Energy, Entropy and Quantum Tunneling of Protons and Electrons in Brain Mitochondria: Relation to Mitochondrial Impairment in Aging-Related Human Brain Diseases and Therapeutic Measures
Abstract
1. Introduction
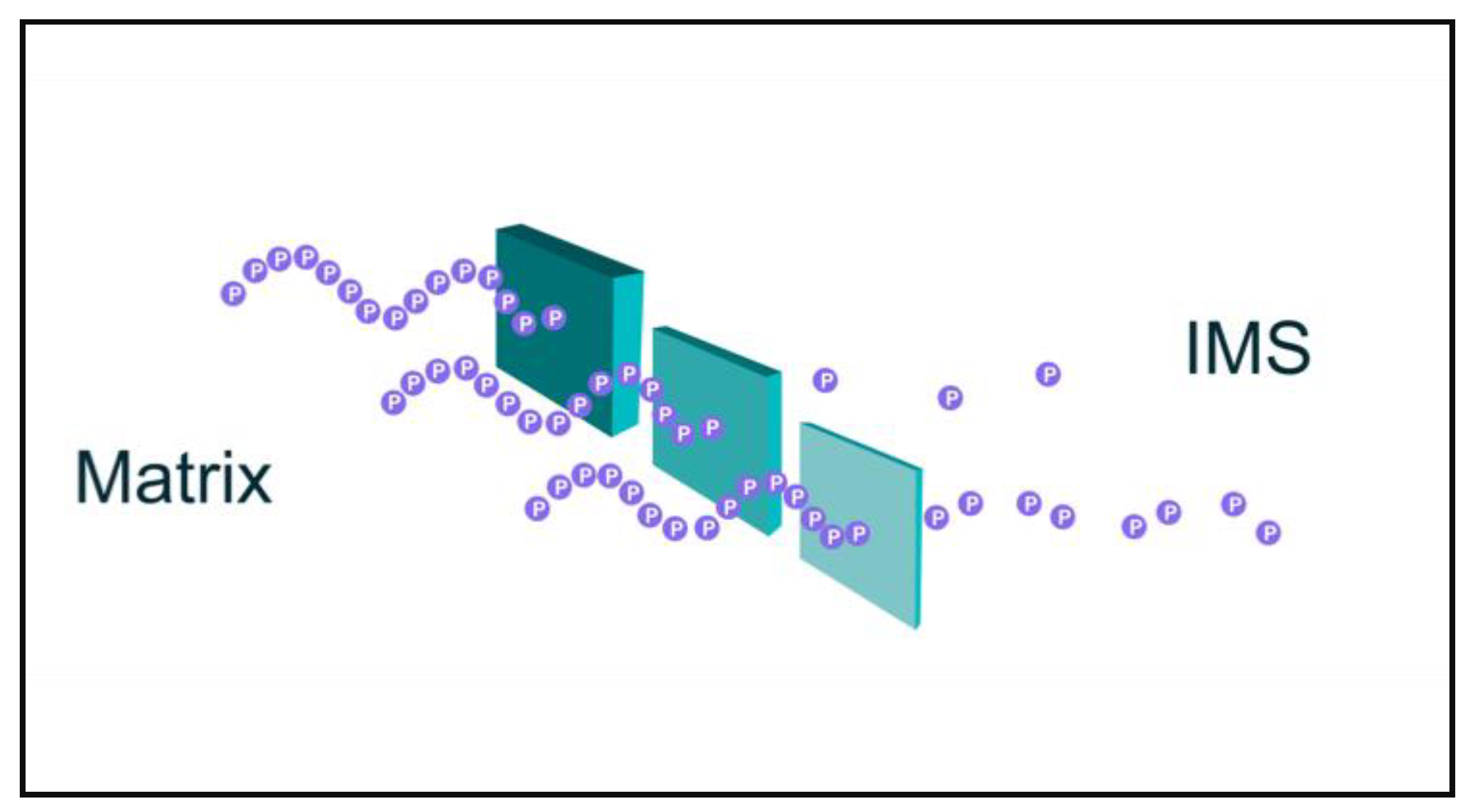
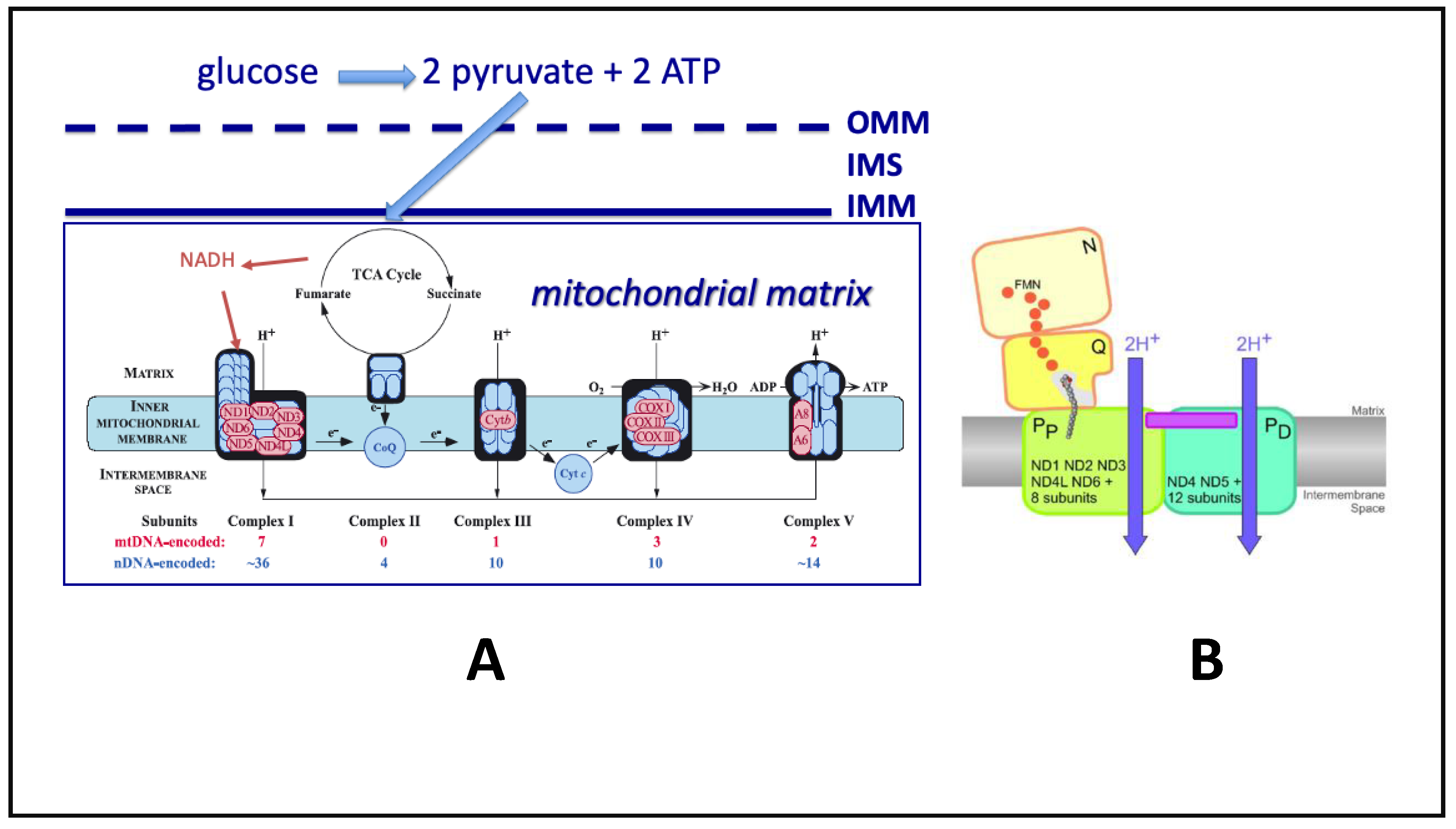
2. Quantum Tunneling of Protons and Electrons in Mitochondria
3. Decoherence
4. Entropy and Neurodegeneration
5. Oxidative Phosphorylation (OXPHOS) Alterations in NDDs
6. Summary of ETC-OXPHOS
7. Brain Mitochondrial Therapeutics
- Correction of mtDNA mutations
- Correction of mtRNA and/or mitochondrial mRNA errors
- Increase in mitochondrial mass leading to increased ETC/OXPHOS capacity to make ATP
- Prevention of epigenetic, nitrative stress (NS) and oxidative stress (OS) damage to ETC/OXPHOS genes or proteins
7.1. Correction of mtDNA Mutations
7.2. Correction of mtRNA and/or Mitochondrial mRNA Errors
7.3. Increase in Mitochondrial Mass Leading to Increased ETC/OXPHOS Capacity to Make ATP
7.4. Prevention of Epigenetic, Nitrative Stress (NS) and Oxidative Stress (OS) Damage to ETC/OXPHOS Genes (Epigenetics) or Proteins (NS and OS)
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blazey, T.; Snyder, A.Z.; Goyal, M.S.; Vlassenko, A.G.; Raichle, M.E. A systematic meta-analysis of oxygen-to-glucose and oxygen-to-carbohydrate ratios in the resting human brain. PLoS ONE 2018, 13, e0204242. [Google Scholar] [CrossRef] [PubMed]
- Von Bartheld, C.S.; Bahney, J.; Herculano-Houzel, S. The search for true numbers of neurons and glial cells in the human brain: A review of 150 years of cell counting. J. Comp. Neurol. 2016, 524, 3865–3895. [Google Scholar] [CrossRef]
- Löwdin, P.-O. Isotope effect in tunneling and its influence on mutation rates. Mutat. Res. Mol. Mech. Mutagen. 1965, 2, 218–221. [Google Scholar] [CrossRef]
- Srivastava, R. The Role of Proton Transfer on Mutations. Front. Chem. 2019, 7, 536. [Google Scholar] [CrossRef]
- Maudlin, T. Philosophy of Physics: Quantum Theory; Princeton Foundations of Contemporary Philosophy Book, 33; Princeton University Press: Princeton, NJ, USA, 2019. [Google Scholar]
- Dröse, S.; Krack, S.; Sokolova, L.; Zwicker, K.; Barth, H.-D.; Morgner, N.; Heide, H.; Steger, M.; Nübel, E.; Zickermann, V.; et al. Functional Dissection of the Proton Pumping Modules of Mitochondrial Complex I. PLoS Biol. 2011, 9, e1001128. [Google Scholar] [CrossRef] [PubMed]
- Leone, V.; Krah, A.; Faraldo-Gómez, J.D. On the Question of Hydronium Binding to ATP-Synthase Membrane Rotors. Biophys. J. 2010, 99, L53–L55. [Google Scholar] [CrossRef][Green Version]
- Schlosshauer, M. Decoherence and the Quantum-to-Classical Transition; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Bennett, J.P. Medical hypothesis: Neurodegenerative diseases arise from oxidative damage to electron tunneling proteins in mitochondria. Med. Hypotheses 2019, 127, 1–4. [Google Scholar] [CrossRef]
- Trixler, F. Quantum Tunnelling to the Origin and Evolution of Life. Curr. Org. Chem. 2013, 17, 1758–1770. [Google Scholar] [CrossRef]
- Yen, F.; Gao, T. Dielectric Anomaly in Ice near 20 K: Evidence of Macroscopic Quantum Phenomena. J. Phys. Chem. Lett. 2015, 6, 2822–2825. [Google Scholar] [CrossRef] [PubMed]
- Cukierman, S. Et tu, Grotthuss! and other unfinished stories. Biochim. Biophys. Acta (BBA) Bioenerg. 2006, 1757, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Gerwert, K.; Freier, E.; Wolf, S. The role of protein-bound water molecules in microbial rhodopsins. Biochim. Biophys. Acta (BBA) Bioenerg. 2014, 1837, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Hammes-Schiffer, S. Proton-Coupled Electron Transfer: Moving Together and Charging Forward. J. Am. Chem. Soc. 2015, 137, 8860–8871. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Belevich, G.; Gamiz-Hernandez, A.P.; Róg, T.; Vattulainen, I.; Verkhovskaya, M.L.; Wikström, M.; Hummer, G.; Kaila, V.R.I. Redox-induced activation of the proton pump in the respiratory complex I. Proc. Natl. Acad. Sci. USA 2015, 112, 11571–11576. [Google Scholar] [CrossRef] [PubMed]
- Wikstrom, M.; Sharma, V.; Kaila, V.R.; Hosler, J.P.; Hummer, G. New Perspectives on Proton Pumping in Cellular Respiration. Chem. Rev. 2015, 115, 2196–2221. [Google Scholar] [CrossRef] [PubMed]
- Ripple, M.O.; Kim, N.; Springett, R. Mammalian Complex I Pumps 4 Protons per 2 Electrons at High and Physiological Proton Motive Force in Living Cells. J. Biol. Chem. 2013, 288, 5374–5380. [Google Scholar] [CrossRef]
- Signes, A.; Fernandez-Vizarra, E. Assembly of Mammalian Oxidative Phosphorylation Complexes I-V and Supercomplexes. Essays Biochem. 2018, 62, 255–270. [Google Scholar] [PubMed]
- Bennett, S.A.; Tanaz, R.; Cobos, S.N.; Torrente, M.P. Epigenetics in amyotrophic lateral sclerosis: A role for histone post-translational modifications in neurodegenerative disease. Transl. Res. 2019, 204, 19–30. [Google Scholar] [CrossRef]
- Consales, C.; Merla, C.; Marino, C.; Benassi, B. The Epigenetic Component of the Brain Response to Electromagnetic Stimulation in Parkinson’s Disease Patients: A Literature Overview. Bioelectromagnetics 2018, 39, 3–14. [Google Scholar] [CrossRef]
- Phillipson, O.T. Alpha-Synuclein, Epigenetics, Mitochondria, Metabolism, Calcium Traffic, & Circadian Dysfunction in Parkinson’s Disease. An Integrated Strategy for Management. Ageing Res. Rev. 2017, 40, 149–167. [Google Scholar]
- Irwin, M.H.; Moos, W.H.; Faller, D.V.; Steliou, K.; Pinkert, C.A. Epigenetic Treatment of Neurodegenerative Disorders: Alzheimer and Parkinson Diseases. Drug Dev. Res. 2016, 77, 109–123. [Google Scholar] [CrossRef]
- Phang, J.M.; Liu, W.; Hancock, C.N.; Fischer, J.W. Proline Metabolism and Cancer: Emerging Links to Glutamine and Collagen. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Parlato, R.; Liss, B. How Parkinson’s Disease Meets Nucleolar Stress. Biochim. Biophys. Acta 2014, 1842, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Valente, A.X.; Das Neves, R.P.; Oliveira, P.J. Epigenetic engineering to reverse the Parkinson’s expression state. Parkinsonism Relat. Disord. 2012, 18, 717–721. [Google Scholar] [CrossRef]
- Hurley, M.J.; Dexter, D.T. Voltage-Gated Calcium Channels and Parkinson’s Disease. Pharmacol. Ther. 2012, 133, 324–333. [Google Scholar] [CrossRef]
- Diederich, N.J.; Parent, A. Parkinson’s Disease: Acquired Frailty of Archaic Neural Networks? J. Neurol. Sci. 2012, 314, 143–151. [Google Scholar] [CrossRef]
- Mendelsohn, A.R.; Larrick, J.W. Rapamycin As an Antiaging Therapeutic?: Targeting Mammalian Target of Rapamycin to Treat Hutchinson–Gilford Progeria and Neurodegenerative Diseases. Rejuvenation Res. 2011, 14, 437–441. [Google Scholar] [CrossRef]
- Dos Santos, S.M.; Romeiro, C.F.R.; Rodrigues, C.A.; Cerqueira, A.R.L.; Monteiro, M.C. Mitochondrial Dysfunction and Alpha-Lipoic Acid: Beneficial or Harmful in Alzheimer’s Disease? Oxidative Med. Cell. Longev. 2019, 2019, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ellison, E.M.; Bradley-Whitman, M.A.; Lovell, M.A. Single-Base Resolution Mapping of 5-Hydroxymethylcytosine Modifications in Hippocampus of Alzheimer’s Disease Subjects. J. Mol. Neurosci. 2017, 63, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Vachharajani, V.T.; Liu, T.; Wang, X.; Hoth, J.J.; Yoza, B.K.; McCall, C.E. Sirtuins Link Inflammation and Metabolism. J. Immunol. Res. 2016, 2016, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kamps, J.J.A.G.; Huang, J.; Poater, J.; Xu, C.; Pieters, B.J.G.E.; Dong, A.; Jasmin, M.; Sherman, W.; Beuming, T.; Bickelhaupt, F.M.; et al. Chemical basis for the recognition of trimethyllysine by epigenetic reader proteins. Nat. Commun. 2015, 6, 8911. [Google Scholar] [CrossRef]
