Current Potential Therapeutic Approaches against SARS-CoV-2: A Review
Abstract
:1. Introduction
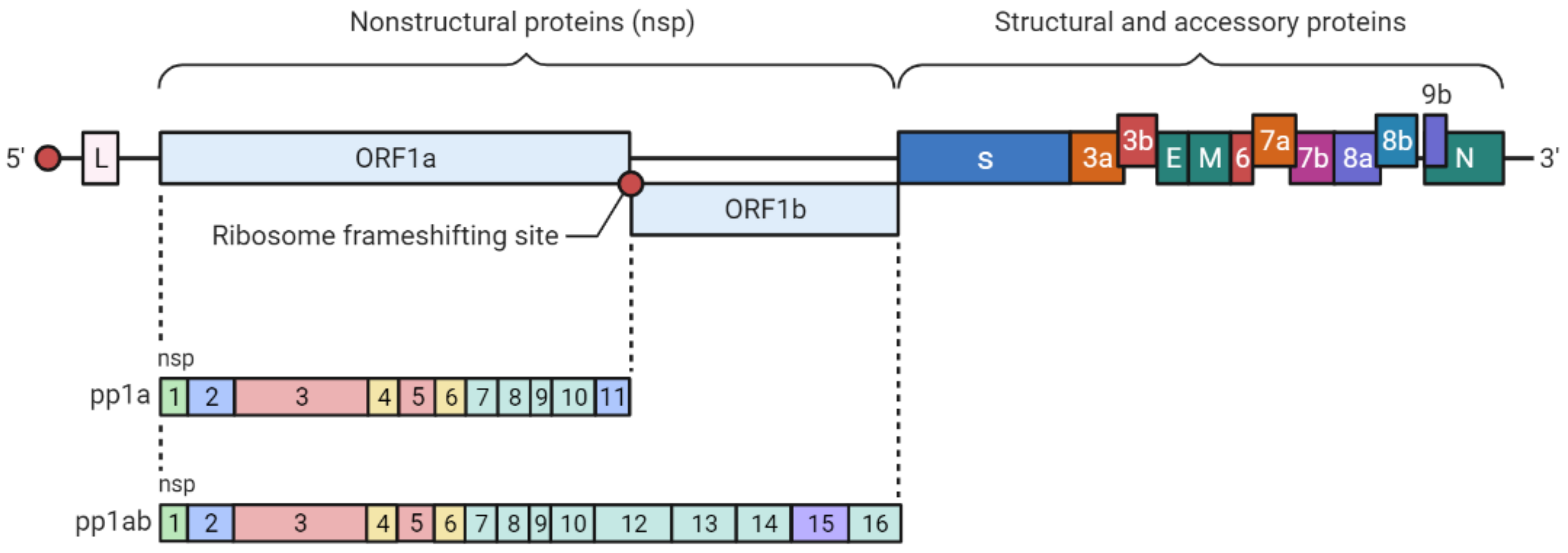
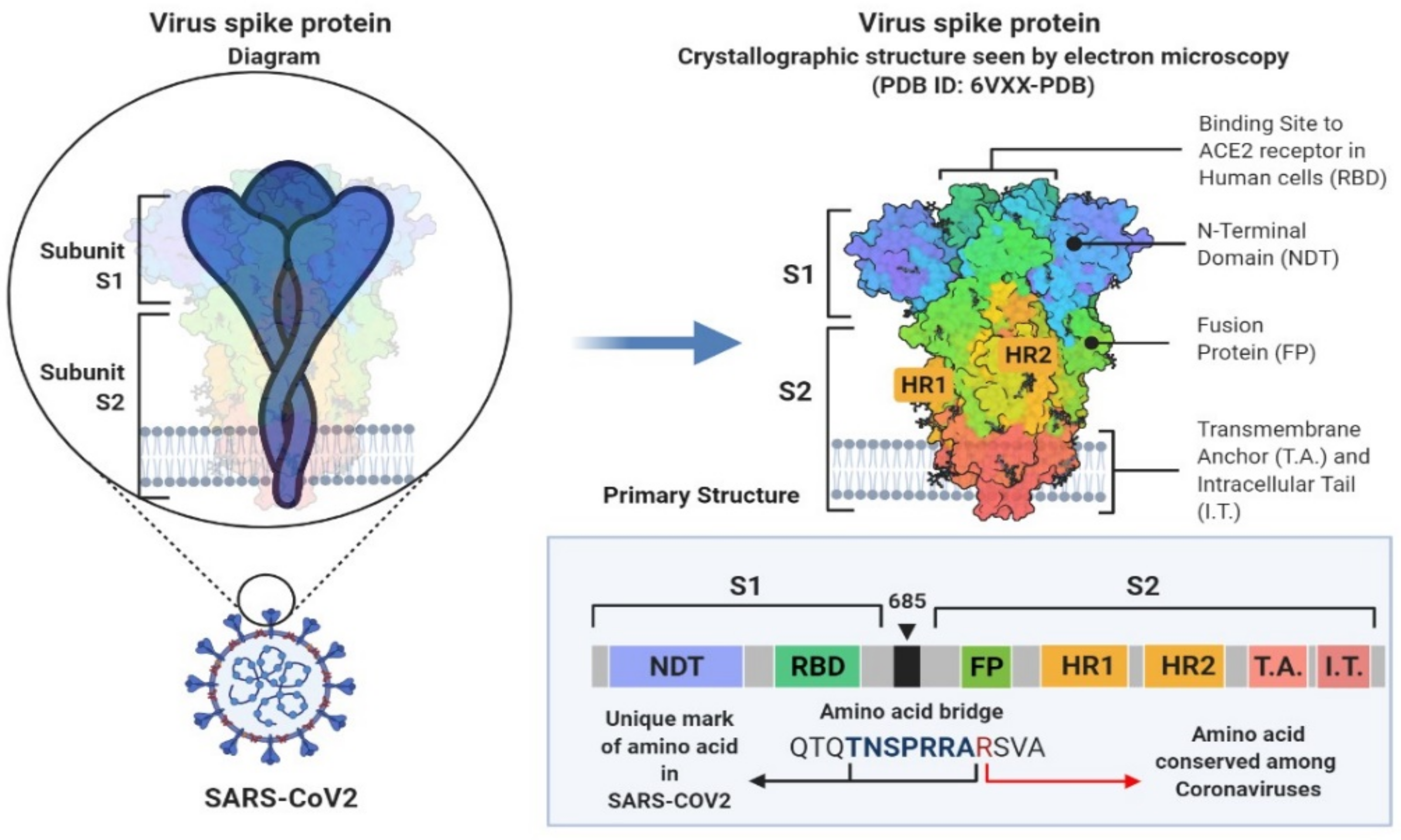
2. SARS-CoV-2 Genetic Structure and Pathogenic Mechanism
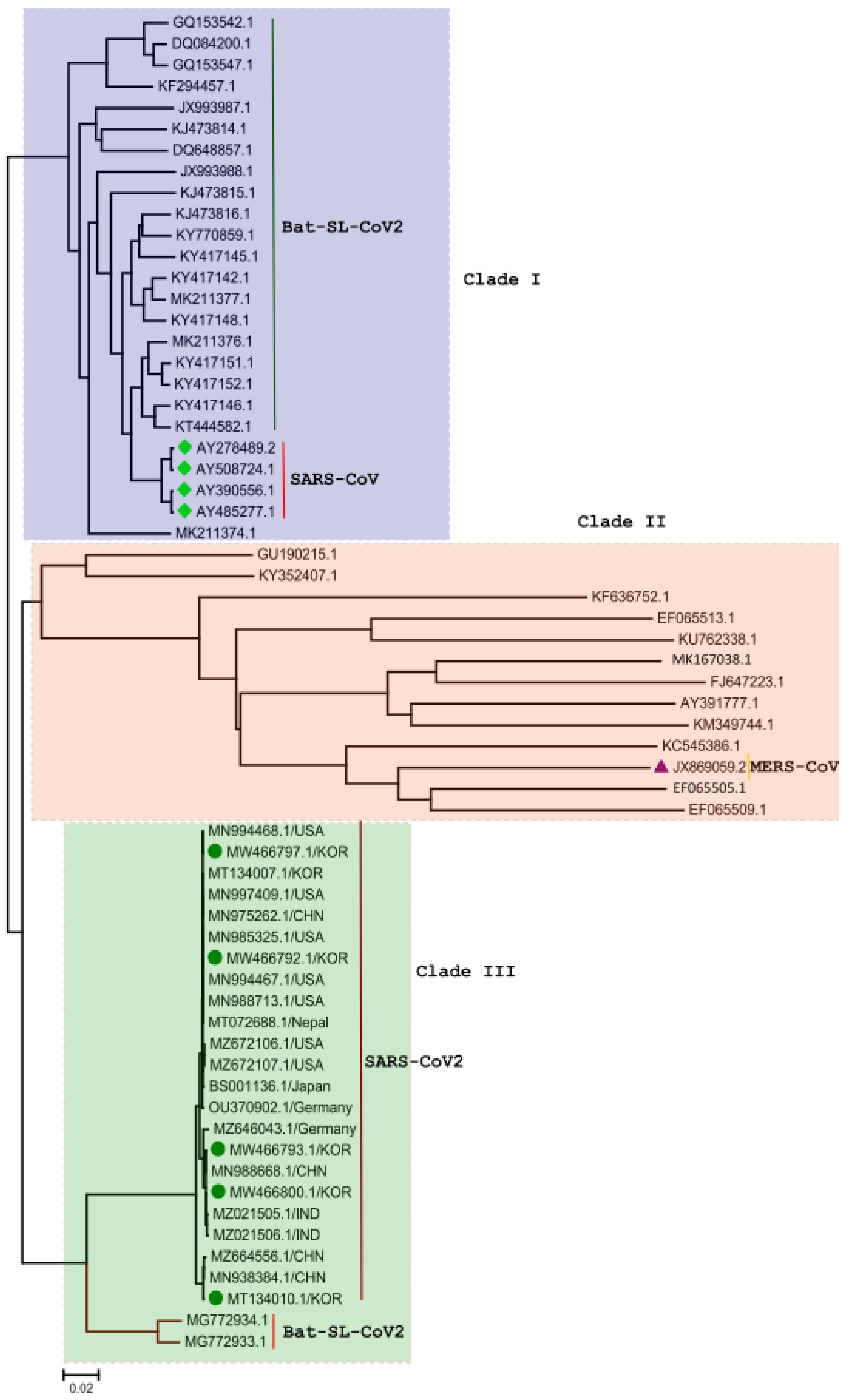
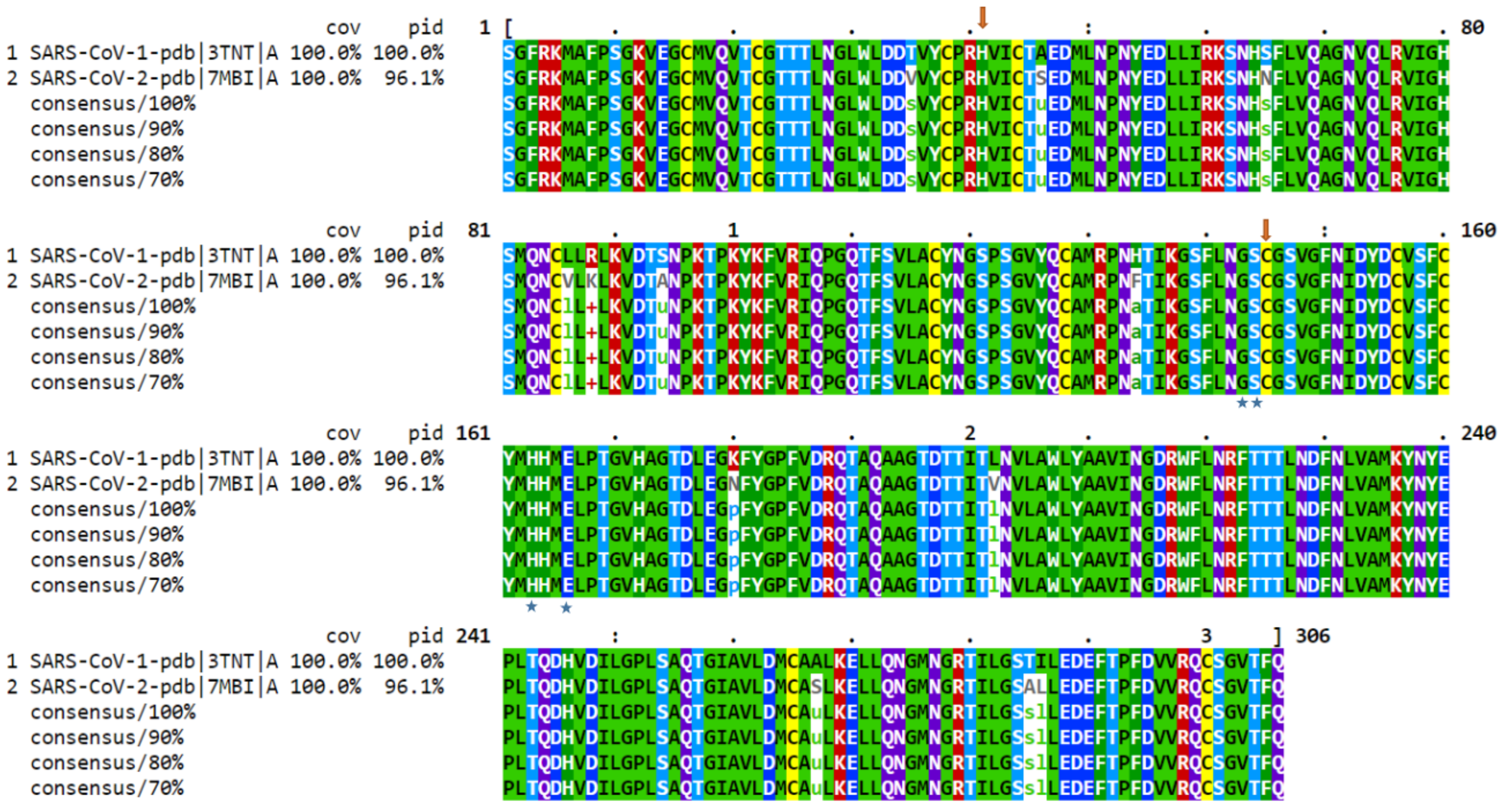
3. Phylogenetic Analysis of SARS-CoV-2 Genome
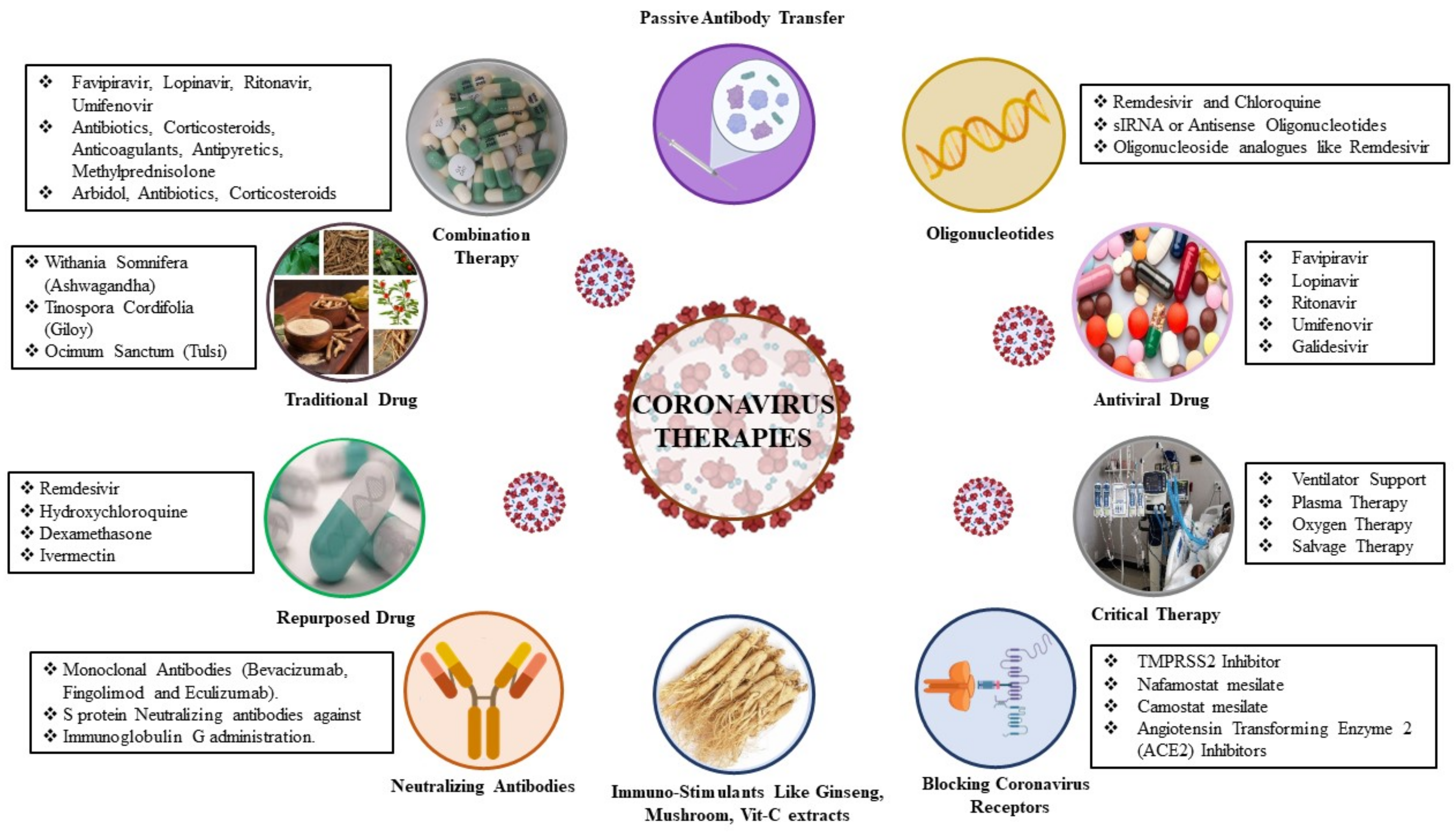
4. Therapeutic Approaches to SAR-COV-2 Infection
4.1. Virus-Based Therapy
4.2. Host-Based Therapy
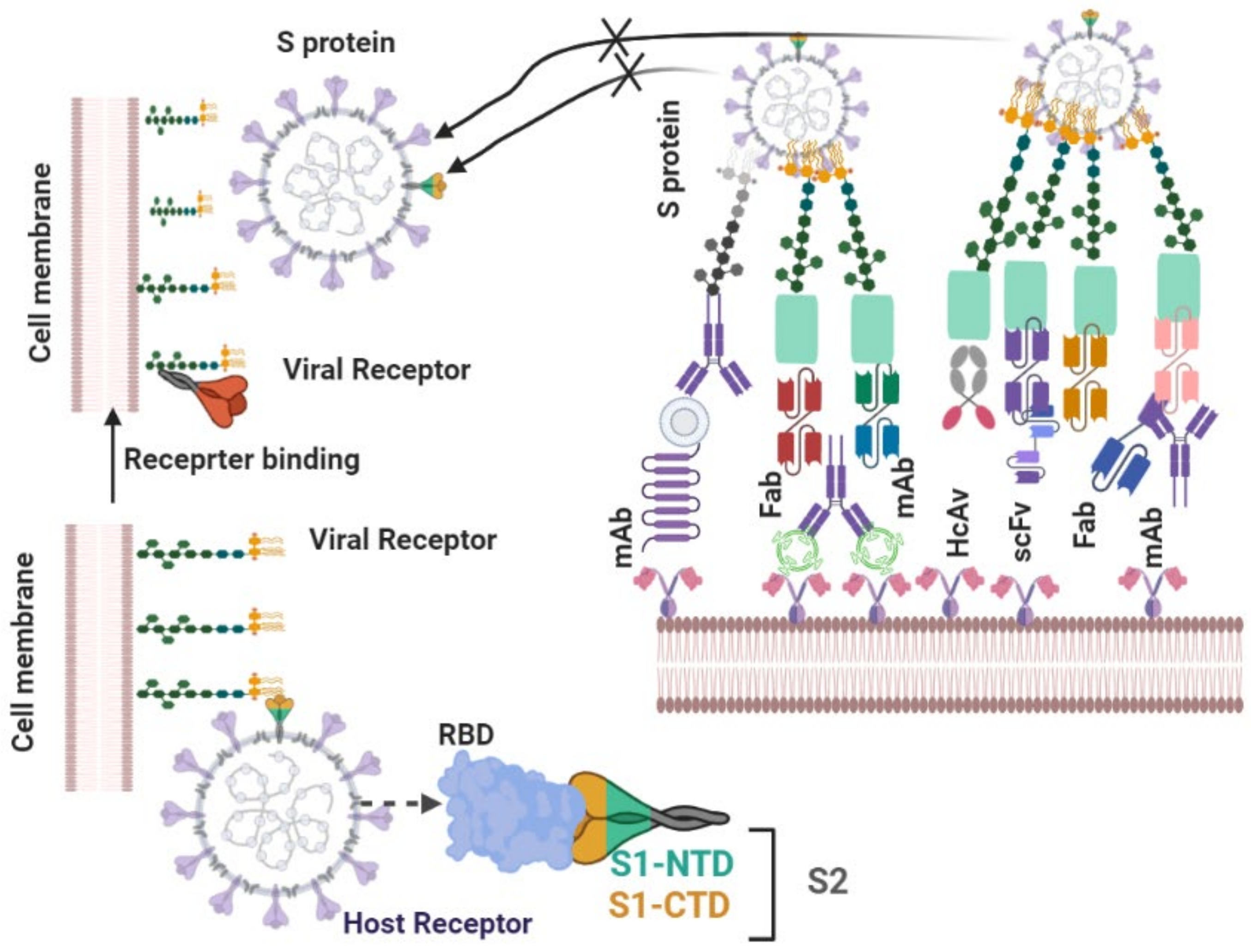
4.2.1. Neutralizing Antibodies
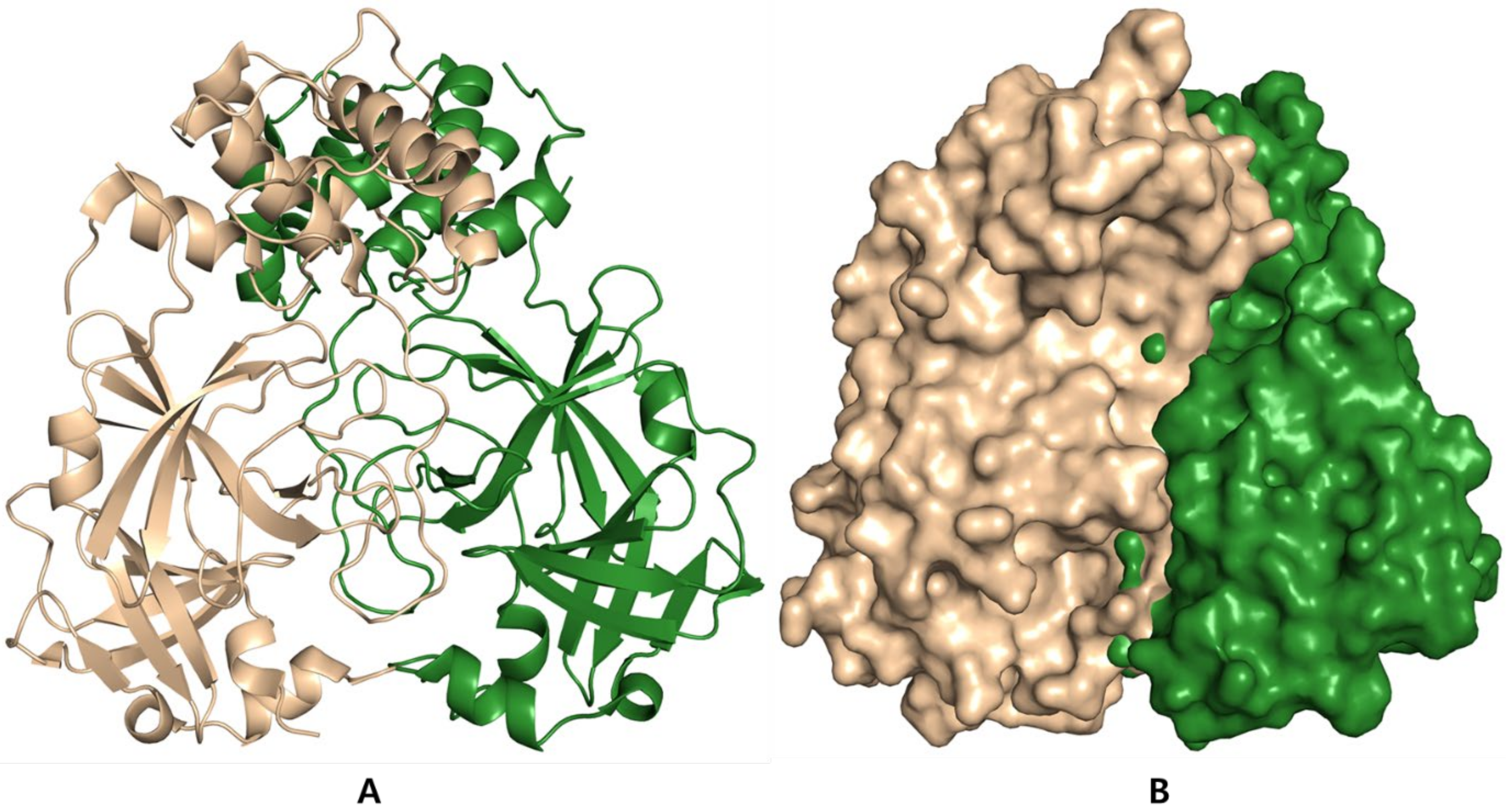
4.2.2. Antiviral Proteases
4.2.3. Combination Therapy
4.2.4. SARS-CoV-2 Cell Entry Inhibitors
Inhibitors of TMPRSS2 Serine Protease
Nafamostat Mesylate
Camostat Mesylate (Foipan™)
Angiotensin Transforming Enzyme 2 (ACE2) Inhibitors and Antimalarial Drugs
Cepharanthine/Selamectin/Mefloquine Hydrochloride
Chloroquine Phosphate and Hydroxychloroquine
4.2.5. Inhibitors of the Replication, Membrane Fusion, and Assembly of SARS-CoV-2
Favipiravir
Umifenovir
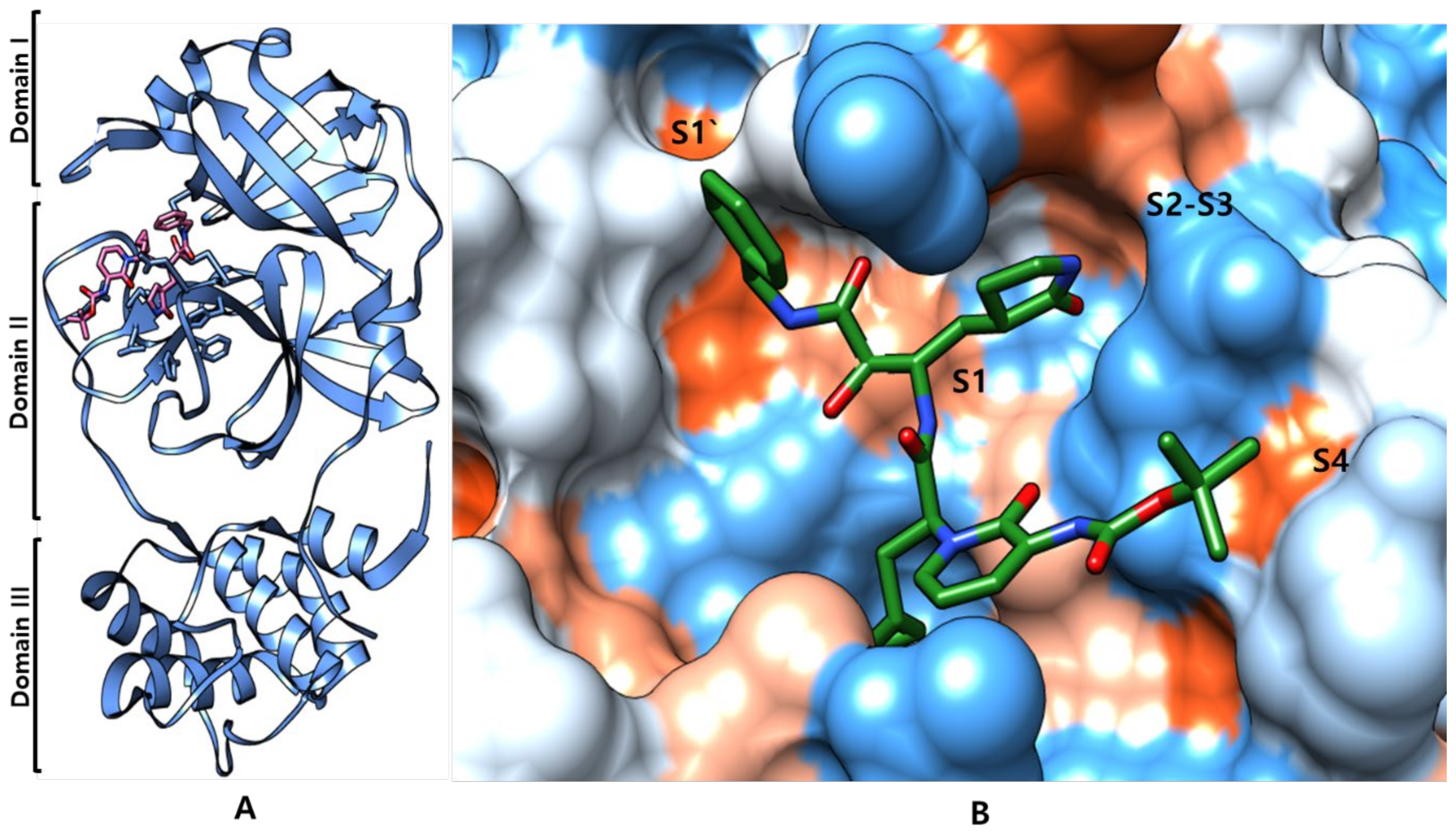
Inhibitors of SARS-CoV-2 3CLpro Protease
SARS-CoV-2 Mpro Inhibitor
5. Nanomaterials Approach for Anti-Coronavirus
| Nanomaterial | Chitosan Nanospheres | CQDsc) Nanocrystals4– | Gold Nanorods | PLGAg) Hollow Nanoparticles | TiO2 Nanoparticles | Ref |
|---|---|---|---|---|---|---|
| Average size | 10 nm–10 µm | 9 nm | 18–54 nm | 114 nm | Not reported | [65,66,67,68,69,70,71,72,73,74,75] |
| Shape | Spherical | Spherical | Rod | Spherical | Predominantly spherical | |
| Strategy | Genipin-crosslinked chitosan | Boronic acid-functionalized CQDs (Carbon quantum dots) | Gold nanorods-based peptides | Viral antigens and STING agonists-loaded hollow nanoparticles | TNPsi)-coated glass coverslips UVC radiation | |
| Coronavirus | HCoV NL63 (Human coronavirus NL63) | HCoV-229E-Luc | MERS-CoV | MERS-CoV | HCoV NL63a) | |
| Potential application | Adsorbents | Antiviral drugs | Antiviral drugs | Vaccine | Self-cleaning surfaces |
| Nanomaterial | Iron Oxide Nanoparticles | Poly Silica Nanoparticles | Gold Nanoparticles | Nano-Sized Formazans | Polylactic-Co-Glycolic Acid (PLGA)-Nanoparticles | Silver Nanoparticles | Ref. |
|---|---|---|---|---|---|---|---|
| Size | N/ra | 210 ± 40 nm | N/ra | 23.75 ± 7.16 nm | N/ra | [65,66,67,68,69,70,71,72,73,74,75,76] | |
| Strategy | Nano-mineral structure of Fe2O3 and Fe3O4 | Optimized polyP encapsulated by SiNPs | Peptide-functionalized gold nanoparticles | Formazan analogs by dithizone and α-haloketones reaction | Optimized Remdesivir-loaded L-PLGA NPs | Artemisinin, Artemether, and Artesunate delivery by silver nanoparticles | |
| Ligand-receptor binding results | Interactions with S1-RBD of SARS-CoV-2 | Inhibition of binding of ACE2 to S-protein SARS-CoV-2, at a physiological solution | More stable complex with RBD of SARS-CoV-2 than ACE2 | Inhibition of SARS-CoV-2 chymotrypsin-like protease, at a physiological solution | Interactions Lisinopril-ACE1 and remdesivir-intracellular targeting protein RdRp | Interactions between negative charges of oxygen atoms of drugs with Ag surface | |
| Potential application | Repurposing medication | Immunologic agents | Antiviral agents | Antiviral agents | Antiviral therapy | Antiviral drugs |
| Nanomaterial | Shape, Size | Strategy | Potential Application | Ref. |
|---|---|---|---|---|
| Graphene sheets | Layers | Modified graphene sheets conjugated with spike antibody of SARS-CoV-2 | Immunodetection | [77] |
| Gold nanoparticles | 2.4 nm Spherical in size | Sulfonated gold nanomaterials | Antiviral agents | [66] |
