MiRNAs as New Tools in Lesion Vitality Evaluation: A Systematic Review and Their Forensic Applications
Abstract
1. Introduction
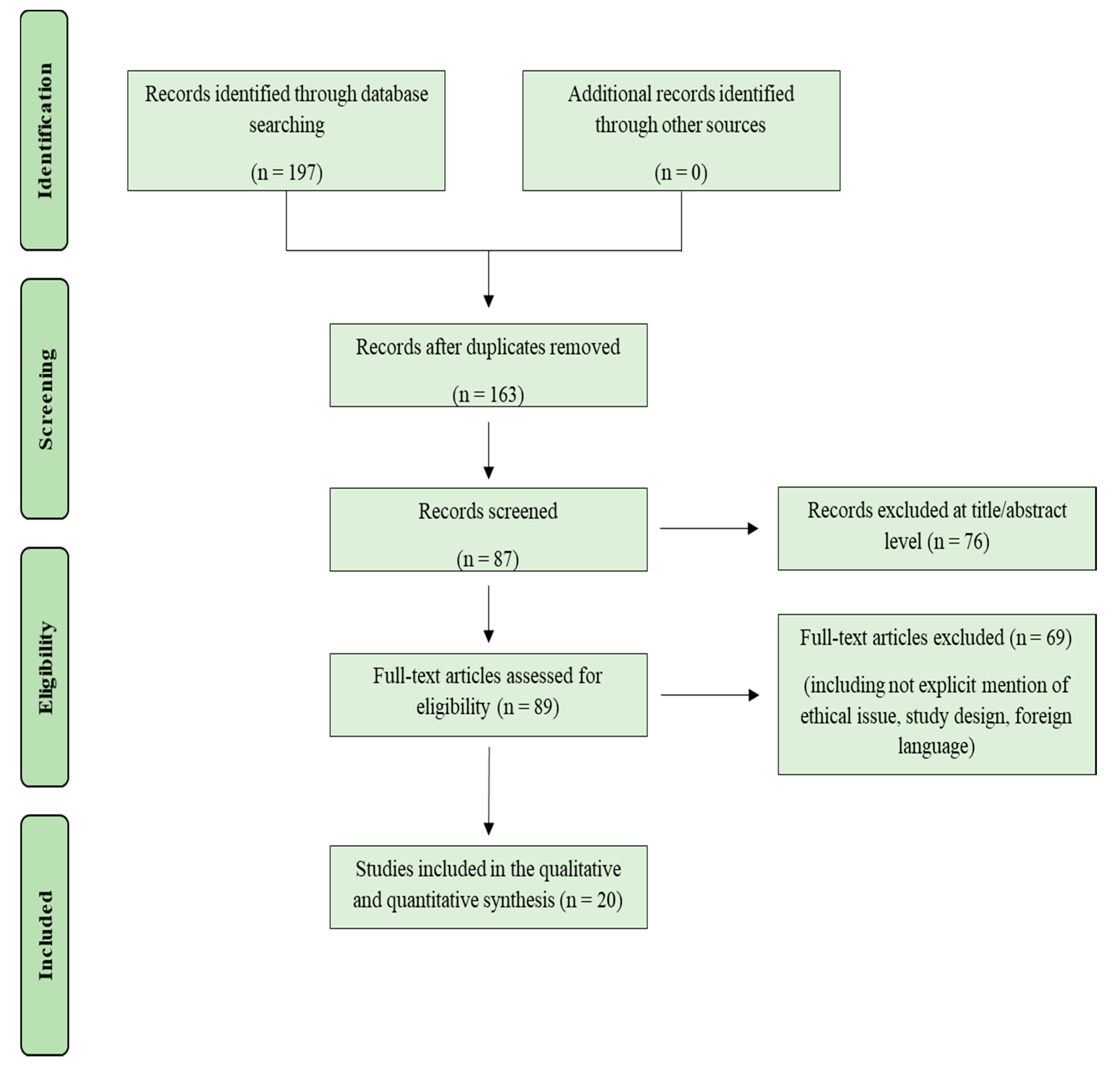
2. Materials and Methods
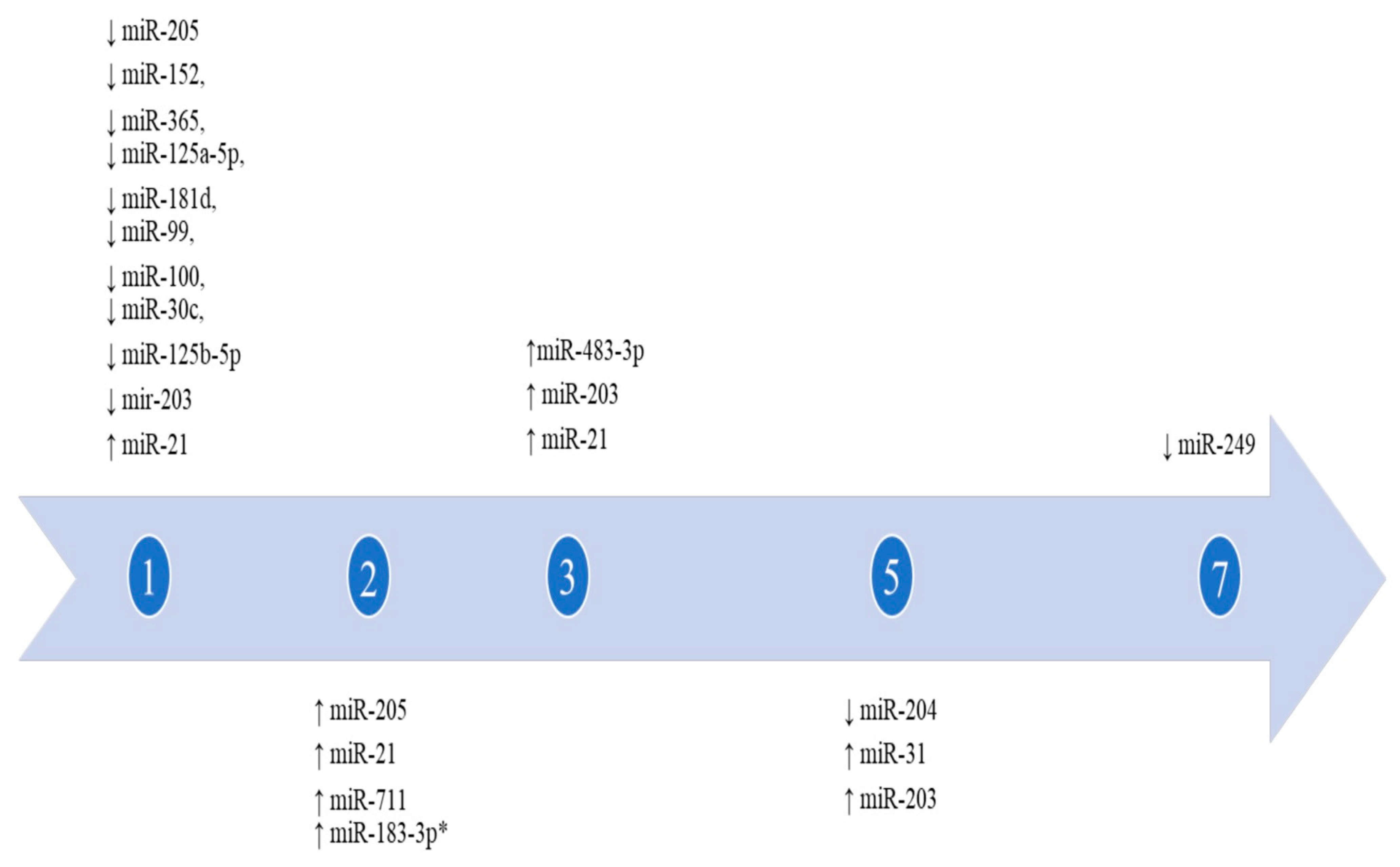
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saukko, P.; Knight, B. Knight’s Forensic Pathology, 4th ed.; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Grellner, W.; Georg, T.; Wilske, J. Quantitative analysis of proinflammatory cytokines (IL-1beta, IL-6, TNF-alpha) in human skin wounds. Forensic Sci. Int. 2000, 113, 251–264. [Google Scholar] [CrossRef]
- Bonelli, A.; Bacci, S.; Norelli, G.A. Affinity cytochemistry analysis of mast cells in skin lesions: A possible tool to assess the timing of lesions after death. Int. J. Legal. Med. 2003, 117, 331–334. [Google Scholar] [CrossRef] [PubMed]
- Casse, J.M.; Martrille, L.; Vignaud, J.M.; Gauchotte, G. Skin wounds vitality markers in forensic pathology: An updated review. Med. Sci. Law. 2016, 56, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Oehmichen, M. Vitality and time course of wounds. Forensic Sci. Int. 2004, 144, 221–231. [Google Scholar] [CrossRef]
- Langlois, N.E.; Gresham, G.A. The ageing of bruises: A review and study of the colour changes with time. Forensic Sci. Int. 1991, 50, 227–238. [Google Scholar] [CrossRef]
- Balandiz, H.; Pehlivan, S.; Çiçek, A.F.; Tuğcu, H. Evaluation of Vitality in the Experimental Hanging Model of Rats by Using Immunohistochemical IL-1β Antibody Staining. Am. J. Forensic Med. Pathol. 2015, 36, 317–322. [Google Scholar] [CrossRef]
- Focardi, M.; Puliti, E.; Grifoni, R.; Palandrini, M.; Bugelli, V.; Pinchi, V.; Norelli, G.A.; Bacci, S. Immunohistochemical localization of Langerhans cells as a tool for vitality in -hanging mark wounds: A pilot study. Aust. J. Forensic Sci. 2019, 52, 393–405. [Google Scholar] [CrossRef]
- Ishida, Y.; Kuninaka, Y.; Nosaka, M.; Shimada, E.; Hata, S.; Yamamoto, H.; Hashizume, Y.; Kimura, A.; Furukawa, F.; Kondo, T. Forensic application of epidermal AQP3 expression to determination of wound vitality in human compressed neck skin. Int. J. Legal. Med. 2018, 132, 1375–1380. [Google Scholar] [CrossRef]
- Turillazzi, E.; Vacchiano, G.; Luna-Maldonado, A.; Neri, M.; Pomara, C.; Rabozzi, R.; Riezzo, I.; Fineschi, V. Tryptase, CD15 and IL-15 as reliable markers for the determination of soft and hard ligature marks vitality. Histol. Histopathol. 2010, 25, 1539–1546. [Google Scholar] [PubMed]
- De Matteis, A.; dell’Aquila, M.; Maiese, A.; Frati, P.; La Russa, R.; Bolino, G.; Fineschi, V. The Troponin-I fast skeletal muscle is reliable marker for the determination of vitality in the suicide hanging. Forensic Sci. Int. 2019, 301, 284–288. [Google Scholar] [CrossRef]
