Embedded Upconversion Nanoparticles in Magnetic Molecularly Imprinted Polymers for Photodynamic Therapy of Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
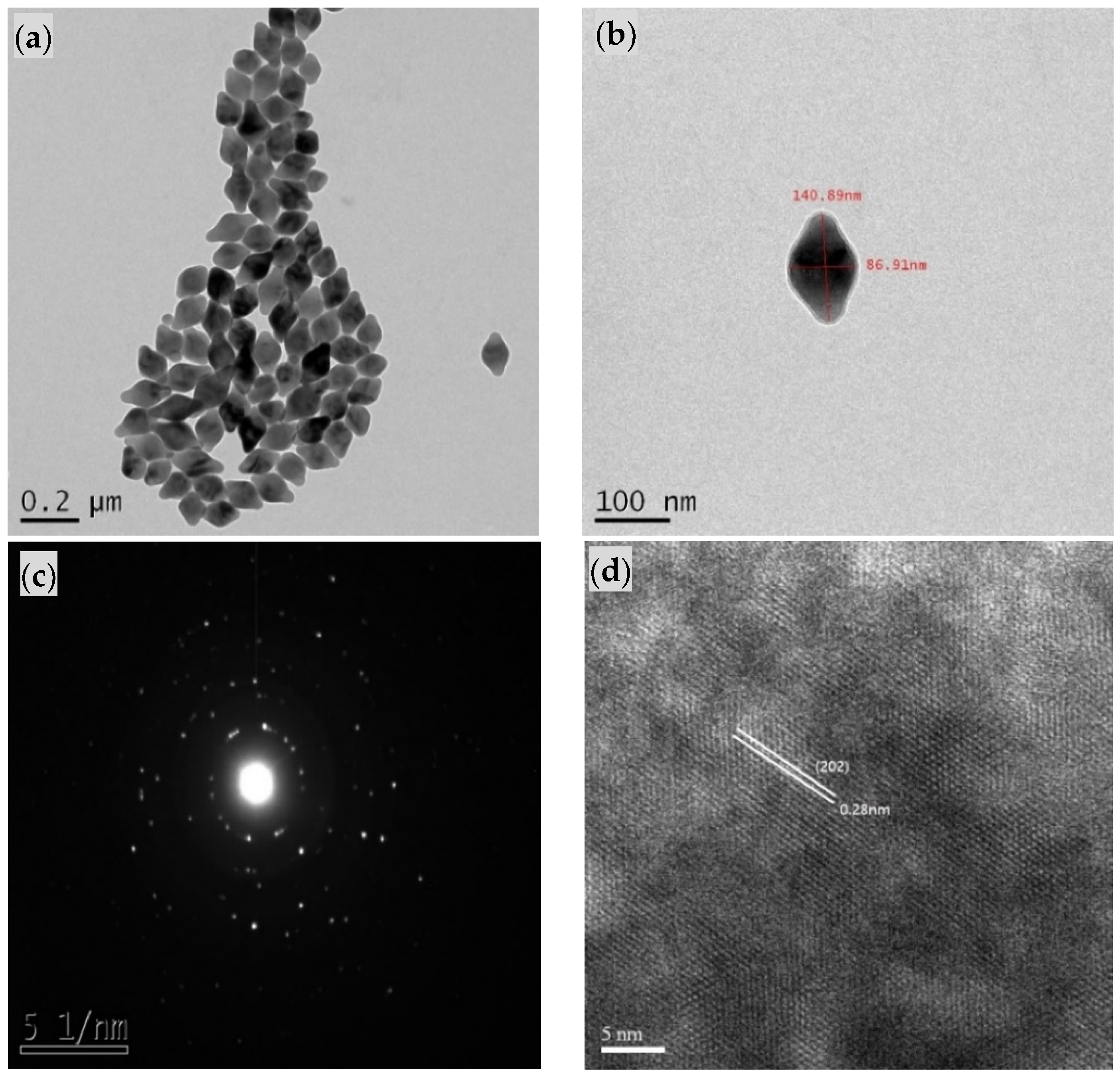
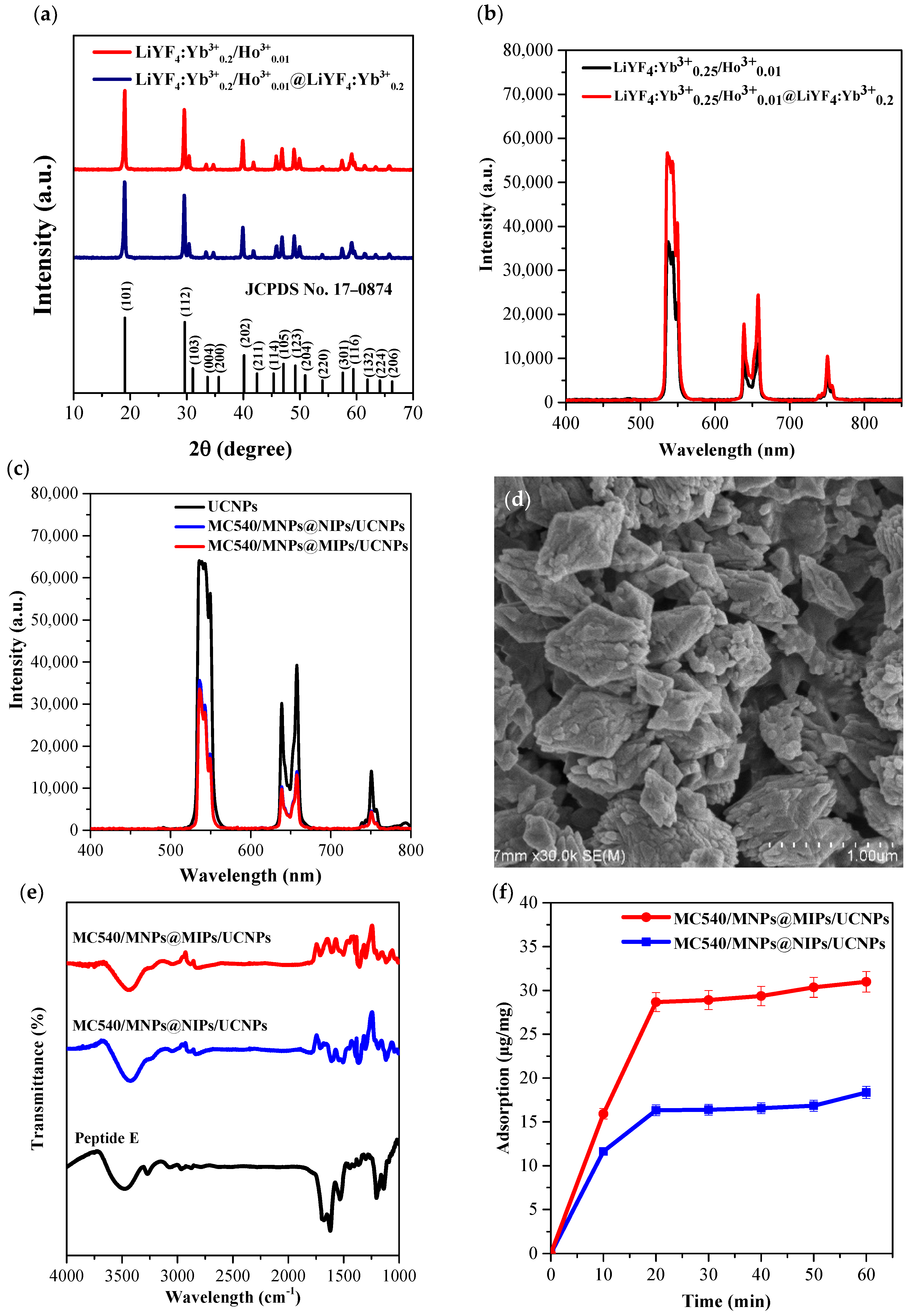
2.2. Synthesis of Core/Shell UCNPs
2.3. Formation of Peptide-Imprinted Composite Nanoparticles
2.4. Characterization of Core/Shell UCNPs and Peptide-Imprinted Composite Nanoparticles
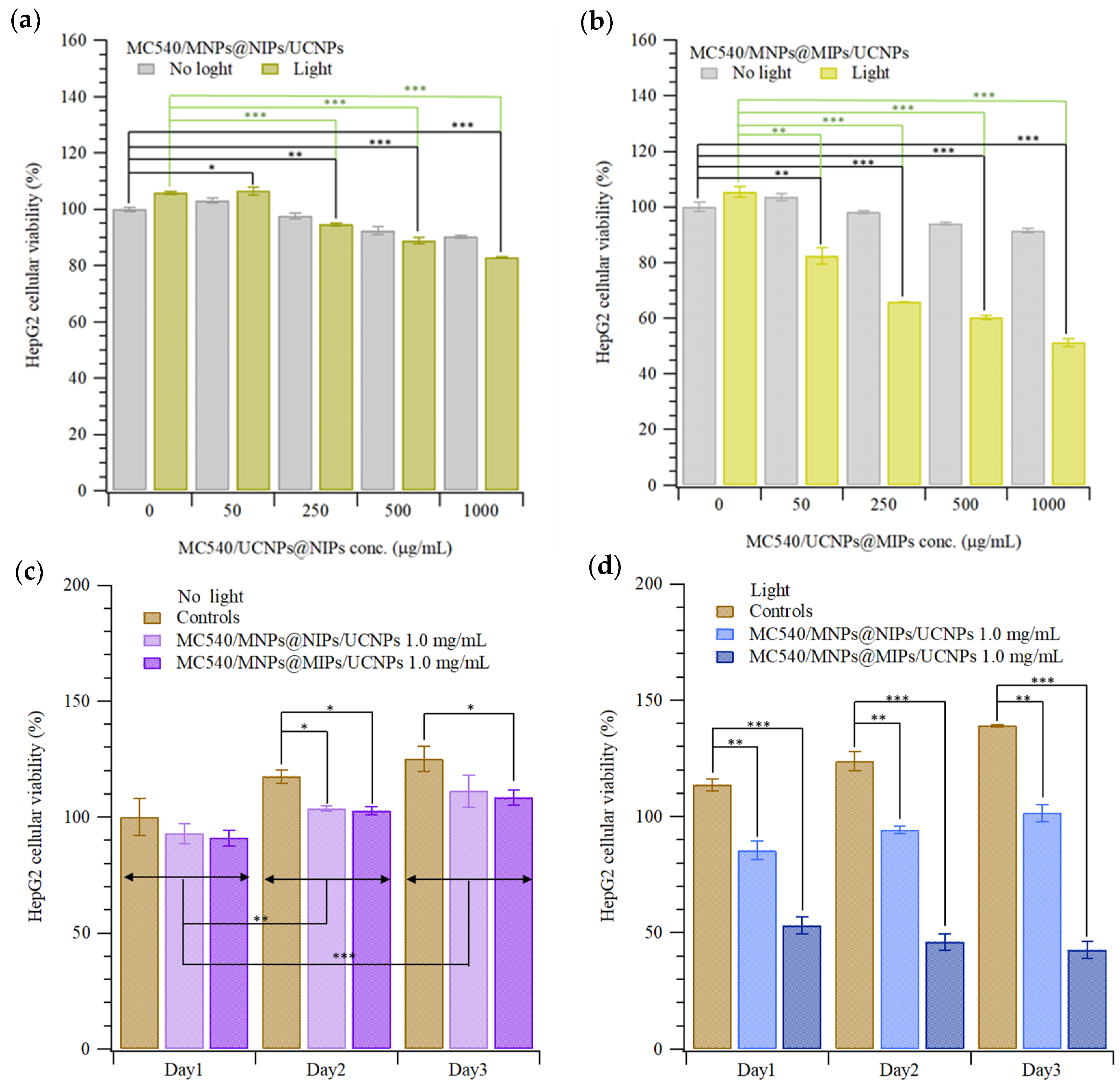
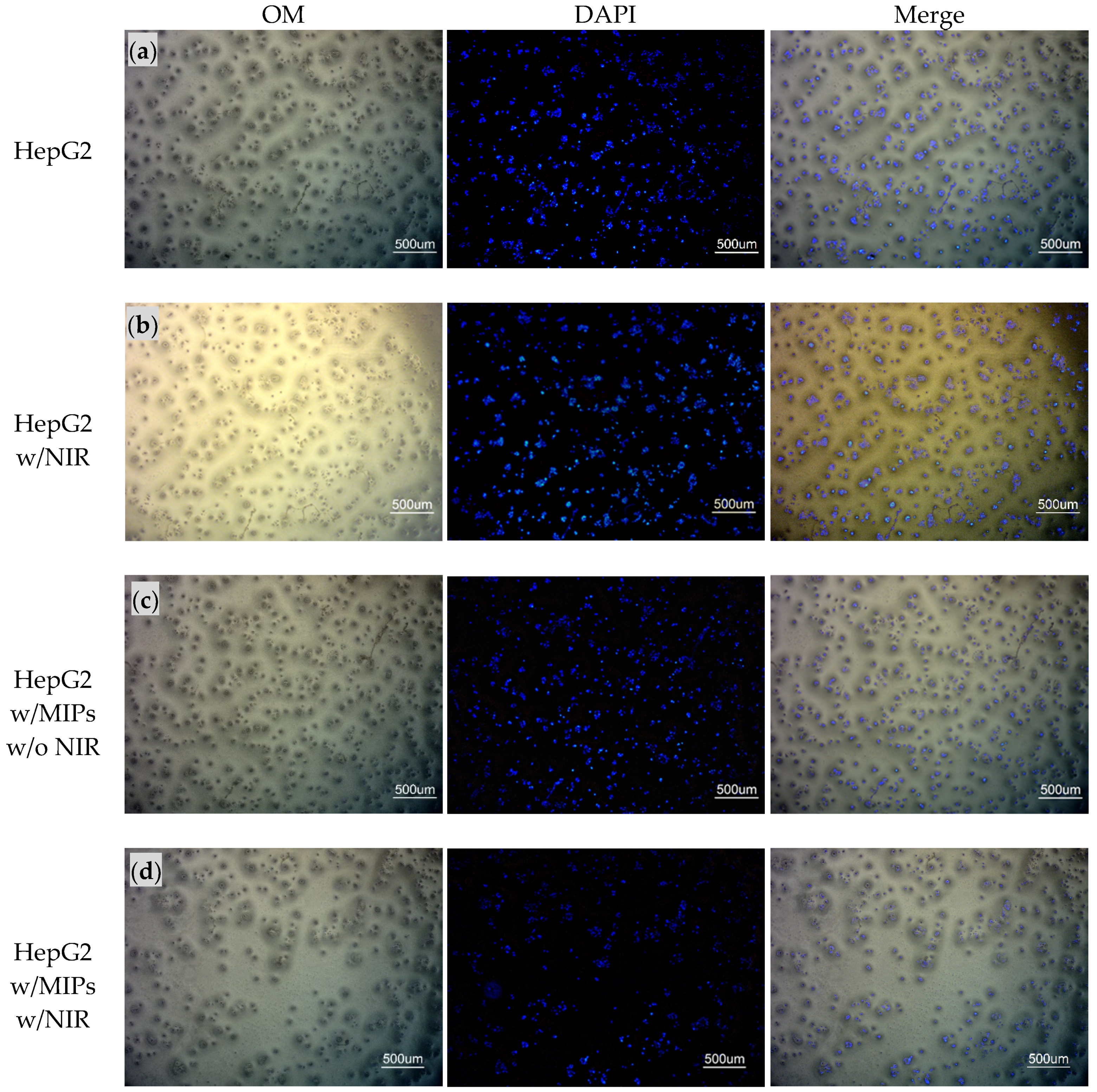
2.5. Cytotoxicity Test and Photodynamic Therapy of HepG2 Cells with MC540/MNPs@MIPs/UCNPs
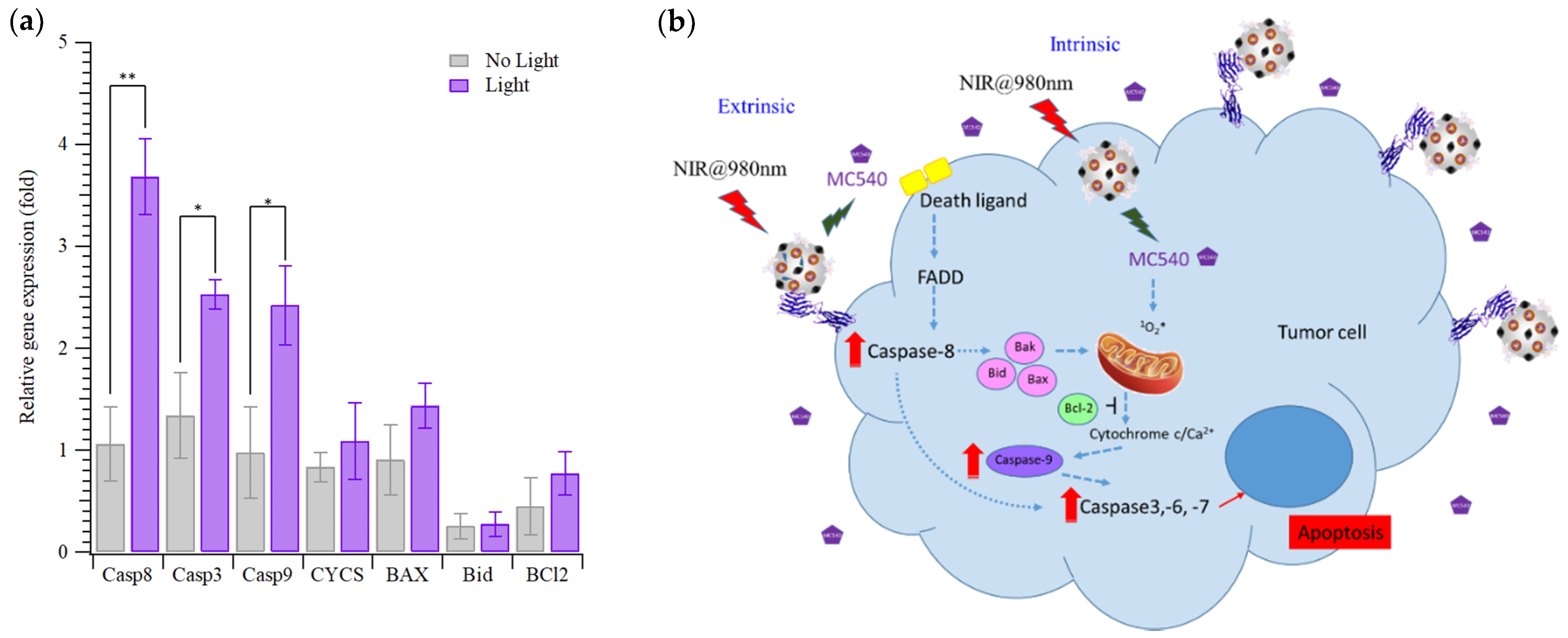
2.6. Gene Expression of HepG2 Cells Treated with MC540/MNPs@MIPs/UCNPs and PDT
| Primer | Sequence | |
|---|---|---|
| Forward | Reverse | |
| GAPDH | ACAGTTGCCATGTAGACC | TTTTTGGTTGAGCACAGG |
| Casp8 | CTACAGGGTCATGCTCTATC | ATTTGGAGATTTCCTCTTGC |
| Casp3 | AAAGCACTGGAATGACATC | CGCATCAATTCCACAATTTC |
| Casp9 | CTCTACTTTCCCAGGTTTTG | TTTCACCGAAACAGCATTAG |
| CYCS | AAGAACAAAGGCATCATCTG | GCTATTAAGTCTGCCCTTTC |
| Bax | AACTGGACAGTAACATGGAG | TTGCTGGCAAAGTAGAAAAG |
| Bid | CTTAGCCAGAAATGGGATG | AGTCACAGCTATCTTCCAG |
| Bcl-2 | GATTGTGGCCTTCCTTTGAG | GTTCCACAAAGGCATCC |
2.7. Data Analysis
3. Results and Discussion
