miR-154 Influences HNSCC Development and Progression through Regulation of the Epithelial-to-Mesenchymal Transition Process and Could Be Used as a Potential Biomarker
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Pathological and Clinical Analysis
2.3. Functional Enrichment Analysis and Prediction of Genes’ Function
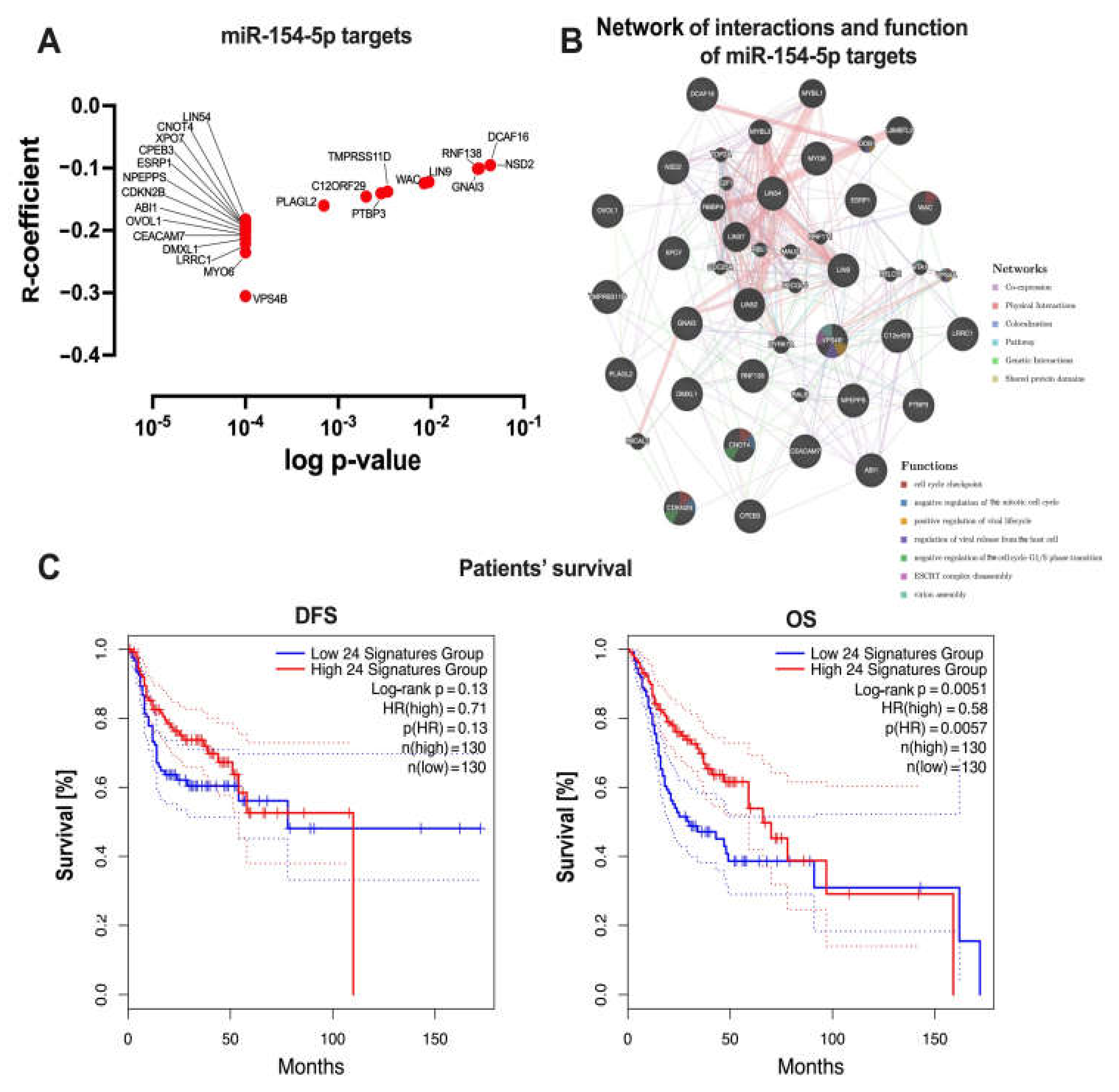
2.4. Target Analysis
2.5. Immune Profile Analysis
2.6. Statistical Analysis
2.7. Availability of Data and Materials
3. Results
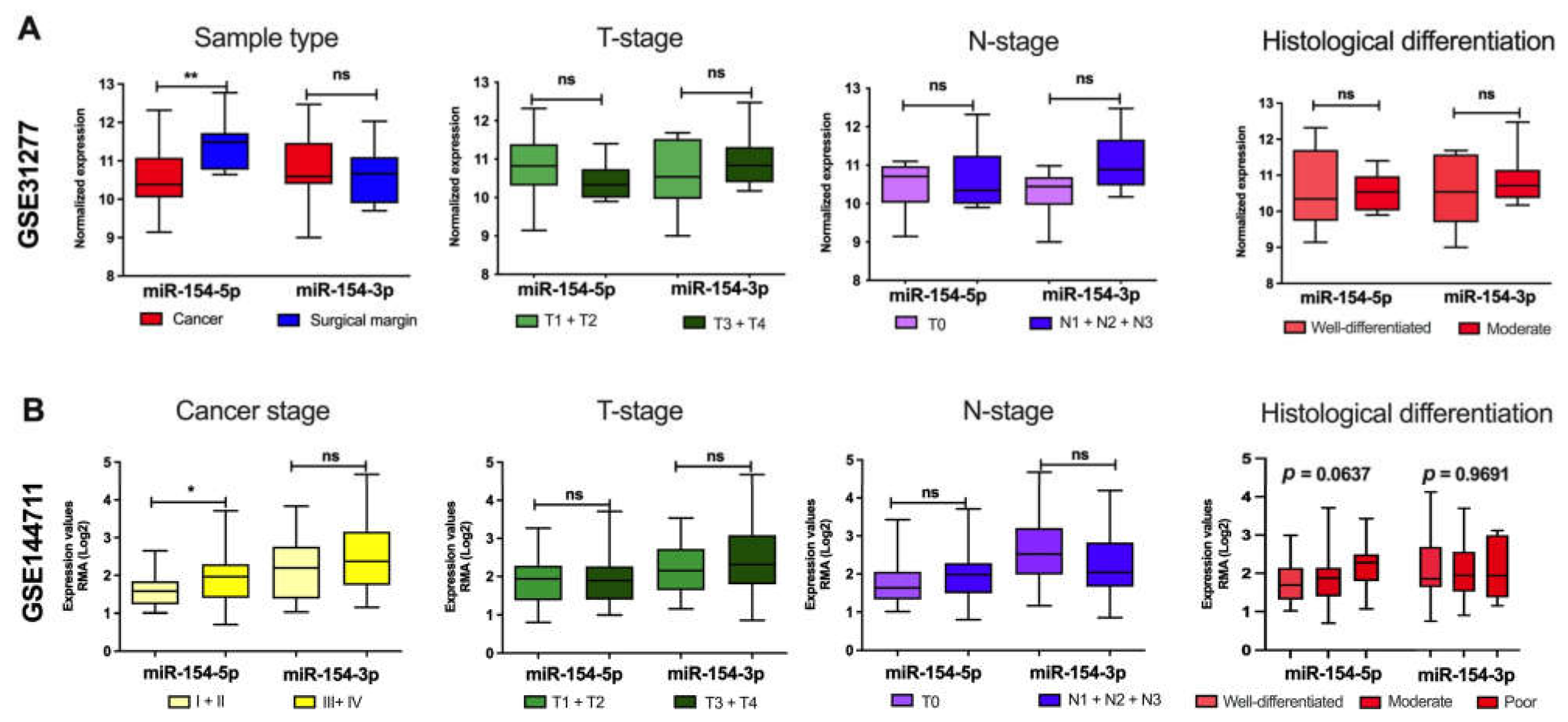
3.1. miR-154-5p and miR-154-3p Levels Differ Depending on the Clinicopathological Parameters
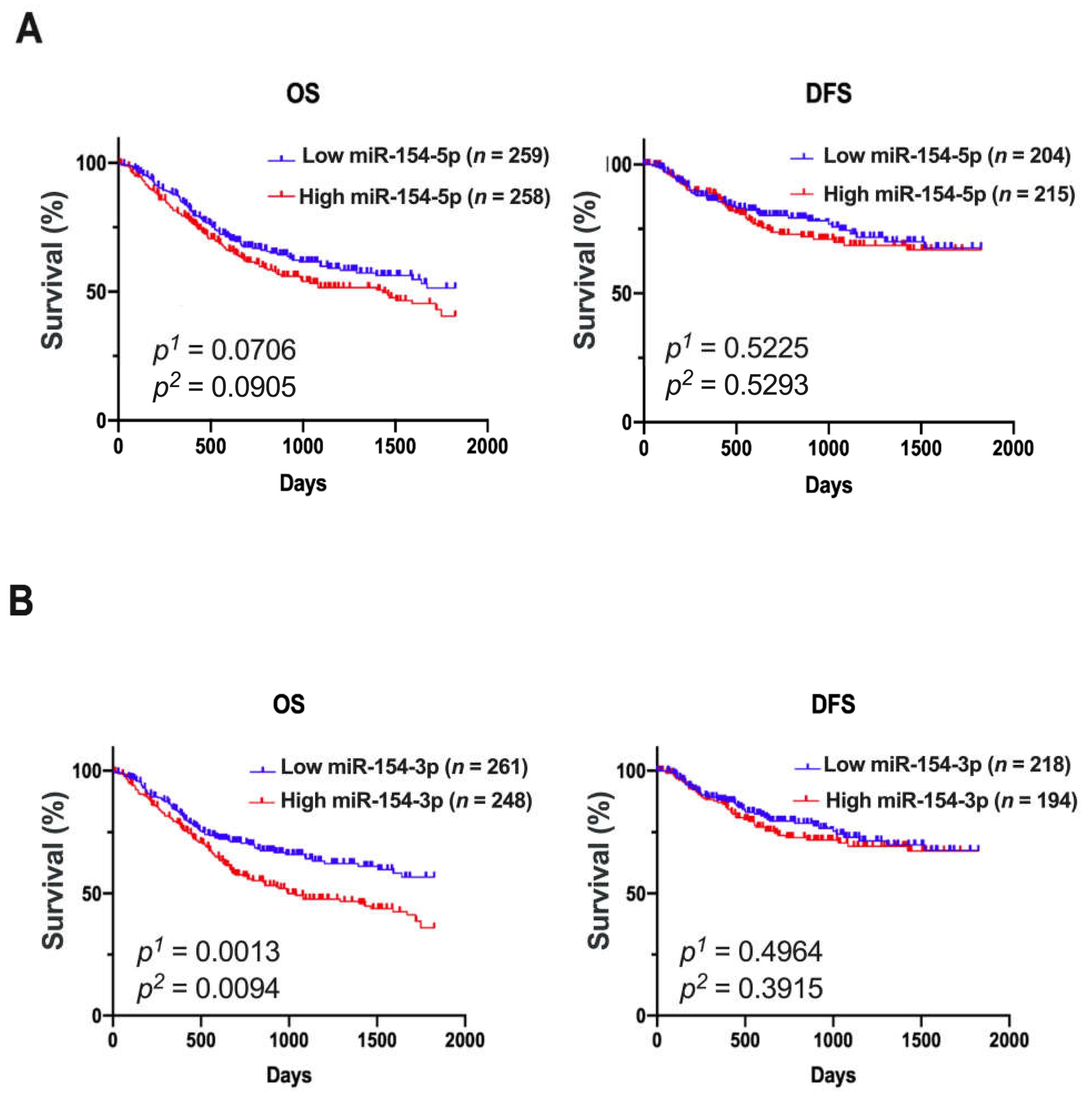
3.2. Patients with Low Expression of miR-154-3p Had a Significantly Extended Overall Survival
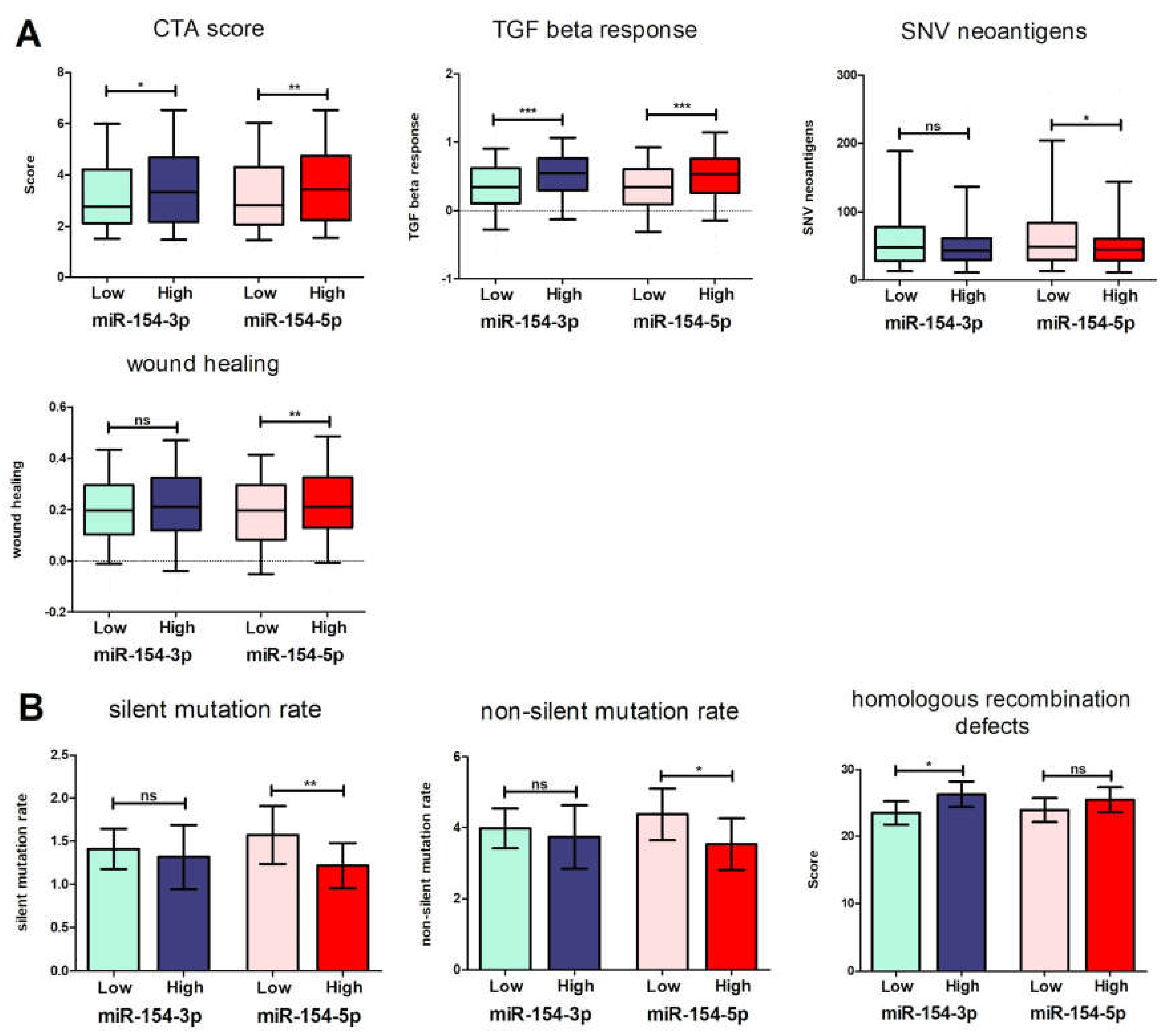
3.3. Higher Expression of miR-154-5p and miR-154-3p Was Associated with the Upregulation of Oncogenic Pathways in the HNSCC Patients
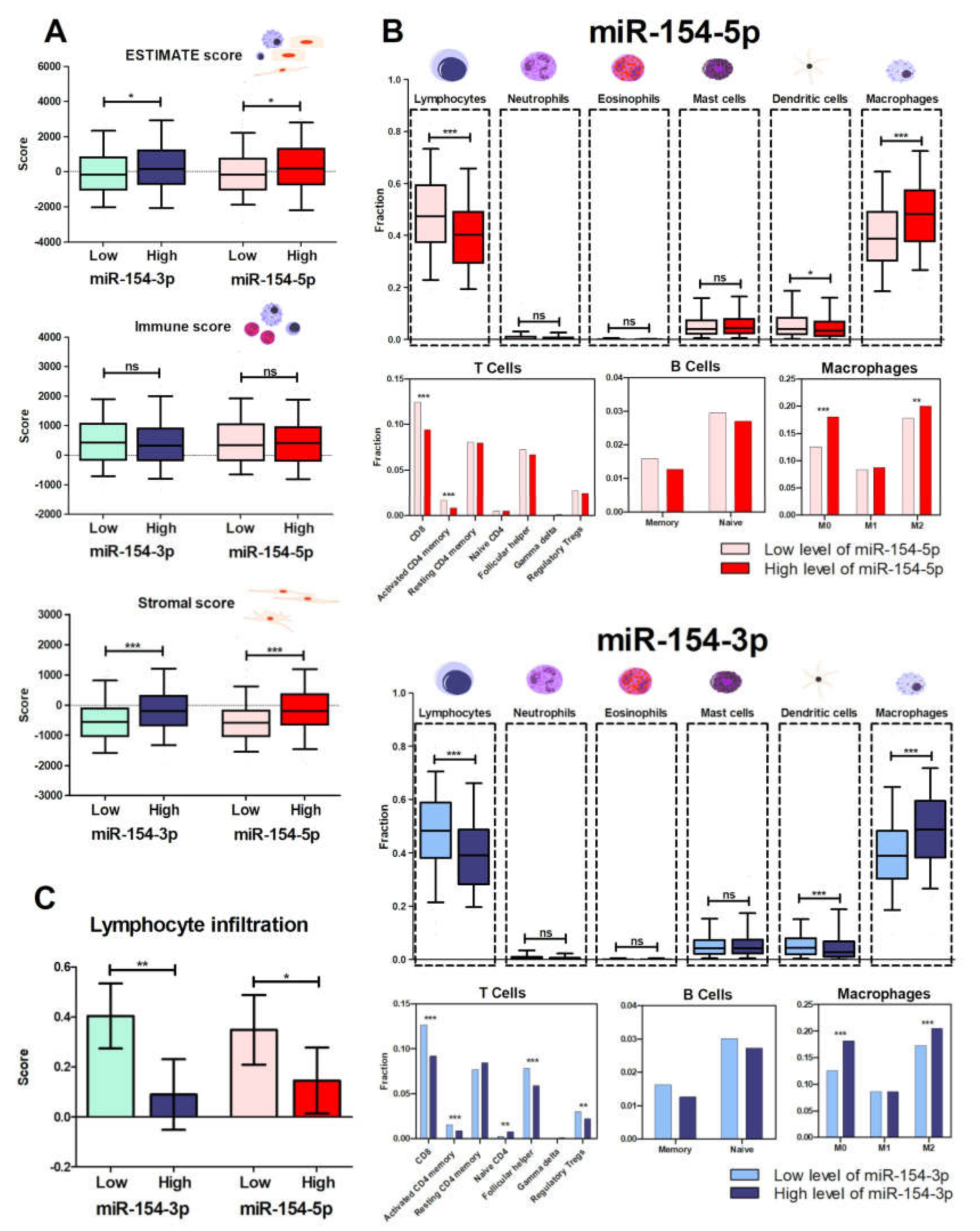
3.4. Patients with Low Levels of Both miR-154 Mature Strands Have a More Favorable Immunological Profile
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marur, S.; Forastiere, A.A. Head and neck cancer: Changing epidemiology, diagnosis, and treatment. Mayo Clin. Proc. 2008, 83, 489–501. [Google Scholar] [CrossRef]
- Kolenda, T.; Guglas, K.; Ryś, M.; Bogaczyńska, M.; Teresiak, A.; Bliźniak, R.; Łasińska, I.; Mackiewicz, J.; Lamperska, K.M. Biological role of long non-coding RNA in head and neck cancers. Rep. Pract. Oncol. Radiother. 2017, 22, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Lubov, J.; Maschietto, M.; Ibrahim, I.; Mlynarek, A.; Hier, M.; Kowalski, L.P.; Alaoui-Jamali, M.A.; Da Silva, S.D. Meta-analysis of microRNAs expression in head and neck cancer: Uncovering association with outcome and mechanisms. Oncotarget 2017, 8, 55511–55524. [Google Scholar] [CrossRef] [Green Version]
- Dok, R.; Nuyts, S. HPV Positive Head and Neck Cancers: Molecular Pathogenesis and Evolving Treatment Strategies. Cancers 2016, 8, 41. [Google Scholar] [CrossRef] [Green Version]
- Jou, A.; Hess, J. Epidemiology and Molecular Biology of Head and Neck Cancer. Oncol. Res. Treat. 2017, 40, 328–332. [Google Scholar] [CrossRef] [PubMed]
- van Kempen, P.M.W.; Noorlag, R.; Braunius, W.W.; Stegeman, I.; Willems, S.M.; Grolman, W. Differences in methylation profiles between HPV-positive and HPV-negative oropharynx squamous cell carcinoma: A systematic review. Epigenetics 2014, 9, 194–203. [Google Scholar] [CrossRef] [Green Version]
- Dahiya, K.; Dhankhar, R. Updated overview of current biomarkers in head and neck carcinoma. World J. Methodol. 2016, 6, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Kalia, M. Personalized oncology: Recent advances and future challenges. Metabolism 2013, 62 (Suppl. 1), S11–S14. [Google Scholar] [CrossRef] [PubMed]
- Fassan, M. Molecular diagnostics in pathology time for a next-generation pathologist? Arch. Pathol. Lab. Med. 2018, 142, 313–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, M.; Liu, L.; Zhang, S.; Yang, X.; Wang, Y. Cancer stem cell biomarkers for head and neck squamous cell carcinoma: A bioinformatic analysis. Oncol. Rep. 2018, 40, 3843–3851. [Google Scholar] [CrossRef] [Green Version]
- Su, N.; Wang, Y.; Qian, M.; Deng, M. Combinatorial regulation of transcription factors and microRNAs. BMC Syst. Biol. 2010, 4, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagadia, R.; Pandit, P.; Coman, W.B.; Cooper-White, J.; Punyadeera, C. miRNAs in head and neck cancer revisited. Cell. Oncol. 2013, 36, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Irani, S. miRNAs Signature in Head and Neck Squamous Cell Carcinoma Metastasis: A Literature Review. J. Dent. (Shiraz Iran.) 2016, 17, 71–83. [Google Scholar]
- Carron, J.; Msc, C.T.; Silva, J.K.; Bsc, G.S.R.Q.; Ortega, M.M.; Lima, C.S.P.; Lourenço, G.J. microRNAs deregulation in head and neck squamous cell carcinoma. Head Neck 2021, 43, 645–667. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Gholipour, M.; Taheri, M.; Farsani, Z.S. MicroRNA profile in the squamous cell carcinoma: Prognostic and diagnostic roles. Heliyon 2020, 6, e05436. [Google Scholar] [CrossRef] [PubMed]
