. Nature 1998, 391, 485–488. [Google Scholar] [CrossRef]
- Bolon, Y.T.; Haun, W.J.; Xu, W.W.; Grant, D.; Stacey, M.G.; Nelson, R.T.; Gerhardt, D.J.; Jeddeloh, J.A.; Stacey, G.; Muehlbauer, G.J.; et al. Phenotypic and Genomic Analyses of a Fast Neutron Mutant Population Resource in Soybean. Plant Physiol. 2011, 156, 240–253. [Google Scholar] [CrossRef] [PubMed]
- Colbert, T.; Till, B.J.; Tompa, R.; Reynolds, S.; Steine, M.N.; Yeung, A.T.; McCallum, C.M.; Comai, L.; Henikoff, S. High-Throughput Screening for Induced Point Mutations. Plant Physiol. 2001, 126, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Jander, G.; Norris, S.R.; Joshi, V.; Fraga, M.; Rugg, A.; Yu, S.; Li, L.; Last, R.L. Application of a High-Throughput HPLC-MS/MS Assay to Arabidopsis Mutant Screening; Evidence That Threonine Aldolase Plays a Role in Seed Nutritional Quality. Plant J. 2004, 39, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Jaganathan, D.; Ramasamy, K.; Sellamuthu, G.; Jayabalan, S.; Venkataraman, G. CRISPR for Crop Improvement: An Update Review. Front. Plant Sci. 2018, 9, 985. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-Frequency off-Target Mutagenesis Induced by CRISPR-Cas Nucleases in Human Cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA Targeting Specificity of RNA-Guided Cas9 Nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Tsai, S.Q.; Nguyen, N.T.; Malagon-Lopez, J.; Topkar, V.V.; Aryee, M.J.; Joung, J.K. CIRCLE-Seq: A Highly Sensitive in Vitro Screen for Genome-Wide CRISPR-Cas9 Nuclease off-Targets. Nat. Methods 2017, 14, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.G.; Kim, S.T.; Choe, S.; Kim, J.S. DNA-Free Genome Editing in Plants with Preassembled CRISPR-Cas9 Ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W.; Tang, X.; Zheng, X.; Voytas, D.F.; Hsieh, T.F.; Zhang, Y.; Qi, Y. A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation. Plant Physiol. 2015, 169, 971–985. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/SgRNA-Mediated Targeted Gene Modification in Arabidopsis, Tobacco, Sorghum and Rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef]
- Feng, Z.; Zhang, B.; Ding, W.; Liu, X.; Yang, D.L.; Wei, P.; Cao, F.; Zhu, S.; Zhang, F.; Mao, Y.; et al. Efficient Genome Editing in Plants Using a CRISPR/Cas System. Cell Res. 2013, 23, 1229–1232. [Google Scholar] [CrossRef]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 System for Plant Genome Editing and Beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Podevin, N.; Davies, H.V.; Hartung, F.; Nogué, F.; Casacuberta, J.M. Site-Directed Nucleases: A Paradigm Shift in Predictable, Knowledge-Based Plant Breeding. Trends Biotechnol. 2013, 31, 375–383. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-Mediated Viral Interference in Plants. Genome Biol. 2015, 16. [Google Scholar] [CrossRef]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 Toolkit for Multiplex Genome Editing in Plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous Editing of Three Homoeoalleles in Hexaploid Bread Wheat Confers Heritable Resistance to Powdery Mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef] [PubMed]
