Comparison of Oxidative and Hypoxic Stress Responsive Genes from Meta-Analysis of Public Transcriptomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Curation of Public Gene Expression Data
2.2. RNA-seq Data Retrieval, Processing, and Quantification
2.3. Calculation of ON_ratio and ON_score
2.4. Analysis and Comparison of Gene Sets
3. Results
3.1. Data Curation/Collection of Oxidative Stress Transcriptome Data
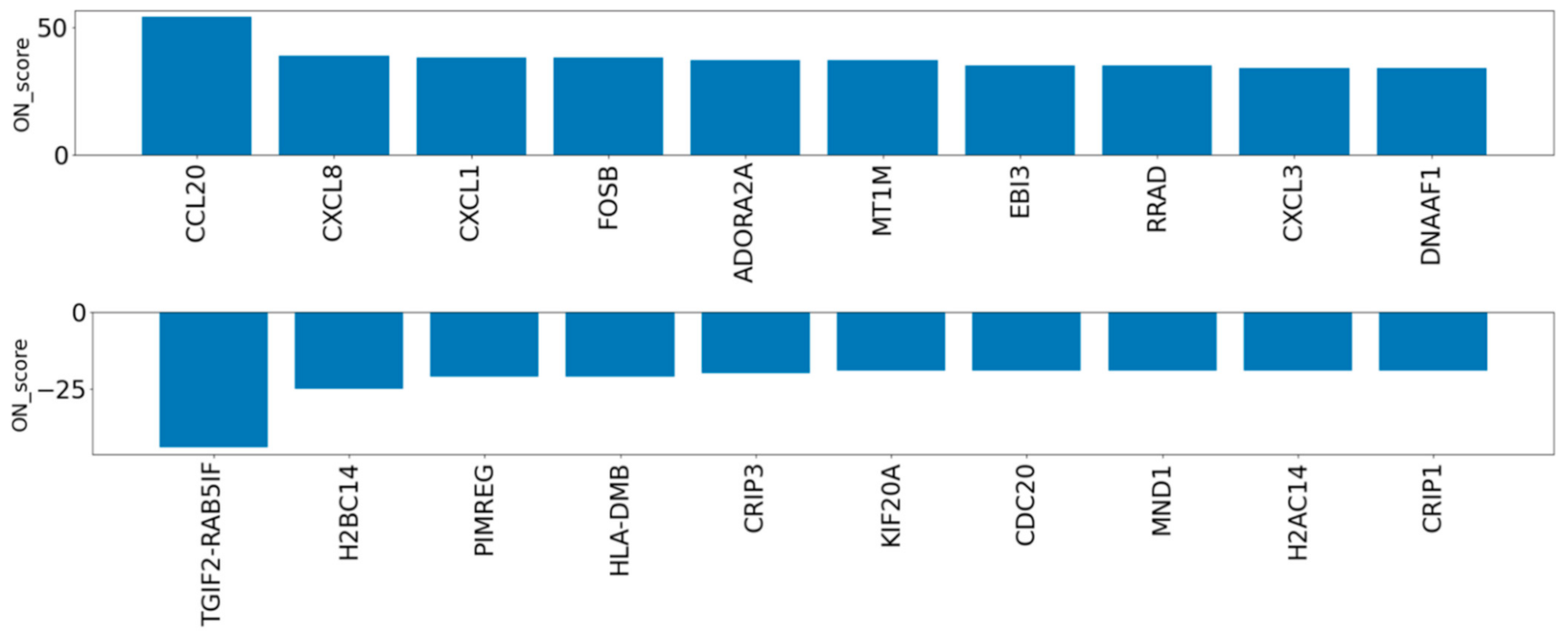
3.2. Verifying the Characteristics of DEGs Using the OS Dataset
3.3. Evaluation of DEGs by Oxidative Stress
3.4. Comparison of the Meta-Analysis Results by OS and Hypoxia
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef] [PubMed]
- Warraich, U.-E.-A.; Hussain, F.; Kayani, H.U.R. Aging—Oxidative stress, antioxidants and computational modeling. Heliyon 2020, 6, e04107. [Google Scholar] [CrossRef]
- Singh, A.; Kukreti, R.; Saso, L.; Kukreti, S. Oxidative stress: A key modulator in neurodegenerative diseases. Molecules 2019, 24, 1583. [Google Scholar] [CrossRef] [Green Version]
- Forman, H.J.; Zhang, H. Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709. [Google Scholar] [CrossRef]
- Li, S.; Hong, M.; Tan, H.-Y.; Wang, N.; Feng, Y. Insights into the role and interdependence of oxidative stress and inflammation in liver diseases. Oxid. Med. Cell. Longev. 2016, 2016, 4234061. [Google Scholar] [CrossRef]
- Cooke, M.S.; Evans, M.D.; Dizdaroglu, M.; Lunec, J. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 2003, 17, 1195–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lushchak, V.I. Free radicals, reactive oxygen species, oxidative stress and its classification. Chem. Biol. Interact. 2014, 224, 164–175. [Google Scholar] [CrossRef]
- McGarry, T.; Biniecka, M.; Veale, D.J.; Fearon, U. Hypoxia, oxidative stress and inflammation. Free Radic. Biol. Med. 2018, 125, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Ono, Y.; Bono, H. Multi-omic meta-analysis of transcriptomes and the bibliome uncovers novel hypoxia-inducible genes. Biomedicines 2021, 9, 582. [Google Scholar] [CrossRef]
- Bono, H. Meta-analysis of oxidative transcriptomes in insects. Antioxidants 2021, 10, 345. [Google Scholar] [CrossRef]
- Bono, H. All of Gene Expression (AOE): An integrated index for public gene expression databases. PLoS ONE 2020, 15, e0227076. [Google Scholar] [CrossRef] [Green Version]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [Green Version]
- Athar, A.; Füllgrabe, A.; George, N.; Iqbal, H.; Huerta, L.; Ali, A.; Snow, C.; Fonseca, N.A.; Petryszak, R.; Papatheodorou, I.; et al. Array express update–from bulk to single-cell expression data. Nucleic Acids Res. 2019, 47, D711–D715. [Google Scholar] [CrossRef]
- Kodama, Y.; Mashima, J.; Kosuge, T.; Ogasawara, O. DDBJ update: The Genomic Expression Archive (GEA) for functional genomics data. Nucleic Acids Res. 2019, 47, D69–D73. [Google Scholar] [CrossRef] [PubMed]