- Emmett, S.; Whiteman, D.C.; Panizza, B.J.; Antonsson, A. An Update on Cellular MicroRNA Expression in Human Papillomavirus-Associated Head and Neck Squamous Cell Carcinoma. Oncology 2018, 95, 193–201. [Google Scholar] [CrossRef]
- Kolenda, T.; Teresiak, A.; Kapałczyńska, M.; Przybyła, W.; Zajączkowska, M.; Bliźniak, R.; Lamperska, K. let-7d and miR-18a as biomarkers of head and neck cancers. Lett. Oncol. Sci. 2015, 12, 37–47. [Google Scholar]
- Wang, J.; Fang, Y.; Liu, Y.-F.; Wang, X.; Wang, R.-Y.; Meng, Z.-D. miR-154 inhibits cells proliferation and metastasis in melanoma by targeting AURKA and serves as a novel prognostic indicator. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4275–4284. [Google Scholar]
- Wang, L.; Wu, L.; Wu, J. Downregulation of miR-154 in human glioma and its clinicopathological and prognostic significance. J. Int. Med. Res. 2016, 44, 994–1001. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Yang, Y.; Chen, L.; Liu, D.; Dong, H. MicroRNA-154 functions as a tumor suppressor in non-small cell lung cancer through directly targeting B-cell-specific Moloney murine leukemia virus insertion site 1. Oncol. Lett. 2018, 15, 10098–10104. [Google Scholar] [CrossRef] [Green Version]
- Qin, C.; Zhao, Y.; Gong, C.; Yang, Z. MicroRNA-154/ADAM9 axis inhibits the proliferation, migration and invasion of breast cancer cells. Oncol. Lett. 2017, 14, 6969–6975. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Ji, Z.; Xie, Y.; Liu, G.; Li, H. MicroRNA-154 as a prognostic factor in bladder cancer inhibits cellular malignancy by targeting RSF1 and RUNX2. Oncol. Rep. 2017, 38, 2727–2734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, W.; Cao, N.; Yang, L. MicroRNA-154 inhibits the growth and metastasis of gastric cancer cells by directly targeting MTDH. Oncol. Lett. 2017, 14, 3268–3274. [Google Scholar] [CrossRef] [Green Version]
- Niu, J.-T.; Zhang, L.-J.; Huang, Y.-W.; Li, C.; Jiang, N.; Niu, Y.-J. miR-154 inhibits the growth of laryngeal squamous cell carcinoma by targeting GALNT7. Biochem. Cell Biol. 2018, 96, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Li, Z.; Chen, P.; Quan, J.; Pan, X.; Zhao, L.; Zhou, L.; Lai, Y.; He, T.; Xu, W.; et al. Oncogene miR-154-5p regulates cellular function and acts as a molecular marker with poor prognosis in renal cell carcinoma. Life Sci. 2018, 209, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Q.; Gao, D. Inhibitory effect of microRNA-154 targeting WHSC1 on cell proliferation of human skin squamous cell carcinoma through mediating the P53 signaling pathway. Int. J. Biochem. Cell Biol. 2018, 100, 22–29. [Google Scholar] [CrossRef]
- Jiang, L.; Huang, Q.; Zhang, S.; Zhang, Q.; Chang, J.; Qiu, X.; Wang, E. Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer 2010, 10, 318. [Google Scholar] [CrossRef] [Green Version]
- Almeida, M.I.; Nicoloso, M.S.; Zeng, L.; Ivan, C.; Spizzo, R.; Gafà, R.; Xiao, L.; Zhang, X.; Vannini, I.; Fanini, F.; et al. Strand-Specific miR-28-5p and miR-28-3p Have Distinct Effects in Colorectal Cancer Cells. Gastroenterology 2012, 142, 886–896.e9. [Google Scholar] [CrossRef] [Green Version]
- Mitra, R.; Adams, C.M.; Jiang, W.; Greenawalt, E.; Eischen, C.M. Pan-cancer analysis reveals cooperativity of both strands of microRNA that regulate tumorigenesis and patient survival. Nat. Commun. 2020, 11, 968. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Pi, J.; Zou, D.; Wang, X.; Xu, J.; Yu, S.; Zhang, T.; Li, F.; Zhang, X.; Zhao, H.; et al. microRNA arm-imbalance in part from complementary targets mediated decay promotes gastric cancer progression. Nat. Commun. 2019, 10, 4397. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.; Craft, B.; Hastie, M.; Repečka, K.; Kamath, A.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; et al. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. bioRxiv 2019, 326470. [Google Scholar] [CrossRef] [Green Version]