- Salminen, A.; Haapasalo, A.; Kauppinen, A.; Kaarniranta, K.; Soininen, H.; Hiltunen, M. Impaired Mitochondrial Energy Metabolism in Alzheimer’s Disease: Impact on Pathogenesis via Disturbed Epigenetic Regulation of Chromatin Landscape. Prog. Neurobiol. 2015, 131, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Daulatzai, M.A. “Boomerang Neuropathology“ of Late-Onset Alzheimer’s Disease Is Shrouded in Harmful “BDDS“: Breathing, Diet, Drinking, and Sleep during Aging. Neurotox. Res. 2015, 28, 55–93. [Google Scholar] [CrossRef] [PubMed]
- Brewer, G.J. Epigenetic oxidative redox shift (EORS) theory of aging unifies the free radical and insulin signaling theories. Exp. Gerontol. 2010, 45, 173–179. [Google Scholar] [CrossRef]
- Hata, Y.; Ma, N.; Yoneda, M.; Morimoto, S.; Okano, H.; Murayama, S.; Kawanishi, S.; Kuzuhara, S.; Kokubo, Y. Nitrative Stress and Tau Accumulation in Amyotrophic Lateral Sclerosis/Parkinsonism-Dementia Complex (Als/Pdc) in the Kii Penin-sula, Japan. Front. Neurosci. 2017, 11, 751. [Google Scholar] [CrossRef] [PubMed]
- Pacher, P.; Mackie, K. Interplay of cannabinoid 2 (CB2) receptors with nitric oxide synthases, oxidative and nitrative stress, and cell death during remote neurodegeneration. J. Mol. Med. 2012, 90, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Nardo, G.; Pozzi, S.; Pignataro, M.; Lauranzano, E.; Spano, G.; Garbelli, S.; Mantovani, S.; Marinou, K.; Papetti, L.; Monteforte, M.; et al. Amyotrophic Lateral Sclerosis Multiprotein Biomarkers in Peripheral Blood Mononuclear Cells. PLoS ONE 2011, 6, e25545. [Google Scholar] [CrossRef]
- Chen, K.; Northington, F.J.; Martin, L.J. Inducible nitric oxide synthase is present in motor neuron mitochondria and Schwann cells and contributes to disease mechanisms in ALS mice. Anat. Embryol. 2010, 214, 219–234. [Google Scholar] [CrossRef]
- Basso, M.; Samengo, G.; Nardo, G.; Massignan, T.; D’Alessandro, G.; Tartari, S.; Cantoni, L.; Marino, M.; Cheroni, C.; De Biasi, S.; et al. Characterization of Detergent-Insoluble Proteins in ALS Indicates a Causal Link between Nitrative Stress and Aggregation in Pathogenesis. PLoS ONE 2009, 4, e8130. [Google Scholar] [CrossRef]
- Martin, L.J.; Gertz, B.; Pan, Y.; Price, A.C.; Molkentin, J.D.; Chang, Q. The mitochondrial permeability transition pore in motor neurons: Involvement in the pathobiology of ALS mice. Exp. Neurol. 2009, 218, 333–346. [Google Scholar] [CrossRef]
- Martin, L.J. Transgenic Mice with Human Mutant Genes Causing Parkinson’s Disease and Amyotrophic Lateral Sclerosis Provide Common Insight into Mechanisms of Motor Neuron Selective Vulnerability to Degeneration. Rev. Neurosci. 2007, 18, 115–136. [Google Scholar] [CrossRef]
- Kochman, A.; Kośka, C.; Metodiewa, D. Submolecular adventures of brain tyrosine: What are we searching for now? Amino Acids 2002, 23, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Li, H.; Gao, Z. Copper Binding Induces Nitration of NPY under Nitrative Stress: Complicating the Role of NPY in Alzheimer’s Disease. Chem. Res. Toxicol. 2018, 31, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Majkutewicz, I.; Kurowska, E.; Podlacha, M.; Myślińska, D.; Grembecka, B.; Ruciński, J.; Pierzynowska, K.; Wrona, D. Age-dependent effects of dimethyl fumarate on cognitive and neuropathological features in the streptozotocin-induced rat model of Alzheimer’s disease. Brain Res. 2018, 1686, 19–33. [Google Scholar] [CrossRef]
- Guivernau, B.; Bonet, J.; Valls-Comamala, V.; Bosch-Morato, M.; Godoy, J.A.; Inestrosa, N.C.; Peralvarez-Marin, A.; Fernan-dez-Busquets, X.; Andreu, D.; Oliva, B.; et al. Amyloid-Beta Peptide Nitrotyrosination Stabilizes Oligomers and En-hances Nmdar-Mediated Toxicity. J. Neurosci. 2016, 36, 11693–11703. [Google Scholar] [CrossRef]
- Lu, N.; Li, J.; Gao, Z. Key Roles of Tyr 10 in Cu Bound Abeta Complexes and Its Relevance to Alzheimer’s Disease. Arch. Biochem. Biophys. 2015, 584, 1–9. [Google Scholar] [CrossRef]
- Lu, N.; Li, J.; Tian, R.; Peng, Y.Y. Key Roles of Arg(5), Tyr(10) and His Residues in Abeta-Heme Peroxidase: Relevance to Alzheimer’s Disease. Biochem. Biophys. Res. Commun. 2014, 452, 676–681. [Google Scholar] [CrossRef]
- Weinreb, O.; Amit, T.; Bar-Am, O.; Youdim, M.B. Ladostigil: A Novel Multimodal Neuroprotective Drug with Cholines-terase and Brain-Selective Monoamine Oxidase Inhibitory Activities for Alzheimer’s Disease Treatment. Curr. Drug. Targets 2012, 13, 483–494. [Google Scholar] [CrossRef]
- Giridharan, V.V.; Thandavarayan, R.A.; Konishi, T. Effect of Shengmai-san on Cognitive Performance and Cerebral Oxidative Damage in BALB/c Mice. J. Med. Food 2011, 14, 601–609. [Google Scholar] [CrossRef]
- Quinn, J.F.; Bussiere, J.R.; Hammond, R.S.; Montine, T.J.; Henson, E.; Jones, R.E.; Stackman, R.W., Jr. Chronic Dietary Alpha-Lipoic Acid Reduces Deficits in Hippocampal Memory of Aged Tg2576 Mice. Neurobiol. Aging 2007, 28, 213–225. [Google Scholar] [CrossRef]
- Calabrese, V.; Sultana, R.; Scapagnini, G.; Guagliano, E.; Sapienza, M.; Bella, R.; Kanski, J.; Pennisi, G.; Mancuso, C.; Stella, A.M.; et al. Nitrosative Stress, Cellular Stress Response, and Thiol Homeostasis in Patients with Alzheimer’s Disease. Antioxid. Redox Signal. 2006, 8, 1975–1986. [Google Scholar] [CrossRef] [PubMed]
- Giasson, B.I.; Ischiropoulos, H.; Lee, V.M.; Trojanowski, J.Q. The Relationship between Oxidative/Nitrative Stress and Pathological Inclusions in Alzheimer’s and Parkinson’s Diseases. Free Radic. Biol. Med. 2002, 32, 1264–1275. [Google Scholar] [CrossRef]
- Williamson, K.S.; Gabbita, S.P.; Mou, S.; West, M.; Pye, Q.N.; Markesbery, W.R.; Cooney, R.V.; Grammas, P.; Reimann-Philipp, U.; Floyd, R.A.; et al. The Nitration Product 5-Nitro-Gamma-Tocopherol Is Increased in the Alzheimer Brain. Nitric Oxide 2002, 6, 221–227. [Google Scholar] [CrossRef]
- Giasson, B.I.; Duda, J.E.; Murray, I.V.; Chen, Q.; Souza, J.M.; Hurtig, H.I.; Ischiropoulos, H.; Trojanowski, J.Q.; Lee, V.M. Oxidative Damage Linked to Neurodegeneration by Selective Alpha-Synuclein Nitration in Synucleinopathy Lesions. Science 2000, 290, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Wen, X.; Jiang, H.; Wang, J.; Song, N.; Xie, J. Interactions between Iron and Alpha-Synuclein Pathology in Parkinson’s Disease. Free Radic. Biol. Med. 2019, 141, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Maki, R.A.; Holzer, M.; Motamedchaboki, K.; Malle, E.; Masliah, E.; Marsche, G.; Reynolds, G.F. Human Myeloperoxidase (Hmpo) Is Expressed in Neurons in the Substantia Nigra in Parkinson’s Disease and in the Hmpo-Alpha-Synuclein-A53t Mouse Model, Correlating with Increased Nitration and Aggregation of Alpha-Synuclein and Exacerbation of Motor Impairment. Free Radic. Biol. Med. 2019, 141, 115–140. [Google Scholar]
- He, Y.; Yu, Z.; Chen, S. Alpha-Synuclein Nitration and Its Implications in Parkinson’s Disease. ACS Chem. Neurosci. 2019, 10, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.Q.; Wang, Y.L.; Yuan, B.S.; Yuan, X.; Hou, X.O.; Bian, Y.S.; Liu, C.F.; Hu, L.F. Impaired Cbs-H2s Signaling Axis Contributes to Mptp-Induced Neurodegeneration in a Mouse Model of Parkinson’s Disease. Brain Behav. Immun. 2018, 67, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Collier, T.J.; Kanaan, N.M.; Kordower, J.H. Aging and Parkinson’s Disease: Different Sides of the Same Coin? Mov. Disord. 2017, 32, 983–990. [Google Scholar] [CrossRef]
- Kleinknecht, A.; Popova, B.; Lazaro, D.F.; Pinho, R.; Valerius, O.; Outeiro, T.F.; Braus, G.H. C-Terminal Tyrosine Residue Modifications Modulate the Protective Phosphorylation of Serine 129 of Alpha-Synuclein in a Yeast Model of Parkinson’s Disease. PLoS Genet. 2016, 12, e1006098. [Google Scholar] [CrossRef]
- Dias, V.; Junn, E.; Mouradian, M.M. The Role of Oxidative Stress in Parkinson’s Disease. J. Parkinson’s Dis. 2013, 3, 461–491. [Google Scholar] [CrossRef]