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, C.; Cheng, L.; Liu, Z. Upconversion nanoparticles for photodynamic therapy and other cancer therapeutics. Theranostics 2013, 3, 317. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Wang, C.; Cheng, L.; He, W.; Cheng, Z.; Liu, Z. Protein modified upconversion nanoparticles for imaging-guided combined photothermal and photodynamic therapy. Biomaterials 2014, 35, 2915–2923. [Google Scholar] [CrossRef]
- Lee, G.; Park, Y.I. Lanthanide-doped upconversion nanocarriers for drug and gene delivery. Nanomaterials 2018, 8, 511. [Google Scholar] [CrossRef] [Green Version]
- Chien, H.-W.; Wu, C.-H.; Yang, C.-H.; Wang, T.-L. Multiple doping effect of LiYF4:Yb3+/Er3+/Ho3+/Tm3+@LiYF4:Yb3+ core/shell nanoparticles and its application in Hg2+ sensing detection. J. Alloy. Compd. 2019, 806, 272–282. [Google Scholar] [CrossRef]
- Xu, J.; Xu, L.; Wang, C.; Yang, R.; Zhuang, Q.; Han, X.; Dong, Z.; Zhu, W.; Peng, R.; Liu, Z. Near-infrared-triggered photodynamic therapy with multitasking upconversion nanoparticles in combination with checkpoint blockade for immunotherapy of colorectal cancer. ACS Nano 2017, 11, 4463–4474. [Google Scholar] [CrossRef]