- Severino, P.; Brüggemann, H.; Andreghetto, F.M.; Camps, C.; Klingbeil, M.D.F.G.; De Pereira, W.O.; Soares, R.M.; Moyses, R.; Wünsch-Filho, V.; Mathor, M.B.; et al. MicroRNA expression profile in head and neck cancer: HOX-cluster embedded microRNA-196a and microRNA-10b dysregulation implicated in cell proliferation. BMC Cancer 2013, 13, 533. [Google Scholar] [CrossRef] [PubMed]
- Holt, J.; Walter, V.; Yin, X.; Marron, D.; Wilkerson, M.D.; Choi, H.Y.; Zhao, X.; Jo, H.; Hayes, D.N.; Ko, Y.H. Integrative analysis of microRNAs identifies clinically relevant epithelial and stromal subtypes of head and neck squamous cell carcinoma. Clin. Cancer Res. 2021, 27, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.-H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.-S.; Tam, W.L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A Pattern-Based Method for the Identification of MicroRNA Binding Sites and Their Corresponding Heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar] [CrossRef] [Green Version]
- Vejnar, C.; Zdobnov, E.M. miRmap: Comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 2012, 40, 11673–11683. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [Green Version]
- Reczko, M.; Maragkakis, M.; Alexiou, P.; Grosse, I.; Hatzigeorgiou, A.G. Functional microRNA targets in protein coding sequences. Bioinformatics 2012, 28, 771–776. [Google Scholar] [CrossRef]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [Green Version]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Trevino, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Eddy, J.A.; Thorsson, V.; Lamb, A.E.; Gibbs, D.L.; Heimann, C.; Yu, J.X.; Chung, V.; Chae, Y.; Dang, K.; Vincent, B.G.; et al. CRI iAtlas: An interactive portal for immuno-oncology research. F1000Research 2020, 9, 1028. [Google Scholar] [CrossRef]
- Cohen, N.; Fedewa, S.; Chen, A.Y. Epidemiology and Demographics of the Head and Neck Cancer Population. Oral Maxillofac. Surg. Clin. N. Am. 2018, 30, 381–395. [Google Scholar] [CrossRef]
- Wu, T.-S.; Lin, B.-R.; Chang, H.-H. Radio Resistance Mechanisms of Cancers: An Overview and Future Perspectives. Biol. Med. 2015, S2, 002. [Google Scholar] [CrossRef] [Green Version]
- Pendleton, K.P.; Grandis, J.R. Cisplatin-Based Chemotherapy Options for Recurrent and/or Metastatic Squamous Cell Cancer of the Head and Neck. Clin. Med. Insights Ther. 2013, 5, CMT.S10409. [Google Scholar] [CrossRef] [Green Version]
- Łasińska, I.; Kolenda, T.; Teresiak, A.; Lamperska, K.M.; Galus, Ł.; Mackiewicz, J. Immunotherapy in Patients with Recurrent and Metastatic Squamous Cell Carcinoma of the Head and Neck. Anticancer Agents Med. Chem. 2019, 19, 290–303. [Google Scholar] [CrossRef]
- Pai, S.I.; Westra, W.H. Molecular Pathology of Head and Neck Cancer: Implications for Diagnosis, Prognosis, and Treatment. Annu. Rev. Pathol. Mech. Dis. 2009, 4, 49–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshpande, A.M.; Wong, D.T. Molecular mechanisms of head and neck cancer. Expert Rev. Anticancer. Ther. 2008, 8, 799–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolenda, T.; Przybyła, W.; Kapałczyńska, M.; Teresiak, A.; Zajączkowska, M.; Bliźniak, R.; Lamperska, K.M. Tumor microenvironment—Unknown niche with powerful therapeutic potential. Rep. Pract. Oncol. Radiother. 2018, 23, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Zhi, X.; Lamperska, K.; Golusinski, P.; Schork, N.J.; Luczewski, L.; Kolenda, T.; Golusiński, W.; Masternak, M.M. Gene expression analysis of head and neck squamous cell carcinoma survival and recurrence. Oncotarget 2014, 6, 547–555. [Google Scholar] [CrossRef]
