Divergent Effects of EZH1 and EZH2 Protein Expression on the Prognosis of Patients with T-Cell Lymphomas
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Sequencing and Mutation Analysis
2.3. Tissue Microarray Construction
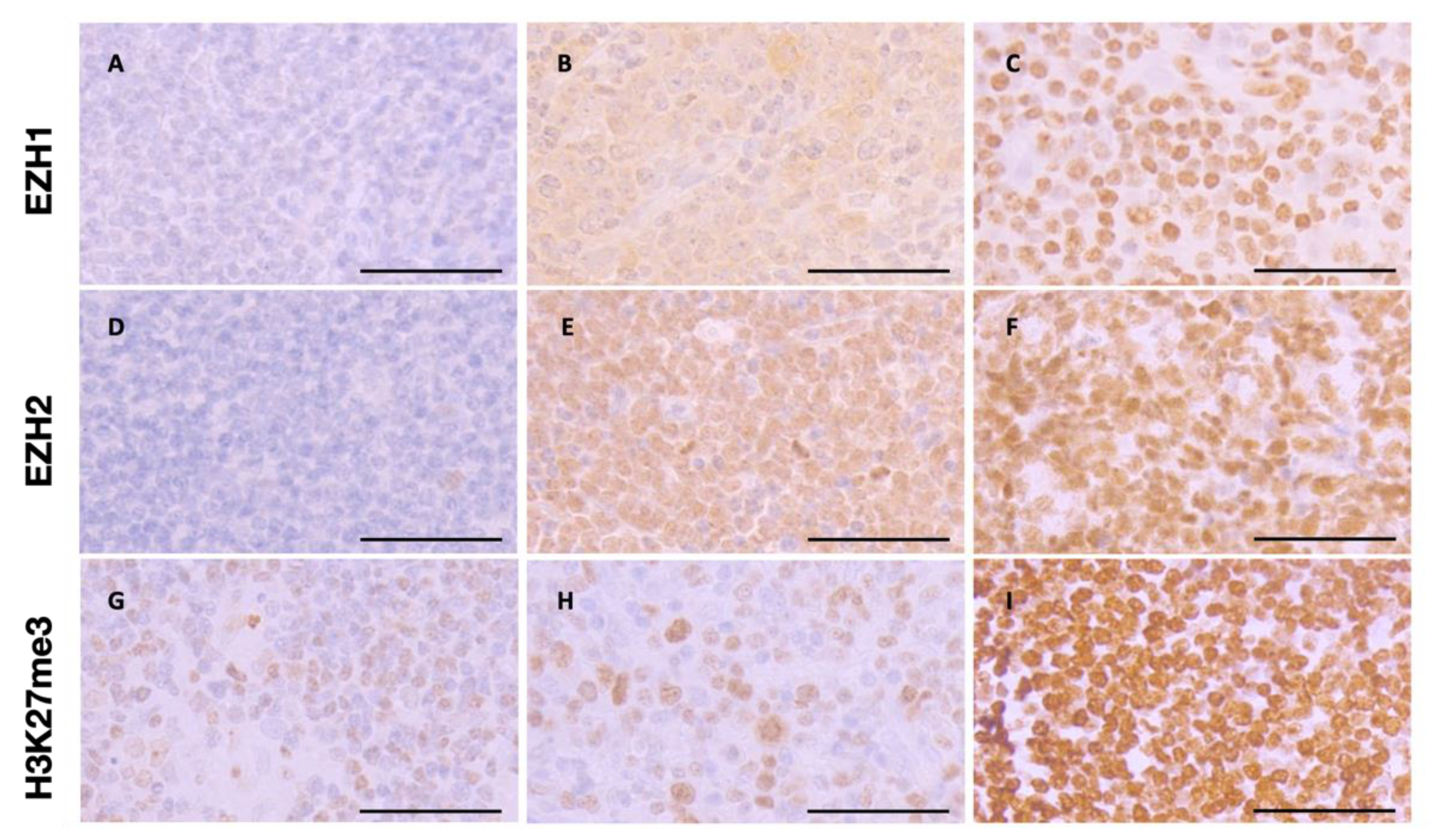
2.4. Immunohistochemistry and Scoring
2.5. Statistical Analysis
3. Results
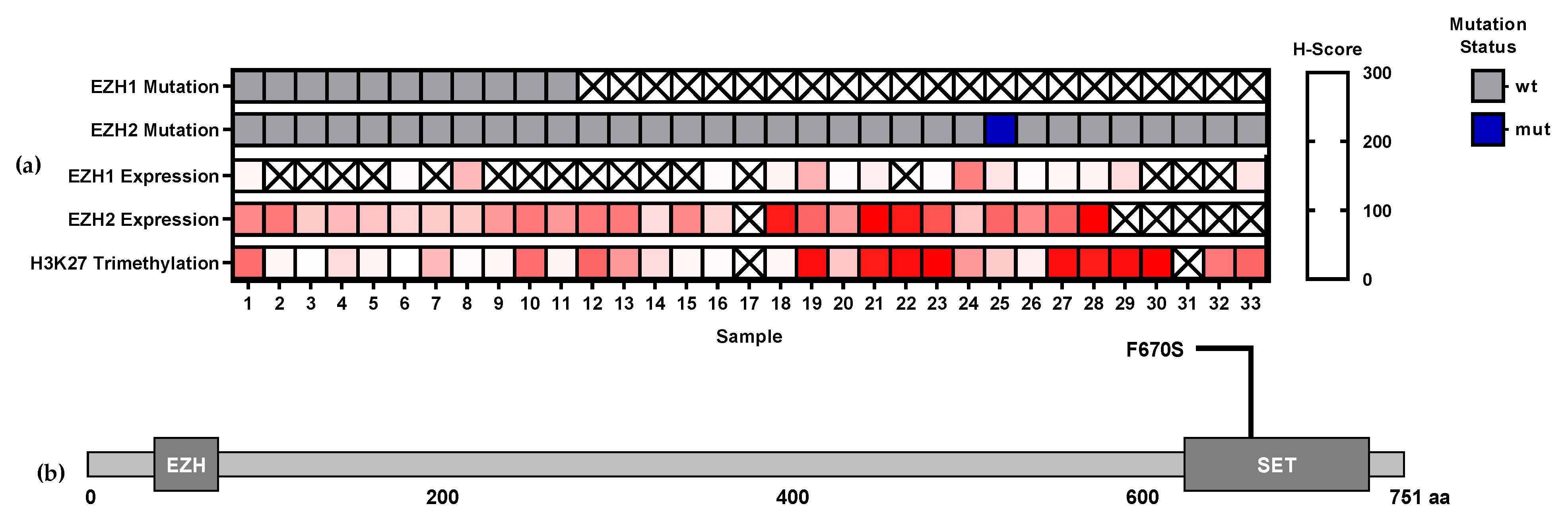
3.1. EZH1 and EZH2 Mutations
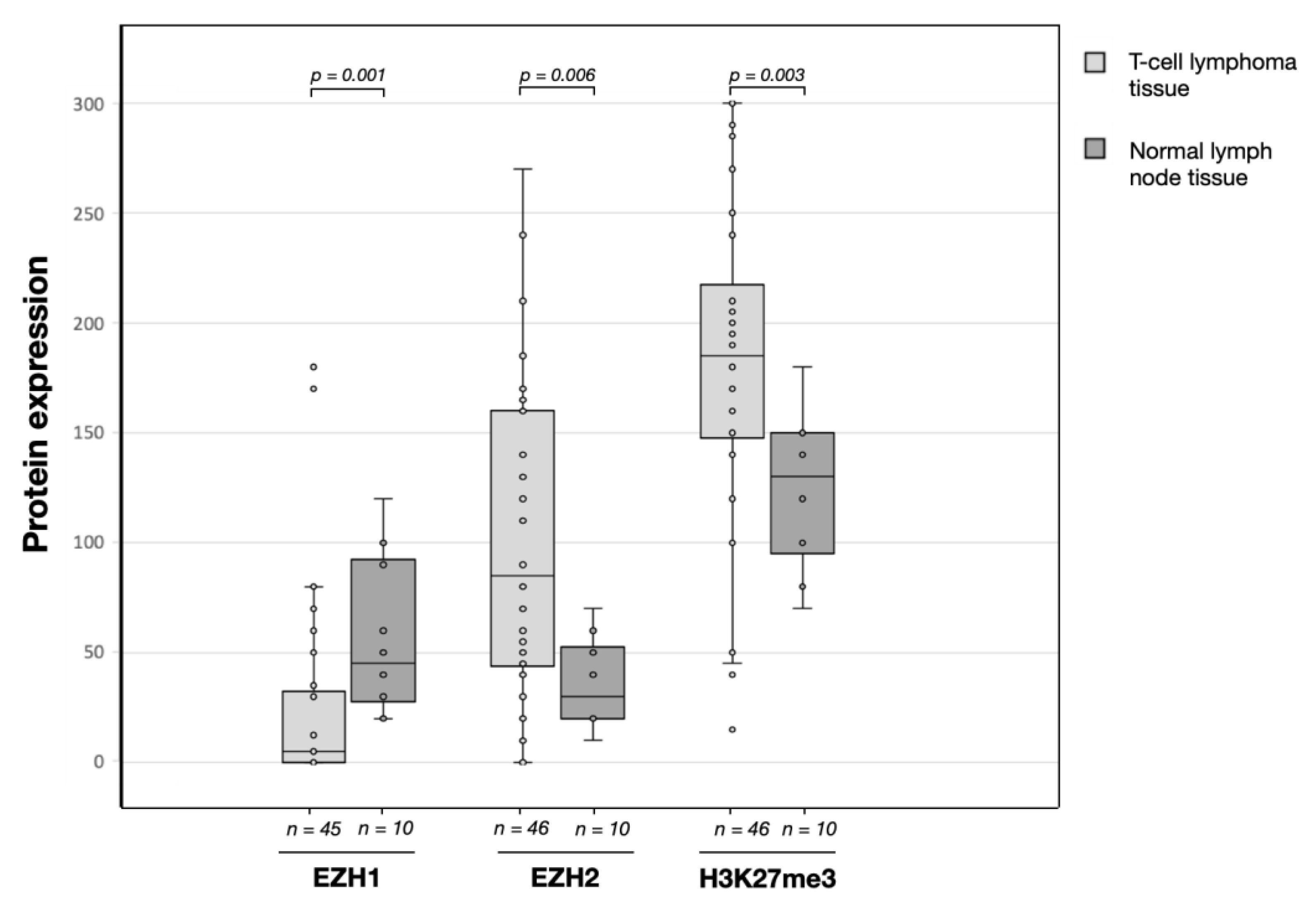
3.2. EZH1, EZH,2 and H3K27me3 Protein Expression Levels in T-Cell Lymphomas
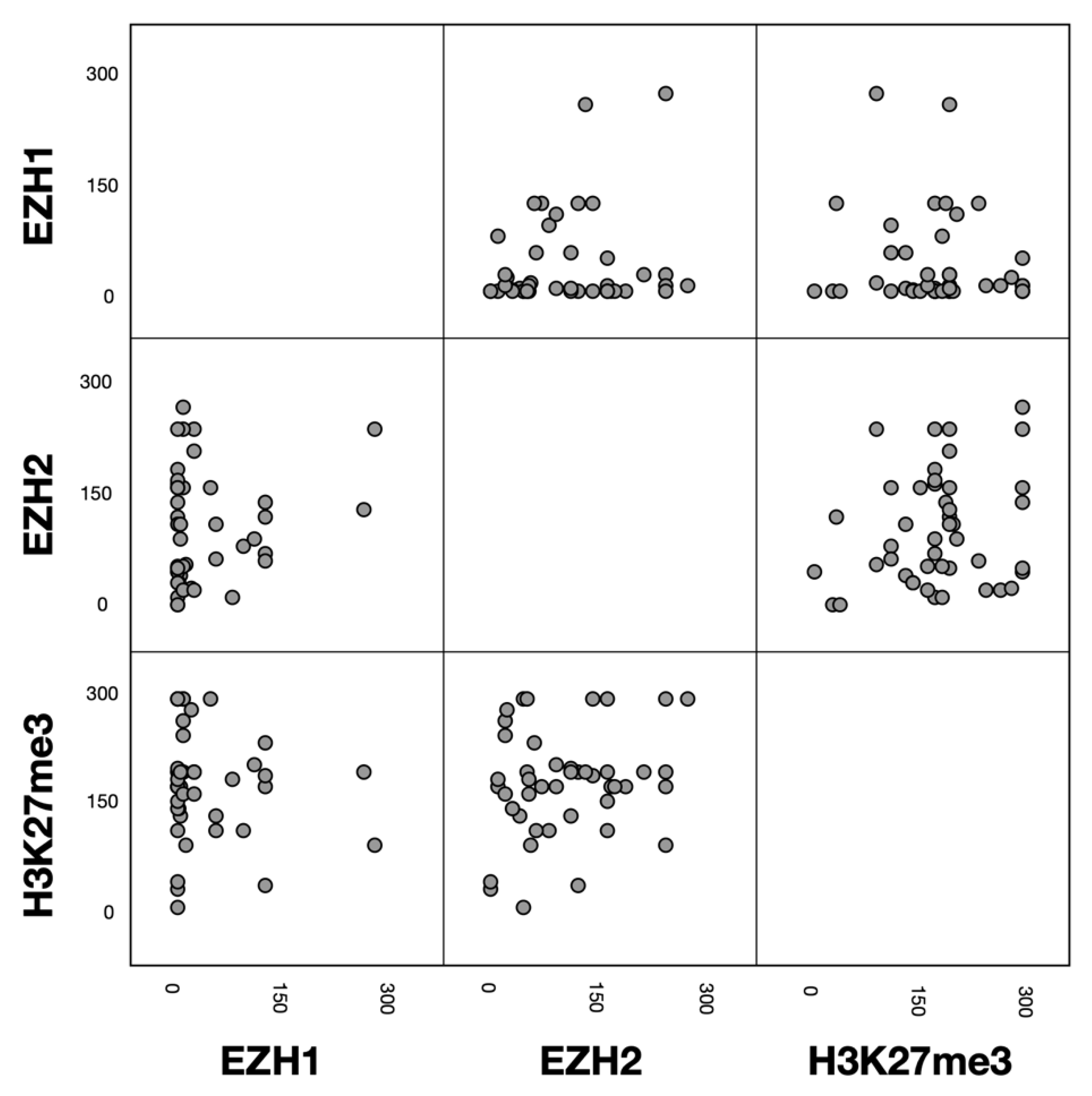
3.3. Associations between EZH1, EZH2 and H3K27me3 Protein Expression
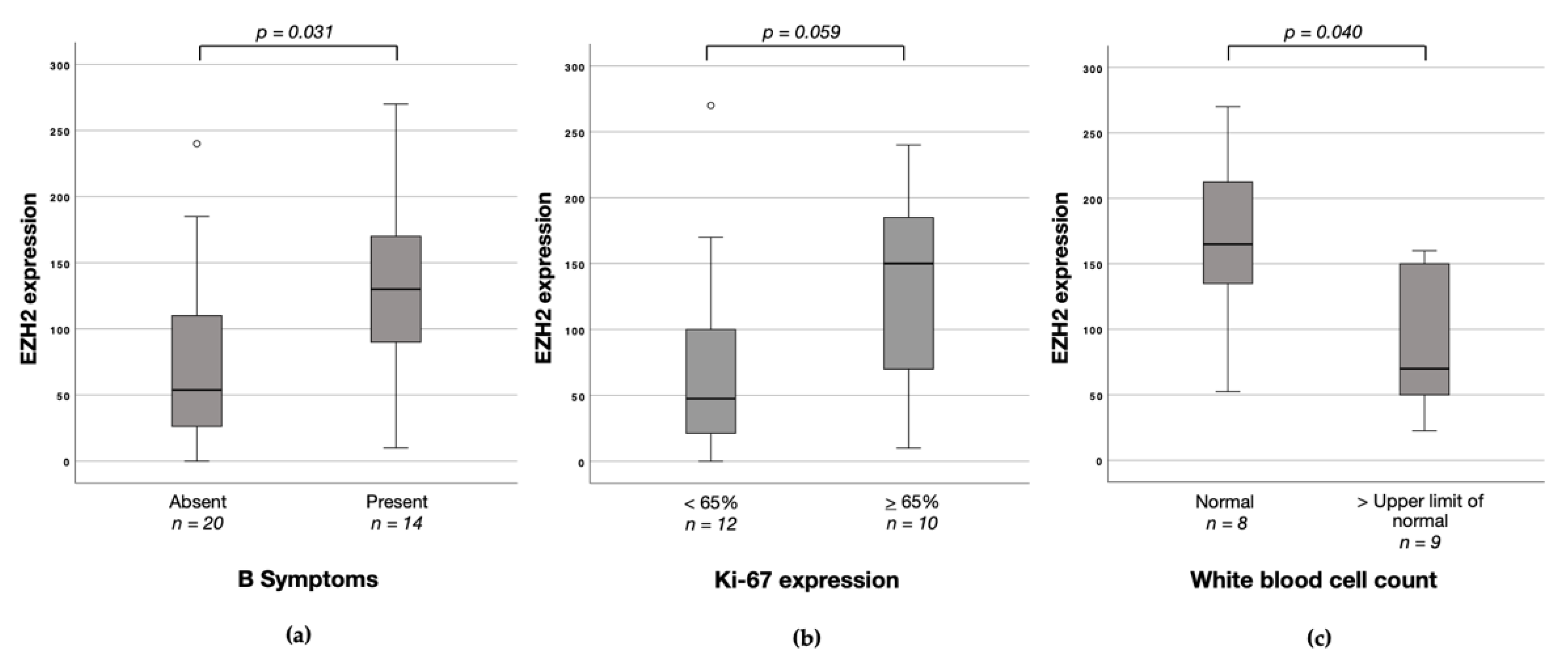
3.4. Associations between EZH1, EZH2 and H3K27me3 Protein Expression and Clinicopathological Characteristics
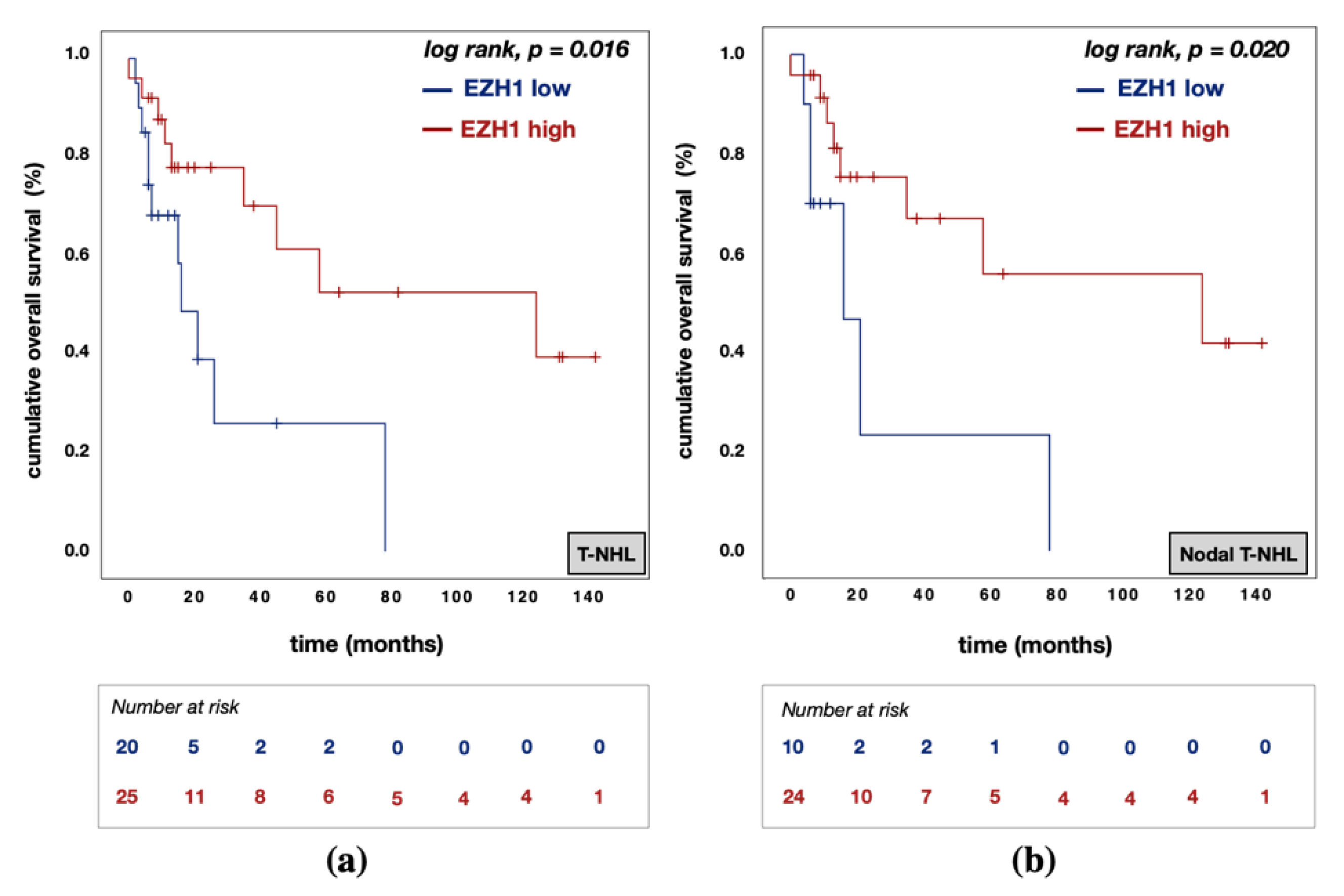
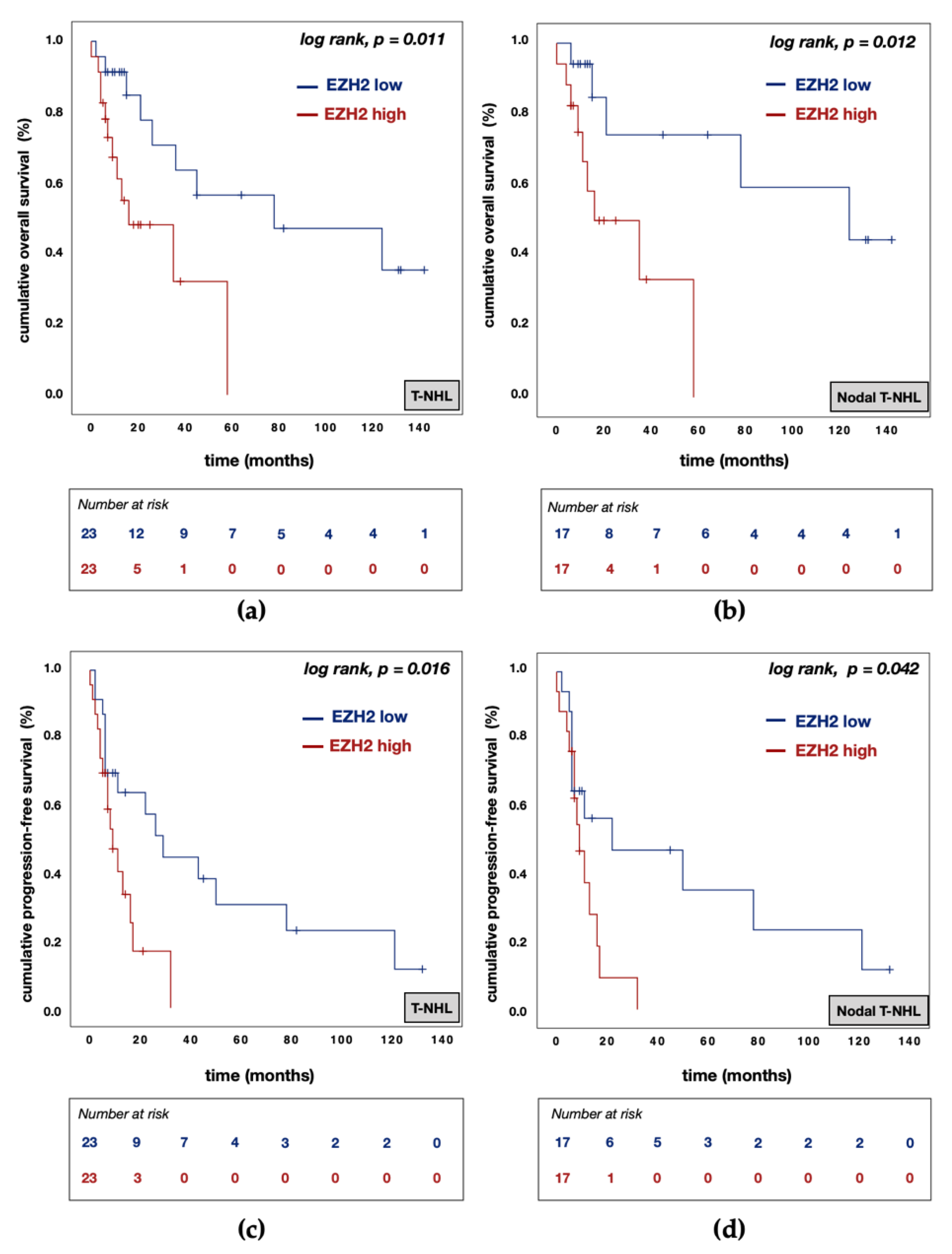
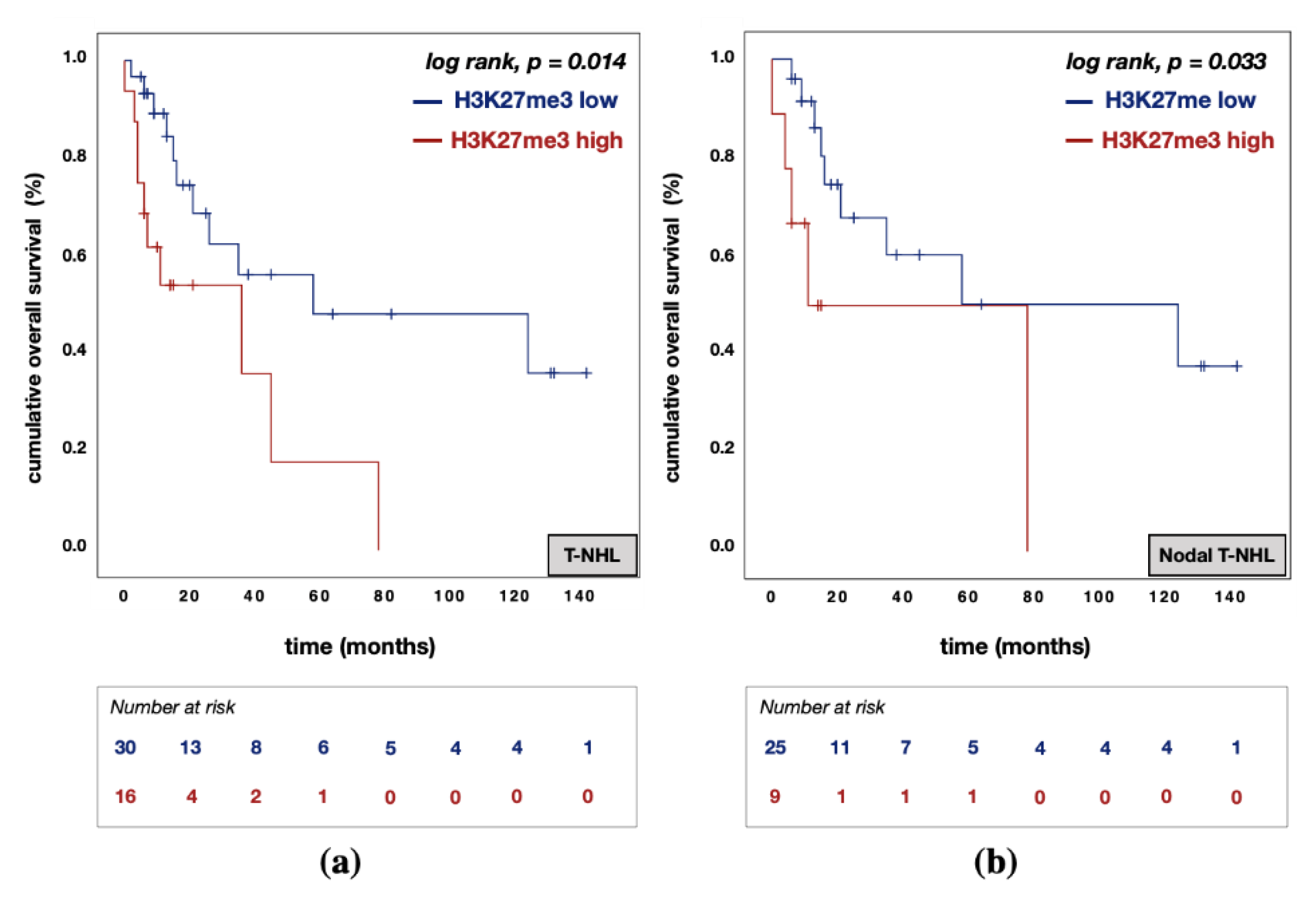
3.5. Divergent Effects of EZH1 and EZH2/H3K27me3 Protein Expression on Patient Prognosis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Swerdlow, S.H.; World Health Organization; International Agency for Research on Cancer. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; International Agency for Research on Cancer: Lyon, France, 2017. [Google Scholar]
- Vose, J.; Armitage, J.; Weisenburger, D.; International, T.C.L.P. International peripheral T-cell and natural killer/T-cell lymphoma study: Pathology findings and clinical outcomes. J. Clin. Oncol. 2008, 26, 4124–4130. [Google Scholar]
- Bellei, M.; Federico, M. The outcome of peripheral T-cell lymphoma patients failing first-line therapy: A report from the prospective International T-Cell Project. Haematologica 2019, 104, e178. [Google Scholar] [CrossRef]
- Czermin, B.; Melfi, R.; McCabe, D.; Seitz, V.; Imhof, A.; Pirrotta, V. Drosophila Enhancer of Zeste/ESC Complexes Have a Histone H3 Methyltransferase Activity that Marks Chromosomal Polycomb Sites. Cell 2002, 111, 185–196. [Google Scholar] [CrossRef] [Green Version]