- Chung, Y.-C.; Yang, C.-H.; Lee, R.-H.; Wang, T.-L. Dual Stimuli-Responsive Block Copolymers for Controlled Release Triggered by Upconversion Luminescence or Temperature Variation. ACS Omega 2019, 4, 3322–3328. [Google Scholar] [CrossRef]
- Lee, M.-H.; Thomas, J.L.; Wang, H.-Y.; Chang, C.-C.; Lin, C.-C.; Lin, H.-Y. Extraction of resveratrol from polygonum cuspidatum with magnetic orcinol-imprinted poly (ethylene-co-vinyl alcohol) composite particles and their in vitro suppression of human osteogenic sarcoma (HOS) cell line. J. Mater. Chem. 2012, 22, 24644–24651. [Google Scholar] [CrossRef]
- Lee, M.-H.; Lin, C.-C.; Thomas, J.L.; Chan, C.-K.; Lin, H.-Y. Epitope recognition of magnetic peptide-imprinted chitosan composite nanoparticles for the extraction of CRISPR/dCas9a proteins from transfected cells. Nanotechnology 2021, 32, 18LT02. [Google Scholar] [CrossRef]
- Lee, M.-H.; Lin, C.-C.; Thomas, J.L.; Li, J.-A.; Lin, H.-Y. Cellular reprogramming with multigene activation by the delivery of CRISPR/dCas9 ribonucleoproteins via magnetic peptide-imprinted chitosan nanoparticles. Mater. Today Bio 2021, 9, 100091. [Google Scholar] [CrossRef]
- Liu, K.-H.; Lin, H.-Y.; Thomas, J.L.; Shih, Y.-P.; Chen, J.-T.; Lee, M.-H. Magnetic analogue-imprinted polymers for the extraction of ginsenosides from the Panax ginseng callus. Ind. Crop. Prod. 2021, 163, 113291. [Google Scholar] [CrossRef]