- Kodama, Y.; Shumway, M.; Leinonen, R. International nucleotide sequence database collaboration the sequence read archive: Explosive growth of sequencing data. Nucleic Acids Res. 2012, 40, D54–D56. [Google Scholar] [CrossRef] [Green Version]
- Yasumizu, Y. Ikra v2.0—RNAseq Pipeline Centered on Salmon. 2021. [Google Scholar] [CrossRef]
- The NCBI SRA (Sequence Read Archive); NCBI—National Center for Biotechnology Information/NLM/NIH: Bethesda, MD, USA, 2021.
- Babraham Bioinformatics—Trim Galore! Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 27 September 2021).
- Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression|Nature Methods. Available online: https://www.nature.com/articles/nmeth.4197 (accessed on 27 September 2021).
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Draw Venn Diagram. Available online: http://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 27 September 2021).
- Suzuki, T. Meta-analysis of human oxidative transcriptomes from public databases. figshare. Collection. 2021. [Google Scholar] [CrossRef]
- Hempe, J.M.; Cousins, R.J. Cysteine-rich intestinal protein binds zinc during transmucosal zinc transport. Proc. Natl. Acad. Sci. USA 1991, 88, 9671–9674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Li, W.; Luo, J.; Wu, Y.; Xu, Y.; Chen, T.; Zhang, W.; Fu, F. Cysteine-rich intestinal protein 1 served as an epithelial ovarian cancer marker via promoting wnt/β-catenin-mediated emt and tumour metastasis. Dis. Markers 2021, 2021, 3566749. [Google Scholar] [CrossRef] [PubMed]
- Marreiro, D.D.N.; Cruz, K.J.C.; Morais, J.B.S.; Beserra, J.B.; Severo, J.S.; De Oliveira, A.R.S. Zinc and oxidative stress: Current mechanisms. Antioxidants 2017, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Takeda, T.-A.; Takagishi, T.; Fukue, K.; Kambe, T.; Fukada, T. Physiological roles of zinc transporters: Molecular and genetic importance in zinc homeostasis. J. Physiol. Sci. 2017, 67, 283–301. [Google Scholar] [CrossRef]
- Choi, S.; Liu, X.; Pan, Z. Zinc deficiency and cellular oxidative stress: Prognostic implications in cardiovascular diseases. Acta Pharmacol. Sin. 2018, 39, 1120–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, H.; Dai, W.; Zhao, L.; Min, J.; Wang, F. The role of zinc and zinc homeostasis in macrophage function. J. Immunol. Res. 2018, 2018, 6872621. [Google Scholar] [CrossRef] [PubMed]
- Du, K.; Liu, M.-Y.; Zhong, X.; Wei, M.-J. Decreased circulating zinc levels in parkinson’s disease: A meta-analysis study. Sci. Rep. 2017, 7, 3902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Read, S.A.; Obeid, S.; Ahlenstiel, C.; Ahlenstiel, G. The role of zinc in antiviral immunity. Adv. Nutr. 2019, 10, 696–710. [Google Scholar] [CrossRef] [Green Version]