- Schildknecht, S.; Gerding, H.R.; Karreman, C.; Drescher, M.; Lashuel, H.A.; Outeiro, T.F.; Di Monte, D.A.; Leist, M. Oxidative and Nitrative Alpha-Synuclein Modifications and Proteostatic Stress: Implications for Disease Mechanisms and Interventions in Synucleinopathies. J. Neurochem. 2013, 125, 491–511. [Google Scholar] [CrossRef]
- Lee, J.; Giordano, S.; Zhang, J. Autophagy, mitochondria and oxidative stress: Cross-talk and redox signalling. Biochem. J. 2011, 441, 523–540. [Google Scholar] [CrossRef] [PubMed]
- Sian-Hulsmann, J.; Mandel, S.; Youdim, M.B.; Riederer, P. The Relevance of Iron in the Pathogenesis of Parkinson’s Disease. J. Neurochem. 2011, 118, 939–957. [Google Scholar] [CrossRef]
- Kupershmidt, L.; Okun, Z.; Amit, T.; Mandel, S.; Saltsman, I.; Mahammed, A.; Bar-Am, O.; Gross, Z.; Youdim, M.B. Metal-locorroles as Cytoprotective Agents against Oxidative and Nitrative Stress in Cellular Models of Neurodegeneration. J. Neurochem. 2010, 113, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Danielson, S.R.; Andersen, J.K. Oxidative and Nitrative Protein Modifications in Parkinson’s Disease. Free Radic. Biol. Med. 2008, 44, 1787–1794. [Google Scholar] [CrossRef]
- Trostchansky, A.; Rubbo, H. Lipid nitration and formation of lipid-protein adducts: Biological insights. Amino Acids 2006, 32, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Weinstock, M.; Luques, L.; Bejar, C.; Shoham, S. Ladostigil, a novel multifunctional drug for the treatment of dementia co-morbid with depression. Focus Extrapyramidal Dysfunct. 2006, 70, 443–446. [Google Scholar] [CrossRef]
- Jenner, P.; Olanow, C.W. The Pathogenesis of Cell Death in Parkinson’s Disease. Neurology 2006, 66 (Suppl. 4), S24–S36. [Google Scholar] [CrossRef]
- Norris, E.H.; Giasson, B.I. Role of Oxidative Damage in Protein Aggregation Associated with Parkinson’s Disease and Related Disorders. Antioxid. Redox Signal. 2005, 7, 672–684. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, A. Postmortem Studies in Parkinson’s Disease. Dialogues Clin. Neurosci. 2004, 6, 281–293. [Google Scholar] [PubMed]
- Yamin, G.; Uversky, V.N.; Fink, A.L. Nitration Inhibits Fibrillation of Human Alpha-Synuclein in Vitro by Formation of Soluble Oligomers. FEBS Lett. 2003, 542, 147–152. [Google Scholar] [CrossRef]
- Bozzo, F.; Mirra, A.; Carri, M.T. Oxidative Stress and Mitochondrial Damage in the Pathogenesis of Als: New Perspectives. Neurosci. Lett. 2017, 636, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Bai, Z.; Qin, X.; Cheng, Y. Aberrations in Oxidative Stress Markers in Amyotrophic Lateral Sclerosis: A Systematic Review and Meta-Analysis. Oxid. Med. Cell. Longev. 2019, 2019, 1712323. [Google Scholar] [CrossRef]
- Butterfield, D.A. Amyloid Beta-Peptide (1-42)-Induced Oxidative Stress and Neurotoxicity: Implications for Neurodegener-ation in Alzheimer’s Disease Brain. A Review. Free Radic. Res. 2002, 36, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, D.A.; Castegna, A.; Drake, J.; Scapagnini, G.; Calabrese, V. Vitamin E and Neurodegenerative Disorders Associated with Oxidative Stress. Nutr. Neurosci. 2002, 5, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, D.A.; Griffin, S.; Munch, G.; Pasinetti, G.M. Amyloid Beta-Peptide and Amyloid Pathology Are Central to the Oxidative Stress and Inflammatory Cascades under Which Alzheimer’s Disease Brain Exists. J. Alzheimer’s Dis. 2002, 4, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, D.A.; Kanski, J. Methionine Residue 35 Is Critical for the Oxidative Stress and Neurotoxic Properties of Alzheimer’s Amyloid Beta-Peptide 1-42. Peptides 2002, 23, 1299–1309. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Lauderback, C.M. Lipid Peroxidation and Protein Oxidation in Alzheimer’s Disease Brain: Potential Causes and Consequences Involving Amyloid Beta-Peptide-Associated Free Radical Oxidative Stress. Free Radic. Biol. Med. 2002, 32, 1050–1060. [Google Scholar] [CrossRef]
- Chang, Y.-T.; Chang, W.-N.; Tsai, N.-W.; Huang, C.-C.; Kung, C.-T.; Su, Y.-J.; Lin, W.-C.; Cheng, B.-C.; Su, C.-M.; Chiang, Y.-F.; et al. The Roles of Biomarkers of Oxidative Stress and Antioxidant in Alzheimer’s Disease: A Systematic Review. BioMed Res. Int. 2014, 2014, 1–14. [Google Scholar] [CrossRef]
- Cheignon, C.; Tomas, M.; Bonnefont-Rousselot, D.; Faller, P.; Hureau, C.; Collin, F. Oxidative stress and the amyloid beta peptide in Alzheimer’s disease. Redox Boil. 2018, 14, 450–464. [Google Scholar] [CrossRef]
- Chen, Z.; Zhong, C. Oxidative Stress in Alzheimer’s Disease. Neurosci. Bull. 2014, 30, 271–281. [Google Scholar] [CrossRef]
- Drake, J.; Kanski, J.; Varadarajan, S.; Tsoras, M.; Butterfield, D.A. Elevation of Brain Glutathione by Gamma-Glutamylcysteine Ethyl Ester Protects against Peroxynitrite-Induced Oxidative Stress. J. Neurosci. Res. 2002, 68, 776–784. [Google Scholar] [CrossRef]
- Kanski, J.; Aksenova, M.; Schoneich, C.; Butterfield, D.A. Substitution of Isoleucine-31 by Helical-Breaking Proline Abol-ishes Oxidative Stress and Neurotoxic Properties of Alzheimer’s Amyloid Beta-Peptide. Free Radic. Biol. Med. 2002, 32, 1205–1211. [Google Scholar] [CrossRef]
- Kanski, J.; Varadarajan, S.; Aksenova, M.; Butterfield, D.A. Role of Glycine-33 and Methionine-35 in Alzheimer’s Amyloid Beta-Peptide 1-42-Associated Oxidative Stress and Neurotoxicity. Biochim. Biophys. Acta 2002, 1586, 190–198. [Google Scholar] [CrossRef]
- LaFontaine, M.A.; Mattson, M.P.; Butterfield, D.A. Oxidative Stress in Synaptosomal Proteins from Mutant Presenilin-1 Knock-in Mice: Implications for Familial Alzheimer’s Disease. Neurochem. Res. 2002, 27, 417–421. [Google Scholar] [CrossRef]
- Tönnies, E.; Trushina, E. Oxidative Stress, Synaptic Dysfunction, and Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 57, 1105–1121. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.; Li, L.; Perry, G.; Lee, H.G.; Zhu, X. Oxidative Stress and Mitochondrial Dysfunction in Alzheimer’s Disease. Biochim. Biophys. Acta 2014, 1842, 1240–1247. [Google Scholar] [CrossRef]
- Guo, J.D.; Zhao, X.; Li, Y.; Li, G.R.; Liu, X.L. Damage to Dopaminergic Neurons by Oxidative Stress in Parkinson’s Disease (Review). Int. J. Mol. Med. 2018, 41, 1817–1825. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.T. Oxidative Stress and Mitochondrial Dysfunction-Linked Neurodegenerative Disorders. Neurol. Res. 2017, 39, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Ali, S.A. Oxidative Stress-Related Biomarkers in Parkinson’s Disease: A Systematic Review and Meta-Analysis. Iran. J. Neurol. 2018, 17, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.; Wong, Y.C.; Ysselstein, D.; Severino, A.; Krainc, D. Synaptic, Mitochondrial, and Lysosomal Dysfunction in Parkinson’s Disease. Trends Neurosci. 2019, 42, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Poewe, W.; Seppi, K.; Tanner, C.M.; Halliday, G.M.; Brundin, P.; Volkmann, J.; Schrag, A.E.; Lang, A.E. Parkinson Disease. Nat. Rev. Dis. Primers 2017, 3, 17013. [Google Scholar] [CrossRef] [PubMed]
- Raza, C.; Anjum, R.; Shakeel, N.U.A. Parkinson’s Disease: Mechanisms, Translational Models and Management Strate-gies. Life Sci. 2019, 226, 77–90. [Google Scholar] [CrossRef]
- Trist, B.G.; Hare, D.J.; Double, K.L. Oxidative Stress in the Aging Substantia Nigra and the Etiology of Parkinson’s Disease. Aging Cell 2019, 18, e13031. [Google Scholar] [CrossRef]
- Wei, Z.; Li, X.; Li, X.; Liu, Q.; Cheng, Y. Oxidative Stress in Parkinson’s Disease: A Systematic Review and Meta-Analysis. Front. Mol. Neurosci. 2018, 11, 236. [Google Scholar] [CrossRef]
- Santibanez-Koref, M.; Griffin, H.; Turnbull, D.M.; Chinnery, P.F.; Herbert, M.; Hudson, F. Assessing Mitochondrial Heteroplasmy Using Next Generation Sequencing: A Note of Caution. Mitochondrion 2019, 46, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Craven, L.; Murphy, J.; Turnbull, D.M.; Taylor, R.W.; Gorman, G.S.; McFarland, R. Scientific and Ethical Issues in Mitochondrial Donation. New Bioeth. 2018, 24, 57–73. [Google Scholar] [CrossRef]
