tRNA Derivatives in Multiple Myeloma: Investigation of the Potential Value of a tRNA-Derived Molecular Signature
Abstract
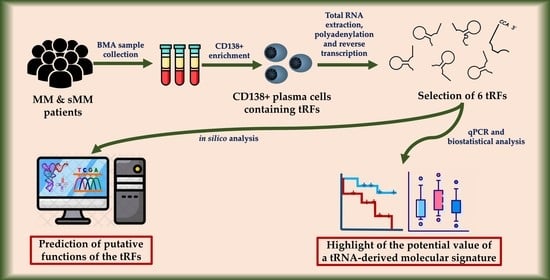
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. CD138+ Plasma Cell Selection
2.3. Total RNA Extraction, In Vitro Polyadenylation, and cDNA Synthesis
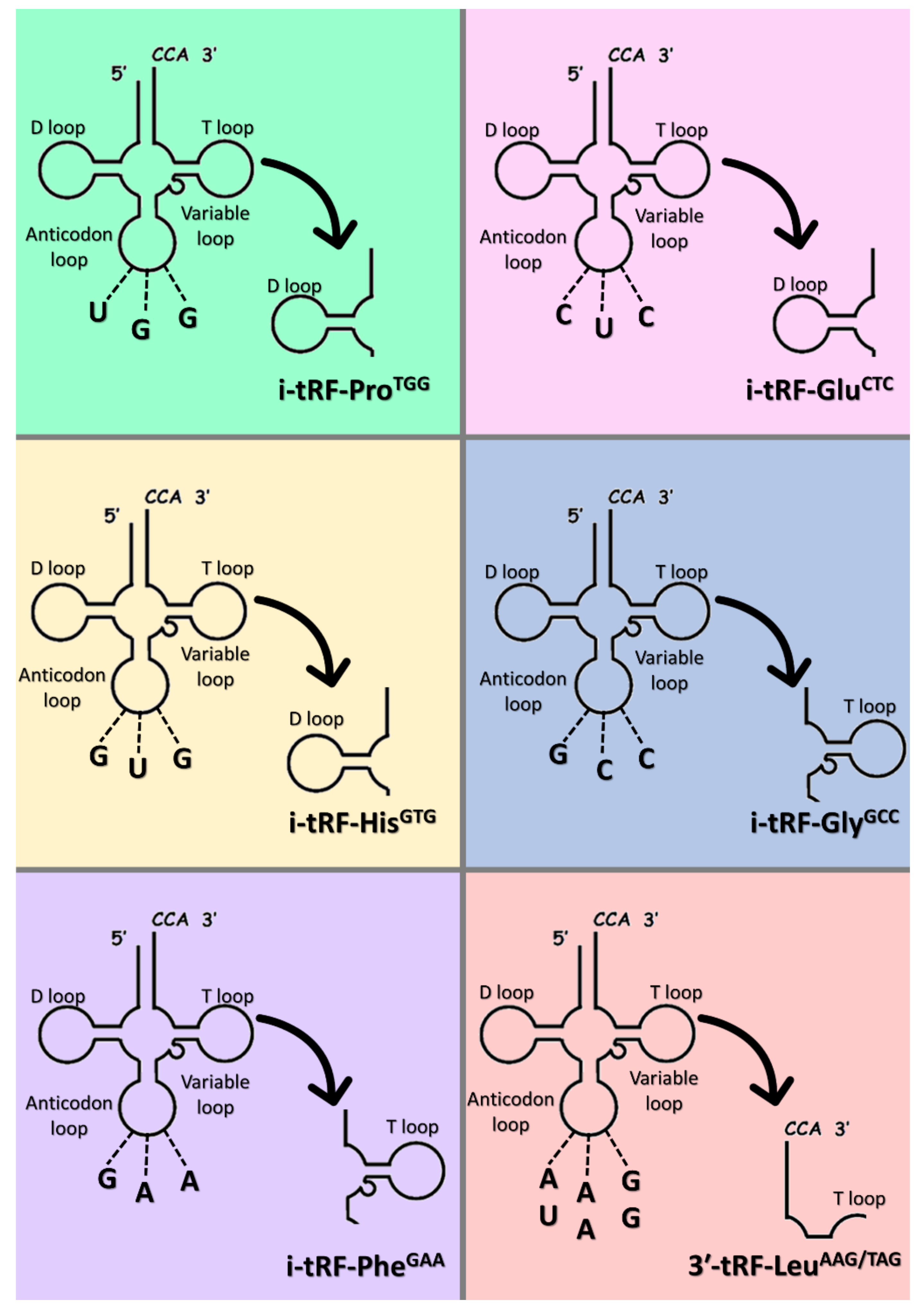
2.4. tRF Selection
2.5. Real-Time qPCR
2.6. Biostatistical Analysis
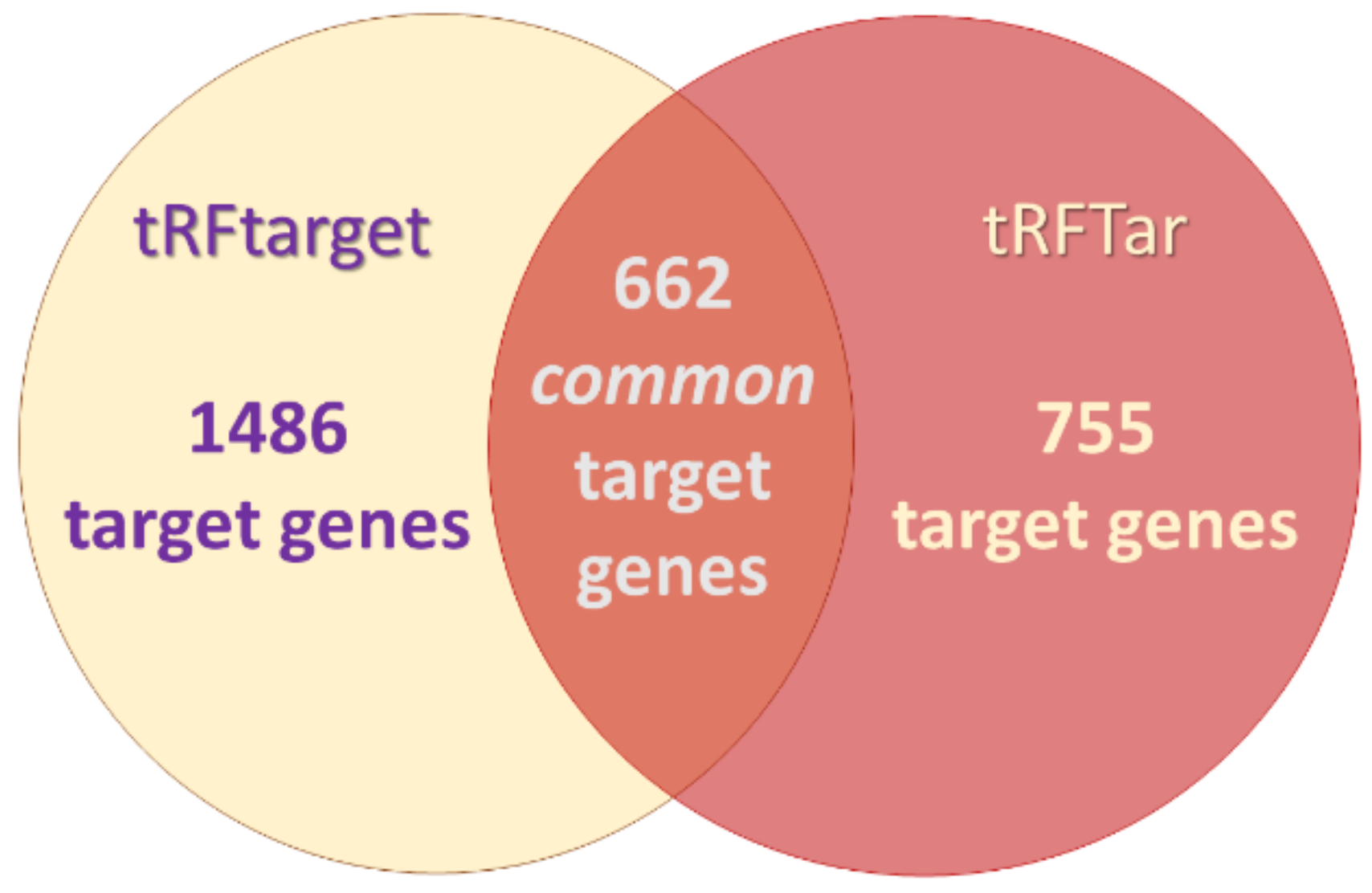
2.7. In Silico Analysis for tRF Target Prediction and Gene Ontology (GO) Enrichment Analysis
3. Results
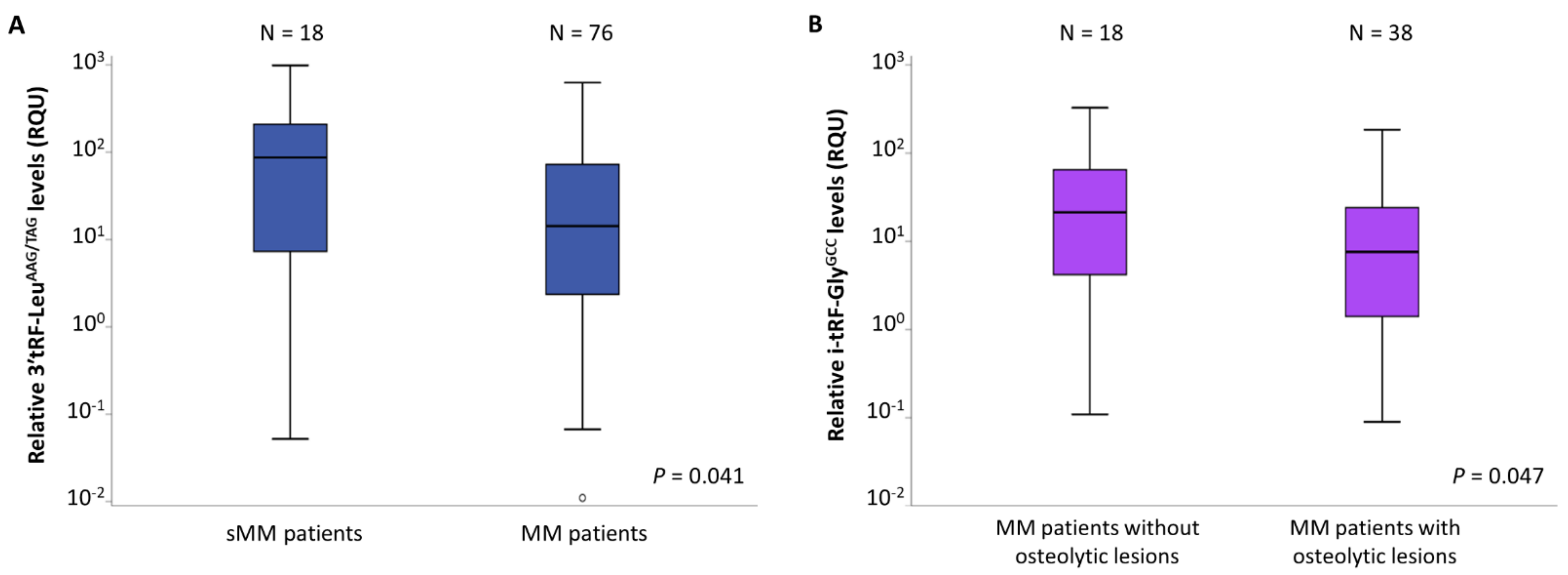
3.1. Differences in tRF Levels of CD138+ Plasma Cells between sMM and MM Patients, as Well as among MM Patients’ Subgroups
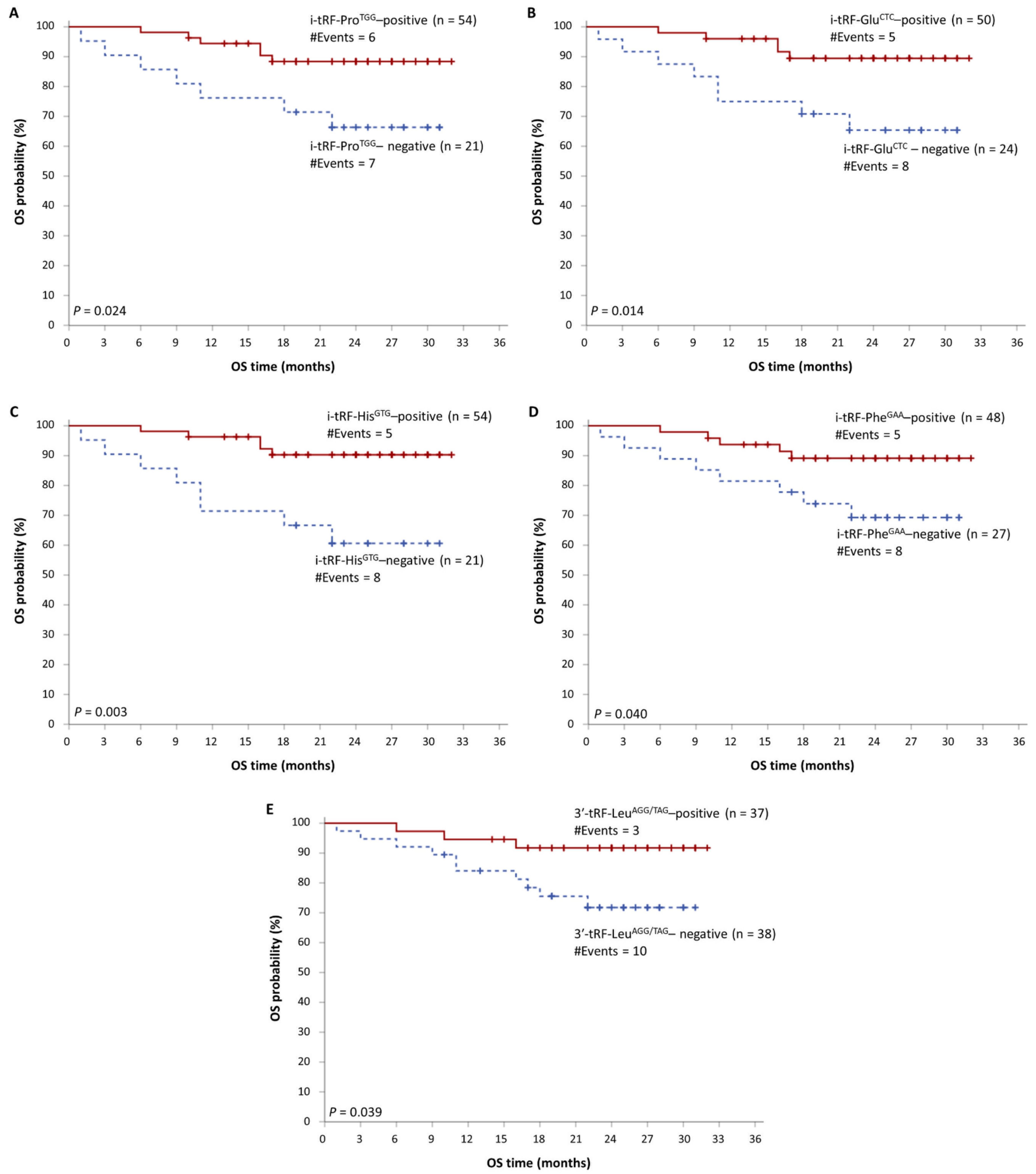
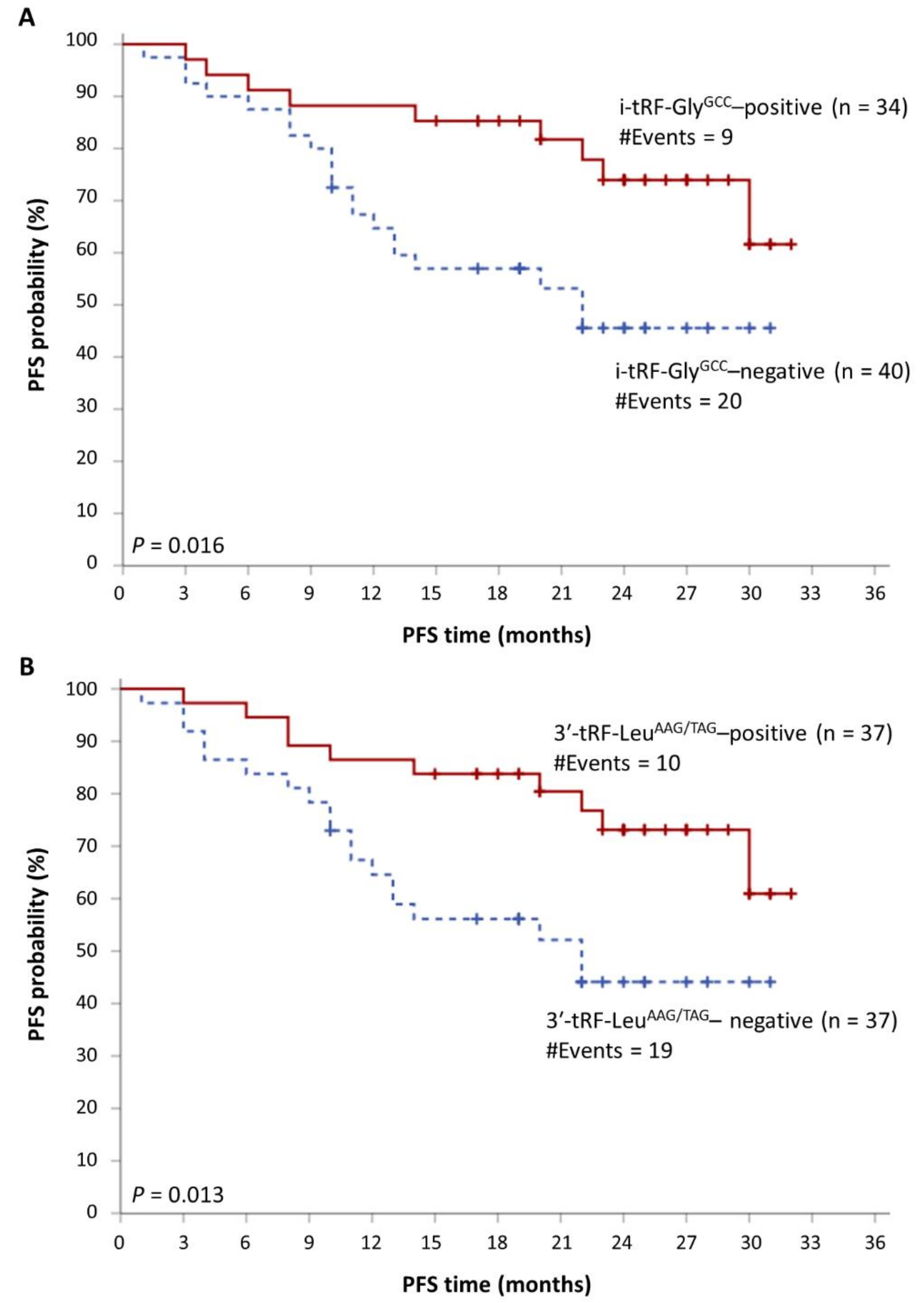
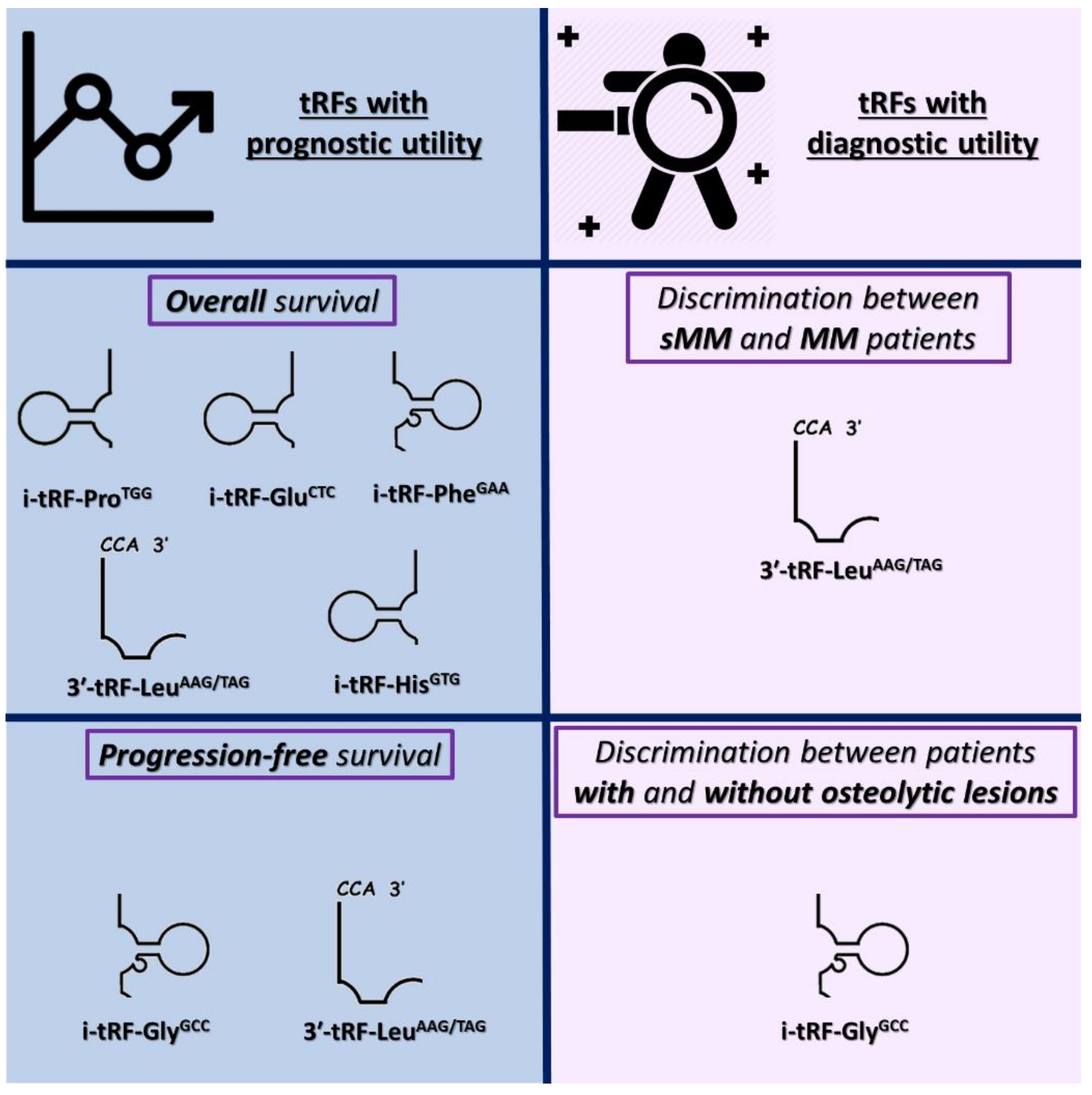
3.2. tRFs as Promising Molecular Indicators of Favorable Prognosis in MM
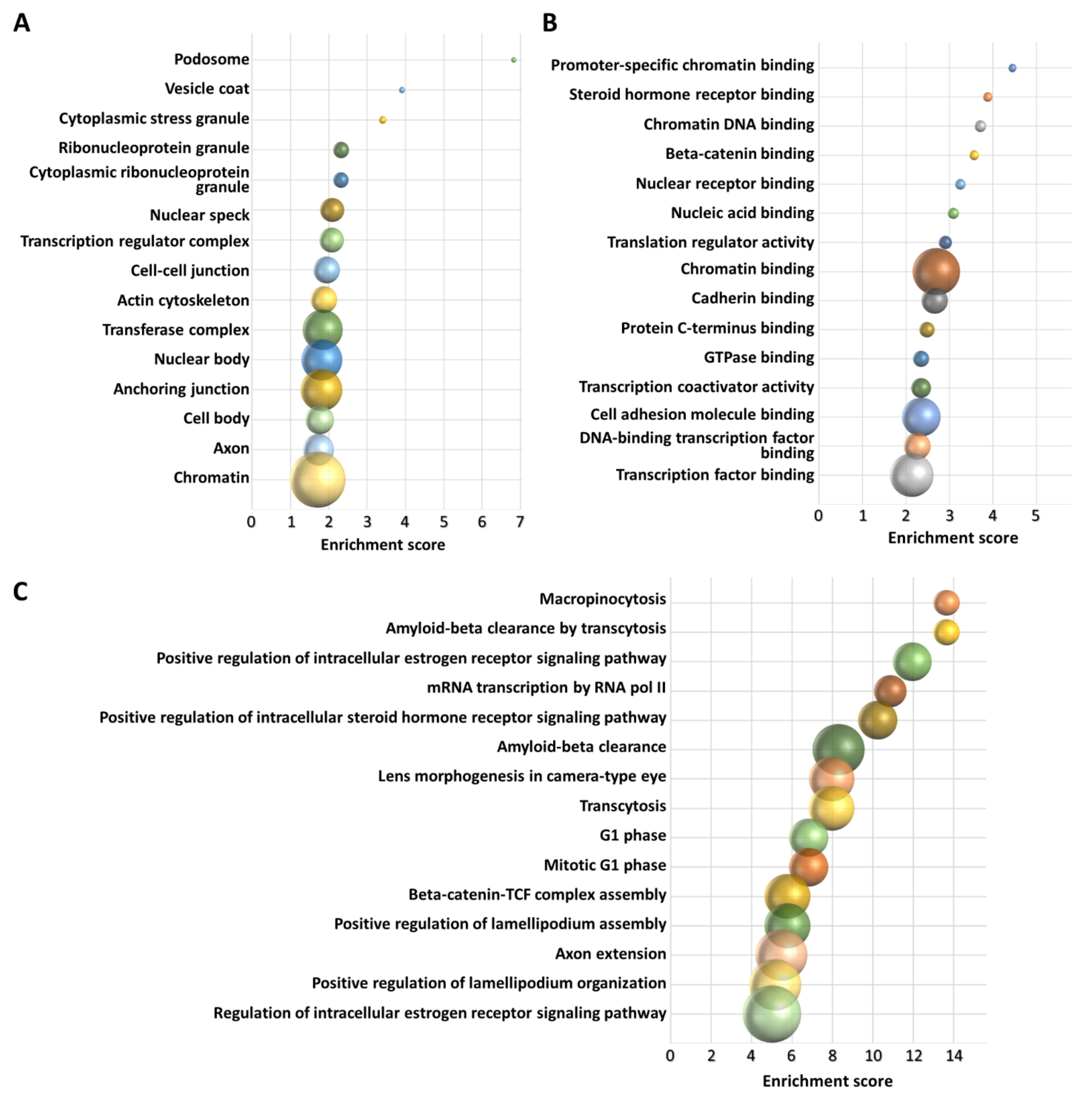
3.3. In Silico Functional Analysis of 3′-tRF-LeuAAG/TAG
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landgren, O.; Kyle, R.A.; Pfeiffer, R.M.; Katzmann, J.A.; Caporaso, N.E.; Hayes, R.B.; Dispenzieri, A.; Kumar, S.; Clark, R.J.; Baris, D.; et al. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: A prospective study. Blood 2009, 113, 5412–5417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajkumar, S.V.; Landgren, O.; Mateos, M.V. Smoldering multiple myeloma. Blood 2015, 125, 3069–3075. [Google Scholar] [CrossRef]
- Rajkumar, S.V. Multiple myeloma: 2018 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2018, 93, 981–1114. [Google Scholar] [CrossRef] [Green Version]
- Terpos, E.; Ntanasis-Stathopoulos, I.; Gavriatopoulou, M.; Dimopoulos, M.A. Pathogenesis of bone disease in multiple myeloma: From bench to bedside. Blood Cancer J. 2018, 8, 7. [Google Scholar] [CrossRef] [Green Version]
- Colombo, M.; Giannandrea, D.; Lesma, E.; Basile, A.; Chiaramonte, R. Extracellular Vesicles Enhance Multiple Myeloma Metastatic Dissemination. Int. J. Mol. Sci. 2019, 20, 3236. [Google Scholar] [CrossRef] [Green Version]
- Matthes, T.; Manfroi, B.; Huard, B. Revisiting IL-6 antagonism in multiple myeloma. Crit. Rev. Oncol. 2016, 105, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Guillerey, C.; Nakamura, K.; Vuckovic, S.; Hill, G.R.; Smyth, M.J. Immune responses in multiple myeloma: Role of the natural immune surveillance and potential of immunotherapies. Cell Mol. Life Sci. 2016, 73, 1569–1589. [Google Scholar] [CrossRef] [PubMed]
- Greipp, P.R.; San Miguel, J.; Durie, B.G.; Crowley, J.J.; Barlogie, B.; Bladé, J.; Boccadoro, M.; Child, J.A.; Avet-Loiseau, H.; Kyle, R.A.; et al. International staging system for multiple myeloma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2005, 23, 3412–3420. [Google Scholar] [CrossRef]