- Lamperska, K.M.; Kozlowski, P.; Kolenda, T.; Teresiak, A.; Blizniak, R.; Przybyla, W.; Masternak, M.M.; Golusinski, P.; Golusinski, W. Unpredictable changes of selected miRNA in expression profile of HNSCC. Cancer Biomark. 2016, 16, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Lamperska, K.M.; Kolenda, T.; Teresiak, A.; Kowalik, A.; Kruszyna-Mochalska, M.; Jackowiak, W.; Bliźniak, R.; Przybyła, W.; Kapałczyńska, M.; Kozlowski, P. Different levels of let-7d expression modulate response of FaDu cells to irradiation and chemotherapeutics. PLoS ONE 2017, 12, e0180265. [Google Scholar] [CrossRef] [PubMed]
- Kolenda, T.; Guglas, K.; Teresiak, A.; Bliźniak, R.; Lamperska, K. Low let-7d and high miR-205 expression levels positively influence HNSCC patient outcome. J. Biomed. Sci. 2019, 26, 17. [Google Scholar] [CrossRef]
- Kolenda, T.; Kopczyńska, M.; Guglas, K.; Teresiak, A.; Bliźniak, R.; Łasińska, I.; Mackiewicz, J.; Lamperska, K. EGOT lncRNA in head and neck squamous cell carcinomas. Pol. J. Pathol. 2018, 69, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Kolenda, T.; Guglas, K.; Kopczyńska, M.; Teresiak, A.; Bliźniak, R.; Mackiewicz, A.; Lamperska, K.; Mackiewicz, J. Oncogenic Role of ZFAS1 lncRNA in Head and Neck Squamous Cell Carcinomas. Cells 2019, 8, 366. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Ma, C.; Zhang, Y.; Pei, S.; Du, M.; Zhang, Y.; Qian, L.-X.; Wang, J.; Yin, L.; He, X. miR-154-5p Suppresses Cell Invasion and Migration Through Inhibiting KIF14 in Nasopharyngeal Carcinoma. OncoTargets Ther. 2020, 13, 2235–2246. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Yang, Z.; Zhang, P.; Shao, G. miR-154 suppresses non-small cell lung cancer growth in vitro and in vivo. Oncol. Rep. 2015, 33, 3053–3060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vojtechova, Z.; Sabol, I.; Salakova, M.; Smahelova, J.; Zavadil, J.; Turek, L.; Grega, M.; Klozar, J.; Prochazka, B.; Tachezy, R. Comparison of the miRNA profiles in HPV-positive and HPV-negative tonsillar tumors and a model system of human keratinocyte clones. BMC Cancer 2016, 16, 382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peta, E.; Sinigaglia, A.; Masi, G.; Di Camillo, B.; Grassi, A.; Trevisan, M.; Messa, L.; Loregian, A.; Manfrin, E.; Brunelli, M.; et al. HPV16 E6 and E7 upregulate the histone lysine demethylase KDM2B through the c-MYC/miR-146a-5p axys. Oncogene 2018, 37, 1654–1668. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, Y.; Zhang, L.; Ding, L.; Li, Y.; Zhang, H.; Wang, T.; Hao, M. MicroRNA-154-5p regulates the HPV16 E7-pRb pathway in Cervical Carcinogenesis by targeting CUL2. J. Cancer 2020, 11, 5379–5389. [Google Scholar] [CrossRef] [PubMed]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kent, L.N.; Leone, G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 2019, 19, 326–338. [Google Scholar] [CrossRef]
- Emmrich, S.; Pützer, B.M. Checks and balances: E2F-microRNA crosstalk in cancer control. Cell Cycle 2010, 9, 2555–2567. [Google Scholar] [CrossRef]
- Xu, H.; Fei, D.; Zong, S.; Fan, Z. MicroRNA-154 inhibits growth and invasion of breast cancer cells through targeting E2F5. Am. J. Transl. Res. 2016, 8, 2620–2630. [Google Scholar] [PubMed]
- Stark, G.R.; Taylor, W.R. Analyzing the G2/M checkpoint. In Checkpoint Controls and Cancer; Methods in Molecular Biology™; Humana Press: Totowa, NJ, USA, 2004; Volume 280, pp. 51–82. [Google Scholar] [CrossRef]
- Löbrich, M.; Jeggo, P.A. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer 2007, 7, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, L.; Liu, M.; Du, R.; Dang, Y.; Bai, M.; Zhang, L.; Ma, F.; Yang, X.; Ning, X.; et al. MicroRNA-493 targets STMN-1 and promotes hypoxia-induced epithelial cell cycle arrest in G2/M and renal fibrosis. FASEB J. 2019, 33, 1565–1577. [Google Scholar] [CrossRef]