| Polymeric nanoparticles | Spherical | Bioinspired DNase-coated melanin-like nanospheres | Sepsis or acute respiratory distress syndrome (ARDS) in sever COVID-19 Patients | [78] |
| Polymeric nanoparticles | 10 nm–1 μm colloidal particles | Ivermectin-delivery by (Poly (lactic-co-glycolic acid)-b-Maleimide PEG (polyethylene glycol) Copolymers | Antiviral drug | [79] |
| Nanostructured lipid carriers | Spherical | Pulmonary delivery of Salinomycin by nanostructured lipid carriers | Drug delivery | [66] |
| Polymeric nanoparticles | Spherical 22.0 nm | DNase-I-coated polydopamine-PEG poly (ethylene glycol) | Sepsis or acute respiratory distress syndrome (ARDS) in sever COVID-19 Patients | [80] |
| Decoy nanoparticles | (Not reported) | Fusing genetically engineered cell membrane nanovesicles (293T/ACE2 and THP-1cells) | Therapeutic vaccines | [81] |
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nadeem, M.S.; Zamzami, M.A.; Choudhry, H.; Murtaza, B.N.; Kazmi, I.; Ahmad, H.; Shakoori, A.R. Origin, potential therapeutic targets and treatment for coronavirus disease (COVID-19). Pathogens 2020, 9, 307. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.Z.; Holmes, E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell 2020, 181, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.C.; Liu, Y.H.; Wang, C.Y.; Wang, Y.H.; Hsueh, S.C.; Yen, M.Y.; Ko, W.C.; Hsueh, P.R. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): Facts and myths. J. Microbiol. Immunol. Infect. 2020, 53, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Q.; Yang, K.; Wang, W.; Jiang, L.; Song, J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020, 46, 846–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Rong, L.; Nian, W.; He, Y. Review article: Gastrointestinal features in COVID-19 and the possibility of faecal transmission. Aliment. Pharmacol. Ther. 2020, 51, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.G.; Klepac, P.; Liu, Y.; Prem, K.; Jit, M.; CMMID COVID-19 Working Group; Eggo, R.M. Age-dependent effects in the transmission and control of COVID-19 epidemics. Nat. Med. 2020, 26, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Siddique, R.; Shereen, M.A.; Ali, A.; Liu, J.; Bai, Q.; Bashir, N.; Xue, M. Emergence of a novel Coronavirus, severe acute respiratory syndrome Coronavirus 2: Biology and therapeutic options. J. Clin. Microbiol. 2020, 58, e00187-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, D.D.; Jain, A. Multipurpose Instantaneous Microarray Detection of Acute Encephalitis Causing Viruses and Their Expression Profiles. Curr. Microbiol. 2012, 65, 290–303. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor recognition by the novel Coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ton, A.T.; Gentile, F.; Hsing, M.; Ban, F.; Cherkasov, A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol. Inform. 2020, 10, 202000028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, H.; Peto, R.; Henao-Restrepo, A.-M.; Karim, Q.A.; Alejandria, M.; García, C.H.; Kieny, M.-P.; Malekzadeh, R.; Murthy, S.; Preziosi, M.-P.; et al. Repurposed antiviral drugs for COVID-19—Interim WHO Solidarity Trial Results. N. Engl. J. Med. 2021, 384, 497–511. [Google Scholar]
- Lam, T.T.-Y.; Shum, M.H.-H.; Zhu, H.-C.; Tong, Y.-G.; Ni, X.-B.; Liao, Y.-S.; Wei, W.; Cheung, W.Y.-M.; Li, W.-J.; Li, L.-F.; et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature 2020, 583, 282–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khateeb, J.; Li, Y.; Zhang, H. Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Crit. Care 2021, 25, 244. [Google Scholar] [CrossRef] [PubMed]