- Palumbo, A.; Avet-Loiseau, H.; Oliva, S.; Lokhorst, H.M.; Goldschmidt, H.; Rosinol, L.; Richardson, P.; Caltagirone, S.; Lahuerta, J.J.; Facon, T.; et al. Revised International Staging System for Multiple Myeloma: A Report From International Myeloma Working Group. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 2863–2869. [Google Scholar] [CrossRef]
- Gupta, N.; Sharma, A.; Sharma, A. Emerging biomarkers in Multiple Myeloma: A review. Clin. Chim. Acta 2020, 503, 45–53. [Google Scholar] [CrossRef]
- Papanota, A.M.; Tsiakanikas, P.; Kontos, C.K.; Malandrakis, P.; Liacos, C.I.; Ntanasis-Stathopoulos, I.; Kanellias, N.; Gavriatopoulou, M.; Kastritis, E.; Avgeris, M.; et al. A Molecular Signature of Circulating MicroRNA Can Predict Osteolytic Bone Disease in Multiple Myeloma. Cancers 2021, 13, 3877. [Google Scholar] [CrossRef]
- Papadimitriou, M.A.; Papanota, A.M.; Adamopoulos, P.G.; Pilala, K.M.; Liacos, C.I.; Malandrakis, P.; Mavrianou-Koutsoukou, N.; Patseas, D.; Eleutherakis-Papaiakovou, E.; Gavriatopoulou, M.; et al. miRNA-seq and clinical evaluation in multiple myeloma: miR-181a overexpression predicts short-term disease progression and poor post-treatment outcome. Br. J. Cancer 2021. [Google Scholar] [CrossRef]
- Artemaki, P.I.; Letsos, P.A.; Zoupa, I.C.; Katsaraki, K.; Karousi, P.; Papageorgiou, S.G.; Pappa, V.; Scorilas, A.; Kontos, C.K. The Multifaceted Role and Utility of MicroRNAs in Indolent B-Cell Non-Hodgkin Lymphomas. Biomedicines 2021, 9, 333. [Google Scholar] [CrossRef]
- Krishna, S.; Raghavan, S.; DasGupta, R.; Palakodeti, D. tRNA-derived fragments (tRFs): Establishing their turf in post-transcriptional gene regulation. Cell Mol. Life Sci 2021, 78, 2607–2619. [Google Scholar] [CrossRef]
- Magee, R.; Rigoutsos, I. On the expanding roles of tRNA fragments in modulating cell behavior. Nucleic Acids Res. 2020, 48, 9433–9448. [Google Scholar] [CrossRef]
- Kuscu, C.; Kumar, P.; Kiran, M.; Su, Z.; Malik, A.; Dutta, A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 2018, 24, 1093–1105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzzi, N.; Bellodi, C. Novel insights into the emerging roles of tRNA-derived fragments in mammalian development. RNA Biol. 2020, 17, 1214–1222. [Google Scholar] [CrossRef] [Green Version]