- Chinnery, P.F.; Craven, L.; Mitalipov, S.; Stewart, J.B.; Herbert, M.; Turnbull, D.M. The challenges of mitochondrial replacement. PLoS Genet. 2014, 10, e1004315. [Google Scholar] [CrossRef]
- Craven, L.; Murphy, J.L.; Turnbull, D.M. Mitochondrial donation—Hope for families with mitochondrial DNA disease. Emerg. Top. Life Sci. 2020, 4, 151–154. [Google Scholar] [CrossRef]
- Hyslop, L.A.; Blakeley, P.; Craven, L.; Richardson, J.; Fogarty, N.M.E.; Fragouli, E.; Lamb, M.; Wamaitha, S.E.; Prathalingam, N.; Zhang, Q.; et al. Towards clinical application of pronuclear transfer to prevent mitochondrial DNA disease. Nature 2016, 534, 383–386. [Google Scholar] [CrossRef]
- Pickett, S.J.; Blain, A.; Ng, Y.S.; Wilson, I.J.; Taylor, R.W.; McFarland, R.; Turnbull, D.M.; Gorman, G.S. Mitochondrial Donation—Which Women Could Benefit? New Engl. J. Med. 2019, 380, 1971–1972. [Google Scholar] [CrossRef]
- Richardson, J.; Irving, L.; Hyslop, L.A.; Choudhary, M.; Murdoch, A.; Turnbull, D.M.; Herbert, M. Concise Reviews: Assisted Reproductive Technologies to Prevent Transmission of Mitochondrial DNA Disease. Stem Cells 2015, 33, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Amato, P.; Tachibana, M.; Sparman, M.; Mitalipov, S. Three-Parent in Vitro Fertilization: Gene Replacement for the Prevention of Inherited Mitochondrial Diseases. Fertil. Steril. 2014, 101, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Byrne, J.A.; Pedersen, D.A.; Clepper, L.L.; Nelson, M.; Sanger, W.G.; Gokhale, S.; Wolf, D.P.; Mitalipov, S.M. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature 2007, 450, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Folmes, C.D.L.; Ma, H.; Mitalipov, S.; Terzic, A. Mitochondria in pluripotent stem cells: Stemness regulators and disease targets. Curr. Opin. Genet. Dev. 2016, 38, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kang, E.; Wu, J.; Gutierrez, N.M.; Koski, A.; Tippner-Hedges, R.; Agaronyan, K.; Platero-Luengo, A.; Martinez-Redondo, P.; Ma, H.; Lee, Y.; et al. Mitochondrial Re-placement in Human Oocytes Carrying Pathogenic Mitochondrial DNA Mutations. Nature 2016, 540, 270–275. [Google Scholar] [CrossRef]
- Ma, H.; Folmes, C.D.L.; Wu, J.; Morey, R.; Mora-Castilla, S.; Ocampo, A.; Ma, L.; Poulton, J.; Wang, X.; Ahmed, R.; et al. Metabolic rescue in pluripotent cells from patients with mtDNA disease. Nat. Cell Biol. 2015, 524, 234–238. [Google Scholar] [CrossRef]
- Ma, H.; Marti Gutierrez, N.; Morey, R.; Van Dyken, C.; Kang, E.; Hayama, T.; Lee, Y.; Li, Y.; Tippner-Hedges, R.; Wolf, D.P.; et al. Incompatibility between Nuclear and Mitochondrial Genomes Contributes to an Interspecies Re-productive Barrier. Cell Metab. 2016, 24, 283–294. [Google Scholar] [CrossRef]
- Ma, H.; O’Neil, R.C.; Gutierrez, N.M.; Hariharan, M.; Zhang, Z.Z.; He, Y.; Cinnioglu, C.; Kayali, R.; Kang, E.; Lee, Y.; et al. Functional Human Oocytes Generated by Transfer of Polar Body Genomes. Cell Stem Cell 2017, 20, 112–119. [Google Scholar] [CrossRef]
- Mitalipov, S.; Wolf, D.P. Clinical and Ethical Implications of Mitochondrial Gene Transfer. Trends Endocrinol. Metab. 2014, 25, 5–7. [Google Scholar] [CrossRef]
- Tachibana, M.; Amato, P.; Sparman, M.; Gutierrez, N.M.; Tippner-Hedges, R.; Ma, H.; Kang, E.; Fulati, A.; Lee, H.-S.; Sritanaudomchai, H.; et al. Human Embryonic Stem Cells Derived by Somatic Cell Nuclear Transfer. Cell 2013, 153, 1228–1238. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, M.; Amato, P.; Sparman, M.; Woodward, J.; Sanchis, D.M.; Ma, H.; Gutierrez, N.M.; Tippner-Hedges, R.; Kang, E.; Lee, H.-S.; et al. Towards germline gene therapy of inherited mitochondrial diseases. Nat. Cell Biol. 2013, 493, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, M.; Sparman, M.; Mitalipov, S. Chromosome transfer in mature oocytes. Fertil. Steril. 2012, 97, e16. [Google Scholar] [CrossRef]
- Tachibana, M.; Sparman, M.; Sritanaudomchai, H.; Ma, H.; Clepper, L.; Woodward, J.; Li, Y.; Ramsey, C.; Kolotushkina, O.; Mitalipov, S. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature 2009, 461, 367–372. [Google Scholar] [CrossRef]
- Wolf, D.P.; Mitalipov, N.; Mitalipov, S. Mitochondrial replacement therapy in reproductive medicine. Trends Mol. Med. 2015, 21, 68–76. [Google Scholar] [CrossRef]
- Wolf, D.P.; Mitalipov, S. Mitochondrial Replacement Therapies Can Circumvent Mtdna-Based Disease Transmission. Cell Metab. 2014, 20, 6–8. [Google Scholar] [CrossRef][Green Version]
- Zhao, M.T.; Chen, H.; Liu, Q.; Shao, N.Y.; Sayed, N.; Wo, H.T.; Zhang, J.Z.; Ong, S.G.; Liu, C.; Kim, Y.; et al. Molecular and Functional Resemblance of Differentiated Cells Derived from Isogenic Human Ipscs and Scnt-Derived Escs. Proc. Natl. Acad. Sci. USA 2017, 114, E11111–E11120. [Google Scholar] [CrossRef] [PubMed]
- Elson, J.L.; Swalwell, H.; Blakely, E.L.; McFarland, R.; Taylor, R.W.; Turnbull, D.M. Pathogenic mitochondrial tRNA mutations—Which mutations are inherited and why? Hum. Mutat. 2009, 30, E984–E992. [Google Scholar] [CrossRef] [PubMed]
- Yarham, J.W.; Elson, J.L.; Blakely, E.L.; McFarland, R.; Taylor, R.W. Mitochondrial tRNA mutations and disease. Wiley Interdiscip. Rev. RNA 2010, 1, 304–324. [Google Scholar] [CrossRef]
- Ribas de Pouplana, L. The Mitochondrial Trna Conundrum. Nat. Rev. Mol. Cell Biol. 2020, 21, 361. [Google Scholar] [CrossRef]
- Richter, U.; Evans, M.E.; Clark, W.C.; Marttinen, P.; Shoubridge, E.A.; Suomalainen, A.; Wredenberg, A.; Wedell, A.; Pan, T.; Battersby, B.J. Rna Modification Landscape of the Human Mitochondrial Trna(Lys) Regulates Protein Synthesis. Nat. Commun. 2018, 9, 3966. [Google Scholar] [CrossRef]
- Karicheva, O.Z.; Kolesnikova, O.A.; Schirtz, T.; Vysokikh, M.Y.; Mager-Heckel, A.-M.; Lombès, A.; Boucheham, A.; Krasheninnikov, I.A.; Martin, R.P.; Entelis, N.; et al. Correction of the consequences of mitochondrial 3243A>G mutation in the MT-TL1 gene causing the MELAS syndrome by tRNA import into mitochondria. Nucleic Acids Res. 2011, 39, 8173–8186. [Google Scholar] [CrossRef]
- Wang, G.; Shimada, E.; Zhang, J.; Hong, J.S.; Smith, G.M.; Teitell, M.A.; Koehler, C.M. Correcting human mitochondrial mutations with targeted RNA import. Proc. Natl. Acad. Sci. USA 2012, 109, 4840–4845. [Google Scholar] [CrossRef]
- Alfadhel, M.; Nashabat, M.; Abu Ali, Q.; Hundallah, K. Mitochondrial iron-sulfur cluster biogenesis from molecular understanding to clinical disease. Neuroscience 2017, 22, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Artika, I.M. Current understanding of structure, function and biogenesis of yeast mitochondrial ATP synthase. J. Bioenerg. Biomembr. 2019, 51, 315–328. [Google Scholar] [CrossRef]
- Babbitt, S.E.; Sutherland, M.C.; San Francisco, B.; Mendez, D.L.; Kranz, R.G. Mitochondrial cytochrome c biogenesis: No longer an enigma. Trends Biochem. Sci. 2015, 40, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Barros, M.H.; McStay, G.P. Modular biogenesis of mitochondrial respiratory complexes. Mitochondrion 2020, 50, 94–114. [Google Scholar] [CrossRef] [PubMed]
- Bishop, D.J.; Botella, J.; Genders, A.J.; Lee, M.J.-C.; Saner, N.J.; Kuang, J.; Yan, X.; Granata, C. High-Intensity Exercise and Mitochondrial Biogenesis: Current Controversies and Future Research Directions. Physiology 2019, 34, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Bouchez, C.; Devin, A. Mitochondrial Biogenesis and Mitochondrial Reactive Oxygen Species (Ros): A Complex Rela-tionship Regulated by the Camp/Pka Signaling Pathway. Cells 2019, 8, 287. [Google Scholar] [CrossRef] [PubMed]
- Bragoszewski, P.; Turek, M.; Chacinska, A. Control of mitochondrial biogenesis and function by the ubiquitin–proteasome system. Open Biol. 2017, 7. [Google Scholar] [CrossRef]