- Jenkins, R.H.; Davies, L.C.; Taylor, P.R.; Akiyama, H.; Cumbes, B.; Beltrami, C.; Carrington, C.P.; Phillips, A.O.; Bowen, T.; Fraser, D.J. miR-192 induces G2/M growth arrest in aristolochic acid nephropathy. Am. J. Pathol. 2014, 184, 996–1009. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Lin, X.; Xu, W.; Lin, X.; Huang, Q.; Shi, L.; Pan, Y.; Zhang, Y.; Zhu, Y.; Li, C.; et al. miR-210-3p protects endometriotic cells from oxidative stress-induced cell cycle arrest by targeting BARD1. Cell Death Dis. 2019, 10, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbie, D.A.; Tamayo, P.; Boehm, J.; Kim, S.Y.; Moody, S.E.; Dunn, I.F.; Schinzel, A.C.; Sandy, P.; Meylan, E.; Scholl, C.; et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nat. Cell Biol. 2009, 462, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Gururajan, M.; Josson, S.; Chu, G.C.-Y.; Lu, C.-L.; Lu, Y.-T.; Haga, C.L.; Zhau, H.E.; Liu, C.; Lichterman, J.; Duan, P.; et al. miR-154* and miR-379 in the DLK1-DIO3 MicroRNA Mega-Cluster Regulate Epithelial to Mesenchymal Transition and Bone Metastasis of Prostate Cancer. Clin. Cancer Res. 2014, 20, 6559–6569. [Google Scholar] [CrossRef] [Green Version]
- Josson, S.; Chung, L.W.K.; Gururajan, M. MicroRNAs and prostate cancer. Adv. Exp. Med. Biol. 2015, 889, 105–118. [Google Scholar]
- Zhang, X.; Huang, Z.; Hu, Y.; Liu, L. Knockdown of Myosin 6 inhibits proliferation of oral squamous cell carcinoma cells. J. Oral Pathol. Med. 2016, 45, 740–745. [Google Scholar] [CrossRef]
- Lei, C.; Du, F.; Sun, L.; Li, T.; Li, T.; Min, Y.; Nie, A.; Wang, X.; Geng, L.; Lu, Y.; et al. miR-143 and miR-145 inhibit gastric cancer cell migration and metastasis by suppressing MYO6. Cell Death Dis. 2017, 8, e3101. [Google Scholar] [CrossRef] [Green Version]
- Murata, M.; Ito, T.; Tanaka, Y.; Yamamura, K.; Furue, K.; Furue, M. OVOL2-Mediated ZEB1 Downregulation May Prevent Promotion of Actinic Keratosis to Cutaneous Squamous Cell Carcinoma. J. Clin. Med. 2020, 9, 618. [Google Scholar] [CrossRef] [Green Version]
- Roca, H.; Hernandez, J.; Weidner, S.; McEachin, R.C.; Fuller, D.; Sud, S.; Schumann, T.; Wilkinson, J.E.; Zaslavsky, A.; Li, H.; et al. Transcription Factors OVOL1 and OVOL2 Induce the Mesenchymal to Epithelial Transition in Human Cancer. PLoS ONE 2013, 8, e76773. [Google Scholar] [CrossRef]
- Jia, D.; Jolly, M.K.; Boareto, M.; Parsana, P.; Mooney, S.M.; Pienta, K.J.; Levine, H.; Ben-Jacob, E. OVOL guides the epithelial-hybrid-mesenchymal transition. Oncotarget 2015, 6, 15436–15448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nath, D.; Li, X.; Mondragon, C.; Post, D.; Chen, M.; White, J.R.; Hryniewicz-Jankowska, A.; Caza, T.; Kuznetsov, V.A.; Hehnly, H.; et al. Abi1 loss drives prostate tumorigenesis through activation of EMT and non-canonical WNT signaling. Cell Commun. Signal. 2019, 17, 120. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.; Liu, J.; Chao, J.; Scheuerman, M.P.; Rahimi, S.A.; Lee, L.Y.; Li, S. PTEN suppresses epithelial–mesenchymal transition and cancer stem cell activity by downregulating Abi1. Sci. Rep. 2020, 10, 12685. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Chen, H.; Zhu, J.Y.; Wang, W.; Teng, Y.; Ding, H.F.; Jing, Q.; Su, S.B.; Huang, S. Epithelial-Mesenchymal Transition of Ovarian Cancer Cells Is Sustained by Rac1 through Simultaneous Activation of MEK1/2 and Src Signaling Pathways. Oncogene 2017, 36, 1546–1558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roshan, V.G.D.; Sinto, M.S.; Vargees, B.T.; Kannan, S. Loss of CDKN2A and CDKN2B expression is associated with disease recurrence in oral cancer. J. Oral Maxillofac. Pathol. 2019, 23, 82–89. [Google Scholar] [CrossRef]
- Li, B.; Li, A.; You, Z.; Xu, J.; Zhu, S. Epigenetic silencing of CDKN1A and CDKN2B by SNHG1 promotes the cell cycle, migration and epithelial-mesenchymal transition progression of hepatocellular carcinoma. Cell Death Dis. 2020, 11, 823. [Google Scholar] [CrossRef]
- Weiße, J.; Rosemann, J.; Krauspe, V.; Kappler, M.; Eckert, A.W.; Haemmerle, M.; Gutschner, T. RNA-Binding Proteins as Regulators of Migration, Invasion and Metastasis in Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2020, 21, 6835. [Google Scholar] [CrossRef]