- Lin, H.-Y.; Ho, M.-S.; Lee, M.-H. Instant formation of molecularly imprinted poly (ethylene-co-vinyl alcohol)/quantum dot composite nanoparticles and their use in one-pot urinalysis. Biosens. Bioelectron. 2009, 25, 579–586. [Google Scholar] [CrossRef]
- Lee, M.-H.; Thomas, J.L.; Ho, M.-H.; Yuan, C.; Lin, H.-Y. Synthesis of magnetic molecularly imprinted poly (ethylene-co-vinyl alcohol) nanoparticles and their uses in the extraction and sensing of target molecules in urine. ACS Appl. Mater. Interfaces 2010, 2, 1729–1736. [Google Scholar] [CrossRef]
- Lee, M.-H.; Chen, Y.-C.; Ho, M.-H.; Lin, H.-Y. Optical recognition of salivary proteins by use of molecularly imprinted poly (ethylene-co-vinyl alcohol)/quantum dot composite nanoparticles. Anal. Bioanal. Chem. 2010, 397, 1457–1466. [Google Scholar] [CrossRef]
- Liu, J.-M.; Cao, F.-Z.; Fang, G.-Z.; Wang, S. Upconversion nanophosphor-involved molecularly imprinted fluorescent polymers for sensitive and specific recognition of sterigmatocystin. Polymers 2017, 9, 299. [Google Scholar] [CrossRef]
- Yan, Z.; Fang, G.-z. Molecularly imprinted polymer based on upconversion nanoparticles for highly selective and sensitive determination of Ochratoxin A. J. Cent. South Univ. 2019, 26, 515–523. [Google Scholar] [CrossRef]