- Zhong, F.; Hu, Z.; Jiang, K.; Lei, B.; Wu, Z.; Yuan, G.; Luo, H.; Dong, C.; Tang, B.; Zheng, C.; et al. Complement C3 activation regulates the production of tRNA-derived fragments Gly-tRFs and promotes alcohol-induced liver injury and steatosis. Cell Res. 2019, 29, 548–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosentino, C.; Toivonen, S.; Diaz Villamil, E.; Atta, M.; Ravanat, J.L.; Demine, S.; Schiavo, A.A.; Pachera, N.; Deglasse, J.P.; Jonas, J.C.; et al. Pancreatic beta-cell tRNA hypomethylation and fragmentation link TRMT10A deficiency with diabetes. Nucleic Acids Res. 2018, 46, 10302–10318. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Yu, J.; Zhou, P. Role of tRNA-derived fragments in cancer: Novel diagnostic and therapeutic targets tRFs in cancer. Am. J. Cancer Res. 2020, 10, 393–402. [Google Scholar] [PubMed]
- Zeng, T.; Hua, Y.; Sun, C.; Zhang, Y.; Yang, F.; Yang, M.; Yang, Y.; Li, J.; Huang, X.; Wu, H.; et al. Relationship between tRNA-derived fragments and human cancers. Int. J. Cancer 2020, 147, 3007–3018. [Google Scholar] [CrossRef]
- Papadimitriou, M.A.; Avgeris, M.; Levis, P.; Papasotiriou, E.C.; Kotronopoulos, G.; Stravodimos, K.; Scorilas, A. tRNA-Derived Fragments (tRFs) in Bladder Cancer: Increased 5’-tRF-LysCTT Results in Disease Early Progression and Patients’ Poor Treatment Outcome. Cancers 2020, 12, 3661. [Google Scholar] [CrossRef]
- Karousi, P.; Adamopoulos, P.G.; Papageorgiou, S.G.; Pappa, V.; Scorilas, A.; Kontos, C.K. A novel, mitochondrial, internal tRNA-derived RNA fragment possesses clinical utility as a molecular prognostic biomarker in chronic lymphocytic leukemia. Clin. Biochem. 2020, 85, 20–26. [Google Scholar] [CrossRef]
- Karousi, P.; Katsaraki, K.; Papageorgiou, S.G.; Pappa, V.; Scorilas, A.; Kontos, C.K. Identification of a novel tRNA-derived RNA fragment exhibiting high prognostic potential in chronic lymphocytic leukemia. Hematol. Oncol. 2019, 37, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Katsaraki, K.; Artemaki, P.I.; Papageorgiou, S.G.; Pappa, V.; Scorilas, A.; Kontos, C.K. Identification of a novel, internal tRNA-derived RNA fragment as a new prognostic and screening biomarker in chronic lymphocytic leukemia, using an innovative quantitative real-time PCR assay. Leuk Res. 2019, 87, 106234. [Google Scholar] [CrossRef]
- Katsaraki, K.; Adamopoulos, P.G.; Papageorgiou, S.G.; Pappa, V.; Scorilas, A.; Kontos, C.K. A 3’ tRNA-derived fragment produced by tRNA(LeuAAG) and tRNA(LeuTAG) is associated with poor prognosis in B-cell chronic lymphocytic leukemia, independently of classical prognostic factors. Eur. J. Haematol. 2021, 106, 821–830. [Google Scholar] [CrossRef]
- Xu, C.; Fu, Y. Expression Profiles of tRNA-Derived Fragments and Their Potential Roles in Multiple Myeloma. OncoTargets Ther. 2021, 14, 2805–2814. [Google Scholar] [CrossRef] [PubMed]
- Rojek, A.E.; Katanski, C.D.; Stefka, A.; Derman, B.A.; Jakubowiak, A.; Pan, T. Abstract 2394: tRNA expression and tRFs in multiple myeloma: Progression from monoclonal gammopathies to relapsed/refractory disease. Cancer Res. 2021, 81, 2394. [Google Scholar] [CrossRef]