- Ishii, H.; Saitoh, M.; Sakamoto, K.; Kondo, T.; Katoh, R.; Tanaka, S.; Motizuki, M.; Masuyama, K.; Miyazawa, K. Epithelial Splicing Regulatory Proteins 1 (ESRP1) and 2 (ESRP2) Suppress Cancer Cell Motility via Different Mechanisms. J. Biol. Chem. 2014, 289, 27386–27399. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Q.; Fang, Y.; Lai, Q.; Wang, S.; He, C.; Li, A.; Liu, S.; Yan, Q. CPEB3 inhibits epithelial-mesenchymal transition by disrupting the crosstalk between colorectal cancer cells and tumor-associated macrophages via IL-6R/STAT3 signaling. J. Exp. Clin. Cancer Res. 2020, 39, 132. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, R.; Li, L. LINC00641 hinders the progression of cervical cancer by targeting miR-378a-3p/CPEB3. J. Gene Med. 2020, 22, e3212. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Han, X.; Feng, Y.; Hao, Y. linc00968 inhibits the tumorigenesis and metastasis of lung adenocarcinoma via serving as a ceRNA against miR-9-5p and increasing CPEB3. Aging 2020, 12, 22582–22598. [Google Scholar] [CrossRef]
- Wu, L.; Zhao, N.; Zhou, Z.; Chen, J.; Han, S.; Zhang, X.; Bao, H.; Yuan, W.; Shu, X. PLAGL2 promotes the proliferation and migration of gastric cancer cells via USP37-mediated deubiquitination of Snail1. Theranostics 2021, 11, 700–714. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, Z.; Han, S.; Chen, J.; Liu, Z.; Zhang, X.; Yuan, W.; Ji, J.; Shu, X. PLAGL2 promotes epithelial–mesenchymal transition and mediates colorectal cancer metastasis via β-catenin-dependent regulation of ZEB1. Br. J. Cancer 2020, 122, 578–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Sun, L.; Liu, R.; Mo, H.; Niu, Y.; Chen, T.; Wang, Y.; Han, S.; Tu, K.; Liu, Q. Long non-coding RNA MAPKAPK5-AS1/PLAGL2/HIF-1α signaling loop promotes hepatocellular carcinoma progression. J. Exp. Clin. Cancer Res. 2021, 40, 72. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Zhang, B.; Li, B.; Cao, X.; Chen, X.; Xue, Q. PTBP3 promotes migration of non-small cell lung cancer through regulating E-cadherin in EMT signaling pathway. Cancer Cell Int. 2020, 20, 172. [Google Scholar] [CrossRef]
- Hou, P.; Li, L.; Chen, F.; Chen, Y.; Liu, H.; Li, J.; Bai, J.; Zheng, J. PTBP3-Mediated Regulation of ZEB1 mRNA Stability Promotes Epithelial–Mesenchymal Transition in Breast Cancer. Cancer Res. 2018, 78, 387–398. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Chi, W.; Cao, H.; Cui, W.; Meng, W.; Guo, W.; Wang, B. Screening and clinical significance of tumor markers in head and neck squamous cell carcinoma through bioinformatics analysis. Mol. Med. Rep. 2018, 19, 143–154. [Google Scholar] [CrossRef] [Green Version]
- Saloura, V.; Vougiouklakis, T.; Sievers, C.; Burkitt, K.; Nakamura, Y.; Hager, G.; van Waes, C. The role of protein methyltransferases as potential novel therapeutic targets in squamous cell carcinoma of the head and neck. Oral Oncol. 2018, 81, 100–108. [Google Scholar] [CrossRef]
- Gameiro, S.F.; Ghasemi, F.; Zeng, P.Y.F.; Mundi, N.; Howlett, C.J.; Plantinga, P.; Barrett, J.W.; Nichols, A.C.; Mymryk, J.S. Low expression of NSD1, NSD2, and NSD3 define a subset of human papillomavirus-positive oral squamous carcinomas with unfavorable prognosis. Infect. Agents Cancer 2021, 16, 13. [Google Scholar] [CrossRef] [PubMed]
- Saloura, V.; Cho, H.-S.; Kiyotani, K.; Alachkar, H.; Zuo, Z.; Nakakido, M.; Tsunoda, T.; Seiwert, T.; Lingen, M.; Licht, J.; et al. WHSC1 Promotes Oncogenesis through Regulation of NIMA-Related Kinase-7 in Squamous Cell Carcinoma of the Head and Neck. Mol. Cancer Res. 2014, 13, 293–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Chen, Y.; Zhao, W.; Fang, C.; Zhou, W.; Yang, X.; Ji, M. The Role of Methyltransferase NSD2 as a Potential Oncogene in Human Solid Tumors. OncoTargets Ther. 2020, 13, 6837–6846. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Zhang, J.; Guo, W. Tumor purity as a prognosis and immunotherapy relevant feature in gastric cancer. Cancer Med. 2020, 9, 9052–9063. [Google Scholar] [CrossRef] [PubMed]
- Kemi, N.; Eskuri, M.; Kauppila, J.H. Tumour-stroma ratio and 5-year mortality in gastric adenocarcinoma: A systematic review and meta-analysis. Sci. Rep. 2019, 9, 16018. [Google Scholar] [CrossRef] [Green Version]