- Kuzmichev, A.; Nishioka, K.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002, 16, 2893–2905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, J.; Hart, C.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.; Kingston, R.E.; Simon, J.A. Histone Methyltransferase Activity of a Drosophila Polycomb Group Repressor Complex. Cell 2002, 111, 197–208. [Google Scholar] [CrossRef] [Green Version]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [Green Version]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.-Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef] [Green Version]
- Margueron, R.; Reinberg, D.; Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Liu, Y.; Hsu, Y.-J.; Fujiwara, Y.; Kim, J.; Mao, X.; Yuan, G.-C.; Orkin, S.H. EZH1 Mediates Methylation on Histone H3 Lysine 27 and Complements EZH2 in Maintaining Stem Cell Identity and Executing Pluripotency. Mol. Cell 2008, 32, 491–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margueron, R.; Li, G.; Sarma, K.; Blais, A.; Zavadil, J.; Woodcock, C.L.; Dynlacht, B.D.; Reinberg, D. Ezh1 and Ezh2 Maintain Repressive Chromatin through Different Mechanisms. Mol. Cell 2008, 32, 503–518. [Google Scholar] [CrossRef] [Green Version]
- Yamagishi, M.; Hori, M.; Fujikawa, D.; Ohsugi, T.; Honma, D.; Adachi, N.; Katano, H.; Hishima, T.; Kobayashi, S.; Nakano, K.; et al. Targeting Excessive EZH1 and EZH2 Activities for Abnormal Histone Methylation and Transcription Network in Malignant Lymphomas. Cell Rep. 2019, 29, 2321–2337.e7. [Google Scholar] [CrossRef]
- Bachmann, I.M.; Halvorsen, O.J.; Collett, K.; Stefansson, I.M.; Straume, O.; Haukaas, S.A.; Salvesen, H.B.; Otte, A.P.; Akslen, L.A. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 2006, 24, 268–273. [Google Scholar] [CrossRef]
- Duan, R.; Du, W.; Guo, W. EZH2: A novel target for cancer treatment. J. Hematol. Oncol. 2020, 13, 104. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Dhanasekaran, S.M.; Zhou, M.; Barrette, T.R.; Kumar, C.; Sanda, M.G.; Ghosh, D.; Pienta, K.; Sewalt, R.G.A.B.; Otte, A.P.; et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nat. Cell Biol. 2002, 419, 624–629. [Google Scholar] [CrossRef]