- Bykov, Y.S.; Rapaport, D.; Herrmann, J.M.; Schuldiner, M. Cytosolic Events in the Biogenesis of Mitochondrial Proteins. Trends Biochem. Sci. 2020, 45, 650–667. [Google Scholar] [CrossRef]
- Cameron, R.B.; Beeson, C.C.; Schnellmann, R.G. Development of Therapeutics that Induce Mitochondrial Biogenesis for the Treatment of Acute and Chronic Degenerative Diseases. J. Med. Chem. 2016, 59, 10411–10434. [Google Scholar] [CrossRef]
- Carelli, V.; Maresca, A.; Caporali, L.; Trifunov, S.; Zanna, C.; Rugolo, M. Mitochondria: Biogenesis and mitophagy balance in segregation and clonal expansion of mitochondrial DNA mutations. Int. J. Biochem. Cell Biol. 2015, 63, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Cherry, A.D.; Piantadosi, C.A. Regulation of Mitochondrial Biogenesis and Its Intersection with Inflammatory Responses. Antioxid. Redox Signal. 2015, 22, 965–976. [Google Scholar] [CrossRef]
- Craig, D.M.; Ashcroft, S.P.; Belew, M.Y.; Stocks, B.; Currell, K.; Baar, K.; Philp, A. Utilizing small nutrient compounds as enhancers of exercise-induced mitochondrial biogenesis. Front. Physiol. 2015, 6, 296. [Google Scholar] [CrossRef] [PubMed]
- Davinelli, S.; De Stefani, D.; De Vivo, I.; Scapagnini, G. Polyphenols as Caloric Restriction Mimetics Regulating Mitochondrial Biogenesis and Mitophagy. Trends Endocrinol. Metab. 2020, 31, 536–550. [Google Scholar] [CrossRef]
- De Oliveira, M.R.; Jardim, F.R.; Setzer, W.N.; Nabavi, S.M. Curcumin, mitochondrial biogenesis, and mitophagy: Exploring recent data and indicating future needs. Biotechnol. Adv. 2016, 34, 813–826. [Google Scholar] [CrossRef]
- De Oliveira, M.R.; Nabavi, S.F.; Manayi, A.; Daglia, M.; Hajheydari, Z. Resveratrol and the mitochondria: From triggering the intrinsic apoptotic pathway to inducing mitochondrial biogenesis, a mechanistic view. Biochim. Biophys. Acta (BBA) Gen. Subj. 2016, 1860, 727–745. [Google Scholar] [CrossRef]
- Doan, K.N.; Ellenrieder, L.; Becker, T. Mitochondrial porin links protein biogenesis to metabolism. Curr. Genet. 2019, 65, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Dorn, G.W., 2nd; Vega, R.B.; Kelly, D.P. Mitochondrial Biogenesis and Dynamics in the Developing and Diseased Heart. Genes Dev. 2015, 29, 1981–1991. [Google Scholar] [CrossRef] [PubMed]
- Drwesh, L.; Rapaport, D. Biogenesis Pathways of Alpha-Helical Mitochondrial Outer Membrane Proteins. Biol. Chem. 2020, 401, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Dutkiewicz, R.; Nowak, M. Molecular chaperones involved in mitochondrial iron–sulfur protein biogenesis. JBIC J. Biol. Inorg. Chem. 2018, 23, 569–579. [Google Scholar] [CrossRef]
- Edens, B.M.; Miller, N.; Ma, Y.-C. Impaired Autophagy and Defective Mitochondrial Function: Converging Paths on the Road to Motor Neuron Degeneration. Front. Cell. Neurosci. 2016, 10, 44. [Google Scholar] [CrossRef]
- Edwards, R.; Gerlich, S.; Tokatlidis, K. The biogenesis of mitochondrial intermembrane space proteins. Biol. Chem. 2020, 401, 737–747. [Google Scholar] [CrossRef]
- Ellenrieder, L.; Martensson, C.U.; Becker, T. Biogenesis of Mitochondrial Outer Membrane Proteins, Problems and Diseases. Biol. Chem. 2015, 396, 1199–1213. [Google Scholar] [CrossRef]
- Erlich, A.T.; Tryon, L.D.; Crilly, M.J.; Memme, J.M.; Moosavi, Z.S.M.; Oliveira, A.N.; Beyfuss, K.; Hood, D.A. Function of specialized regulatory proteins and signaling pathways in exercise-induced muscle mitochondrial biogenesis. Integr. Med. Res. 2016, 5, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Finsterer, J.; Scorza, F.A. Effects of antiepileptic drugs on mitochondrial functions, morphology, kinetics, biogenesis, and survival. Epilepsy Res. 2017, 136, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Fontecha-Barriuso, M.; Martin-Sanchez, D.; Martinez-Moreno, J.M.; Monsalve, M.; Ramos, A.M.; Sanchez-Niño, M.D.; Ruiz-Ortega, M.; Ortiz, A.; Sanz, A.B. The Role of PGC-1α and Mitochondrial Biogenesis in Kidney Diseases. Biomolecules 2020, 10, 347. [Google Scholar] [CrossRef] [PubMed]
- Franz, A.; Kevei, É.; Hoppe, T. Double-edged alliance: Mitochondrial surveillance by the UPS and autophagy. Curr. Opin. Cell Biol. 2015, 37, 18–27. [Google Scholar] [CrossRef]
- Fu, W.; Liu, Y.; Yin, H. Mitochondrial Dynamics: Biogenesis, Fission, Fusion, and Mitophagy in the Regulation of Stem Cell Behaviors. Stem Cells Int. 2019, 2019, 1–15. [Google Scholar] [CrossRef]
- Golpich, M.; Amini, E.; Mohamed, Z.; Ali, R.A.; Ibrahim, N.M.; Ahmadiani, A. Mitochondrial Dysfunction and Biogenesis in Neurodegenerative diseases: Pathogenesis and Treatment. CNS Neurosci. Ther. 2017, 23, 5–22. [Google Scholar] [CrossRef]
- Gomes, J.V.P.; Rigolon, T.C.B.; da Silveira Souza, M.S.; Alvarez-Leite, J.I.; Lucia, C.M.D.; Martino, H.S.D.; Rosa, C.D.O.B. Anti-obesity Effects of Anthocyanins on Mitochondrial Biogenesis, Inflammation, and Oxidative Stress: A Systematic Review. Nutrition 2019, 66, 192–202. [Google Scholar] [CrossRef]
- Granata, C.; Jamnick, N.A.; Bishop, D.J. Principles of Exercise Prescription, and How They Influence Exercise-Induced Changes of Transcription Factors and Other Regulators of Mitochondrial Biogenesis. Sports Med. 2018, 48, 1541–1559. [Google Scholar] [CrossRef]
- Grevel, A.; Pfanner, N.; Becker, T. Coupling of import and assembly pathways in mitochondrial protein biogenesis. Biol. Chem. 2019, 401, 117–129. [Google Scholar] [CrossRef]
- Groennebaek, T.; Vissing, K. Impact of Resistance Training on Skeletal Muscle Mitochondrial Biogenesis, Content, and Function. Front. Physiol. 2017, 8, 713. [Google Scholar] [CrossRef]
- Gupta, A.; Becker, T. Mechanisms and pathways of mitochondrial outer membrane protein biogenesis. Biochim. Biophys. Acta (BBA) Bioenerg. 2021, 1862, 148323. [Google Scholar] [CrossRef] [PubMed]
- Gureev, A.P.; Shaforostova, E.A.; Popov, V.N. Regulation of Mitochondrial Biogenesis as a Way for Active Longevity: Interaction between the Nrf2 and PGC-1α Signaling Pathways. Front. Genet. 2019, 10, 435. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Duan, Y.; Yao, K.; Li, F.; Hou, Y.; Wu, G.; Yin, Y. Beta-Hydroxy-Beta-Methylbutyrate, Mitochondrial Biogenesis, and Skeletal Muscle Health. Amino Acids 2016, 48, 653–664. [Google Scholar] [CrossRef]
- Herrmann, J.M.; Riemer, J. Apoptosis inducing factor and mitochondrial NADH dehydrogenases: Redox-controlled gear boxes to switch between mitochondrial biogenesis and cell death. Biol. Chem. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hey-Mogensen, M.; Clausen, T.R. Targeting Mitochondrial Biogenesis and Mitochondrial Substrate Utilization to Treat Obesity and Insulin Resistance, Respectively—Two Data-Driven Hypotheses. Curr. Diabetes Rev. 2017, 13, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Hiraumi, Y.; Huang, C.; Andres, A.M.; Xiong, Y.; Ramil, J.; Gottlieb, R.A. Myogenic Progenitor Cell Differentiation Is Dependent on Modulation of Mitochondrial Biogenesis through Autophagy. In Etiology and Morphogenesis of Congenital Heart Disease: From Gene Function and Cellular Interaction to Morphology; Nakanishi, T., Markwald, R.R., Baldwin, H.S., Keller, B.B., Srivastava, D., Yamagishi, H., Eds.; Springer: Tokyo, Japan, 2016. [Google Scholar]
- Hood, D.A.; Tryon, L.D.; Carter, H.N.; Kim, Y.; Chen, C.C. Unravelling the Mechanisms Regulating Muscle Mitochondrial Biogenesis. Biochem. J. 2016, 473, 2295–2314. [Google Scholar] [CrossRef]
- Horten, P.; Colina-Tenorio, L.; Rampelt, H. Biogenesis of Mitochondrial Metabolite Carriers. Biomolecules 2020, 10, 1008. [Google Scholar] [CrossRef]