- Pérez, I.L.; Falcón, M.; Gimenez, M.; Diaz, F.M.; Pérez-Cárceles, M.D.; Osuna, E.; Nuno-Vieira, D.; Luna, A. Diagnosis of Vitality in Skin Wounds in the Ligature Marks Resulting From Suicide Hanging. Am. J. Forensic Med. Pathol. 2017, 38, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Maiese, A.; Scatena, A.; Costantino, A.; Di Paolo, M.; La Russa, R.; Turillazzi, E.; Frati, P.; Fineschi, V. MicroRNAs as Useful Tools to Estimate Time Since Death. A Systematic Review of Current Literature. Diagnostics 2021, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Rocchi, A.; Chiti, E.; Maiese, A.; Turillazzi, E.; Spinetti, I. MicroRNAs: An Update of Applications in Forensic Science. Diagnostics 2020, 11, 32. [Google Scholar] [CrossRef]
- Lyu, H.P.; Cheng, M.; Liu, J.C.; Ye, M.Y.; Xu, D.; He, J.T.; Xie, X.L.; Wang, Q. Differentially expressed microRNAs as potential markers for vital reaction of burned skin. Forensic Sci. Med. 2018, 4, 135–141. [Google Scholar]
- Ibrahim, S.F.; Ali, M.M.; Basyouni, H.; Laila, A.; Rashed, E.; Amer, A.E.; Abd El-Kareem, D. Histological and miRNAs postmortem changes in incisional wound. Egypt J. Forensic Sci. 2019, 9, 37. [Google Scholar] [CrossRef]
- Neri, M.; Fabbri, M.; D’Errico, S.; Di Paolo, M.; Frati, P.; Gaudio, R.M.; La Russa, R.; Maiese, A.; Marti, M.; Pinchi, E.; et al. Regulation of miRNAs as new tool for cutaneous vitality lesions demonstration in ligature marks in deaths by hanging. Sci. Rep. 2019, 9, 20011. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: Explanation and elaboration. BMJ. 2009, 339, b2700. [Google Scholar] [CrossRef]
- Bertero, T.; Gastaldi, C.; Bourget-Ponzio, I.; Imbert, V.; Loubat, A.; Selva, E.; Busca, R.; Mari, B.; Hofman, P.; Barbry, P.; et al. miR-483-3p controls proliferation in wounded epithelial cells. FASEB J. 2011, 25, 3092–3105. [Google Scholar] [CrossRef]
- Cao, G.; Chen, B.; Zhang, X.; Chen, H. Human Adipose-Derived Mesenchymal Stem Cells-Derived Exosomal microRNA-19b Promotes the Healing of Skin Wounds Through Modulation of the CCL1/TGF-β Signaling Axis. Clin. Cosmet. Investig. Dermatol. 2020, 13, 957–971. [Google Scholar] [CrossRef]
- Etich, J.; Bergmeier, V.; Pitzler, L.; Brachvogel, B. Identification of a reference gene for the quantification of mRNA and miRNA expression during skin wound healing. Connect. Tissue. Res. 2017, 58, 196–207. [Google Scholar] [CrossRef]
- He, L.; Zhu, C.; Jia, J.; Hao, X.Y.; Yu, X.Y.; Liu, X.Y.; Shu, M.G. ADSC-Exos containing MALAT1 promotes wound healing by targeting miR-124 through activating Wnt/β-catenin pathway. Biosci. Rep. 2020, 40, BSR20192549. [Google Scholar] [CrossRef]
- Jiang, Z.; Wei, J.; Yang, W.; Li, W.; Liu, F.; Yan, X.; Yan, X.; Hu, N.; Li, J. MicroRNA-26a inhibits wound healing through decreased keratinocytes migration by regulating ITGA5 through PI3K/AKT signaling pathway. Biosci. Rep. 2020, 40, BSR20201361. [Google Scholar] [CrossRef]
- Jin, Y.; Tymen, S.D.; Chen, D.; Fang, Z.J.; Zhao, Y.; Dragas, D.; Dai, Y.; Marucha, P.T.; Zhou, X. MicroRNA-99 family targets AKT/mTOR signaling pathway in dermal wound healing. PLoS ONE 2013, 8, e64434. [Google Scholar] [CrossRef]