- Wang, C.-G.; Ye, Y.-J.; Yuan, J.; Liu, F.-F.; Zhang, H.; Wang, S. EZH2 and STAT6 expression profiles are correlated with colorectal cancer stage and prognosis. World J. Gastroenterol. 2010, 16, 2421–2427. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, S.; Chuang, S.-S.; Ashton-Key, M.; Ochoa, E.; Bolli, N.; Vassiliou, G.; Gao, Z.; Du, M.-Q. Angioimmunoblastic T cell lymphoma: Novel molecular insights by mutation profiling. Oncotarget 2017, 8, 17763–17770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palomero, T.; Couronné, L.; Khiabanian, H.; Kim, M.-Y.; Ambesi-Impiombato, A.; Perez-Garcia, A.; Carpenter, Z.; Abate, F.; Allegretta, M.; Haydu, J.E.; et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat. Genet. 2014, 46, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Sakata-Yanagimoto, M.; Enami, T.; Yoshida, K.; Shiraishi, Y.; Ishii, R.; Miyake, Y.; Muto, H.; Tsuyama, N.; Sato-Otsubo, A.; Okuno, Y.; et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat. Genet. 2014, 46, 171–175. [Google Scholar] [CrossRef]
- Crescenzo, R.; Abate, F.; Lasorsa, E.; Gaudiano, M.; Chiesa, N.; Di Giacomo, F.; Spaccarotella, E.; Barbarossa, L.; Ercole, E.; Todaro, M.; et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell 2015, 27, 516–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maura, F.; Dodero, A.; Carniti, C.; Bolli, N.; Magni, M.; Monti, V.; Cabras, A.; Leongamornlert, D.; Abascal, F.; Diamond, B.; et al. CDKN2A deletion is a frequent event associated with poor outcome in patients with peripheral T-cell lymphoma not otherwise specified (PTCL-NOS). Haematologica 2021, 106, 2918. [Google Scholar] [CrossRef]
- Roberti, A.; Dobay, M.P.; Bisig, B.; Vallois, D.; Boéchat, C.; Lanitis, E.; Bouchindhomme, B.; Parrens, M.-C.; Bossard, C.; Quintanilla-Martinez, L.; et al. Type II enteropathy-associated T-cell lymphoma features a unique genomic profile with highly recurrent SETD2 alterations. Nat. Commun. 2016, 7, 12602. [Google Scholar] [CrossRef]