- Monteil, V.; Kwon, H.; Patricia, P.; Prado, P.; Hagelkrüys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Hurtado del Pozo, C.; et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell Press 2020, 181, 905–913. [Google Scholar] [CrossRef]
- Zhou, Y.; Hou, Y.; Shen, J.; Huang, Y.; Martin, W.; Cheng, F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020, 6, 14–18. [Google Scholar] [CrossRef] [Green Version]
- Rabaan, A.A.; Al-Ahmed, S.H.; Haque, S.; Sah, R.; Tiwari, R.; Malik, Y.S.; Dhama, K.; Yatoo, M.I.; Bonilla-Aldana, D.K.; RodriguezMorales, A.J.; et al. SARS-CoV-2, SARS-CoV, and MERS-CoV: A comparative overview. Infez. Med. 2020, 28, 174–184. [Google Scholar] [PubMed]
- Ko, M.; Chang, S.; Byun, S.; Ianevski, A.; Choi, I.; D’Orengiani, A.-L.P.H.D.; Ravlo, E.; Wang, W.; Bjørås, M.; Kainov, D.; et al. Screening of FDA-Approved Drugs Using a MERS-CoV Clinical isolate from South korea identifies potential therapeutic options for COVID-19. Viruses 2021, 13, 651. [Google Scholar] [CrossRef]
- Yadav, R.; Chaudhary, J.; Jain, N.; Chaudhary, P.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821. [Google Scholar] [CrossRef]
- Finkelstein, M.T.; Mermelstein, A.G.; Parker Miller, E.; Seth, P.C.; Stancofski, E.D.; Fera, D. Structural analysis of neutralizing epitopes of the SARS-CoV-2 spike to guide therapy and vaccine design strategies. Viruses 2021, 13, 134. [Google Scholar] [CrossRef]
- Lai, C.-C.; Chen, C.-H.; Wang, C.-Y.; Chen, K.-H.; Wang, Y.-H.; Hsueh, P.-R. Clinical efficacy and safety of remdesivir in patients with COVID-19: A systematic review and network meta-analysis of randomized controlled trials. J. Antimicrob. Chemother. 2021, 76, 1962–1968. [Google Scholar] [CrossRef]
- Kocayiğit, H.; Süner, K.Ö.; Tomak, Y.; Demir, G.; Yaylacı, S.; Dheir, H.; Güçlü, E.; Erdem, A.F. Observational study of the effects of Favipiravir vs Lopinavir/Ritonavir on clinical outcomes in critically Ill patients with COVID-19. J. Clin. Pharm. Ther. 2021, 46, 454–459. [Google Scholar] [CrossRef]
- Saw, P.E.; Song, E.-W. siRNA therapeutics: A clinical reality. Sci. China Life Sci. 2020, 63, 485–500. [Google Scholar] [CrossRef] [PubMed]
- Zanella, I.; Zizioli, D.; Castelli, F.; Quiros-Roldan, E. Tenofovir, another inexpensive, well-known and widely available old drug repurposed for SARS-CoV-2 infection. Pharmaceuticals 2021, 14, 454. [Google Scholar] [CrossRef]
- Dejmek, M.; Konkol’ová, E.; Eyer, L.; Straková, P.; Svoboda, P.; Šála, M.; Krejčová, K.; Růžek, D.; Boura, E.; Nencka, R. Non-Nucleotide RNA-Dependent RNA Polymerase Inhibitor That Blocks SARS-CoV-2 Replication. Viruses 2021, 13, 1585. [Google Scholar] [CrossRef]
- Singh, D.D.; Han, I.; Choi, E.H.; Yadav, D.K. Immunopathology, host-virus genome interactions, and effective vaccine development in SARS-CoV-2. Comput. Struct. Biotechnol. J. 2020, 18, 3774–3787. [Google Scholar] [CrossRef]
- Hurt, A.; Wheatley, A. Neutralizing antibody therapeutics for COVID-19. Viruses 2021, 13, 628. [Google Scholar] [CrossRef]
- Li, X.; Zhang, L.; Chen, S.; Ouyang, H.; Ren, L. Possible targets of Pan-coronavirus antiviral strategies for emerging or re-emerging Coronaviruses. Microorganisms 2021, 9, 1479. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, S.V.; Wallace, F.E.; Stoddard, S.D.; Cheng, Q.; Acosta, D.; Barzani, S.; Bobay, M.; Briant, J.; Cisneros, C.; Feinstein, S.; et al. In silico design of peptide-based SARS-CoV-2 fusion inhibitors that target wt and mutant versions of SARS-CoV-2 HR1 Domains. Biophysica 2021, 1, 311–327. [Google Scholar] [CrossRef]
- Yamamoto, M.; Kiso, M.; Sakai-Tagawa, Y.; Iwatsuki-Horimoto, K.; Imai, M.; Takeda, M.; Kinoshita, N.; Ohmagari, N.; Gohda, J.; Semba, K.; et al. The anticoagulant nafamostat potently inhibits SARS-CoV-2 s protein-mediated fusion in a cell fusion assay system and viral infection in vitro in a cell-type-dependent manner. Viruses 2020, 12, 629. [Google Scholar] [CrossRef]
