Chick Embryo Experimental Platform for Micrometastases Research in a 3D Tissue Engineering Model: Cancer Biology, Drug Development, and Nanotechnology Applications
Abstract
1. Introduction
2. Materials and Methods
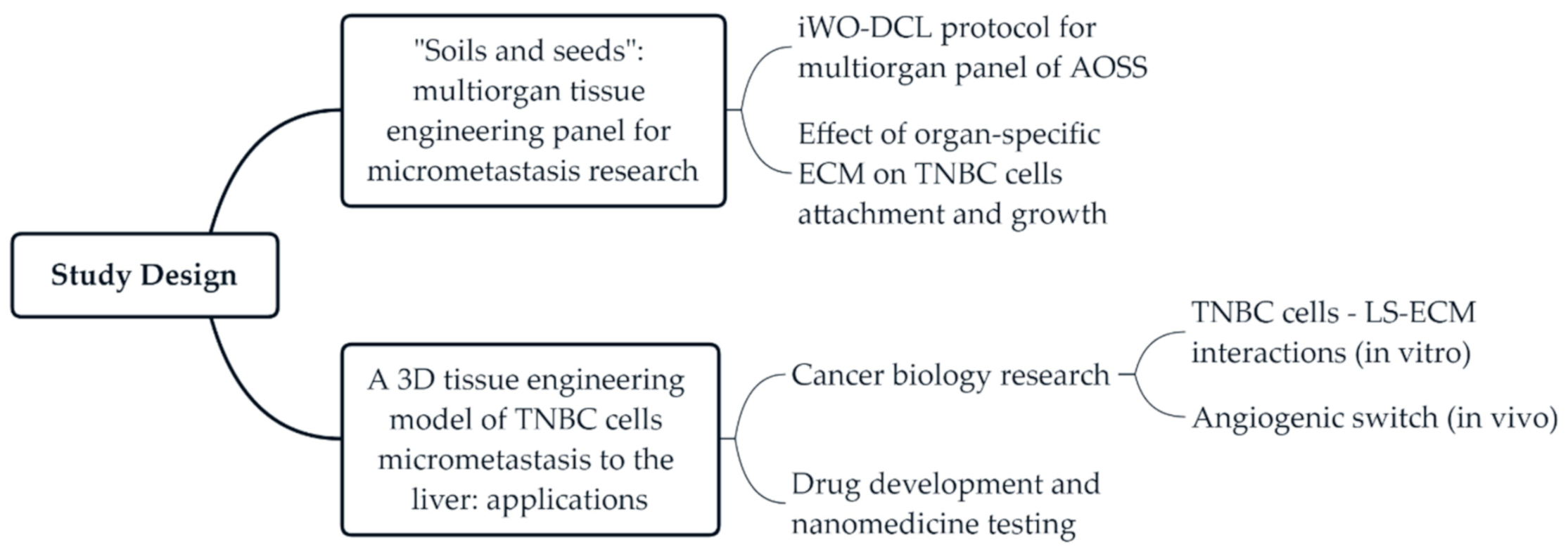
2.1. Study Design
2.2. Chick Embryo Incubation and Organ Collection
2.3. Immersion Whole-Organ Decellularization (iWO-DCL)
2.4. Cell Culture
2.5. Recellularization of CE Acellular Organ-Sepcific Scaffolds (AOSSs) with MDA-MB- 231 Cells
2.6. Analysis of the Structural Evolution of TECs
2.6.1. Histology, Histomorphometry, and Fluorescence Microscopy
2.6.2. Scanning Electron Microscopy
2.6.3. Atomic Force Microscopy
2.6.4. Image Analysis for Histological Morphometry
2.6.5. MTT Assay for Evaluation of the Population Evolution of MDA-MB-231 Cells in 2D and 3D TECs
2.6.6. Modeling Cell Growth Dynamics
2.7. Evaluation of Angiogenic Potential of Liver TNBC TECs in Vivo (CAM Assay)
2.7.1. Grafting and Imaging Procedures
2.7.2. Angiogenesis Quantification
2.8. Cytotoxicity and Cellular Uptake of Doxorubicin (Dox) and Mesoporous Silica Nanoparticles Loaded with Dox in 3D Liver TNBC TECs and in Matching 2D Cultures of TNBC Cells
2.8.1. Preparation of Mesoporous Silica Nanoparticles and Their Loading with Dox
2.8.2. Characterization of Mesoporous Silica Nanoparticles
2.8.3. MTT Viability Assay of 2D and 3D TEC Cell Cultures Treated with Free and Nanoformulated Dox
2.8.4. Pharmacodynamics Modeling
2.8.5. Confocal Microscopy Study of the Uptake of Free and Nanoformulated Dox in 3D TECs and 2D Cultures
2.9. Statistical Analysis
3. Results
3.1. Soils and Seeds: Development of a Chick Embryo-Based Multiorgan Tissue Engineering Platform for Modeling Cancer Micrometastases
3.2. Three-Dimensional Tissue-Engineered Model of TNBC Micrometastatsis to the Liver for Cancer Biology Research
3.2.1. IWO-DCL Reveals Compartmental Organization of LS-ECM
3.2.2. TNBC Cells Differentially Colonize Parenchymal and Stromal LS-ECM, While ECMs Stimulate Phenotypic Plasticity of the Cells