- Mao, Y.; Feng, Q.; Zheng, P.; Yang, L.; Liu, T.; Xu, Y.; Zhu, D.; Chang, W.; Ji, M.; Ren, L.; et al. Low tumor purity is associated with poor prognosis, heavy mutation burden, and intense immune phenotype in colon cancer. Cancer Manag. Res. 2018, 10, 3569–3577. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Cheng, W.; Ren, X.; Wang, Z.; Liu, X.; Li, G.; Han, S.; Jiang, T.; Wu, A. Tumor Purity as an Underlying Key Factor in Glioma. Clin. Cancer Res. 2017, 23, 6279–6291. [Google Scholar] [CrossRef] [Green Version]
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. Curr. Biol. CB 2020, 30, R921–R925. [Google Scholar] [CrossRef]
- Mandal, R.; Şenbabaoğlu, Y.; Desrichard, A.; Havel, J.J.; Dalin, M.G.; Riaz, N.; Lee, K.-W.; Ganly, I.; Hakimi, A.A.; Chan, T.A.; et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight 2016, 1, e89829. [Google Scholar] [CrossRef] [Green Version]
- Balermpas, P.; Michel, Y.; Wagenblast, J.; Seitz, O.; Weiss, C.; Rödel, F.; Rödel, C.; Fokas, E. Tumour-infiltrating lymphocytes predict response to definitive chemoradiotherapy in head and neck cancer. Br. J. Cancer 2013, 110, 501–509. [Google Scholar] [CrossRef]
- Kong, C.S.; Narasimhan, B.; Cao, H.; Kwok, S.; Erickson, J.P.; Koong, A.; Pourmand, N.; Le, Q.-T. The Relationship Between Human Papillomavirus Status and Other Molecular Prognostic Markers in Head and Neck Squamous Cell Carcinomas. Int. J. Radiat. Oncol. 2009, 74, 553–561. [Google Scholar] [CrossRef] [Green Version]
- Ba, D.W.; Light, E.; Thomas, D.; Worden, F.; Prince, M.; Urba, S.; Chepeha, D.; Kumar, B.; Cordell, K.; Eisbruch, A.; et al. Infiltrating lymphocytes and human papillomavirus-16-associated oropharyngeal cancer. Laryngoscope 2012, 122, 121–127. [Google Scholar] [CrossRef] [Green Version]
- da Silva, V.L.; Fonseca, A.F.; Fonseca, M.; da Silva, T.E.; Coelho, A.C.; Kroll, J.E.; de Souza, J.; Stransky, B.; de Souza, G.A.; de Souza, S.J. Genome-wide identification of cancer/testis genes and their association with prognosis in a pan-cancer analysis. Oncotarget 2017, 8, 92966–92977. [Google Scholar] [CrossRef]
- Simpson, A.J.G.; Caballero, O.L.; Jungbluth, A.; Chen, Y.-T.; Old, L.J. Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer 2005, 5, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.T.; Knops, A.; Swendseid, B.; Martinez-Outschoom, U.; Harshyne, L.; Philp, N.; Rodeck, U.; Luginbuhl, A.; Cognetti, D.; Johnson, J.; et al. Prognostic Significance of Tumor-Associated Macrophage Content in Head and Neck Squamous Cell Carcinoma: A Meta-Analysis. Front. Oncol. 2019, 9, 656. [Google Scholar] [CrossRef]
- She, L.; Qin, Y.; Wang, J.; Liu, C.; Zhu, G.; Li, G.; Wei, M.; Chen, C.; Liu, G.; Zhang, D.; et al. Tumor-associated macrophages derived CCL18 promotes metastasis in squamous cell carcinoma of the head and neck. Cancer Cell Int. 2018, 18, 120. [Google Scholar] [CrossRef]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, C.J.; Leibovich, S.J. Regulation of Macrophage Polarization and Wound Healing. Adv. Wound Care 2012, 1, 10–16. [Google Scholar] [CrossRef] [Green Version]
- Ellisen, L.W. A wound-healing program is hijacked to promote cancer metastasis. J. Exp. Med. 2017, 214, 2813–2815. [Google Scholar] [CrossRef]
- Song, D.; Lyu, H.; Feng, Q.; Luo, J.; Li, L.; Wang, X. Subtyping of head and neck squamous cell cancers based on immune signatures. Int. Immunopharmacol. 2021, 99, 108007. [Google Scholar] [CrossRef]
- House, R.; Majumder, M.; Janakiraman, H.; Ogretmen, B.; Kato, M.; Erkul, E.; Hill, E.; Atkinson, C.; Barth, J.; Day, T.A.; et al. Smoking-induced control of miR-133a-3p alters the expression of EGFR and HuR in HPV-infected oropharyngeal cancer. PLoS ONE 2018, 13, e0205077. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Wu, J.; Li, Y.; Li, X.; Yang, T.; Yang, Q.; Jiang, Y. Deregulation of Serum MicroRNA Expression Is Associated with Cigarette Smoking and Lung Cancer. BioMed Res. Int. 2014, 2014, 364316. [Google Scholar] [CrossRef] [PubMed]