- Li, J.F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and Homologous Recombination-Mediated Genome Editing in Arabidopsis and Nicotiana Benthamiana Using Guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef] [PubMed]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Patron, N.J.; Nekrasov, V. Editing Plant Genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 2015, 32, 76–84. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Z.B.; Xing, A.; Moon, B.P.; Koellhoffer, J.P.; Huang, L.; Ward, R.T.; Clifton, E.; Falco, S.C.; Cigan, A.M. Cas9-Guide RNA Directed Genome Editing in Soybean. Plant Physiol. 2015, 169, 960–970. [Google Scholar] [CrossRef]
- Mackinnon, L.; McDougall, G.; Aziz, N.; Millam, S. Progress Towards Transformation of Fibre Hemp; Scottish Crop Research Institute Annual Report 2000/2001; Scottish Crop Research Institute: Dundee, UK, 2000; pp. 84–86. [Google Scholar]
- Feeney, M.; Punja, Z.K. Tissue Culture and Agrobacterium-Mediated Transformation of Hemp (Cannabis sativa L.). Vitr. Cell. Dev. Biol. Plant 2003, 39, 578–585. [Google Scholar] [CrossRef]
- Wahby, I.; Caba, J.M.; Ligero, F. Agrobacterium Infection of Hemp (Cannabis sativa L.): Establishment of Hairy Root Cultures. J. Plant Interact. 2013, 8, 312–320. [Google Scholar] [CrossRef]
- Srivastava, S.; Srivastava, A.K. Hairy Root Culture for Mass-Production of High-Value Secondary Metabolites. Crit. Rev. Biotechnol. 2007, 27, 29–43. [Google Scholar] [CrossRef]
- Ślusarkiewicz-Jarzina, A.; Ponitka, A.; Kaczmarek, Z. Influence of Cultivar, Explant Source and Plant Growth Regulator on Callus Induction and Plant Regeneration of Cannabis sativa L. Acta Biol. Cracoviensia Ser. Bot. 2005, 47, 145–151. [Google Scholar]
- Carvalho, Â.; Hansen, E.H.; Kayser, O.; Carlsen, S.; Stehle, F. Designing Microorganisms for Heterologous Biosynthesis of Cannabinoids. FEMS Yeast Res. 2017, 17, Fox037. [Google Scholar] [CrossRef] [PubMed]
- Zirpel, B.; Stehle, F.; Kayser, O. Production of Δ9-Tetrahydrocannabinolic Acid from Cannabigerolic Acid by Whole Cells of Pichia (Komagataella) Pastoris Expressing Δ9-Tetrahydrocannabinolic Acid Synthase from Cannabis sativa L. Biotechnol. Lett. 2015, 37, 1869–1875. [Google Scholar] [CrossRef] [PubMed]
- Ohto, C.; Muramatsu, M.; Obata, S.; Sakuradani, E.; Shimizu, S. Overexpression of the Gene Encoding HMG-CoA Reductase in Saccharomyces Cerevisiae for Production of Prenyl Alcohols. Appl. Microbiol. Biotechnol. 2009, 82, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Zirpel, B.; Degenhardt, F.; Martin, C.; Kayser, O.; Stehle, F. Engineering Yeasts as Platform Organisms for Cannabinoid Biosynthesis. J. Biotechnol. 2017, 259, 204–212. [Google Scholar] [CrossRef]