- Watatani, Y.; Sato, Y.; Miyoshi, H.; Sakamoto, K.; Nishida, K.; Gion, Y.; Nagata, Y.; Shiraishi, Y.; Chiba, K.; Tanaka, H.; et al. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia 2019, 33, 2867–2883. [Google Scholar] [CrossRef]
- Laginestra, M.A.; Cascione, L.; Motta, G.; Fuligni, F.; Agostinelli, C.; Rossi, M.; Sapienza, M.R.; Righi, S.; Broccoli, A.; Indio, V.; et al. Whole exome sequencing reveals mutations in FAT1 tumor suppressor gene clinically impacting on peripheral T-cell lymphoma not otherwise specified. Mod. Pathol. 2020, 33, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdalkader, L.; Oka, T.; Takata, K.; Sato, H.; Murakami, I.; Otte, A.P.; Yoshino, T. Aberrant differential expression of EZH1 and EZH2 in Polycomb repressive complex 2 among B- and T/NK-cell neoplasms. Pathology 2016, 48, 467–482. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Yang, W.I.; Min, Y.H.; Ko, Y.H.; Yoon, S.O. The role of the polycomb repressive complex pathway in T and NK cell lymphoma: Biological and prognostic implications. Tumor Biol. 2015, 37, 2037–2047. [Google Scholar] [CrossRef]
- Yi, S.; Sun, J.; Qiu, L.; Fu, W.; Wang, A.; Liu, X.; Yang, Y.; Kadin, M.E.; Tu, P.; Wang, Y. Dual Role of EZH2 in Cutaneous Anaplastic Large Cell Lymphoma: Promoting Tumor Cell Survival and Regulating Tumor Microenvironment. J. Investig. Dermatol. 2018, 138, 1126–1136. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Ng, S.-B.; Tay, J.L.-S.; Lin, B.; Koh, T.L.; Tan, J.; Selvarajan, V.; Liu, S.-C.; Bi, C.; Wang, S.; et al. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood 2013, 121, 4512–4520. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Shahsafaei, A.; Liu, C.; Yu, H.; Dorfman, D.M. Enhancer of zeste homolog 2 is widely expressed in T-cell neoplasms, is associated with high proliferation rate and correlates with MYC and pSTAT3 expression in a subset of cases. Leuk. Lymphoma 2014, 56, 2087–2091. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lv, H.; Jia, X.; Hu, G.; Kong, L.; Zhang, T.; Li, L.; Pan, Y.; Zhai, Q.; Meng, B.; et al. Clinical significance of enhancer of zeste homolog 2 and histone deacetylases 1 and 2 expression in peripheral T-cell lymphoma. Oncol. Lett. 2019, 18, 1415–1423. [Google Scholar] [CrossRef]