- Xu, C.; Liang, T.; Zhang, F.; Liu, J.; Fu, Y. tRNA-derived fragments as novel potential biomarkers for relapsed/refractory multiple myeloma. BMC Bioinform. 2021, 22, 238. [Google Scholar] [CrossRef]
- Ridley, R.C.; Xiao, H.; Hata, H.; Woodliff, J.; Epstein, J.; Sanderson, R.D. Expression of syndecan regulates human myeloma plasma cell adhesion to type I collagen. Blood 1993, 81, 767–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawano, Y.; Fujiwara, S.; Wada, N.; Izaki, M.; Yuki, H.; Okuno, Y.; Iyama, K.; Yamasaki, H.; Sakai, A.; Mitsuya, H.; et al. Multiple myeloma cells expressing low levels of CD138 have an immature phenotype and reduced sensitivity to lenalidomide. Int. J. Oncol. 2012, 41, 876–884. [Google Scholar] [CrossRef] [Green Version]
- Wijdenes, J.; Vooijs, W.C.; Clement, C.; Post, J.; Morard, F.; Vita, N.; Laurent, P.; Sun, R.X.; Klein, B.; Dore, J.M. A plasmocyte selective monoclonal antibody (B-B4) recognizes syndecan-1. Br. J. Haematol. 1996, 94, 318–323. [Google Scholar] [CrossRef]
- Pliatsika, V.; Loher, P.; Magee, R.; Telonis, A.G.; Londin, E.; Shigematsu, M.; Kirino, Y.; Rigoutsos, I. MINTbase v2.0: A comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all The Cancer Genome Atlas projects. Nucleic Acids Res. 2018, 46, D152–D159. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Camp, R.L.; Dolled-Filhart, M.; Rimm, D.L. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 2004, 10, 7252–7259. [Google Scholar] [CrossRef] [Green Version]
- Papageorgiou, S.G.; Kontos, C.K.; Kotsianidis, I.; Karousi, P.; Symeonidis, A.; Galanopoulos, A.; Bouchla, A.; Hatzimichael, E.; Repousis, P.; Zikos, P.; et al. Effectiveness of 5-Azacytidine in older patients with high-risk myelodysplastic syndromes and oligoblastic acute myeloid leukemia: A retrospective analysis of the Hellenic (Greek) MDS Study Group. J. Geriatr. Oncol. 2020, 11, 121–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Peng, H.; Cui, Q.; Zhou, Y. tRFTar: Prediction of tRF-target gene interactions via systemic re-analysis of Argonaute CLIP-seq datasets. Methods 2021, 187, 57–67. [Google Scholar] [CrossRef]
- Li, N.; Shan, N.; Lu, L.; Wang, Z. tRFtarget: A database for transfer RNA-derived fragment targets. Nucleic Acids Res. 2021, 49, D254–D260. [Google Scholar] [CrossRef]
- Yu, M.; Lu, B.; Zhang, J.; Ding, J.; Liu, P.; Lu, Y. tRNA-derived RNA fragments in cancer: Current status and future perspectives. J. Hematol. Oncol. 2020, 13, 121. [Google Scholar] [CrossRef]
- Huang, S.Q.; Sun, B.; Xiong, Z.P.; Shu, Y.; Zhou, H.H.; Zhang, W.; Xiong, J.; Li, Q. The dysregulation of tRNAs and tRNA derivatives in cancer. J. Exp. Clin. Cancer Res. 2018, 37, 101. [Google Scholar] [CrossRef] [PubMed]