- Mills, E. The Carbon Footprint of Indoor Cannabis Production. Energy Policy 2012, 46, 58–67. [Google Scholar] [CrossRef]
- Borthwick, H.A.; Scully, N.J. Photoperiodic Responses of Hemp. Bot. Gaz. 1954, 116, 14–29. [Google Scholar] [CrossRef]
- Schaffner, J.H. The Influence of Relative Length of Daylight on the Reversal of Sex in Hemp. Ecology 1923, 4, 323–334. [Google Scholar] [CrossRef]
- Potter, D.J.; Duncombe, P. The Effect of Electrical Lighting Power and Irradiance on Indoor-Grown Cannabis Potency and Yield. J. Forensic Sci. 2012, 57, 618–622. [Google Scholar] [CrossRef]
- Spitzer-Rimon, B.; Duchin, S.; Bernstein, N.; Kamenetsky, R. Architecture and Florogenesis in Female Cannabis sativa Plants. Front. Plant Sci. 2019, 10, 350. [Google Scholar] [CrossRef]
- Vanhove, W.; Van Damme, P.; Meert, N. Factors Determining Yield and Quality of Illicit Indoor Cannabis (Cannabis Spp.) Production. Forensic Sci. Int. 2011, 212, 1. [Google Scholar] [CrossRef] [PubMed]
- Viršile, A.; Olle, M.; Duchovskis, P. LED Lighting in Horticulture. In Light Emitting Diodes for Agriculture: Smart Lighting; Springer: Singapore, 2017; pp. 113–147. [Google Scholar] [CrossRef]
- Nelson, J.A.; Bugbee, B. Economic Analysis of Greenhouse Lighting: Light Emitting Diodes vs. High Intensity Discharge Fixtures. PLoS ONE 2014, 9, e99010. [Google Scholar] [CrossRef]
- Tamulaitis, G.; Duchovskis, P.; Bliznikas, Z.; Breive, K.; Ulinskaite, R.; Brazaityte, A.; Novičkovas, A.; Žukauskas, A. High-Power Light-Emitting Diode Based Facility for Plant Cultivation. J. Phys. D. Appl. Phys. 2005, 38, 3182–3187. [Google Scholar] [CrossRef]
- Hogewoning, S.W.; Douwstra, P.; Trouwborst, G.; Van Ieperen, W.; Harbinson, J. An Artificial Solar Spectrum Substantially Alters Plant Development Compared with Usual Climate Room Irradiance Spectra. J. Exp. Bot. 2010, 61, 1267–1276. [Google Scholar] [CrossRef]
- Backer, R.; Schwinghamer, T.; Rosenbaum, P.; McCarty, V.; Eichhorn Bilodeau, S.; Lyu, D.; Ahmed, M.B.; Robinson, G.; Lefsrud, M.; Wilkins, O.; et al. Closing the Yield Gap for Cannabis: A Meta-Analysis of Factors Determining Cannabis Yield. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Chandra, S.; Lata, H.; Khan, I.A.; Elsohly, M.A. Photosynthetic Response of Cannabis sativa L. to Variations in Photosynthetic Photon Flux Densities, Temperature and CO2 Conditions. Physiol. Mol. Biol. Plants 2008, 14, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Lata, H.; Mehmedic, Z.; Khan, I.A.; ElSohly, M.A. Light Dependence of Photosynthesis and Water Vapor Exchange Characteristics in Different High Δ9-THC Yielding Varieties of Cannabis sativa L. J. Appl. Res. Med. Aromat. Plants 2015, 2, 39–47. [Google Scholar] [CrossRef]