- Chien, H.-W.; Tsai, M.-T.; Yang, C.-H.; Lee, R.-H.; Wang, T.-L. Interaction of LiYF4:Yb3+/Er3+/Ho3+/Tm3+@LiYF4:Yb3+ upconversion nanoparticles, molecularly imprinted polymers, and templates. RSC Adv. 2020, 10, 35600–35610. [Google Scholar] [CrossRef]
- Piletsky, S.; Canfarotta, F.; Poma, A.; Bossi, A.M.; Piletsky, S. Molecularly Imprinted Polymers for Cell Recognition. Trends Biotechnol. 2020, 38, 368–387. [Google Scholar] [CrossRef]
- Peng, H.; Qin, Y.-T.; He, X.-W.; Li, W.-Y.; Zhang, Y.-K. Epitope molecularly imprinted polymer nanoparticles for chemo-/photodynamic synergistic cancer therapy guided by targeted fluorescence imaging. ACS Appl. Mater. Interfaces 2020, 12, 13360–13370. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Su, Z.-C.; He, X.-W.; Li, W.-Y.; Zhang, Y.-K. H2O2 self-supplying degradable epitope imprinted polymers for targeted fluorescence imaging and chemodynamic therapy. Nanoscale 2021, 13, 12553–12564. [Google Scholar] [CrossRef]
- Lee, M.-H.; Thomas, J.L.; Li, J.-A.; Chen, J.-R.; Wang, T.-L.; Lin, H.-Y. Synthesis of Multifunctional Nanoparticles for the Combination of Photodynamic Therapy and Immunotherapy. Pharmaceuticals 2021, 14, 508. [Google Scholar] [CrossRef]