- Liu, J.; Liang, L.; Huang, S.; Nong, L.; Li, D.; Zhang, B.; Li, T. Aberrant differential expression of EZH2 and H3K27me3 in extranodal NK/T-cell lymphoma, nasal type, is associated with disease progression and prognosis. Hum. Pathol. 2019, 83, 166–176. [Google Scholar] [CrossRef]
- Cai, M.-Y.; Hou, J.-H.; Rao, H.-L.; Luo, R.-Z.; Li, M.; Pei, X.-Q.; Lin, M.C.; Guan, X.-Y.; Kung, H.-F.; Zeng, Y.-X.; et al. High Expression of H3K27me3 in Human Hepatocellular Carcinomas Correlates Closely with Vascular Invasion and Predicts Worse Prognosis in Patients. Mol. Med. 2010, 17, 12–20. [Google Scholar] [CrossRef]
- He, L.-R.; Liu, M.-Z.; Li, B.-K.; Rao, H.-L.; Liao, Y.-J.; Guan, X.-Y.; Zeng, Y.-X.; Xie, D. Prognostic impact of H3K27me3 expression on locoregional progression after chemoradiotherapy in esophageal squamous cell carcinoma. BMC Cancer 2009, 9, 461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Xia, W.; Zhang, Z.; Liu, J.; Wang, H.; Adsay, V.; Albarracin, C.; Yu, D.; Abbruzzese, J.L.; Mills, G.B.; et al. Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Mol. Carcinog. 2008, 47, 701–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holm, K.; Grabau, D.; Lövgren, K.; Aradottir, S.; Gruvberger-Saal, S.; Howlin, J.; Saal, L.H.; Ethier, S.P.; Bendahl, P.-O.; Stål, O.; et al. Global H3K27 trimethylation and EZH2 abundance in breast tumor subtypes. Mol. Oncol. 2012, 6, 494–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandmann, S.; Karimi, M.; De Graaf, A.O.; Rohde, C.; Göllner, S.; Varghese, J.; Ernsting, J.; Walldin, G.; Van Der Reijden, B.A.; Müller-Tidow, C.; et al. appreci8: A pipeline for precise variant calling integrating 8 tools. Bioinformatics 2018, 34, 4205–4212. [Google Scholar] [CrossRef]