- Sardanelli, A.M.; Isgrò, C.; Palese, L.L. SARS-CoV-2 main protease active site ligands in the human metabolome. Molecules 2021, 26, 1409. [Google Scholar] [CrossRef]
- Citarella, A.; Scala, A.; Piperno, A.; Micale, N. SARS-CoV-2 Mpro: A potential target for peptidomimetics and small-molecule inhibitors. Biomolecules 2021, 11, 607. [Google Scholar] [CrossRef] [PubMed]
- Riva, A.; Conti, F.; Bernacchia, D.; Pezzati, L.; Sollima, S.; Merli, S.; Siano, M.; Lupo, A.; Rusconi, S.; Cattaneo, D.; et al. Da-runavir does not prevent SARS-CoV-2 infection in HIV patients. Pharmacol. Res. 2020, 157, 104826. [Google Scholar] [CrossRef] [PubMed]
- Nersisyan, S.; Shkurnikov, M.; Turchinovich, A.; Knyazev, E.; Tonevitsky, A. Integrative analysis of miRNA and mRNA sequencing data reveals potential regulatory mechanisms of ACE2 and TMPRSS2. PLoS ONE 2020, 15, e0235987. [Google Scholar] [CrossRef] [PubMed]
- Gatti, M.; Turrini, E.; Raschi, E.; Sestili, P.; Fimognari, C. Janus kinase inhibitors and Coronavirus dDisease (COVID)-19: Rationale, clinical evidence and safety issues. Pharmaceuticals 2021, 14, 738. [Google Scholar] [CrossRef] [PubMed]
- Arisan, E.D.; Dart, A.; Grant, G.H.; Arisan, S.; Cuhadaroglu, S.; Lange, S.; Uysal-Onganer, P. The prediction of miRNAs in SARS-CoV-2 genomes: Hsa-miR databases identify 7 Key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses 2020, 12, 614. [Google Scholar] [CrossRef]
- Chien, M.; Anderson, T.K.; Jockusch, S.; Tao, C.; Li, X.; Kumar, S.; Russo, J.J.; Kirchdoerfer, R.N.; Ju, J. Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase, a Key Drug Target for COVID-19. J. Proteome Res. 2020, 19, 4690–4697. [Google Scholar] [CrossRef]
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of Coronavirus cell entry mediated by the viral spike protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef] [Green Version]
- Gil Martínez, V.; Avedillo Salas, A.; Santander Ballestín, S. Antiviral therapeutic approaches for SARS-CoV-2 infection: A systematic review. Pharmaceuticals 2021, 14, 736. [Google Scholar] [CrossRef] [PubMed]
- Janik, E.; Niemcewicz, M.; Podogrocki, M.; Saluk-Bijak, J.; Bijak, M. Existing drugs considered as promising in COVID-19 therapy. Int. J. Mol. Sci. 2021, 22, 5434. [Google Scholar] [CrossRef] [PubMed]
- Malhani, A.A.; Enani, M.A.; Sharif-Askari, F.S.; Alghareeb, M.R.; Bin-Brikan, R.T.; AlShahrani, S.A.; Halwani, R.; Tleyjeh, I.M. Combination of (interferon beta-1b, lopinavir/ritonavir and ribavirin) versus favipiravir in hospitalized patients with non-critical COVID-19: A cohort study. PLoS ONE 2021, 16, e0252984. [Google Scholar] [CrossRef] [PubMed]
- Jonsdottir, H.R.; Bielecki, M.; Siegrist, D.; Buehrer, T.W.; Züst, R.; Deuel, J.W. Titers of neutralizing antibodies against SARS-CoV-2 are independent of symptoms of non-severe COVID-19 in young adults. Viruses 2021, 13, 284. [Google Scholar] [CrossRef] [PubMed]
- Magro, P.; Zanella, I.; Pescarolo, M.; Castelli, F.; Quiros-Roldan, E. Lopinavir/ritonavir: Repurposing an old drug for HIV infection in COVID-19 treatment. Biomed. J. 2021, 44, 43–53. [Google Scholar] [CrossRef]
- Tampere, M.; Pettke, A.; Salata, C.; Wallner, O.; Koolmeister, T.; Cazares-Körner, A.; Visnes, T.; Hesselman, M.C.; Kunold, E.; Wiita, E.; et al. Novel broad-spectrum antiviral inhibitors targeting host factors essential for replication of pathogenic RNA viruses. Viruses 2020, 12, 1423. [Google Scholar] [CrossRef] [PubMed]