- Lee, M.-H.; Thomas, J.L.; Liao, C.-L.; Jurcevic, S.; Crnogorac-Jurcevic, T.; Lin, H.-Y. Epitope recognition of peptide-imprinted polymers for Regenerating protein 1 (REG1). Sep. Purif. Technol. 2018, 192, 213–219. [Google Scholar] [CrossRef]
- Lee, M.-H.; Liu, K.-H.; Thomas, J.L.; Chen, J.-R.; Lin, H.-Y. Immunotherapy of Hepatocellular Carcinoma with Magnetic PD-1 Peptide-Imprinted Polymer Nanocomposite and Natural Killer Cells. Biomolecules 2019, 9, 651. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, G.; Yu, Y.; Zhang, H.; Gao, J.; Sun, Z.; Lu, Y.; Zou, H. NIR-Responsive Copolymer Upconversion Nanocomposites for Triggered Drug Release in Vitro and in Vivo. ACS Appl. Bio Mater. 2019, 2, 495–503. [Google Scholar] [CrossRef]
- Tang, G.; He, J.; Liu, J.; Yan, X.; Fan, K. Nanozyme for tumor therapy: Surface modification matters. Exploration 2021, 1, 75–89. [Google Scholar] [CrossRef]
- Zhang, R.; Chen, L.; Liang, Q.; Xi, J.; Zhao, H.; Jin, Y.; Gao, X.; Yan, X.; Gao, L.; Fan, K. Unveiling the active sites on ferrihydrite with apparent catalase-like activity for potentiating radiotherapy. Nano Today 2021, 41, 101317. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-C.; Lin, H.