- Dimopoulos, M.A.; Moreau, P.; Terpos, E.; Mateos, M.V.; Zweegman, S.; Cook, G.; Delforge, M.; Hajek, R.; Schjesvold, F.; Cavo, M.; et al. Multiple Myeloma: EHA-ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-up. Hemasphere 2021, 5, e528. [Google Scholar] [CrossRef]
- Zhu, L.; Li, J.; Gong, Y.; Wu, Q.; Tan, S.; Sun, D.; Xu, X.; Zuo, Y.; Zhao, Y.; Wei, Y.Q.; et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer 2019, 18, 74. [Google Scholar] [CrossRef]
- Gu, X.; Wang, L.; Coates, P.J.; Boldrup, L.; Fahraeus, R.; Wilms, T.; Sgaramella, N.; Nylander, K. Transfer-RNA-Derived Fragments Are Potential Prognostic Factors in Patients with Squamous Cell Carcinoma of the Head and Neck. Genes 2020, 11, 1344. [Google Scholar] [CrossRef]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef]
- Murphy, D.A.; Courtneidge, S.A. The ‘ins’ and ‘outs’ of podosomes and invadopodia: Characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 2011, 12, 413–426. [Google Scholar] [CrossRef] [Green Version]
- Georgess, D.; Machuca-Gayet, I.; Blangy, A.; Jurdic, P. Podosome organization drives osteoclast-mediated bone resorption. Cell Adh. Migr. 2014, 8, 191–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Tu, C.; Zhang, J.; Wang, J. Inhibition of multiple myelomaderived exosomes uptake suppresses the functional response in bone marrow stromal cell. Int J. Oncol. 2019, 54, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- De Luca, L.; Laurenzana, I.; Trino, S.; Lamorte, D.; Caivano, A.; Musto, P. An update on extracellular vesicles in multiple myeloma: A focus on their role in cell-to-cell cross-talk and as potential liquid biopsy biomarkers. Expert Rev. Mol. Diagn. 2019, 19, 249–258. [Google Scholar] [CrossRef]
- Commisso, C. The pervasiveness of macropinocytosis in oncological malignancies. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180153. [Google Scholar] [CrossRef] [Green Version]
- Masoumi Moghaddam, S.; Spencer, A. KRAS-Mutated Myeloma Cells Exploit Autophagy and Macropinocytosis to Tolerate Glutamine-Deprived Conditions. Blood 2017, 130, 1786. [Google Scholar] [CrossRef]
- Alexanian, R.; Dimopoulos, M.A.; Delasalle, K.; Barlogie, B. Primary dexamethasone treatment of multiple myeloma. Blood 1992, 80, 887–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gehring, U.; Mohit, B.; Tomkins, G.M. Glucocorticoid action on hybrid clones derived from cultured myeloma and lymphoma cell lines. Proc. Natl. Acad. Sci. USA 1972, 69, 3124–3127. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, D.; Auclair, D.; Robinson, E.K.; Hideshima, T.; Li, G.; Podar, K.; Gupta, D.; Richardson, P.; Schlossman, R.L.; Krett, N.; et al. Identification of genes regulated by dexamethasone in multiple myeloma cells using oligonucleotide arrays. Oncogene 2002, 21, 1346–1358. [Google Scholar] [CrossRef] [Green Version]