- Jeong, G.U.; Song, H.; Yoon, G.Y.; Kim, D.; Kwon, Y.-C. Therapeutic strategies against COVID-19 and structural characterization of SARS-CoV-2: A Review. Front. Microbiol. 2020, 11, 1723. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.D.; Han, I.; Choi, E.H.; Yadav, D.K. Recent advances in pathophysiology, drug development and future perspectives of SARS-CoV-2. Front. Cell Dev. Biol. 2020, 8, 580202. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.D. Assessment of antimicrobial activity of hundreds extract of twenty Indian medicinal plants. Biomed. Res. 2018, 29, 1797–1814. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Zhi, K.; Mukherji, A.; Gerth, K. Repurposing antiviral protease inhibitors using extracellular vesicles for potential therapy of COVID-19. Viruses 2020, 12, 486. [Google Scholar] [CrossRef]
- Xiao, L.; Sakagami, H.; Miwa, N. ACE2: The key molecule for understanding the pathophysiology of severe and critical conditions of COVID-19: Demon or Angel? Viruses 2020, 12, 491. [Google Scholar] [CrossRef] [PubMed]
- Amraei, R.; Rahimi, N. COVID-19, renin-angiotensin system and endothelial dysfunction. Cells 2020, 9, 1652. [Google Scholar] [CrossRef]
- Eze, P.; Mezue, K.N.; Nduka, C.U.; Obianyo, I.; Egbuche, O. Efficacy and safety of chloroquine and hydroxychloroquine for treatment of COVID-19 patients-a systematic review and meta-analysis of randomized controlled trials. Am. J. Cardiovasc. Dis. 2021, 11, 93–107. [Google Scholar]
- Huang, X.; Pearce, R.; Omenn, G.; Zhang, Y. Identification of 13 Guanidinobenzoyl- or Aminidinobenzoyl-containing drugs to potentially inhibit TMPRSS2 for COVID-19 treatment. Int. J. Mol. Sci. 2021, 22, 7060. [Google Scholar] [CrossRef]
- Hoffmann, M.; Hofmann-Winkler, H.; Smith, J.C.; Kruger, N.; Arora, P.; Sorensen, L.K.; Sogaard, O.S.; Hasselstrom, J.B.; Winkler, M.; Hempel, T.; et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine 2021, 65, 103255. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Shah, B.; Modi, P.; Sagar, S.R. In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sci. 2020, 252, 117652. [Google Scholar] [CrossRef]
- Zandi, K.; Amblard, F.; Musall, K.; Downs-Bowen, J.; Kleinbard, R.; Oo, A.; Cao, D.; Liang, B.; Russell, O.O.; McBrayer, T.; et al. Repurposing nucleoside analogs for human Coronaviruses. Antimicrob. Agents Chemother. 2020, 65, e01652-20. [Google Scholar] [CrossRef] [PubMed]
- Touret, F.; de Lamballerie, X. Of chloroquine and COVID-19. Antiv. Res. 2020, 177, 104762. [Google Scholar] [CrossRef]
- Ho, T.-C.; Wang, Y.-H.; Chen, Y.-L.; Tsai, W.-C.; Lee, C.-H.; Chuang, K.-P.; Chen, Y.-M.; Yuan, C.-H.; Ho, S.-Y.; Yang, M.-H.; et al. Chloroquine and hydroxychloroquine: Efficacy in the treatment of the COVID-19. Pathogens 2021, 10, 217. [Google Scholar] [CrossRef]
- Marcianò, G.; Roberti, R.; Palleria, C.; Mirra, D.; Rania, V.; Casarella, A.; De Sarro, G.; Gallelli, L. SARS-CoV-2 Treatment: Current therapeutic options and the pursuit of tailored therapy. Appl. Sci. 2021, 11, 7457. [Google Scholar] [CrossRef]
- Asselah, T.; Durantel, D.; Pasmant, E.; Lau, G.; Schinazi, R.F. COVID-19: Discovery, diagnostics and drug development. J. Hepatol. 2021, 74, 168–184. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [Green Version]
- Kneller, D.W.; Phillips, G.; O’Neill, H.M.; Jedrzejczak, R.; Stols, L.; Langan, P.; Joachmiak, A.; Coates, L.; Kovalevsky, A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020, 11, 3202. [Google Scholar] [CrossRef] [PubMed]
- Kumar, Y.; Singh, H.; Patel, C.N. In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation-based drug-repurposing. J. Infect. Public Health 2020, 13, 1210–1223. [Google Scholar] [CrossRef] [PubMed]