- Lydon, J.; Teramura, A.H.; Coffman, C.B. UV-B Radiation Effects on Photosynthesis, Growth and Cannabinoid Production of Two Cannabis sativa Chemotypes. Photochem. Photobiol. 1987, 46, 201–206. [Google Scholar] [CrossRef]
- Berry, J.; Bjorkman, O. Photosynthetic Response and Adaptation to Temperature in Higher Plants. Annu. Rev. Plant Physiol. 1980, 31, 491–543. [Google Scholar] [CrossRef]
- Larcher, W. Photosynthesis as a Tool for Indicating Temperature Stress Events. In Ecophysiology of Photosynthesis; Schulze, E., Caldwell, M.M., Eds.; Springer: Berlin/Heidelberg, Germany, 1995; pp. 261–277. [Google Scholar] [CrossRef]
- Hikosaka, K.; Ishikawa, K.; Borjigidai, A.; Muller, O.; Onoda, Y. Temperature Acclimation of Photosynthesis: Mechanisms Involved in the Changes in Temperature Dependence of Photosynthetic Rate. J. Exp. Bot. 2006, 57, 291–302. [Google Scholar] [CrossRef]
- Chandra, S.; Lata, H.; Khan, I.A.; ElSohly, M.A. Temperature Response of Photosynthesis in Different Drug and Fiber Varieties of Cannabis sativa L. Physiol. Mol. Biol. Plants 2011, 17, 297–303. [Google Scholar] [CrossRef]
- Van der Werf, H.M.G.; Brouwer, K.; Wijlhuizen, M.; Withagen, J.C.M. The Effect of Temperature on Leaf Appearance and Canopy Establishment in Fibre Hemp (Cannabis sativa L.). Ann. Appl. Biol. 1995, 126, 551–561. [Google Scholar] [CrossRef]
- Bernstein, N.; Gorelick, J.; Zerahia, R.; Koch, S. Impact of N, P, K, and Humic Acid Supplementation on the Chemical Profile of Medical Cannabis (Cannabis sativa L). Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Maļceva, M.; Vikmane, M.; Stramkale, V. Changes of Photosynthesis-Related Parameters and Productivity of Cannabis sativa under Different Nitrogen Supply. Environ. Exp. Biol. 2011, 9, 61–69. [Google Scholar]
- Mansouri, H.; Asrar, Z. Effects of Abscisic Acid on Content and Biosynthesis of Terpenoids in Cannabis sativa at Vegetative Stage. Biol. Plant. 2012, 56, 153–156. [Google Scholar] [CrossRef]
- Mansouri, H.; Asrar, Z.; Mehrabani, M. Effects of Gibberellic Acid on Primary Terpenoids and Δ9-Tetrahydrocannabinol in Cannabis sativa at Flowering Stage. J. Integr. Plant Biol. 2009, 51, 553–561. [Google Scholar] [CrossRef]
- Larkin, J.C.; Oppenheimer, D.G.; Lloyd, A.M.; Paparozzi, E.T.; Marks, M.D. Roles of the GLABROUS1 and TRANSPARENT TESTA GLABRA Genes in Arabidopsis Trichome Development. Plant Cell 1994, 6, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Payne, C.T.; Zhang, F.; Lloyd, A.M. GL3 Encodes a BHLH Protein That Regulates Trichome Development in Arabidopsis through Interaction with GL1 and TTG1. Genetics 2000, 156, 1349–1362. [Google Scholar]
- Rerie, W.G.; Feldmann, K.A.; Marks, M.D. The GLABRA2 Gene Encodes a Homeo Domain Protein Required for Normal Trichome Development in Arabidopsis. Genes Dev. 1994, 8, 1388–1399. [Google Scholar] [CrossRef]