- Lang, H.; Zhao, F.; Zhang, T.; Liu, X.; Wang, Z.; Wang, R.; Shi, P.; Pang, X. MicroRNA-149 contributes to scarless wound healing by attenuating inflammatory response. Mol. Med. Rep. 2017, 16, 2156–2162. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, D.; Halilovic, A.; Yue, P.; Bellner, L.; Wang, K.; Wang, L.; Zhang, C. Inhibition of miR-205 impairs the wound-healing process in human corneal epithelial cells by targeting KIR4.1 (KCNJ10). Investig. Ophthalmol. Vis. Sci. 2013, 54, 6167–6178. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; He, Q.; Luo, C.; Qian, L. Differentially expressed miRNAs in acute wound healing of the skin: A pilot study. Medicine 2015, 94, 458. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shu, B.; Zhou, Z.; Xu, Y.; Liu, Y.; Wang, P.; Xiong, K.; Xie, J. Involvement of miRNA203 in the proliferation of epidermal stem cells during the process of DM chronic wound healing through Wnt signal pathways. Stem. Cell Res. Ther. 2020, 11, 348. [Google Scholar] [CrossRef]
- Long, S.; Zhao, N.; Ge, L.; Wang, G.; Ran, X.; Wang, J.; Su, Y.; Wang, T. MiR-21 ameliorates age-associated skin wound healing defects in mice. J. Gene Med. 2018, 20, e3022. [Google Scholar] [CrossRef]
- Pastar, I.; Khan, A.A.; Stojadinovic, O.; Lebrun, E.A.; Medina, M.C.; Brem, H.; Kirsner, R.S.; Jimenez, J.J.; Leslie, C.; Tomic-Canic, M. Induction of specific microRNAs inhibits cutaneous wound healing. J. Biol. Chem. 2012, 287, 29324–29335. [Google Scholar] [CrossRef] [PubMed]
- Viticchiè, G.; Lena, A.M.; Cianfarani, F.; Odorisio, T.; Annicchiarico-Petruzzelli, M.; Melino, G.; Candi, E. MicroRNA-203 contributes to skin re-epithelialization. Cell Death Dis. 2012, 3, e435. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Feng, Y.; Sun, H.; Zhang, L.; Hao, L.; Shi, C.; Wang, J.; Li, R.; Ran, X.; Su, Y.; et al. miR-21 regulates skin wound healing by targeting multiple aspects of the healing process. Am. J. Pathol. 2012, 181, 1911–1920. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, J.; Guo, S.L.; Fan, K.J.; Li, J.; Wang, Y.L.; Teng, Y.; Yang, X. miR-21 promotes keratinocyte migration and re-epithelialization during wound healing. Int. J. Biol. Sci. 2011, 7, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Peng, H.; Ruan, Q.; Fatima, A.; Getsios, S.; Lavker, R.M. MicroRNA-205 promotes keratinocyte migration via the lipid phosphatase SHIP2. FASEB J. 2010, 24, 3950–3959. [Google Scholar] [CrossRef]
- Zhang, K.; Cheng, M.; Xu, J.; Chen, L.; Li, J.; Li, Q.; Xie, X.; Wang, Q. MiR-711 and miR-183-3p as potential markers for vital reaction of burned skin. Forensic Sci. Res. 2020. [Google Scholar] [CrossRef]
- Pinchi, E.; Frati, A.; Cantatore, S.; D’Errico, S.; La Russa, R.; Maiese, A.; Palmieri, M.; Pesce, A.; Viola, R.V.; Frati, P.; et al. Acute Spinal Cord Injury: A Systematic Review Investigating miRNA Families Involved. Int. J. Mol. Sci. 2019, 20, 1841. [Google Scholar] [CrossRef]
- Pinchi, E.; Frati, P.; Aromatario, M.; Cipolloni, L.; Fabbri, M.; La Russa, R.; Maiese, A.; Neri, M.; Santurro, A.; Scopetti, M.; et al. miR-1, miR-499 and miR-208 are sensitive markers to diagnose sudden death due to early acute myocardial infarction. J. Cell. Mol. Med. 2019, 23, 6005–6016. [Google Scholar] [CrossRef]