- Thomas, A.L.; Coarfa, C.; Qian, J.; Wilkerson, J.J.; Rajapakshe, K.; Krett, N.L.; Gunaratne, P.H.; Rosen, S.T. Identification of potential glucocorticoid receptor therapeutic targets in multiple myeloma. Nucl. Recept. Signal. 2015, 13, e006. [Google Scholar] [CrossRef]
- Wang, L.H.; Yang, X.Y.; Mihalic, K.; Xiao, W.; Li, D.; Farrar, W.L. Activation of estrogen receptor blocks interleukin-6-inducible cell growth of human multiple myeloma involving molecular cross-talk between estrogen receptor and STAT3 mediated by co-regulator PIAS3. J. Biol. Chem. 2001, 276, 31839–31844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papanota, A.M.; Karousi, P.; Kontos, C.K.; Ntanasis-Stathopoulos, I.; Scorilas, A.; Terpos, E. Multiple Myeloma Bone Disease: Implication of MicroRNAs in Its Molecular Background. Int. J. Mol. Sci. 2021, 22, 2375. [Google Scholar] [CrossRef]
- Van Andel, H.; Kocemba, K.A.; Spaargaren, M.; Pals, S.T. Aberrant Wnt signaling in multiple myeloma: Molecular mechanisms and targeting options. Leukemia 2019, 33, 1063–1075. [Google Scholar] [CrossRef]
- Kumar, S.; Kimlinger, T.; Morice, W. Immunophenotyping in multiple myeloma and related plasma cell disorders. Best Pract. Res. Clin. Haematol. 2010, 23, 433–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, S.; Yang, S.; Brown, R.; Kabani, K.; Aklilu, E.; Ho, P.J.; Woodland, N.; Joshua, D. Characterisation and relevance of CD138-negative plasma cells in plasma cell myeloma. Int. J. Lab. Hematol. 2010, 32, e190–e196. [Google Scholar] [CrossRef]
- Harada, H.; Kawano, M.M.; Huang, N.; Harada, Y.; Iwato, K.; Tanabe, O.; Tanaka, H.; Sakai, A.; Asaoku, H.; Kuramoto, A. Phenotypic difference of normal plasma cells from mature myeloma cells. Blood 1993, 81, 2658–2663. [Google Scholar] [CrossRef] [PubMed]
- Paiva, B.; Almeida, J.; Perez-Andres, M.; Mateo, G.; Lopez, A.; Rasillo, A.; Vidriales, M.B.; Lopez-Berges, M.C.; Miguel, J.F.; Orfao, A. Utility of flow cytometry immunophenotyping in multiple myeloma and other clonal plasma cell-related disorders. Cytom. B Clin. Cytom. 2010, 78, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Rawstron, A.C.; Orfao, A.; Beksac, M.; Bezdickova, L.; Brooimans, R.A.; Bumbea, H.; Dalva, K.; Fuhler, G.; Gratama, J.; Hose, D.; et al. Report of the European Myeloma Network on multiparametric flow cytometry in multiple myeloma and related disorders. Haematologica 2008, 93, 431–438. [Google Scholar] [CrossRef] [Green Version]