- Szymanski, D.B.; Jilk, R.A.; Pollock, S.M.; Marks, M.D. Control of GL2 Expression in Arabidopsis Leaves and Trichomes. Development 1998, 125, 1161–1171. [Google Scholar]
- Hülskamp, M.; Miséra, S.; Jürgens, G. Genetic Dissection of Trichome Cell Development in Arabidopsis. Cell 1994, 76, 555–566. [Google Scholar] [CrossRef]
- Perazza, D.; Herzog, M.; Hülskamp, M.; Brown, S.; Dorne, A.M.; Bonneville, J.M. Trichome Cell Growth in Arabidopsis thaliana Can Be Derepressed by Mutations in at Least Five Genes. Genetics 1999, 152, 461–476. [Google Scholar]
- Liu, Y.; Liu, D.; Hu, R.; Hua, C.; Ali, I.; Zhang, A.; Liu, B.; Wu, M.; Huang, L.; Gan, Y. AtGIS, a C2H2 Zinc-Finger Transcription Factor from Arabidopsis Regulates Glandular Trichome Development through GA Signaling in Tobacco. Biochem. Biophys. Res. Commun. 2017, 483, 209–215. [Google Scholar] [CrossRef]
- Luo, M.; Wang, Z.; Li, H.; Xia, K.F.; Cai, Y.; Xu, Z.F. Overexpression of a Weed (Solanum Americanum) Proteinase Inhibitor in Transgenic Tobacco Results in Increased Glandular Trichome Density and Enhanced Resistance to Helicoverpa Armigera and Spodoptera Litura. Int. J. Mol. Sci. 2009, 10, 1896–1910. [Google Scholar] [CrossRef] [PubMed]
- Paetzold, H.; Garms, S.; Bartram, S.; Wieczorek, J.; Urós-Gracia, E.M.; Rodríguez-Concepción, M.; Boland, W.; Strack, D.; Hause, B.; Walter, M.H. The Isogene 1-Deoxy-D-Xylulose 5-Phosphate Synthase 2 Controls Isoprenoid Profiles, Precursor Pathway Allocation, and Density of Tomato Trichomes. Mol. Plant 2010, 3, 904–916. [Google Scholar] [CrossRef]
- Ma, D.; Hu, Y.; Yang, C.; Liu, B.; Fang, L.; Wan, Q.; Liang, W.; Mei, G.; Wang, L.; Wang, H.; et al. Genetic Basis for Glandular Trichome Formation in Cotton. Nat. Commun. 2016, 7, 10456. [Google Scholar] [CrossRef] [PubMed]
- Salas Fernandez, M.G.; Becraft, P.W.; Yin, Y.; Lübberstedt, T. From Dwarves to Giants? Plant Height Manipulation for Biomass Yield. Trends Plant Sci. 2009, 14, 454–461. [Google Scholar] [CrossRef]
- Campiglia, E.; Radicetti, E.; Mancinelli, R. Plant Density and Nitrogen Fertilization Affect Agronomic Performance of Industrial Hemp (Cannabis sativa L.) in Mediterranean Environment. Ind. Crop. Prod 2017, 100, 246–254. [Google Scholar] [CrossRef]
- Van der Werf, H.M.G.; Wijlhuizen, M.; de Schutter, J.A.A. Plant Density and Self-Thinning Affect Yield and Quality of Fibre Hemp (Cannabis sativa L.). Field Crop. Res. 1995, 40, 153–164. [Google Scholar] [CrossRef]
- Small, E. Dwarf Germplasm: The Key to Giant Cannabis Hempseed and Cannabinoid Crops. Genet. Resour. Crop Evol. 2018, 65, 1071–1107. [Google Scholar] [CrossRef]
- Graham, L.A.; Besser, K.; Blumer, S.; Branigan, C.A.; Czechowski, T.; Elias, L.; Guterman, I.; Harvey, D.; Isaac, P.G.; Khan, A.M.; et al. The Genetic Map of Artemisia Annua L Identifies Loci Affecting Yield of the Antimalarial Drug Artemisinin. Science 2010, 327, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Fairbairn, J.; Kapoor, L. The Lactiferous Vessels of Papaver Somniferum L. Planta Med. 1960, 8, 49–61. [Google Scholar] [CrossRef]