- Manetti, A.C.; Maiese, A.; Di Paolo, M.; De Matteis, A.; La Russa, R.; Turillazzi, E.; Frati, P.; Fineschi, V. MicroRNAs and Sepsis-Induced Cardiac Dysfunction: A Systematic Review. Int. J. Mol. Sci. 2020, 22, 321. [Google Scholar] [CrossRef]
- Chiti, E.; Di Paolo, M.; Turillazzi, E.; Rocchi, A. MicroRNAs in Hypertrophic, Arrhythmogenic and Dilated Cardiomyopathy. Diagnostics 2021, 11, 1720. [Google Scholar] [CrossRef]
- Tahamtan, A.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. Anti-Inflammatory MicroRNAs and Their Potential for Inflammatory Diseases Treatment. Front. Immunol. 2018, 9, 1377. [Google Scholar] [CrossRef]
- Marques-Rocha, J.L.; Samblas, M.; Milagro, F.I.; Bressan, J.; Martínez, J.A.; Marti, A. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J. 2015, 29, 3595–3611. [Google Scholar] [CrossRef]
- Brudecki, L.; Ferguson, D.A.; McCall, C.E.; El Gazzar, M. MicroRNA-146a and RBM4 form a negative feed-forward loop that disrupts cytokine mRNA translation following TLR4 responses in human THP-1 monocytes. Immunol. Cell Biol. 2013, 91, 532–540. [Google Scholar] [CrossRef]
- Sessa, F.; Salerno, M.; Bertozzi, G.; Cipolloni, L.; Messina, G.; Aromatario, M.; Polo, L.; Turillazzi, E.; Pomara, C. miRNAs as Novel Biomarkers of Chronic Kidney Injury in Anabolic-Androgenic Steroid Users: An Experimental Study. Front. Pharmacol. 2020, 11, 563756. [Google Scholar] [CrossRef]
- Chang, R.; Yi, S.; Tan, X.; Huang, Y.; Wang, Q.; Su, G.; Zhou, C.; Cao, Q.; Yuan, G.; Kijlstra, A.; et al. MicroRNA-20a-5p suppresses IL-17 production by targeting OSM and CCL1 in patients with Vogt-Koyanagi-Harada disease. Br. J. Ophthalmol. 2018, 102, 282–290. [Google Scholar] [CrossRef]
- Pasparakis, M. Regulation of tissue homeostasis by NF-kappaB signalling: Implications for inflammatory diseases. Nat. Rev. Immunol. 2009, 9, 778–788. [Google Scholar] [CrossRef]
- Balkwill, F. TNF-alpha in promotion and progression of cancer. Cancer Metastasis Rev. 2006, 25, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Pullen, N.; Thomas, G. The modular phosphorylation and activation of p70s6k. FEBS Lett. 1997, 410, 78–82. [Google Scholar] [CrossRef]
- Suwa, A.; Kurama, T.; Shimokawa, T. SHIP2 and its involvement in various diseases. Expert Opin. Ther. Targets 2010, 14, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Liu, X.; Kolokythas, A.; Yu, J.; Wang, A.; Heidbreder, C.E.; Shi, F.; Zhou, X. Downregulation of the Rho GTPase signaling pathway is involved in the microRNA-138-mediated inhibition of cell migration and invasion in tongue squamous cell carcinoma. Int. J. Cancer 2010, 127, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Su, N.; Qiu, H.; Chen, Y.; Yang, T.; Yan, Q.; Wan, X. miR-205 promotes tumor proliferation and invasion through targeting ESRRG in endometrial carcinoma. Oncol. Rep. 2013, 29, 2297–2302. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, X. miR-122-5p promotes aggression and epithelial-mesenchymal transition in triple-negative breast cancer by suppressing charged multivesicular body protein 3 through mitogen-activated protein kinase signaling. J. Cell. Physiol. 