- Islam, H.; Edgett, B.A.; Gurd, B.J. Coordination of mitochondrial biogenesis by PGC-1α in human skeletal muscle: A reevaluation. Metabolism 2018, 79, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Islam, H.; Hood, D.A.; Gurd, B.J. Looking beyond PGC-1α: Emerging regulators of exercise-induced skeletal muscle mitochondrial biogenesis and their activation by dietary compounds. Appl. Physiol. Nutr. Metab. 2020, 45, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Jamwal, S.; Blackburn, J.K.; Elsworth, J.D. PPARgamma/Pgc1alpha Signaling as a Potential Therapeutic Target for Mitochondrial Biogenesis in Neurodegenerative Disorders. Pharmacol. Ther. 2020, 107705. [Google Scholar] [CrossRef]
- Kandezi, N.; Mohammadi, M.; Ghaffari, M.; Gholami, M.; Motaghinejad, M.; Safari, S. Novel Insight to Neuroprotective Potential of Curcumin: A Mechanistic Review of Possible Involvement of Mitochondrial Biogenesis and PI3/Akt/ GSK3 or PI3/Akt/CREB/BDNF Signaling Pathways. Int. J. Mol. Cell. Med. 2020, 9, 1–32. [Google Scholar]
- Kiyama, T.; Chen, C.K.; Wang, S.W.; Pan, P.; Ju, Z.; Wang, J.; Takada, S.; Klein, W.H.; Mao, C.A. Essential Roles of Mitochondrial Biogenesis Regulator Nrf1 in Retinal Development and Homeostasis. Mol. Neurodegener. 2018, 13, 56. [Google Scholar] [CrossRef] [PubMed]
- Kondadi, A.K.; Anand, R.; Reichert, A.S. Functional Interplay between Cristae Biogenesis, Mitochondrial Dynamics and Mitochondrial DNA Integrity. Int. J. Mol. Sci. 2019, 20, 4311. [Google Scholar] [CrossRef]
- Krämer, L.; Groh, C.; Herrmann, J.M. The proteasome: Friend and foe of mitochondrial biogenesis. FEBS Lett. 2020. [Google Scholar] [CrossRef]
- Li, P.A.; Hou, X.; Hao, S. Mitochondrial biogenesis in neurodegeneration. J. Neurosci. Res. 2017, 95, 2025–2029. [Google Scholar] [CrossRef]
- Lill, R.; Freibert, S.-A. Mechanisms of Mitochondrial Iron-Sulfur Protein Biogenesis. Annu. Rev. Biochem. 2020, 89, 471–499. [Google Scholar] [CrossRef]
- Lima, T.I.; Araujo, H.N.; Menezes, E.S.; Sponton, C.H.; Araújo, M.B.; Bomfim, L.H.; Queiroz, A.L.; Passos, M.A.; DE Sousa, T.A.; Hirabara, S.M.; et al. Role of microRNAs on the Regulation of Mitochondrial Biogenesis and Insulin Signaling in Skeletal Muscle. J. Cell. Physiol. 2016, 232, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Widlund, H.R.; Puigserver, P. PGC-1 Coactivators: Shepherding the Mitochondrial Biogenesis of Tumors. Trends Cancer 2016, 2, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Manganas, P.; MacPherson, L.; Tokatlidis, K. Oxidative protein biogenesis and redox regulation in the mitochondrial intermembrane space. Cell Tissue Res. 2017, 367, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Mansilla, N.; Racca, S.; Gras, D.E.; Gonzalez, D.H.; Welchen, E. The Complexity of Mitochondrial Complex IV: An Update of Cytochrome c Oxidase Biogenesis in Plants. Int. J. Mol. Sci. 2018, 19, 662. [Google Scholar] [CrossRef]
- McKie, G.L.; Wright, D.C. Biochemical Adaptations in White Adipose Tissue Following Aerobic Exercise: From Mitochondrial Biogenesis to Browning. Biochem. J. 2020, 477, 1061–1081. [Google Scholar] [CrossRef] [PubMed]
- Melber, A.; Winge, D.R. Steps toward Understanding Mitochondrial Fe/S Cluster Biogenesis. Methods Enzym. 2018, 599, 265–292. [Google Scholar] [CrossRef]
- Miao, L.; Shen, X.; Whiteman, M.; Xin, H.; Shen, Y.; Xin, X.; Moore, P.K.; Zhu, Y.-Z. Hydrogen Sulfide Mitigates Myocardial Infarction via Promotion of Mitochondrial Biogenesis-Dependent M2 Polarization of Macrophages. Antioxid. Redox Signal. 2016, 25, 268–281. [Google Scholar] [CrossRef]
- Moulin, C.; Caumont-Sarcos, A.; Ieva, R. Mitochondrial Presequence Import: Multiple Regulatory Knobs Fine-Tune Mitochondrial Biogenesis and Homeostasis. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 930–944. [Google Scholar] [CrossRef]
- Ndi, M.; Marin-Buera, L.; Salvatori, R.; Singh, A.P.; Ott, M. Biogenesis of the bc1 Complex of the Mitochondrial Respiratory Chain. J. Mol. Biol. 2018, 430, 3892–3905. [Google Scholar] [CrossRef]
- Nirwane, A.; Majumdar, A. Understanding Mitochondrial Biogenesis through Energy Sensing Pathways and Its Translation in Cardio-Metabolic Health. Arch. Physiol. Biochem. 2018, 124, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Nowinski, S.M.; Van Vranken, J.G.; Dove, K.K.; Rutter, J. Impact of Mitochondrial Fatty Acid Synthesis on Mitochondrial Biogenesis. Curr. Biol. 2018, 28, R1212–R1219. [Google Scholar] [CrossRef] [PubMed]
- Ogunbona, O.B.; Claypool, S.M. Emerging Roles in the Biogenesis of Cytochrome C Oxidase for Members of the Mitochondrial Carrier Family. Front. Cell. Dev. Biol. 2019, 7, 3. [Google Scholar] [CrossRef]
- Ortega, S.P.; Chouchani, E.T.; Boudina, S. Stress turns on the heat: Regulation of mitochondrial biogenesis and UCP1 by ROS in adipocytes. Adipocyte 2016, 6, 56–61. [Google Scholar] [CrossRef]
- Panajatovic, M.; Singh, F.; Duthaler, U.; Krahenbuhl, S.; Bouitbir, J. Role of Pgc-1-Alpha-Associated Mitochondrial Biogenesis in Statin-Induced Myotoxicity. Eur. Cardiol. 2020, 15, e35. [Google Scholar] [CrossRef] [PubMed]
- Perry, C.G.; Hawley, J.A. Molecular Basis of Exercise-Induced Skeletal Muscle Mitochondrial Biogenesis: Historical Advances, Current Knowledge, and Future Challenges. Cold Spring Harb. Perspect. Med. 2017, 8, a029686. [Google Scholar] [CrossRef]
- Pfanner, N.; Warscheid, B.; Wiedemann, N. Mitochondrial proteins: From biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 2019, 20, 267–284. [Google Scholar] [CrossRef]
- Picca, A.; Lezza, A.M. Regulation of Mitochondrial Biogenesis through Tfam-Mitochondrial DNA Interactions: Useful Insights from Aging and Calorie Restriction Studies. Mitochondrion 2015, 25, 67–75. [Google Scholar] [CrossRef]
- Ploumi, C.; Daskalaki, I.; Tavernarakis, N. Mitochondrial biogenesis and clearance: A balancing act. FEBS J. 2016, 284, 183–195. [Google Scholar] [CrossRef]
- Popov, L. Mitochondrial biogenesis: An update. J. Cell. Mol. Med. 2020, 24, 4892–4899. [Google Scholar] [CrossRef] [PubMed]
- Praharaj, P.P.; Panigrahi, D.P.; Bhol, C.S.; Patra, S.; Mishra, S.R.; Mahapatra, K.K.; Behera, B.P.; Singh, A.; Patil, S.; Bhutia, S.K. Mitochondrial rewiring through mitophagy and mitochondrial biogenesis in cancer stem cells: A potential target for anti-CSC cancer therapy. Cancer Lett. 2021, 498, 217–228. [Google Scholar] [CrossRef]
- Rainey, N.E.; Moustapha, A.; Petit, P.X. Curcumin, a Multifaceted Hormetic Agent, Mediates an Intricate Crosstalk between Mitochondrial Turnover, Autophagy, and Apoptosis. Oxidative Med. Cell. Longev. 2020, 2020, 1–23. [Google Scholar] [CrossRef]
- Scholpa, N.E.; Schnellmann, R.G. Mitochondrial-Based Therapeutics for the Treatment of Spinal Cord Injury: Mitochondrial Biogenesis as a Potential Pharmacological Target. J. Pharmacol. Exp. Ther. 2017, 363, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Sheremet, N.L.; Andreeva, N.A.; Shmel’kova, M.S.; Tsigankova, P.G. Mitochondrial biogenesis in hereditary optic neuropathies. Вестник Офтальмологии 2019, 135, 85–91. [Google Scholar] [CrossRef]
- Simmons, E.C.; Scholpa, N.E.; Schnellmann, R.G. Mitochondrial biogenesis as a therapeutic target for traumatic and neurodegenerative CNS diseases. Exp. Neurol. 2020, 329, 113309. [Google Scholar] [CrossRef]
- Skuratovskaia, D.; Komar, A.; Vulf, M.; Litvinova, L. Mitochondrial destiny in type 2 diabetes: The effects of oxidative stress on the dynamics and biogenesis of mitochondria. PeerJ 2020, 8, e9741. [Google Scholar] [CrossRef] [PubMed]
- Smiles, W.J.; Camera, D.M. The Guardian of the Genome P53 Regulates Exercise-Induced Mitochondrial Plasticity Beyond Organelle Biogenesis. Acta Physiol. 2018, 222, e13004. [Google Scholar] [CrossRef]
- Tamura, Y.; Endo, T. Role of Intra- and Inter-mitochondrial Membrane Contact Sites in Yeast Phospholipid Biogenesis. Single Mol. Single Cell Seq. 2017, 997, 121–133. [Google Scholar] [CrossRef]