- Nessler, C.L.; Mahlberg, P.G. Laticifers in Stamens of Papaver Somniferum L. Planta 1976, 129, 83–85. [Google Scholar] [CrossRef]
- Weid, M.; Ziegler, J.; Kutchan, T.M. The Roles of Latex and the Vascular Bundle in Morphine Biosynthesis in the Opium Poppy, Papaver Somniferum. Proc. Natl. Acad. Sci. USA 2004, 101, 13957–13962. [Google Scholar] [CrossRef] [PubMed]
- Fuller, J.G.; McMorland, G.H.; Douglas, M.J.; Palmer, L. Epidural Morphine for Analgesia after Caesarean Section: A Report of 4880 Patients. Can. J. Anaesth. 1990, 37, 636–640. [Google Scholar] [CrossRef] [PubMed]
- Stenseth, K.; Sellevold, O.; Breivik, H. Epidural Morphine for Postoperative Pain: Experience with 1085 Patients. Acta Anaesthesiol. Scand. 1985, 29, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Walder, B.; Schafer, M.; Henzi, I.; Tramèr, M.R. Efficacy and Safety of Patient-Controlled Opioid Analgesia for Acute Postoperative Pain. Acta Anaesthesiol. Scand. 2001, 45, 795–804. [Google Scholar] [CrossRef]
- Goldsack, C.; Scuplak, S.M.; Smith, M. A Double-Blind Comparison of Codeine and Morphine for Postoperative Analgesia Following Intracranial Surgery. Anaesthesia 1996, 51, 1029–1032. [Google Scholar] [CrossRef]
- Walker, D.J.; Zacny, J.P. Subjective, Psychomotor, and Analgesic Effects of Oral Codeine and Morphine in Healthy Volunteers. Psychopharmacology 1998, 140, 191–201. [Google Scholar] [CrossRef]
- Sevelius, H.; McCoy, J.F.; Colmore, J.P. Dose Response to Codeine in Patients with Chronic Cough. Clin. Pharmacol. Ther. 1971, 12, 449–455. [Google Scholar] [CrossRef]
- Bolser, D.C.; Davenport, P.W. Codeine and Cough: An Ineffective Gold Standard. Curr. Opin. Allergy Clin. Immunol. 2007, 7, 32–36. [Google Scholar] [CrossRef]
- Freestone, C.; Eccles, R. Assessment of the Antitussive Efficacy of Codeine in Cough Associated with Common Cold. J. Pharm. Pharmacol. 1997, 49, 1045–1049. [Google Scholar] [CrossRef]
- Takahama, K.; Shirasaki, T. Central and Peripheral Mechanisms of Narcotic Antitussives: Codeine-Sensitive and -Resistant Coughs. Cough 2007, 152, 349–355. [Google Scholar] [CrossRef]
- Jackson, T.; Chougule, M.B.; Ichite, N.; Patlolla, R.R.; Singh, M. Antitumor Activity of Noscapine in Human Non-Small Cell Lung Cancer Xenograft Model. Cancer Chemother. Pharmacol. 2008, 63, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Joshi, H.C.; Zhou, J. Noscapine and Analogues as Potential Chemotherapeutic Agents. Drug News Perspect. 2000, 13, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Rida, P.C.G.; Livecche, D.; Ogden, A.; Zhou, J.; Aneja, R. The Noscapine Chronicle: A Pharmaco-Historic Biography of the Opiate Alkaloid Family and Its Clinical Applications. Med. Res. Rev. 2015, 35, 1072–1096. [Google Scholar] [CrossRef]