2020, 235, 2825–2835. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.H.; Caricilli, A.M.; de Abreu, L.L.; Araújo, E.P.; Pelegrinelli, F.F.; Thirone, A.C.; Tsukumo, D.M.; Pessoa, A.F.; dos Santos, M.F.; de Moraes, M.A.; et al. Topical insulin accelerates wound healing in diabetes by enhancing the AKT and ERK pathways: A double-blind placebo-controlled clinical trial. PLoS ONE 2012, 7, e36974. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Chi, Y.J.; Lin, G.Q.; Luo, S.H.; Jiang, Q.Y.; Chen, Y.K. MiRNA-26a promotes angiogenesis in a rat model of cerebral infarction via PI3K/AKT and MAPK/ERK pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3485–3492. [Google Scholar] [PubMed]
- Eniafe, J.; Jiang, S. MicroRNA-99 family in cancer and immunity. Wiley Interdiscip. Rev. RNA 2021, 12, e1635. [Google Scholar] [CrossRef]
- Clevers, H.; Nusse, R. Wnt/β-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef]
- Gorostizaga, A.; Brion, L.; Maloberti, P.; Cornejo Maciel, F.; Podestá, E.J.; Paz, C. Heat shock triggers MAPK activation and MKP-1 induction in Leydig testicular cells. Biochem. Biophys. Res. Commun. 2005, 327, 23–28. [Google Scholar] [CrossRef]
- Song, B.; Wang, C.; Liu, J.; Wang, X.; Lv, L.; Wei, L.; Xie, L.; Zheng, Y.; Song, X. MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J. Exp. Clin. Cancer Res. 2010, 29, 29. [Google Scholar] [CrossRef]
- Manetti, A.C.; Baronti, A.; Bosetti, C.; Costantino, A.; Di Paolo, M.; Turillazzi, E.; Maiese, A. Bleeding varicose veins’ ulcer as a cause of death: A case report and review of the current literature. Clin. Ter. 2021, 172, 395–406. [Google Scholar]
- Whyte, J.L.; Smith, A.A.; Helms, J.A. Wnt signaling and injury repair. Cold. Spring Harb. Perspect. Biol. 2012, 4, a008078. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, L.; Shi, C.; Sun, H.; Wang, J.; Li, R.; Zou, Z.; Ran, X.; Su, Y. TGF-β-induced miR-21 negatively regulates the antiproliferative activity but has no effect on EMT of TGF-β in HaCaT cells. Int. J. Biochem. Cell Biol. 2012, 44, 366–376. [Google Scholar] [CrossRef]
- Vanbokhoven, H.; Melino, G.; Candi, E.; Declercq, W. p63, a story of mice and men. J. Investig. Dermatol. 2011, 131, 1196–1207. [Google Scholar] [CrossRef] [PubMed]
- Bonni, A.; Brunet, A.; West, A.E.; Datta, S.R.; Takasu, M.A.; Greenberg, M.E. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 1999, 286, 1358–1362. [Google Scholar] [CrossRef] [PubMed]
- Grellner, W.; Madea, B. Demands on scientific studies: Vitality of wounds and wound age estimation. Forensic Sci. Int. 2007, 165, 150–154. [Google Scholar] [CrossRef]
- Grellner, W.; Vieler, S.; Madea, B. Transforming growth factors (TGF-alpha and TGF-beta1) in the determination of vitality and wound age: Immunohistochemical study on human skin wounds. Forensic Sci. Int. 2005, 153, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Madea, B.; Doberentz, E.; Jackowski, C. Vital reactions—An updated overview. Forensic Sci. Int. 2019, 305, 110029. [Google Scholar] [CrossRef] [PubMed]