- Tang, J.X.; Thompson, K.; Taylor, R.W.; Olahova, M. Mitochondrial Oxphos Biogenesis: Co-Regulation of Protein Synthesis, Import, and Assembly Pathways. Int. J. Mol. Sci. 2020, 21, 3820. [Google Scholar] [CrossRef] [PubMed]
- Thornton, C.; Jones, A.; Nair, S.; Aabdien, A.; Mallard, C.; Hagberg, H. Mitochondrial dynamics, mitophagy and biogenesis in neonatal hypoxic-ischaemic brain injury. FEBS Lett. 2017, 592, 812–830. [Google Scholar] [CrossRef]
- Timón-Gómez, A.; Nývltová, E.; Abriata, L.A.; Vila, A.J.; Hosler, J.; Barrientos, A. Mitochondrial cytochrome c oxidase biogenesis: Recent developments. Semin. Cell Dev. Biol. 2018, 76, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Tong, M.; Sadoshima, J. Mitochondrial autophagy in cardiomyopathy. Curr. Opin. Genet. Dev. 2016, 38, 8–15. [Google Scholar] [CrossRef]
- Van Haute, L.; Powell, C.A.; Minczuk, M. Dealing with an Unconventional Genetic Code in Mitochondria: The Biogenesis and Pathogenic Defects of the 5-Formylcytosine Modification in Mitochondrial tRNAMet. Biomolecules 2017, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- VanderVeen, B.N.; Fix, D.K.; Carson, J.A. Disrupted Skeletal Muscle Mitochondrial Dynamics, Mitophagy, and Biogenesis during Cancer Cachexia: A Role for Inflammation. Oxid. Med. Cell. Longev. 2017, 2017, 3292087. [Google Scholar] [CrossRef] [PubMed]
- Vega, R.B.; Horton, J.L.; Kelly, D.P. Maintaining Ancient Organelles: Mitochondrial Biogenesis and Maturation. Circ. Res. 2015, 116, 1820–1834. [Google Scholar] [CrossRef]
- Paz, M.V.; Cotán, D.; Garrido-Maraver, J.; Cordero, M.D.; Oropesa-Ávila, M.; De La Mata, M.; Pavón, A.D.; De Lavera, I.; Alcocer-Gómez, E.; Sánchez-Alcázar, J.A. Targeting autophagy and mitophagy for mitochondrial diseases treatment. Expert Opin. Ther. Targets 2015, 20, 487–500. [Google Scholar] [CrossRef]
- Wang, J.; Wen, X.; Liu, J.; Sun, H. Mitochondrial biogenesis Inhibitors for Anticancer Therapy: A Review of Recent Patents. Recent Pat. Anti-Cancer Drug Discov. 2016, 11, 332–341. [Google Scholar] [CrossRef]
- Whitaker, R.M.; Corum, D.; Beeson, C.C.; Schnellmann, R.G. Mitochondrial Biogenesis as a Pharmacological Target: A New Approach to Acute and Chronic Diseases. Annu. Rev. Pharmacol. Toxicol. 2016, 56, 229–249. [Google Scholar] [CrossRef]
- Wideman, J.G.; Muñoz-Gómez, S.A. The evolution of ERMIONE in mitochondrial biogenesis and lipid homeostasis: An evolutionary view from comparative cell biology. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2016, 1861, 900–912. [Google Scholar] [CrossRef]
- Williams, J.A.; Zhao, K.; Jin, S.; Ding, W.-X. New methods for monitoring mitochondrial biogenesis and mitophagy in vitro and in vivo. Exp. Biol. Med. 2017, 242, 781–787. [Google Scholar] [CrossRef]
- Dos Santos, T.W.; Pereira, Q.C.; Teixeira, L.; Gambero, A.; Villena, J.A.; Ribeiro, M.L. Effects of Polyphenols on Thermogenesis and Mitochondrial Biogenesis. Int. J. Mol. Sci. 2018, 19, 2757. [Google Scholar] [CrossRef]
- Yambire, K.F.; Fernandez-Mosquera, L.; Steinfeld, R.; Mühle, C.; Ikonen, E.; Milosevic, I.; Raimundo, N. Mitochondrial biogenesis is transcriptionally repressed in lysosomal lipid storage diseases. eLife 2019, 8. [Google Scholar] [CrossRef]
- Yuan, Y.; Cruzat, V.F.; Newsholme, P.; Cheng, J.; Chen, Y.; Lu, Y. Regulation of SIRT1 in aging: Roles in mitochondrial function and biogenesis. Mech. Ageing Dev. 2016, 155, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Zamora, M.; Pardo, R.; Villena, J.A. Pharmacological induction of mitochondrial biogenesis as a therapeutic strategy for the treatment of type 2 diabetes. Biochem. Pharmacol. 2015, 98, 16–28. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, H. Translational regulation of mitochondrial biogenesis. Biochem. Soc. Trans. 2016, 44, 1717–1724. [Google Scholar] [CrossRef] [PubMed]
- Andhavarapu, S.; Katuri, A.; Bryant, J.; Patel, V.; Gupta, U.; Asemu, G.; Makar, T.K. Intersecting roles of ER stress, mitochondrial dysfunction, autophagy, and calcium homeostasis in HIV-associated neurocognitive disorder. J. NeuroVirol. 2020, 26, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Baquero, P.; Dawson, A.; Helgason, G.V. Autophagy and mitochondrial metabolism: Insights into their role and therapeutic potential in chronic myeloid leukaemia. FEBS J. 2019, 286, 1271–1283. [Google Scholar] [CrossRef]
- Chen, S.Y.; Gao, Y.; Sun, J.Y.; Meng, X.L.; Yang, D.; Fan, L.H.; Xiang, L.; Wang, P. Traditional Chinese Medicine: Role in Reducing Beta-Amyloid, Apoptosis, Autophagy, Neuroinflammation, Oxidative Stress, and Mitochondrial Dysfunction of Alzheimer’s Disease. Front. Pharmacol. 2020, 11, 497. [Google Scholar] [CrossRef]
- Cheng, J.; Wei, L.; Li, M. Progress in Regulation of Mitochondrial Dynamics and Mitochondrial Autophagy. Sheng Li Xue Bao 2020, 72, 475–487. [Google Scholar]
- Furukawa, K.; Innokentev, A.; Kanki, T. Regulatory Mechanisms of Mitochondrial Autophagy: Lessons from Yeast. Front. Plant. Sci. 2019, 10, 1479. [Google Scholar] [CrossRef]
- Gafar, A.A.; Draz, H.M.; Goldberg, A.A.; Bashandy, M.A.; Bakry, S.; Khalifa, M.A.; AbuShair, W.; Titorenko, V.I.; Sanderson, J.T. Lithocholic acid induces endoplasmic reticulum stress, autophagy and mitochondrial dysfunction in human prostate cancer cells. PeerJ 2016, 4, e2445. [Google Scholar] [CrossRef] [PubMed]
- Go, K.L.; Lee, S.; Zendejas, I.; Behrns, K.E.; Kim, J.S. Mitochondrial Dysfunction and Autophagy in Hepatic Ischemia/Reperfusion Injury. Biomed. Res. Int. 2015, 2015, 183469. [Google Scholar] [CrossRef]
- Hamacher-Brady, A.; Brady, N.R. Mitophagy programs: Mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell. Mol. Life Sci. 2016, 73, 775–795. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.; Shi, Y. Regulation of autophagy by mitochondrial phospholipids in health and diseases. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2017, 1862, 114–129. [Google Scholar] [CrossRef]
- Huang, M.L.-H.; Chiang, S.; Kalinowski, D.S.; Bae, D.-H.; Sahni, S.; Richardson, D.R. The Role of the Antioxidant Response in Mitochondrial Dysfunction in Degenerative Diseases: Cross-Talk between Antioxidant Defense, Autophagy, and Apoptosis. Oxidative Med. Cell. Longev. 2019, 2019, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Katuri, A.; Bryant, J.L.; Patel, D.; Patel, V.; Andhavarapu, S.; Asemu, G.; Davis, H.; Makar, T.K. HIVAN associated tubular pathology with reference to ER stress, mitochondrial changes, and autophagy. Exp. Mol. Pathol. 2019, 106, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Kubli, D.A.; Gustafsson, Å.B. Unbreak My Heart: Targeting Mitochondrial Autophagy in Diabetic Cardiomyopathy. Antioxid. Redox Signal. 2015, 22, 1527–1544. [Google Scholar] [CrossRef]
- Madhavi, Y.V.; Gaikwad, N.; Yerra, V.G.; Kalvala, A.K.; Nanduri, S.; Kumar, A. Targeting AMPK in Diabetes and Diabetic Complications: Energy Homeostasis, Autophagy and Mitochondrial Health. Curr. Med. Chem. 2019, 26, 5207–5229. [Google Scholar] [CrossRef]
- Miyamoto, S. Hexokinase 2 mediated cellular protection: Interaction with Akt/mTORC1 to regulate mitochondrial protection and autophagy. Seikagaku. J. Jpn. Biochem. Soc. 2017, 89, 199–210. [Google Scholar]
- Ney, P.A. Mitochondrial Autophagy: Origins, Significance, and Role of Bnip3 and Nix. Biochim. Biophys. Acta 2015, 1853, 2775–2783. [Google Scholar] [CrossRef]
- Roca-Agujetas, V.; De Dios, C.; Lestón, L.; Marí, M.; Morales, A.; Colell, A. Recent Insights into the Mitochondrial Role in Autophagy and Its Regulation by Oxidative Stress. Oxidative Med. Cell. Longev. 2019, 2019, 1–16. [Google Scholar] [CrossRef]
- Saito, T.; Sadoshima, J. Molecular Mechanisms of Mitochondrial Autophagy/Mitophagy in the Heart. Circ. Res. 2015, 116, 1477–1490. [Google Scholar] [CrossRef]
- Shah, S.Z.A.; Zhao, D.; Hussain, T.; Sabir, N.; Mangi, M.H.; Yang, L. p62-Keap1-NRF2-ARE Pathway: A Contentious Player for Selective Targeting of Autophagy, Oxidative Stress and Mitochondrial Dysfunction in Prion Diseases. Front. Mol. Neurosci. 2018, 11, 310. [Google Scholar] [CrossRef]