- Ye, K.; Ke, Y.; Keshava, N.; Shanks, J.; Kapp, J.A.; Tekmal, R.R.; Petros, J.; Joshi, H.C. Opium Alkaloid Noscapine Is an Antitumor Agent That Arrests Metaphase and Induces Apoptosis in Dividing Cells. Proc. Natl. Acad. Sci. USA 1998, 95, 1601–1606. [Google Scholar] [CrossRef] [PubMed]
- Carroll, R.J.; Leisch, H.; Rochon, L.; Hudlicky, T.; Cox, D.P. One-Pot Conversion of Thebaine to Hydrocodone and Synthesis of Neopinone Ketal. J. Org. Chem. 2009, 74, 747–752. [Google Scholar] [CrossRef]
- Endoma-Arias, M.A.A.; Cox, D.P.; Hudlicky, T. General Method of Synthesis for Naloxone, Naltrexone, Nalbuphone, and Nalbuphine by the Reaction of Grignard Reagents with an Oxazolidine Derived from Oxymorphone. Adv. Synth. Catal. 2013, 355, 1869–1873. [Google Scholar] [CrossRef]
- MacHara, A.; Werner, L.; Endoma-Arias, M.A.; Cox, D.P.; Hudlicky, T. Improved Synthesis of Buprenorphine from Thebaine and/or Oripavine via Palladium-Catalyzed N-Demethylation/Acylation and/or Concomitant O-Demethylation. Adv. Synth. Catal. 2012, 354, 613–626. [Google Scholar] [CrossRef]
- Murphy, B.; Šnajdr, I.; Machara, A.; Endoma-Arias, M.A.A.; Stamatatos, T.C.; Cox, D.P.; Hudlický, T. Conversion of Thebaine to Oripavine and Other Useful Intermediates for the Semisynthesis of Opiate-Derived Agents: Synthesis of Hydromorphone. Adv. Synth. Catal. 2014, 356, 2679–2687. [Google Scholar] [CrossRef]
- Orman, J.S.; Keating, G.M. Buprenorphine/Naloxone: A Review of Its Use in the Treatment of Opioid Dependence. Drugs 2009, 69, 577–607. [Google Scholar] [CrossRef] [PubMed]
- Werner, L.; Wernerova, M.; MacHara, A.; Endoma-Arias, M.A.; Duchek, J.; Adams, D.R.; Cox, D.P.; Hudlicky, T. Unexpected N-Demethylation of Oxymorphone and Oxycodone N-Oxides Mediated by the Burgess Reagent: Direct Synthesis of Naltrexone, Naloxone, and Other Antagonists from Oxymorphone. Adv. Synth. Catal. 2012, 354, 2706–2712. [Google Scholar] [CrossRef]
- Millgate, A.G.; Pogson, B.J.; Wilson, I.W.; Kutchan, T.M.; Zenk, M.H.; Gerlach, W.L.; Fist, A.J.; Larkin, P.J. Morphine-Pathway Block in Top1 Poppies. Nature 2004, 431, 413–414. [Google Scholar] [CrossRef]
- Hagel, J.M.; Facchini, P.J. Dioxygenases Catalyze the O-Demethylation Steps of Morphine Biosynthesis in Opium Poppy. Nat. Chem. Biol. 2010, 6, 273–275. [Google Scholar] [CrossRef] [PubMed]
- Fist, A.J.; Miller, J.A.C.; Gregory, D. Papaver Somniferum with High Concentration of Codeine. WO2009143574, 3 December 2009. [Google Scholar]
- Winzer, T.; Walker, T.C.; Meade, F.; Larson, T.R.; Graham, I.A. Modified Plant. WO2017122011, 20 July 2017. [Google Scholar]
- Winzer, T.; Gazda, V.; He, Z.; Kaminski, F.; Kern, M.; Larson, T.R.; Li, Y.; Meade, F.; Teodor, R.; Vaistij, F.E.; et al. A Papaver Somniferum 10-Gene Cluster for Synthesis of the Anticancer Alkaloid Noscapine. Science 2012, 336, 1704–1708. [Google Scholar] [CrossRef]