- Courts, C.; Madea, B. Micro-RNA—A potential for forensic science? Forensic Sci. Int. 2010, 203, 106–111. [Google Scholar] [CrossRef]
| References | N. Cases | Model | Cells and/or Animals | Kind of Lesions | Controls | miRNAs | miRNAs Expression | Performed Analysis | RNA Isolation/cDNA Synthesis |
|---|---|---|---|---|---|---|---|---|---|
| Bertero et al., 2011 [21] | 6 | In vitro + in vivo | NHKs, HaCat + Mice | Scratch wound + Excisional wound | Normal skin | miR-483-3p | ↑ 3d after injury infliction Peak: 6–7 days Normalization: at day 10 In epidermis (PM) | H + IHC + WB + RT-PCR | TRIzol reagent/NP |
| Cao et al., 2020 [22] | 15 | In vitro + in vivo | HaCaT, HSF + Mice | H2O2-induced wound + Excisional wound | Normal skin | miR-19b | ↓ In epidermis if ↑ H2O2 | H + IHC + WB + RT-PCR | TRIzol reagent/NP |
| Etich et al., 2017 [23] | 15 | In vivo | Mice | Excisional wound | Normal skin | miR-204 | ↓ from day 5 to day 10 | H + RT-PCR | miRNeasy mini kit/miScript II RT kit |
| miR-205 | ↓ from day 1 to day 7 | ||||||||
| miR-31 | ↑ In epidermis from day 5 to 14 after injury infliction (PM samples) | ||||||||
| He et al., 2020 [24] | NS | In vitro | HADSCs exosome, MALAT1 knockdown HaCaT and HSF | H2O2 induced scratch wound | Normal HaCaT and HSF | miR-124 | ↓ In epidermis if ↑ H2O2 | WB + RT-PCR | TRIzol reagent/cDNA Synthesis SuperMix |
| Ibrahim et al., 2019 [18] | 18 | In vivo | Rats | Incisional wound | Normal skin | miR-205, miR-21 | No statistical significance In epidermis 0, 24, and 48 h after death | H + RT-PCR | mirVana PARIS kit/NP |
| Jiang et al., 2020 [25] | 3 | In vitro | TGF-β1-treated HaCaT | Scratch wound | HaCaT | miR-26a | ↓ In keratinocytes | WB + RT-PCR | EZ-Magna RIP kit/miScript II RT kit |
| Jin et al., 2013 [26] | 5 | In vivo + in vitro | HaCaT + Mice | Scratch wound + Excisional wound | Normal skin | miR-152, miR-365, miR-125a/b-5p, miR-181d, miR-99, miR-100, miR-30c | ↓ In keratinocytes Peak: day 1 Normalization: day 5 | WB + RT-PCR | TRIzol reagent/CyQUANT assay |
| Lang et al., 2017 [27] | 9 | In vitro + In vivo | HaCaT, HSF + Rats | Scratch wound + Excisional wound | No- TNFα-exposed cells | miR-149 | ↓ In epidermis | H + IHC + WB | mirVana miRNA isolation kit/NP |
| Lin et al., 2013 [28] | NS | In vitro | HCECs and HEKs | Scratch wound | Normal miR-16 levels | miR-205 | ↑ In corneal epithelial cells 24h after incision | WB + RT-PCR | TRIzol reagent/cDNA Synthesis SuperMix |
| Li et al., 2015 [29] | 9 | In vivo | Human (surgical samples) | Hypertrophic scar | Normal skin | miR-149, miR-203a, miR-222, miR-122 | ↓ In epidermis | ISH | miRcute RNA Isolation kit/NP |
| Liu et al., 2020 [30] | 24 | In vivo | Rats and DM rats | Excisional wound | No-diabetic rats | mir-203 | ↓ Over the course of the first 4 days in non-diabetic rats, over the course of the first 6 days in DM rats, in epidermis, dermis, and subcutaneous fat | H + IHC+ IHF + WB + RT-PCR | TRIzol rea-gent/RevertAid H minus first-strand cDNA synthesis kit |