- Schümann, F.L.; Bauer, M.; Groß, E.; Terziev, D.; Wienke, A.; Wickenhauser, C.; Binder, M.; Weber, T. RBMX Protein Expression in T-Cell Lymphomas Predicts Chemotherapy Response and Prognosis. Cancers 2021, 13, 4788. [Google Scholar] [CrossRef] [PubMed]
- Detre, S.; Jotti, G.S.; Dowsett, M. A “quickscore” method for immunohistochemical semiquantitation: Validation for oestrogen receptor in breast carcinomas. J. Clin. Pathol. 1995, 48, 876–878. [Google Scholar] [CrossRef] [Green Version]
- Kagiyama, Y.; Fujita, S.; Shima, Y.; Yamagata, K.; Katsumoto, T.; Nakagawa, M.; Honma, D.; Adachi, N.; Araki, K.; Kato, A.; et al. CDKN1C-mediated growth inhibition by an EZH1/2 dual inhibitor overcomes resistance of mantle cell lymphoma to ibrutinib. Cancer Sci. 2021, 112, 2314–2324. [Google Scholar] [CrossRef] [PubMed]
- Ishitsuka, K.; Izutsu, K.; Maruyama, D.; Makita, S.; Jacobsen, E.D.; Horwitz, S.; Kusumoto, S.; Allen, P.; Porcu, P.; Imaizumi, Y.; et al. First-in-human Study of the EZH1 and EZH2 Dual Inhibitor Valemetostat Tosylate (Ds-3201b) in Patients with Relapsed or Refractory Non-Hodgkin Lymphomas. Hematol. Oncol. 2021, 39, 1–520. [Google Scholar] [CrossRef]
| Characteristics | MEITL n = 33 | |
|---|---|---|
| n (%) | ||
| Sex | Female | 16 (49) |
| Male | 12 (36) | |
| Not evaluable | 5 (15) | |
| Age | Median (years) (range) | 62 (38–92) |
| Histopathology | CD3positive | 33 (100) |
| CD4negative | 31(94) | |
| CD8positive | 27 (82) | |
| CD58positive | 31 (94) | |
| T-cell Receptor | αβpositive | 10 (30) |
| γδpositive | 9 (27) | |
| Not evaluable | 14 (42) | |
| Characteristics | T-NHL n = 46 | Nodal T-NHL n = 34 | |
|---|---|---|---|
| n (%) | n (%) | ||
| Sex | Female | 15 (33) | 12 (35) |
| Male | 31 (67) | 22 (65) | |
| Age | Median (years) (range) | 64.1 (36–92) | 64.4 (51–92) |
| B symptoms | Absent | 27 (59) | 20 (59) |
| Present | 18 (39) | 14 (41) | |
| Not evaluable | 1 (2) | 0 (0) | |
| Bone marrow involvement | Absent | 32 (70) | 25 (74) |
| Present | 10 (22) | 7 (21) | |
| Not evaluable | 4 (9) | 2 (6) | |
| Ann Arbor stage | Stages I and II | 8 (17) | 6 (18) |