-Y.; Thomas, J.L.; Yu, J.-X.; Lin, C.-Y.; Chang, Y.-H.; Lee, M.-H.; Wang, T.-L. Embedded Upconversion Nanoparticles in Magnetic Molecularly Imprinted Polymers for Photodynamic Therapy of Hepatocellular Carcinoma. Biomedicines 2021, 9, 1923. https://doi.org/10.3390/biomedicines9121923
Lin C-C, Lin H-Y, Thomas JL, Yu J-X, Lin C-Y, Chang Y-H, Lee M-H, Wang T-L. Embedded Upconversion Nanoparticles in Magnetic Molecularly Imprinted Polymers for Photodynamic Therapy of Hepatocellular Carcinoma. Biomedicines. 2021; 9(12):1923. https://doi.org/10.3390/biomedicines9121923
Chicago/Turabian StyleLin, Cheng-Chih, Hung-Yin Lin, James L. Thomas, Jia-Xin Yu, Chien-Yu Lin, Yu-Hua Chang, Mei-Hwa Lee, and Tzong-Liu Wang. 2021. "Embedded Upconversion Nanoparticles in Magnetic Molecularly Imprinted Polymers for Photodynamic Therapy of Hepatocellular Carcinoma" Biomedicines 9, no. 12: 1923. https://doi.org/10.3390/biomedicines9121923
APA StyleLin, C.-C., Lin, H.-Y., Thomas, J. L., Yu, J.-X., Lin, C.-Y., Chang, Y.-H., Lee, M.-H., & Wang, T.-L. (2021). Embedded Upconversion Nanoparticles in Magnetic Molecularly Imprinted Polymers for Photodynamic Therapy of Hepatocellular Carcinoma. Biomedicines, 9(12), 1923. https://doi.org/10.3390/biomedicines9121923