| Lyu et al., 2018 [17] | 22 | In vivo | Mice | Burned skin | Normal skin | miR-135a-1-3p miR-183-3p miR-188-5p miR-3081-5p miR-5103 miR-6378 miR-6385 miR-6391 miR-6769b-5p miR-6969-5p miR-7005-5p miR-7036a-5p miR-7044-5p miR-710 miR-711 miR-7118-5p miR-7668-3p miR-8090 mmu-miR-874-3p | ↑ | H + RT-PCR | NS/PrimeScript RT reagent |
| miR-155-5p miR-28a-3p miR-467b-3p miR-5132-5p miR-6924-3p | ↓ | ||||||||
| Long et al., 2018 [31] | NS | In vivo | miR-21 knock-in mice | Excisional wound | Normal skin | miR-21 | ↑ | H + MP + ISH + RT-PCR | TaqMan microR-NA assay kit/NP |
| Neri et al., 2019 [19] | 64 | Autopsy casuistry | Human | Ligature marks | Normal skin | miR125a/b-5p miR130a-3p miR214-3p miR92a-3p | ↑ | H + IHC + RT-PCR | miScript miRNA PCR Array- Human Cell Differentiation/miScript II RT Kit |
| Pastar et al., 2012 [32] | 21 | In vivo + in vitro | HEK, HSF + Rats (6) + Humans (15) | Chronic ulcers + Excisional wound | Normal skin | miR-16, miR-20a, miR-21, miR-106a, miR-130a, miR-203 | ↑ In epidermis (miR-21: in epidermis, dermis, and blood vessels) | H + ISH + WB + RT-PCR | TaqMan Mi-croRNA As-says/NP |
| Viticchiè et al., 2012 [33] | NS | In vitro + in vivo | HEK and MEK + Mice | Excisional wound | Normal skin | miR-203 | ↑ In epidermis surrounding the wound and wound margin ↓ In keratinocytes at the migratory front (day 3 and 5 after wounding) | H + ISH + WB + RT-PCR | mirVana miRNA isolation kit/NP |
| Wang et al., 2012 [34] | NS | In vivo | Mice | Excisional wound | Normal skin | miR-31, miR-21, miR-203 | ↑ In epithelial cells | H + IHC + ISH + RT-PCR | TaqMan Mi-croRNA As-says/NP |
| miR-249 | ↓ (day 7 after wounding) | ||||||||
| Yang et al., 2011 [35] | 9 | In vivo | Mice | Excisional wound | miR-21 knockdown mice | miR-21 | ↑ In epidermis after 0, 1, 2 and 3 days | H + IHC + IHF + RT-PCR | NS/NS |
| Yu et al., 2010 [36] | NS | In vitro | HCECs and HEKs (miR-205 and miR-184 downregulated) | Scratch wound | Normal cells | miR-205, miR-184 | ↑ In epithelial cells | IHC + PCM + WB | Quik-Change Site-Directed Muta-genesis kit/NP |
| Zhang et al., 2020 [37] | 35 | In vivo + autopsy cases | Mice (9) + autoptic human samples (26) | Excisional wound in burned skin | Mice unburned skin + mice PM burned skin | miR-711, miR-183-3p | ↑ 48 h after excision in human burned skin (PM samples), 120 h in mice burned skin (AM samples) | RT-PCR | NS/PrimeScript RT reagent kit |
| Tot: 20 articles | Tot: 255 (at least) | Tot: 51 different miRNAs |
| Study’s Characteristics | N. of Studies (Tot. 20) | References | ||
|---|---|---|---|---|
| Model | In Vivo | 8 | [17,18,23,29,30,31,34,35] | |
| In Vitro | 4 | [24,25,28,36] | ||
| In Vivo + In Vitro | 6 | [21,22,26,27,32,33] | ||
| Human samples | Autoptic | 2 | [19,37] | |
| Surgical | 1 | [29] | ||
| Type of lesion * | Excisional wound | 10 | [21,22,23,26,27,30,31,33,34,35] | |
| Incisional wound | 1 | [18] | ||
| Burned skin | 1 | [37] | ||
| Ligature mark | 1 | [19] | ||