- Ianevski, A.; Yao, R.; Biza, S.; Zusinaite, E.; Mannik, A.; Kivi, G.; Planken, A.; Kurg, K.; Tombak, E.-M.; Ustav, M.; et al. Identification and tracking of antiviral drug combinations. Viruses 2020, 12, 1178. [Google Scholar] [CrossRef] [PubMed]
- Stepniowski, W.J.; Misiolek, W.Z. Review of fabrication methods, physical properties, and applications of nanostructured copper oxides formed via electrochemical oxidation. Nanomaterials 2018, 8, 379. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, J.P.F.; Silva, A.C.Q.; Silvestre, A.J.D.; Freire, C.S.R.; Vilela, C. Spherical Cellulose Micro and Nanoparticles: A Review of Recent Developments and Applications. Nanomaterials 2021, 11, 2744. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, S.K.; Bhardwaj, N.; Kumar, V.; Bhatt, D.; Azzouz, A.; Bhaumik, J.; Kim, K.-H.; Deep, A. Recent progress in nanomaterial-based sensing of airborne viral and bacterial pathogens. Environ. Int. 2021, 146, 106183. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.; Zhang, Y. Colloidal nanoparticle inks for printing functional devices: Emerging trends and future prospects. J. Mater. Chem. A 2019, 7, 23301–23336. [Google Scholar] [CrossRef]
- Zeng, M.; Chen, M.; Huang, D.; Lei, S.; Zhang, X.; Wang, L.; Cheng, Z. Engineered two-dimensional nanomaterials: An emerging paradigm for water purification and monitoring. Mater. Horiz. 2021, 8, 758–802. [Google Scholar] [CrossRef]
- De la Sota, P.G.; Lorente, E.; Notario, L.; Mir, C.; Zaragoza, O.; López, D. Mitoxantrone Shows in vitro, but not in vivo antiviral activity against human respiratory syncytial virus. Biomedicines 2021, 9, 1176. [Google Scholar] [CrossRef]
- Mosselhy, D.; Virtanen, J.; Kant, R.; He, W.; Elbahri, M.; Sironen, T. COVID-19 pandemic: What about the safety of Anti-Coronavirus nanoparticles? Nanomaterials 2021, 11, 796. [Google Scholar] [CrossRef]
- Sivasankarapillai, V.S.; Pillai, A.M.; Rahdar, A.; Sobha, A.P.; Das, S.S.; Mitropoulos, A.C.; Mokarrar, M.H.; Kyzas, G.Z. On facing the SARS-CoV-2 (COVID-19) with combination of nanomaterials and medicine: Possible strategies and first challenges. Nanomaterials 2020, 10, 852. [Google Scholar] [CrossRef] [PubMed]
- Tolksdorf, B.; Nie, C.; Niemeyer, D.; Röhrs, V.; Berg, J.; Lauster, D.; Adler, J.M.; Haag, R.; Trimpert, J.; Kaufer, B.; et al. Inhibition of SARS-CoV-2 replication by a small interfering RNA targeting the leader sequence. Viruses 2021, 13, 2030. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Ma, Q.; Chen, X.; Chen, T.; Ying, Y.; Xi, X.; Wang, L.; Ma, C.; Shaw, C.; Zhou, M. Recent advances and challenges in nanodelivery systems for antimicrobial peptides (AMPs). Antibiotics 2021, 10, 990. [Google Scholar] [CrossRef] [PubMed]
- Nasrollahzadeh, M.; Sajjadi, M.; Soufi, G.J.; Iravani, S.; Varma, R.S. Nanomaterials and nanotechnology-associated innovations against viral infections with a focus on Coronaviruses. Nanomaterials 2020, 10, 1072. [Google Scholar] [CrossRef] [PubMed]
- Perrella, F.; Coppola, F.; Petrone, A.; Platella, C.; Montesarchio, D.; Stringaro, A.; Ravagnan, G.; Fuggetta, M.; Rega, N.; Musumeci, D. Interference of Polydatin/Resveratrol in the ACE2: Spike recognition during COVID-19 infection. A focus on their potential mechanism of action through computational and biochemical assays. Biomolecules 2021, 11, 1048. [Google Scholar] [CrossRef]
- Cordaro, A.; Neri, G.; Sciortino, M.T.; Scala, A.; Piperno, A. Graphene-based strategies in liquid biopsy and in viral diseases diagnosis. Nanomaterials 2020, 10, 1014. [Google Scholar] [CrossRef]
- Kassirian, S.; Taneja, R.; Mehta, S. Diagnosis and management of acute respiratory distress syndrome in a time of COVID-19. Diagnostics 2020, 10, 1053. [Google Scholar] [CrossRef]