| Stages III and IV | 28 (61) | 25 (74) | |
| Not evaluable | 10(22) | 3 (9) | |
| IPI | 0–2 | 18 (39) | 15 (44) |
| 3–5 | 19 (41) | 17 (50) | |
| Not evaluable | 9 (20) | 2 (6) | |
| ECOG | 0–1 | 20 (43) | 16 (47) |
| 2–5 | 6 (13) | 5 (15) | |
| Not evaluable | 20 (43) | 13 (38) | |
| WBC | Normal | 13 (28) | 8 (24) |
| Upper limit of normal | 11 (24) | 7 (21) | |
| Not evaluable | 22 (48) | 19 (56) | |
| LDH | Normal | 6 (13) | 2 (6) |
| Upper limit of normal | 19 (41) | 17 (50) | |
| Not evaluable | 21 (46) | 15 (44) | |
| Ki-67 expression | <65% | 16 (35) | 12 (35) |
| ≥65% | 14 (30) | 10 (29) | |
| Not evaluable | 19 (35) | 12 (35) | |
| Relapse | Absent | 23 (50) | 14 (41) |
| Present | 23 (50) | 20 (59) | |
| First-line treatment | (R)-CHO(E)P | 37 (80) | 33 (97) |
| Others | 9 (20) | 1 (1) | |
| T-NHL (n = 41) | Nodal T-NHL (n = 30) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | Categories | HR | 95% CI | p-Value | HR | 95% CI | p-Value | ||
| LL | UL | LL | UL | ||||||
| Overall Survival | |||||||||
| Sex | female vs. male | 3.816 | 0.990 | 14.699 | 0.052 | 2.900 | 0.462 | 18.198 | 0.256 |
| Age in years | 1.053 | 0.998 | 1.111 | 0.059 | 1.070 | 0.979 | 1.171 | 0.137 | |
| Ann Arbor stage | III–IV vs. I–II | 0.148 | 0.021 | 1.042 | 0.055 | ||||
| B Symptoms | present vs. absent | 1.182 | 0.411 | 3.397 | 0.756 | 1.066 | 0.166 | 6.822 | 0.947 |
| Bone marrow involvement | present vs. absent | 2.148 | 0.614 | 7.513 | 0.232 | 1.312 | 0.220 | 7.814 | 0.766 |
| EZH1 expression | high vs. low | 0.183 | 0.044 | 0.767 | 0.020 | 0.085 | 0.008 | 0.859 | 0.037 |
| EZH2 expression | high vs. low | 8.245 | 1.898 | 35.826 | 0.005 | 28.398 | 2.166 | 372.334 | 0.011 |
| H3K27me3 expression | high vs. low | 2.322 | 0.688 | 7.836 | 0.175 | 3.500 | 0.735 | 16.652 | 0.115 |
| Progression-Free Survival | |||||||||
| Sex | female vs. male | 1.607 | 0.630 | 4.097 | 0.321 | 0.959 | 0.289 | 3.179 | 0.945 |
| Age in years | 1.008 | 0.966 | 1.051 | 0.728 | 0.987 | 0.932 | 1.046 | 0.660 | |