| Chronic ulcer | 1 | [32] | ||
| Hypertrophic scar | 1 | [29] | ||
| Performed analysis | RT-PCR | 17 | [17,18,19,21,22,23,24,25,26,28,30,31,32,33,34,35,37] | |
| Histology (HE) | 13 | [18,19,21,22,23,24,27,30,31,32,33,34,35] | ||
| WB | 11 | [21,22,24,25,26,27,28,30,32,33,36] | ||
| IHC | 8 | [19,21,22,27,30,34,35,36] | ||
| ISH | 5 | [29,31,32,33,34] | ||
| IHF | 2 | [30,35] | ||
| MP | 1 | [31] | ||
| PMC | 1 | [36] | ||
| miRNAs | Target Genes and/or Proteins | References |
|---|---|---|
| miR19-b | CCL1, TGFβ | [22] |
| miR-21 | Factor 3, vinculin, LepR, EGR3, Collagen, TGF-β, TIMP3, TIAM1, TP53 | [29,32,34,35] |
| miR-26a | ITGA5, PI3K/ AKT, SMAD1, GSK3β | [25] |
| miR-30c | PI3K/AKT, mTOR, IGF1R | [26] |
| miR-31 | IL-1b, PTPRC/CD45, SHIP2, RNU6B, Col1a1 | [23] |
| miR-99 | PI3K/AKT, mTOR, IGF1R | [26] |
| miR-100 | PI3K/AKT, mTOR, IGF1R | [26] |
| miR-122 | MAPK, lysosome, insulin signaling pathway, focal adhesion | [29] |
| miR-125a-5p, miR-125b-5p | PI3K/AKT, mTOR, IGF1R | [26] |
| miR-130a | EGR3 | [32] |
| miR-149 | IL-1a, IL-1b, IL-6, TGF-β3, collagen III, Nf-Kb, RelB, Rel, MAPK | [27,29] |
| miR-152 | PI3K/AKT, mTOR, IGF1R | [26] |
| miR-181d | PI3K/AKT, mTOR, IGF1R | [26] |
| miR-184 | SHIP2, PI3K-Akt, actin filaments, p-cofilin (via Rho) | [36] |
| miR-203 | MAPK, lysosome, insulin signaling pathway, focal adhesion, K15, P63, integrin-β1, TCF-4, ID-2, CD44, VEGFA, NRCAM, C-MET, Wnt, Notch, Factor 3, vinculin, LepR, EGR3, p63, LASP1, RAN, RAPH1 | [29,30,32,33] |
| miR-204 | IL-1b, PTPRC/CD45, SHIP2, RNU6B, Col1a1 | [23] |
| miR-205 | IL-1b, PTPRC/CD45, SHIP2, RNU6B, Col1a1, KCNJ10, SHIP2, PI3K-Akt, actin filaments, p-cofilin (via Rho), p-ERM | [23,28,36] |
| miR-222 | DDK2, AXIN2, FRAT2, MAPK | [29] |
| miR-365 | PI3K/AKT, mTOR, IGF1R | [26] |
| miR-483-3p | MK2, YAP1, ASH2, MKI67 | [21] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manetti, A.C.; Maiese, A.; Baronti, A.; Mezzetti, E.; Frati, P.; Fineschi, V.; Turillazzi, E. MiRNAs as New Tools in Lesion Vitality Evaluation: A Systematic Review and Their Forensic Applications. Biomedicines 2021, 9, 1731. https://doi.org/10.3390/biomedicines9111731
Manetti AC, Maiese A, Baronti A, Mezzetti E, Frati P, Fineschi V, Turillazzi E. MiRNAs as New Tools in Lesion Vitality Evaluation: A Systematic Review and Their Forensic Applications. Biomedicines. 2021; 9(11):1731. https://doi.org/10.3390/biomedicines9111731
Chicago/Turabian StyleManetti, Alice Chiara, Aniello Maiese, Arianna Baronti, Eleonora Mezzetti, Paola Frati, Vittorio Fineschi, and Emanuela Turillazzi. 2021. "MiRNAs as New Tools in Lesion Vitality Evaluation: A Systematic Review and Their Forensic Applications" Biomedicines 9, no. 11: 1731. https://doi.org/10.3390/biomedicines9111731
APA StyleManetti, A. C., Maiese, A., Baronti, A., Mezzetti, E., Frati, P., Fineschi, V., & Turillazzi, E. (2021). MiRNAs as New Tools in Lesion Vitality Evaluation: A Systematic Review and Their Forensic Applications. Biomedicines, 9(11), 1731. https://doi.org/10.3390/biomedicines9111731