- Sifuentes-Franco, S.; Pacheco-Moisés, F.P.; Rodríguez-Carrizalez, A.D.; Miranda-Díaz, A.G. The Role of Oxidative Stress, Mitochondrial Function, and Autophagy in Diabetic Polyneuropathy. J. Diabetes Res. 2017, 2017, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.A.; Yen, P.M. Thyroid hormone-mediated autophagy and mitochondrial turnover in NAFLD. Cell Biosci. 2016, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Tagaya, M.; Arasaki, K. Regulation of Mitochondrial Dynamics and Autophagy by the Mitochondria-Associated Membrane. Adv. Exp. Med. Biol. 2017, 997, 33–47. [Google Scholar] [PubMed]
- Tricarico, P.M.; Crovella, S.; Celsi, F. Mevalonate Pathway Blockade, Mitochondrial Dysfunction and Autophagy: A Possible Link. Int. J. Mol. Sci. 2015, 16, 16067–16084. [Google Scholar] [CrossRef]
- Van Houten, B.; Hunter, S.E.; Meyer, J.N. Mitochondrial DNA Damage Induced Autophagy, Cell Death, and Disease. Front. Biosci. (Landmark Ed.) 2016, 21, 42–54. [Google Scholar] [CrossRef]
- Wang, B.; Abraham, N.; Gao, G.; Yang, Q. Dysregulation of Autophagy and Mitochondrial Function in Parkinson’s Disease. Transl. Neurodegener. 2016, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, G. Autophagy in Mitochondrial Quality Control. In Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Swtizerland, 2019; Volume 1206, pp. 421–434. [Google Scholar]
- Yamano, K.; Matsuda, N.; Tanaka, K. The Ubiquitin Signal and Autophagy: An Orchestrated Dance Leading to Mitochondrial Degradation. EMBO Rep. 2016, 17, 300–316. [Google Scholar] [CrossRef]
- Yamashita, S.I.; Kanki, T. How Autophagy Eats Large Mitochondria: Autophagosome Formation Coupled with Mitochondrial Fragmentation. Autophagy 2017, 13, 980–981. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Gao, G.; Mao, Z.; Yang, Q. Chaperone-Mediated Autophagy and Mitochondrial Homeostasis in Parkinson’s Disease. Park. Dis. 2016, 2016, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Culp, M.L.; Craver, J.G.; Darley-Usmar, V. Mitochondrial Function and Autophagy: Integrating Proteotoxic, Redox, and Metabolic Stress in Parkinson’s Disease. J. Neurochem. 2018, 144, 691–709. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zheng, Y.; Chen, Z. Autophagy and Mitochondrial Encephalomyopathies. In Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2020; pp. 103–110. [Google Scholar]
- Kopinski, P.K.; Janssen, K.A.; Schaefer, P.M.; Trefely, S.; Perry, C.E.; Potluri, P.; Tintos-Hernandez, J.A.; Singh, L.N.; Karch, K.R.; Campbell, S.L.; et al. Regulation of nuclear epigenome by mitochondrial DNA heteroplasmy. Proc. Natl. Acad. Sci. USA 2019, 116, 16028–16035. [Google Scholar] [CrossRef]
- Matilainen, O.; Quirós, P.M.; Auwerx, J. Mitochondria and Epigenetics—Crosstalk in Homeostasis and Stress. Trends Cell Biol. 2017, 27, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Schell, J.C.; Rutter, J. Mitochondria link metabolism and epigenetics in haematopoiesis. Nat. Cell Biol. 2017, 19, 589–591. [Google Scholar] [CrossRef]
- Sharma, N.; Pasala, M.S.; Prakash, A. Mitochondrial DNA: Epigenetics and environment. Environ. Mol. Mutagen. 2019, 60, 668–682. [Google Scholar] [CrossRef]
- Danzeisen, R.; Schwalenstoecker, B.; Gillardon, F.; Buerger, E.; Krzykalla, V.; Klinder, K.; Schild, L.; Hengerer, B.; Ludolph, A.C.; Dorner-Ciossek, C.; et al. Targeted Antioxidative and Neuroprotective Properties of the Dopamine Agonist Pramipexole and Its Nondopaminergic Enantiomer SND919CL2x [(+)2-Amino-4,5,6,7-tetrahydro-6-lpropylamino-benzathiazole Dihydrochloride]. J. Pharmacol. Exp. Ther. 2005, 316, 189–199. [Google Scholar] [CrossRef]
- Ferrari-Toninelli, G.; Maccarinelli, G.; Uberti, D.L.; Buerger, E.; Memo, M. Mitochondria-targeted antioxidant effects of S(-) and R(+) pramipexole. BMC Pharmacol. 2010, 10, 2. [Google Scholar] [CrossRef]
- Akhmadishina, R.A.; Garifullin, R.; Petrova, N.V.; Kamalov, M.I.; Abdullin, T.I. Triphenylphosphonium Moiety Modulates Proteolytic Stability and Potentiates Neuroprotective Activity of Antioxidant Tetrapeptides in Vitro. Front. Pharmacol. 2018, 9, 115. [Google Scholar] [CrossRef]
- Battogtokh, G.; Choi, Y.S.; Kang, D.S.; Park, S.J.; Shim, M.S.; Huh, K.M.; Cho, Y.-Y.; Lee, J.Y.; Lee, H.S.; Kang, H.C. Mitochondria-targeting drug conjugates for cytotoxic, anti-oxidizing and sensing purposes: Current strategies and future perspectives. Acta Pharm. Sin. B 2018, 8, 862–880. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Liaw, K.; Sharma, R.; Zhang, Z.; Kannan, S.; Kannan, R.M. Targeting Mitochondrial Dysfunction and Oxidative Stress in Activated Microglia Using Dendrimer-Based Therapeutics. Theranostics 2018, 8, 5529–5547. [Google Scholar] [CrossRef]
- Titova, E.; Shagieva, G.; Ivanova, O.; Domnina, L.; Domninskaya, M.; Strelkova, O.; Khromova, N.; Kopnin, P.; Chernyak, B.; Skulachev, V.; et al. Mitochondria-targeted antioxidant SkQ1 suppresses fibrosarcoma and rhabdomyosarcoma tumour cell growth. Cell Cycle 2018, 17, 1797–1811. [Google Scholar] [CrossRef]
- Apostolova, N.; Victor, V.M. Molecular Strategies for Targeting Antioxidants to Mitochondria: Therapeutic Implications. Antioxid. Redox Signal. 2015, 22, 686–729. [Google Scholar] [CrossRef]
- Fetisova, E.K.; Antoschina, M.M.; Cherepanynets, V.D.; Izumov, D.S.; Kireev, I.I.; Kireev, R.I.; Lyamzaev, K.G.; Riabchenko, N.I.; Chernyak, B.V.; Skulachev, V.P. Radioprotective Effects of Mitochondria-Targeted Antioxidant SkQR. Radiat. Res. 2015, 183, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Genrikhs, E.E.; Stelmashook, E.V.; Popova, O.V.; Kapay, N.A.; Korshunova, G.A.; Sumbatyan, N.V.; Skrebitsky, V.G.; Skulachev, V.P.; Isaev, N.K. Mitochondria-Targeted Antioxidant Skqt1 Decreases Trauma-Induced Neurological Deficit in Rat and Prevents Amyloid-Beta-Induced Impairment of Long-Term Potentiation in Rat Hippocampal Slices. J. Drug Target. 2015, 23, 347–352. [Google Scholar] [CrossRef]
- Silachev, D.N.; Plotnikov, E.Y.; Zorova, L.D.; Pevzner, I.B.; Sumbatyan, N.V.; Korshunova, G.A.; Gulyaev, M.V.; Pirogov, Y.A.; Skulachev, V.P.; Zorov, D.B. Neuroprotective Effects of Mitochondria-Targeted Plastoquinone and Thymoquinone in a Rat Model of Brain Ischemia/Reperfusion Injury. Molecules 2015, 20, 14487–14503. [Google Scholar] [CrossRef]
- Margulis, L. Symbiotic theory of the origin of eukaryotic organelles; criteria for proof. Symp. Soc. Exp. Biol. 1975, 29, 21–38. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bennett, J.P., Jr.; Onyango, I.G. Energy, Entropy and Quantum Tunneling of Protons and Electrons in Brain Mitochondria: Relation to Mitochondrial Impairment in Aging-Related Human Brain Diseases and Therapeutic Measures. Biomedicines 2021, 9, 225. https://doi.org/10.3390/biomedicines9020225
Bennett JP Jr., Onyango IG. Energy, Entropy and Quantum Tunneling of Protons and Electrons in Brain Mitochondria: Relation to Mitochondrial Impairment in Aging-Related Human Brain Diseases and Therapeutic Measures. Biomedicines. 2021; 9(2):225. https://doi.org/10.3390/biomedicines9020225
Chicago/Turabian StyleBennett, James P., Jr., and Isaac G. Onyango. 2021. "Energy, Entropy and Quantum Tunneling of Protons and Electrons in Brain Mitochondria: Relation to Mitochondrial Impairment in Aging-Related Human Brain Diseases and Therapeutic Measures" Biomedicines 9, no. 2: 225. https://doi.org/10.3390/biomedicines9020225
APA StyleBennett, J. P., Jr., & Onyango, I. G. (2021). Energy, Entropy and Quantum Tunneling of Protons and Electrons in Brain Mitochondria: Relation to Mitochondrial Impairment in Aging-Related Human Brain Diseases and Therapeutic Measures. Biomedicines, 9(2), 225. https://doi.org/10.3390/biomedicines9020225