| Ann Arbor stage | III–IV vs. I–II | 0.933 | 0.249 | 3.495 | 0.918 | ||||
| B Symptoms | present vs. absent | 1.299 | 0.512 | 3.298 | 0.582 | 0.888 | 0.250 | 3.159 | 0.855 |
| Bone marrow involvement | present vs. absent | 4.621 | 1.706 | 12.515 | 0.003 | 3.750 | 1.252 | 11.231 | 0.018 |
| EZH1 expression | high vs. low | 0.668 | 0.246 | 1.814 | 0.428 | 0.391 | 0.104 | 1.471 | 0.165 |
| EZH2 expression | high vs. low | 3.754 | 1.233 | 11.426 | 0.020 | 5.147 | 1.472 | 17.998 | 0.010 |
| H3K27me3 expression | high vs. low | 1.326 | 0.546 | 3.218 | 0.533 | 1.537 | 0.469 | 5.038 | 0.478 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schümann, F.L.; Groß, E.; Bauer, M.; Rohde, C.; Sandmann, S.; Terziev, D.; Müller, L.P.; Posern, G.; Wienke, A.; Fend, F.; et al. Divergent Effects of EZH1 and EZH2 Protein Expression on the Prognosis of Patients with T-Cell Lymphomas. Biomedicines 2021, 9, 1842. https://doi.org/10.3390/biomedicines9121842
Schümann FL, Groß E, Bauer M, Rohde C, Sandmann S, Terziev D, Müller LP, Posern G, Wienke A, Fend F, et al. Divergent Effects of EZH1 and EZH2 Protein Expression on the Prognosis of Patients with T-Cell Lymphomas. Biomedicines. 2021; 9(12):1842. https://doi.org/10.3390/biomedicines9121842
Chicago/Turabian StyleSchümann, Franziska Lea, Elisabeth Groß, Marcus Bauer, Christian Rohde, Sarah Sandmann, Denis Terziev, Lutz P. Müller, Guido Posern, Andreas Wienke, Falko Fend, and et al. 2021. "Divergent Effects of EZH1 and EZH2 Protein Expression on the Prognosis of Patients with T-Cell Lymphomas" Biomedicines 9, no. 12: 1842. https://doi.org/10.3390/biomedicines9121842
APA StyleSchümann, F. L., Groß, E., Bauer, M., Rohde, C., Sandmann, S., Terziev, D., Müller, L. P., Posern, G., Wienke, A., Fend, F., Hansmann, M.-L., Klapper, W., Rosenwald, A., Stein, H., Dugas, M., Müller-Tidow, C., Wickenhauser, C., Binder, M., & Weber, T. (2021). Divergent Effects of EZH1 and EZH2 Protein Expression on the Prognosis of Patients with T-Cell Lymphomas. Biomedicines, 9(12), 1842. https://doi.org/10.3390/biomedicines9121842