- Cazzoli, R.; Buttitta, F.; DI Nicola, M.; Malatesta, S.; Marchetti, A.; Rom, W.; Pass, H.I. microRNAs Derived from Circulating Exosomes as Noninvasive Biomarkers for Screening and Diagnosing Lung Cancer. J. Thorac. Oncol. 2013, 8, 1156–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Parameter | Group | miR-154-5p | miR-154-3p | ||||
|---|---|---|---|---|---|---|---|
| Cases | Mean ± SEM | p | Cases | Mean ± SEM | p | ||
| Age | ≤ 60 | n = 250 | 1.919 ± 0.8643 | 0.2086 | n = 247 | 1.491 ± 0.7910 | 0.2127 |
| > 61 | n = 269 | 2.013 ± 0.8272 | n = 264 | 1.582 ± 0.8511 | |||
| Gender | Female | n = 147 | 1.950 ± 0.7729 | 0.7350 | n = 43 | 1.490 ± 0.7226 | 0.3860 |
| Male | n = 373 | 1.978 ± 0.8747 | n = 369 | 1.560 ± 0.8606 | |||
| Alcohol | Yes | n = 337 | 1.959 ± 0.8285 | 0.9619 | n = 331 | 1.539 ± 0.8437 | 0.8766 |
| No | n = 173 | 1.962 ± 0.8560 | n = 171 | 1.527 ± 0.7874 | |||
| Smoking | No/ex | n = 203 | 1.925 ± 0.8307 | 0.2880 | n = 201 | 1.529 ± 0.8028 | 0.8000 |
| Yes | n = 302 | 2.006 ± 0.8551 | n = 296 | 1.549 ± 0.8442 | |||
| Cancer stage | I + II | n = 125 | 2.145 ± 0.8002 | 0.0077 | n = 123 | 1.625 ± 0.7244 | 0.1674 |
| III + IV | n = 382 | 1.913 ± 0.8548 | n = 376 | 1.510 ± 0.8291 | |||
| T-stage | T1 + T2 | n = 189 | 2.001 ± 0.8369 | 0.5924 | n = 189 | 1.511 ± 0.7406 | 0.5106 |
| T3 + T4 | n = 316 | 1.959 ± 0.8497 | n = 308 | 1.560 ± 0.8428 | |||
| N-stage | N0 | n = 252 | 2.071 ± 0.8426 | 0.0066 | n = 245 | 1.623 ± 0.8086 | 0.0208 |
| N1 + N2 + N3 | n = 248 | 1.866 ± 0.8409 | n = 246 | 1.455 ± 0.7938 | |||
| Grade | G1 + G2 | n = 370 | 1.913 ± 0.8247 | 0.0017 | n = 361 | 1.507 ± 0.7755 | 0.0589 |
| G3 + G4 | n = 126 | 2.182 ± 0.8299 | n = 127 | 1.667 ± 0.9413 | |||
| Perineural invasion | Positive | n = 171 | 2.090 ± 0.7819 | 0.0777 | n = 169 | 1.682 ± 0.8359 | 0.1173 |
| Negative | n = 192 | 1.936 ± 0.8610 | n = 188 | 1.538 ± 0.8889 | |||
| Lymph node neck dissection | Positive | n = 416 | 1.986 ± 0.8395 | 0.4054 | n = 407 | 1.564 ± 0.8272 | 0.2810 |
| Negative | n = 102 | 1.908 ± 0.8814 | n = 102 | 1.466 ± 0.8081 | |||
| HPV p16 status | Positive | n = 38 | 1.492 ± 0.9784 | 0.0102 | n = 36 | 1.208 ± 0.7566 | 0.0277 |
| Negative | n = 65 | 1.943 ± 0.7552 | n = 65 | 1.534 ± 0.6708 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomaszewska, W.; Kozłowska-Masłoń, J.; Baranowski, D.; Perkowska, A.; Szałkowska, S.; Kazimierczak, U.; Severino, P.; Lamperska, K.; Kolenda, T. miR-154 Influences HNSCC Development and Progression through Regulation of the Epithelial-to-Mesenchymal Transition Process and Could Be Used as a Potential Biomarker. Biomedicines 2021, 9, 1894. https://doi.org/10.3390/biomedicines9121894
Tomaszewska W, Kozłowska-Masłoń J, Baranowski D, Perkowska A, Szałkowska S, Kazimierczak U, Severino P, Lamperska K, Kolenda T. miR-154 Influences HNSCC Development and Progression through Regulation of the Epithelial-to-Mesenchymal Transition Process and Could Be Used as a Potential Biomarker. Biomedicines. 2021; 9(12):1894. https://doi.org/10.3390/biomedicines9121894
Chicago/Turabian StyleTomaszewska, Weronika, Joanna Kozłowska-Masłoń, Dawid Baranowski, Anna Perkowska, Sandra Szałkowska, Urszula Kazimierczak, Patricia Severino, Katarzyna Lamperska, and Tomasz Kolenda. 2021. "miR-154 Influences HNSCC Development and Progression through Regulation of the Epithelial-to-Mesenchymal Transition Process and Could Be Used as a Potential Biomarker" Biomedicines 9, no. 12: 1894. https://doi.org/10.3390/biomedicines9121894
APA StyleTomaszewska, W., Kozłowska-Masłoń, J., Baranowski, D., Perkowska, A., Szałkowska, S., Kazimierczak, U., Severino, P., Lamperska, K., & Kolenda, T. (2021). miR-154 Influences HNSCC Development and Progression through Regulation of the Epithelial-to-Mesenchymal Transition Process and Could Be Used as a Potential Biomarker. Biomedicines, 9(12), 1894. https://doi.org/10.3390/biomedicines9121894