- Beasley, A.B.; Acheampong, E.; Lin, W.; Gray, E.S. Multi-Marker Immunomagnetic Enrichment of Circulating Melanoma Cells. Methods Mol. Biol. 2021, 2265, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Van Dongen, J.J.; Lhermitte, L.; Böttcher, S.; Almeida, J.; van der Velden, V.H.; Flores-Montero, J.; Rawstron, A.; Asnafi, V.; Lécrevisse, Q.; Lucio, P.; et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia 2012, 26, 1908–1975. [Google Scholar] [CrossRef] [Green Version]
| Variable | Number of Patients (%) |
|---|---|
| Gender (76/76 patients) | |
| Male | 44 (57.9%) |
| Female | 32 (42.1%) |
| M-protein isotype (75/76 patients) | |
| IgG | 44 (58.7%) |
| IgA | 17 (22.7%) |
| IgD | 2 (2.7%) |
| Kappa light chain | 7 (9.2%) |
| Lambda light chain | 3 (4.0%) |
| Not typed | 2 (2.7%) |
| del(17p) (71/76 patients) | |
| Absence | 59 (83.1%) |
| Presence | 12 (16.9%) |
| t(4;14) (70/76 patients) | |
| Absence | 62 (88.6%) |
| Presence | 8 (11.4%) |
| t(14;16) (68/76 patients) | |
| Absence | 67 (98.5%) |
| Presence | 1 (1.5%) |
| (+1q) (54/76 patients) | |
| Absence | 30 (55.6%) |
| Presence | 24 (44.4%) |
| ISS 1 stage (74/76 patients) | |
| I | 15 (20.3%) |
| II | 25 (33.8%) |
| III | 34 (45.9%) |
| R-ISS 2 stage (69/76 patients) | |
| I | 11 (15.9%) |
| II | 40 (58.0%) |
| III | 18 (26.1%) |
| Bone disease (72/76 patients) | |
| No | 22 (30.6%) |
| Yes | 50 (69.4%) |
| WBLDCT 3 osteolysis (56/76 patients) | |
| No | 18 (32.1%) |
| Yes | 38 (67.9%) |
| tRF | Fragment Sequence | Anticodon | Localization | Accession Number | MINTbase Unique ID |
|---|---|---|---|---|---|
| i-tRF-ProTGG | 5′-GUUGGUCUAGGGGUAUGAUUCUCGG-3′ | UGG | Nucleus | MK671729 | tRF-25-78WPRLXN48 |
| i-tRF-GluCTC | 5′-GUCUAGUGGUUAGGAUUCGGCG-3′ | CUC | Nucleus | MK671728 | tRF-22-SX73V2Y8K |
| i-tRF-HisGTG | 5′-UGAUCGUAUAGUGGUUAGUACUCUGCG-3′ | GUG | Nucleus | MW650833 | tRF-27-XMSL73VL4YK |
| i-tRF-GlyGCC | 5′-GAGGCCCGGGUUCGAUUC-3′ | GCC | Nucleus | MK642309 | tRF-18-5J3KYU05 |
| i-tRF-PheGAA | 5′-UUUAGACGGGCUCACAUCACC-3′ | GAA | Mitochondrion | MK671731 | tRF-21-ZPEK45H5D |
| 3’-tRF-LeuAAG/TAG | 5′-AUCCCACCGCUGCCACCA-3′ | AAG, UAG | Nucleus | MK671733 | tRF-18-HR0VX6D2 |
| Amplified Molecule | Primer Sequence (5′→3′) | Direction | Length (nt 1) | Tm (°C) |
|---|---|---|---|---|
| i-tRF-ProTGG | GTTGGTCTAGGGGTATGATTCTCGGA | Forward | 26 | 62 |
| i-tRF-GluCTC | GTCTAGTGGTTAGGATTCGGCGA | 23 | 61 | |
| i-tRF-HisGTG | TGATCGTATAGTGGTTAGTACTCTGCG | 27 | 59 | |
| i-tRF-GlyGCC | GAGGCCCGGGTTCGATTC | 18 | 62 | |
| i-tRF-PheGAA | TTTAGACGGGCTCACATCACC | 21 | 59 | |
| 3’-tRF-LeuAAG/TAG | ATCCCACCGCTGCCACCA | 18 | 66 | |
| SNORD43 | ACTTATTGACGGGCGGACA | 19 | 59 | |
| SNORD48 | TGATGATGACCCCAGGTAACTCT | 23 | 59 | |
| Universal reverse | GCGAGCACAGAATTAATACGAC | Reverse | 22 | 56 |
| Covariate | HR 1 | 95% CI 2 | p Value 3 | BCa 4 Bootstrap 5 95% CI 2 | Bootstrap 5 p Value 3 | |
|---|---|---|---|---|---|---|