- Begines, B.; Ortiz, T.; Pérez-Aranda, M.; Martínez, G.; Merinero, M.; Argüelles-Arias, F.; Alcudia, A. Polymeric nanoparticles for drug delivery: Recent developments and future prospects. Nanomaterials 2020, 10, 1403. [Google Scholar] [CrossRef]
- Rasmi, Y.; Saloua, K.; Nemati, M.; Choi, J. Recent progress in nanotechnology for COVID-19 prevention, diagnostics and treatment. Nanomaterials 2021, 11, 1788. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Sahu, R.; Singh, D.D.; Egbo, T.E. A CRISPR/Cas9 based polymeric nanoparticles to treat/inhibit microbial infections. Semin. Cell Dev. Biol. 2019, 96, 44–52. [Google Scholar] [CrossRef] [PubMed]
| Antiviral Agent | Drug Target | Mechanism of Action | Infectious Disease | References |
|---|---|---|---|---|
| Remdesivir | RdRp | Terminates the non-obligate chain | SARS-CoV-2, MERS-CoV, SARS-CoV | [20] |
| Favipiravir | RdRp | Inhibits RdRp | SARS-CoV-2, Influenza | [21] |
| siRNA | RdRp | Short chains of dsRNA that interfere | SARS-CoV, MERS-CoVWu | [22] |
| Galidesivir | RdRp | Inhibits viral RNA polymerase function by | Galidesivir SARS-CoV-2, | [23] |
| Ribavirin | RdRp | Inhibits viral RNA synthesis and mRNA capping | SARS-CoV-2, MERS-CoV, SARS-CoV, | [24] |
| LJ001 and JL103 | Lipid membrane | Membrane-binding photosensitizers that induce | Enveloped viruses (IAV, filoviruses, poxviruses, arenaviruses, bunyaviruses, paramyxoviruses, flaviviruses and HIV-1) | [25] |
| CR3022 | Spike glycoprotein | Immunogenic antigen against Spike protein | SARS-CoV-2, SARS-CoV | [26] |
| Griffithsin | Spike glycoprotein | Griffithsin binds to the SARSCoV-2 spike | SARS-CoV-2 | [27] |
| Peptide (P9) | Spike glycoprotein | Inhibits spike protein-mediated cell-cell entry or | Broad-spectrum (SARS-CoV, MERS-CoV, influenza) | [28] |
| Nafamostat | Spike glycoprotein | Inhibits spike-mediated membrane fusion A | SARS-CoV-2, MERS-CoV | [29] |
| Ritonavir | 3CLpro | Inhibits 3CLpro | SARS-CoV-2, MERS-CoV | [30] |
| Lopinavir | 3CLpro | Inhibits 3CLpro | SARS-CoV-2, MERS-CoV, SARS-CoV, HCoV-229E, HIV, HPV | [31] |
| Darunavir and cobicistat | 3CLpro | Inhibits 3CLpro | SARS-CoV-2 | [32] |
| Antiviral Agent | Drug Target | Mechanism of Action | Infectious Disease | References |
|---|---|---|---|---|
| Baricitinib | Clathrin-mediated endocytosis | Baricitinib | Clathrin-mediated endocytosis | [34] |
| Chloroquine | Endosomal acidification | A lysosomatropic base that appears to disrupt intracellular trafficking and viral fusion events | SARS-CoV-2, SARS-CoV, MERS-CoV | [33] |
| Convalescent plasma | - | Inhibits virus entry to the target cells | SARS-CoV, SARS-CoV-2, Influenza | [35,36] |
| Camostat Mesylate | Surface protease | Potent serine protease inhibitor | SARS-CoV, MERS-CoV, HcoV-229E | [33] |
| Corticosteroids | Pulsed methylprednisolone | Patients with severe MERS who were treated with systemic corticosteroid with or without antivirals and interferons had no favorable response | SARS-CoV, MERS-CoVL | [35] |
| Nitazoxanide | Interferon response | Induces the host innate immune response | Coronaviruses, SARS-CoV-2 | [19] |
| Recombinant interferons | Interferon response | Exogenous interferons | SARS-CoV-2, SARS-CoV, MERS-CoV | [37] |
| S.N. | Antibody Name | Antibody Type | Origin | PDB ID | Epitopes | Neutralizing Mechanism | Cross Neutralizing Activity | Protective Efficacy | Ref |
|---|---|---|---|---|---|---|---|---|---|
| 1 | CV30 | Human IgG | Infected COVID-19 patients | 6XE1 | D420-Y421, Y453, L455-N460, Y473-S477, F486-N487, Y489, Q493, T500, G502, Y505 | Block hACE2-RBD interaction | no | IC50 value of 0.03 µg/mL | [35] |