- Guo, L.; Winzer, T.; Yang, X.; Li, Y.; Ning, Z.; He, Z.; Teodor, R.; Lu, Y.; Bowser, T.A.; Graham, I.A.; et al. The Opium Poppy Genome and Morphinan Production. Science 2018, 362, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Winzer, T.; Graham, I.A.; Walker, T.C. Genes Involved in Noscapine Production. WO2013136057, 19 September 2013. [Google Scholar]
- Winzer, T.; Kern, M.; King, A.J.; Larson, T.R.; Teodor, R.I.; Donninger, S.L.; Li, Y.; Dowle, A.A.; Cartwright, J.; Bates, R.; et al. Morphinan Biosynthesis in Opium Poppy Requires a P450-Oxidoreductase Fusion Protein. Science 2015, 349, 309–3012. [Google Scholar] [CrossRef] [PubMed]
- Winzer, T.; Graham, I.A.; Walker, T.C. Production of Noscapine. WO2016207643, 29 December 2016. [Google Scholar]
- Wijekoon, C.P.; Facchini, P.J. Systematic Knockdown of Morphine Pathway Enzymes in Opium Poppy Using Virus-Induced Gene Silencing. Plant J. 2012, 69, 1052–1063. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.S.; Millgate, A.G.; Chitty, J.A.; Thisleton, J.; Miller, J.A.C.; Fist, A.J.; Gerlach, W.L.; Larkin, P.J. RNAi-Mediated Replacement of Morphine with the Nonnarcotic Alkaloid Reticuline in Opium Poppy. Nat. Biotechnol. 2004, 22, 1559–1566. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.R.; Lal, R.K.; Gupta, A.P.; Misra, H.O.; Pant, V.; Singh, N.K.; Pandey, V. Development of Non-Narcotic (Opiumless and Alkaloid-Free) Opium Poppy, Papaver Somniferum. Plant Breed. 1999, 118, 449–452. [Google Scholar] [CrossRef]

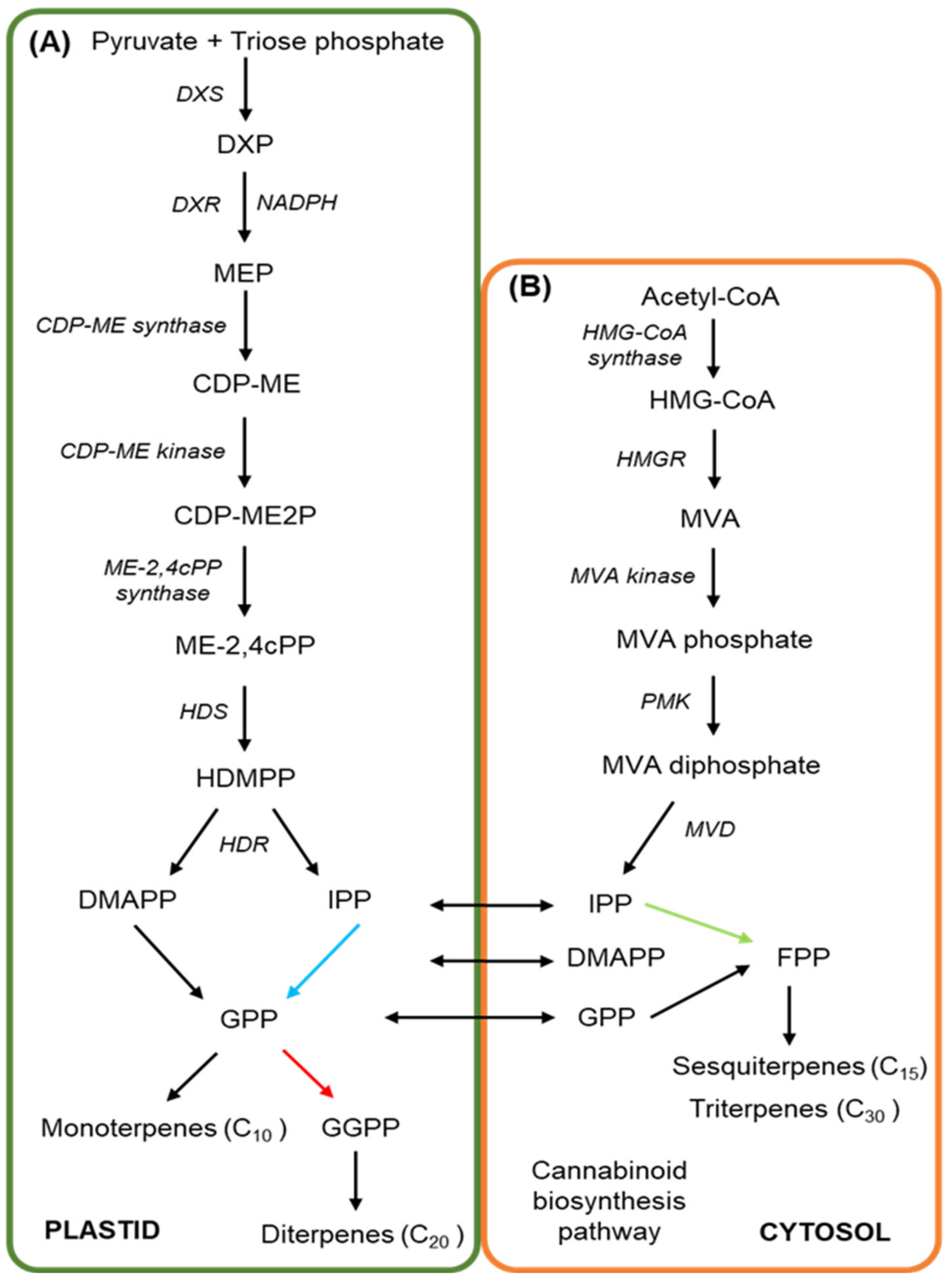
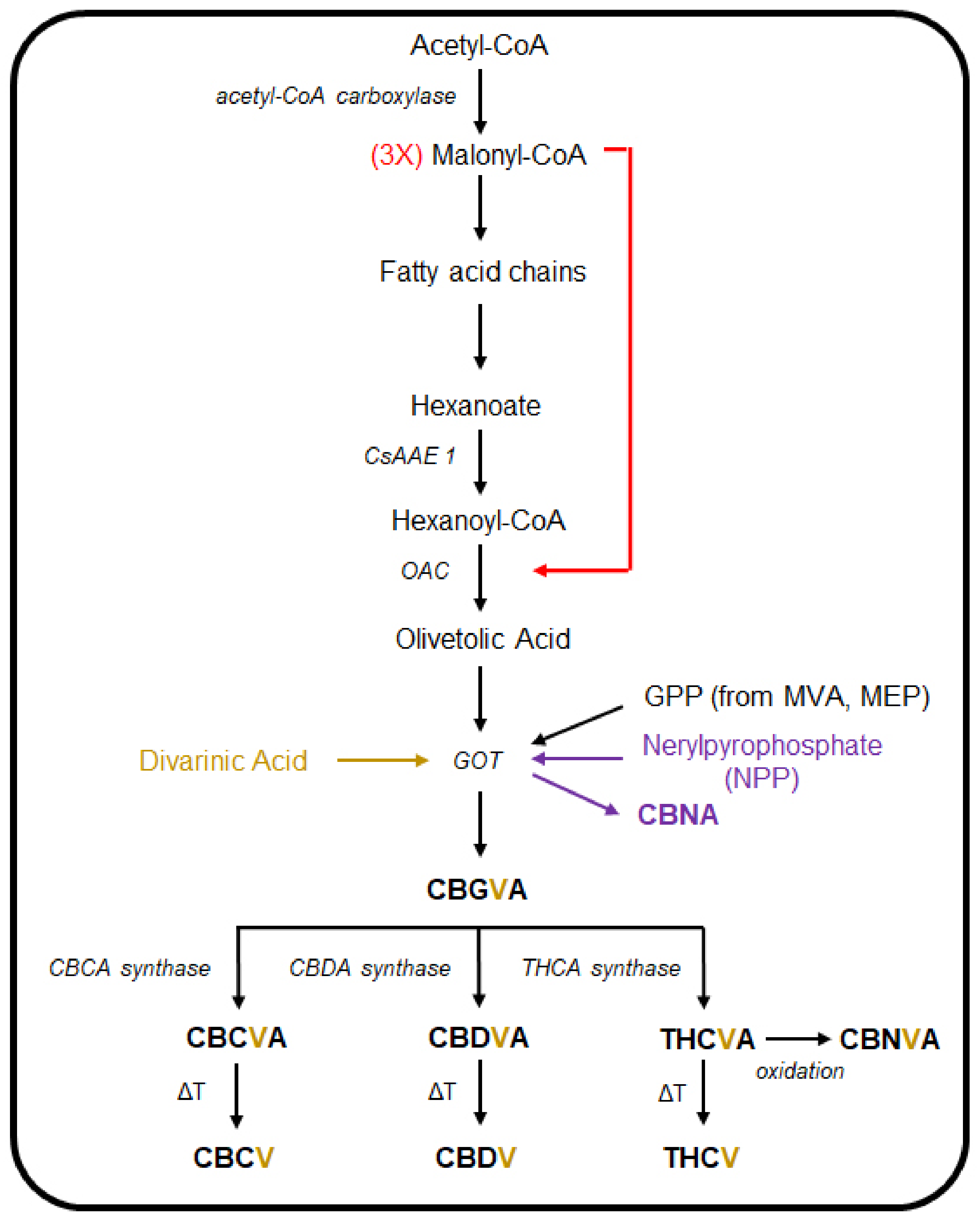
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oultram, J.M.J.; Pegler, J.L.; Bowser, T.A.; Ney, L.J.; Eamens, A.L.; Grof, C.P.L. Cannabis sativa: Interdisciplinary Strategies and Avenues for Medical and Commercial Progression Outside of CBD and THC. Biomedicines 2021, 9, 234. https://doi.org/10.3390/biomedicines9030234
Oultram JMJ, Pegler JL, Bowser TA, Ney LJ, Eamens AL, Grof CPL. Cannabis sativa: Interdisciplinary Strategies and Avenues for Medical and Commercial Progression Outside of CBD and THC. Biomedicines. 2021; 9(3):234. https://doi.org/10.3390/biomedicines9030234
Chicago/Turabian StyleOultram, Jackson M. J., Joseph L. Pegler, Timothy A. Bowser, Luke J. Ney, Andrew L. Eamens, and Christopher P. L. Grof. 2021. "Cannabis sativa: Interdisciplinary Strategies and Avenues for Medical and Commercial Progression Outside of CBD and THC" Biomedicines 9, no. 3: 234. https://doi.org/10.3390/biomedicines9030234
APA StyleOultram, J. M. J., Pegler, J. L., Bowser, T. A., Ney, L. J., Eamens, A. L., & Grof, C. P. L. (2021). Cannabis sativa: Interdisciplinary Strategies and Avenues for Medical and Commercial Progression Outside of CBD and THC. Biomedicines, 9(3), 234. https://doi.org/10.3390/biomedicines9030234