- Skrajnowska, D.; Bobrowska-Korczak, B. Role of zinc in immune system and anti-cancer defense mechanisms. Nutrients 2019, 11, 2273. [Google Scholar] [CrossRef] [Green Version]
- Shackelford, R.E.; Kaufmann, W.K.; Paules, R.S. Oxidative stress and cell cycle checkpoint function. Free Radic. Biol. Med. 2000, 28, 1387–1404. [Google Scholar] [CrossRef]
- Klein, J.A.; Ackerman, S.L. Oxidative stress, cell cycle, and neurodegeneration. J. Clin. Invest. 2003, 111, 785–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuhrmann, D.C.; Brüne, B. Mitochondrial composition and function under the control of hypoxia. Redox Biol. 2017, 12, 208–215. [Google Scholar] [CrossRef]
- Gerber, P.A.; Rutter, G.A. The role of oxidative stress and hypoxia in pancreatic beta-cell dysfunction in diabetes mellitus. Antioxid. Redox Signal. 2017, 26, 501–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, T.; Hirakawa, Y.; Nangaku, M. The role of oxidative stress and hypoxia in renal disease. Kidney Res. Clin. Pract. 2019, 38, 414–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Zhao, J.-J.; Zhu, Y.; Xiong, W.; Lin, J.-Y.; Ma, X. Identification of candidate cancer genes involved in human retinoblastoma by Data Mining. Childs Nerv. Syst. 2008, 24, 893–900. [Google Scholar] [CrossRef] [PubMed]



| Source of OS | Number of Data Pairs |
|---|---|
| Hydrogen peroxide (H2O2) | 98 (25%) |
| Ultra-Violet rays (UV) | 59 (15%) |
| Rotenone | 45 (12%) |
| Lipopolysaccharide (LPS) | 38 (10%) |
| Arsenite | 33 (9%) |
| Infra-Red rays (Radiation) | 24 (6%) |
| NRF2 knockdown/KO, BRD4 KO | 22 (6%) |
| Deoxynivalenol | 10 (3%) |
| Palmitate/high fat/high glucose | 10 (3%) |
| Cadmium, Methylmercury, Zinc dimethyldithiocarbamate | 8 (2%) |
| Aging | 6 (2%) |
| Paraquat | 5 (1%) |
| Others (Senescence, Menadione, entinostat, etc.) | 28 (7%) |
| Total | 386 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suzuki, T.; Ono, Y.; Bono, H. Comparison of Oxidative and Hypoxic Stress Responsive Genes from Meta-Analysis of Public Transcriptomes. Biomedicines 2021, 9, 1830. https://doi.org/10.3390/biomedicines9121830
Suzuki T, Ono Y, Bono H. Comparison of Oxidative and Hypoxic Stress Responsive Genes from Meta-Analysis of Public Transcriptomes. Biomedicines. 2021; 9(12):1830. https://doi.org/10.3390/biomedicines9121830
Chicago/Turabian StyleSuzuki, Takayuki, Yoko Ono, and Hidemasa Bono. 2021. "Comparison of Oxidative and Hypoxic Stress Responsive Genes from Meta-Analysis of Public Transcriptomes" Biomedicines 9, no. 12: 1830. https://doi.org/10.3390/biomedicines9121830
APA StyleSuzuki, T., Ono, Y., & Bono, H. (2021). Comparison of Oxidative and Hypoxic Stress Responsive Genes from Meta-Analysis of Public Transcriptomes. Biomedicines, 9(12), 1830. https://doi.org/10.3390/biomedicines9121830