| Overall survival (OS) | i-tRF-ProTGG status | |||||
| Positive | 1.00 | |||||
| Negative | 4.06 | 1.28–12.82 | 0.017 | 0.98–46.70 | 0.011 | |
| R-ISS 6 (ordinal) | 3.39 | 1.28–8.96 | 0.014 | 0.93–38.24 | 0.024 | |
| i-tRF-GluCTC status | ||||||
| Positive | 1.00 | |||||
| Negative | 5.87 | 1.75–19.63 | 0.004 | 1.02–5.58 × 105 | 0.001 | |
| R-ISS 6 (ordinal) | 3.98 | 1.50–10.57 | 0.006 | 0.58 –7.39 × 105 | 0.010 | |
| i-tRF-HisGTG status | ||||||
| Positive | 1.00 | |||||
| Negative | 6.49 | 1.94–21.74 | 0.002 | 0.97–7.32 × 105 | 0.001 | |
| R-ISS 6 (ordinal) | 3.77 | 1.44–9.91 | 0.007 | 0.87–4.42 × 105 | 0.008 | |
| Progression-free survival (PFS) | i-tRF-GlyGCC status | |||||
| Positive | 1.00 | |||||
| Negative | 3.06 | 1.33–7.00 | 0.008 | 1.12–12.63 | 0.007 | |
| ISS 7 (ordinal) | 2.22 | 1.25–3.95 | 0.007 | 1.20–7.71 | 0.008 | |
| 3′-tRF-LeuAAG/TAG status | ||||||
| Positive | 1.00 | |||||
| Negative | 2.94 | 1.32–6.55 | 0.008 | 1.28–8.76 | 0.005 | |
| ISS 7 (ordinal) | 2.16 | 1.22–3.80 | 0.008 | 1.21–6.37 | 0.008 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karousi, P.; Papanota, A.-M.; Artemaki, P.I.; Liacos, C.-I.; Patseas, D.; Mavrianou-Koutsoukou, N.; Liosi, A.-A.; Kalioraki, M.-A.; Ntanasis-Stathopoulos, I.; Gavriatopoulou, M.; et al. tRNA Derivatives in Multiple Myeloma: Investigation of the Potential Value of a tRNA-Derived Molecular Signature. Biomedicines 2021, 9, 1811. https://doi.org/10.3390/biomedicines9121811
Karousi P, Papanota A-M, Artemaki PI, Liacos C-I, Patseas D, Mavrianou-Koutsoukou N, Liosi A-A, Kalioraki M-A, Ntanasis-Stathopoulos I, Gavriatopoulou M, et al. tRNA Derivatives in Multiple Myeloma: Investigation of the Potential Value of a tRNA-Derived Molecular Signature. Biomedicines. 2021; 9(12):1811. https://doi.org/10.3390/biomedicines9121811
Chicago/Turabian StyleKarousi, Paraskevi, Aristea-Maria Papanota, Pinelopi I. Artemaki, Christine-Ivy Liacos, Dimitrios Patseas, Nefeli Mavrianou-Koutsoukou, Aikaterini-Anna Liosi, Maria-Anna Kalioraki, Ioannis Ntanasis-Stathopoulos, Maria Gavriatopoulou, and et al. 2021. "tRNA Derivatives in Multiple Myeloma: Investigation of the Potential Value of a tRNA-Derived Molecular Signature" Biomedicines 9, no. 12: 1811. https://doi.org/10.3390/biomedicines9121811
APA StyleKarousi, P., Papanota, A.-M., Artemaki, P. I., Liacos, C.-I., Patseas, D., Mavrianou-Koutsoukou, N., Liosi, A.-A., Kalioraki, M.-A., Ntanasis-Stathopoulos, I., Gavriatopoulou, M., Kastritis, E., Dimopoulos, M.-A., Scorilas, A., Terpos, E., & Kontos, C. K. (2021). tRNA Derivatives in Multiple Myeloma: Investigation of the Potential Value of a tRNA-Derived Molecular Signature. Biomedicines, 9(12), 1811. https://doi.org/10.3390/biomedicines9121811