| 2 | REGN10933 Recombinant | full-human antibodies | Humanized mice and COVID-19-convalescent patients | 6XDG | R403, K417, Y421, Y453, L455-F456, A475-G476, E484-Y489, Q493 | Block hACE2-RBD interaction, ADCC & ADCP | no | IC50 value of 37.4 pM | |
| 3 | B38 | Human IgG | COVID-19-convalescent patient | 7BZ5 | R403, D405-E406, Q409, D420-Y421, Y452, L454-N460, Y473-S477, F486-N487, Y489-F490, Q493-G496, Q498, T500-V503, Y505 | Block hACE2-RBD interaction | no | A single dose of B38 (25 mg/kg) | [35] |
| 4 | CC12.1 | Human IgG | COVID-19-convalescent patient | 6XC3 | R403, D405-E406, R408-Q409, D420-Y421, Y453, L455-N460, Y473-S477, F486-N487, Y489, Q493-G496, Q498, T500-V503, Y505 | Block hACE2-RBD interaction | no | IC50 value of 0.019 µg/mL | [36] |
| 5 | CB6 | Human IgG | COVID-19-convalescent patient | 7C01 | R403, D405-E406, R408-Q409, D420-Y421, L455-N460, Y473-S477, F486-N487, Y489, Q493, Y495, N501-G502, G504-Y505 | Block hACE2-RBD interaction | no | A single dose of CB6-LALA (50 mg/kg) | [37] |
| 6 | C105 | Human IgG | COVID-19-convalescent patient | 6XCN, 6XCM | R403, D405, R408, D420-Y421, Y453, L455-N460, Y473, A475-G476, F486-N487, G502, Y505 | Block hACE2-RBD interaction | no | IC50 value of 26.1 ng/mL | [41] |
| 7 | CC12.3 | Human IgG | COVID-19-convalescent patient | 6XC7 | R403, D405, D420-Y421, Y453, L455-N460, Y473-S477, F486-N487, Y489, Q493, G496, N501, Y505 | Block hACE2-RBD interaction | no | IC50 value of 0.018 µg/mL | [42] |
| 8 | CR3022 | Human IgG | SARS-convalescent patient | 6YOR, 6 W41 | Y369-N370, F374-K386, L390, F392, D428, T430, F515-L517 | Trapping RBD in the less stable up conformation while leading to destabilization of S | SARS-CoV, SARS-CoV-2 | ND50 value of 0.114 µg/mL | [19] |
| 9 | EY6A | Human IgG | Late-stage COVID-19 patient | 6ZDH, 6ZER, 6ZCZ | Y369, F374-S375, F377-K386, N388, L390, P412-G413, D427-F429, L517 | destabilization of S | SARS-CoV, SARS-CoV-2 | ND50 value of ~10.8 µg/mL | [26] |
| 10 | VHH-72 | Llama single domain antibody | llama immunized with prefusionstabilized betacoronavirus spikes | 6WAQ | Y356-T359, F361-C366, A371-T372, G391-D392, R395, N424, I489, Y494 | Trapping RBD in the less stable up conformation while leading to destabilization of S, Block hACE2_RBD interaction | SARS-CoV, SARS-Co-V-2 | IC50 values of 0.14 µg/mL and 0.2 mg/mL. | [19] |
| 11 | BD23 | Human IgG | COVID-19-convalescent patient | 7BYR | G446, Y449, L452, T470, E484-F486, Y489-F490, L492-S494, G496, Q498, T500-N501, Y505 | Block hACE-RBD2 interaction | no | IC50 value of 8.5 µg/mL | [26] |
| 12 | Fab 2–4 | Human IgG | Infected COVID-19 patients | 6XEY | Y449, Y453, L455-F456, E484-F486, Y489-F490, L492-S494, G496 | Locking RBD in the down conformation while occluding access to ACE2 | no | Neutralizing SARS-CoV-2 live virus with IC50 value of 0.057 µg/mL | [41] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, D.K.; Singh, D.D.; Han, I.; Kumar, Y.; Choi, E.-H. Current Potential Therapeutic Approaches against SARS-CoV-2: A Review. Biomedicines 2021, 9, 1620. https://doi.org/10.3390/biomedicines9111620
Yadav DK, Singh DD, Han I, Kumar Y, Choi E-H. Current Potential Therapeutic Approaches against SARS-CoV-2: A Review. Biomedicines. 2021; 9(11):1620. https://doi.org/10.3390/biomedicines9111620
Chicago/Turabian StyleYadav, Dharmendra Kumar, Desh Deepak Singh, Ihn Han, Yogesh Kumar, and Eun-Ha Choi. 2021. "Current Potential Therapeutic Approaches against SARS-CoV-2: A Review" Biomedicines 9, no. 11: 1620. https://doi.org/10.3390/biomedicines9111620
APA StyleYadav, D. K., Singh, D. D., Han, I., Kumar, Y., & Choi, E.-H. (2021). Current Potential Therapeutic Approaches against SARS-CoV-2: A Review. Biomedicines, 9(11), 1620. https://doi.org/10.3390/biomedicines9111620