3.2.3. Cell Growth Dynamics in 3D TECs Is Different vs. 2D Cultures
3.2.4. 3D TECs Are Angiogenic In Vivo
3.3. The 3D TNBC-Liver TECs as Testbeds for Drug and Nanoparticles Testing
3.3.1. TNBC Cells in 3D TECs Are Less Sensitive to the Cytotoxic Action of Dox and AMS-6-Dox Than in 2D Cultures of MDA-MB-231 Cells
3.3.2. Uptake of Dox and AMS-6-Dox in 3D TECs and 2D Cultures of MDA-MB-231 Cells
4. Discussion
4.1. Methodological Advancements
4.2. Cancer Biology Findings
4.2.1. Organ-Specific ECMs Differentially Modulate TNBC Cell Attachment and Growth
4.2.2. Liver ECMs as a Component of the Organ-Specific Metastatic Niche
4.2.3. Colonization of the Liver ECM by TNBC Cells
4.2.4. MDA-MB-231 Cell Growth in 2D Monolayer and 3D TEC Cultures in Vitro
4.2.5. Results of the CAM Assay
4.3. Feasibility of 3D Engineered Models of Cancer Micrometastases for the Testing of Drugs and Nanoparticles
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Design of the Study on the Feasibility of the 3D Tissue Engineered Model of TNBC Micrometastases to the Liver

Appendix A.2. Decellularization of Multiple CE Organs
| Organs | Average DCL Time, h | Minimal Washing Time, h | Recommended Shaking Speed during DCL3, rpm |
|---|---|---|---|
| Brain 1 | ~48 | 48 | 100 (first 24 h), 90 (second 24 h) |
| Heart | ~180–200 | ~48–60 | 150 (first 24 h), 120 (second 24 h) |
| Lungs | ~48 | 48 | 100 (first 24 h), 90 (second 24 h) |
| Liver | ~240–280 | 48 | 120 (first 5–6 days), 50 (the rest of the time) |
| SI 2 | ~48 | 48 | 150 (first 24 h), 120 (second 24 h) |
| PV 2 | ~90–120 | 48 | 150 (first 24 h), 120 (the rest of the time) |
| Ve 2 | ~250–280 | ~48–60 | 150 (first 24 h), 120 (the rest of the time) |
| Spleen | ~36–48 | 48 | 150 (first 24 h), 120 (the rest of the time) |
| BMu2 | ~90–120 | 48 | 150 (first 24 h), 120 (the rest of the time) |
| Skin | ~90–120 | 48 | 150 (first 24 h), 120 (the rest of the time) |
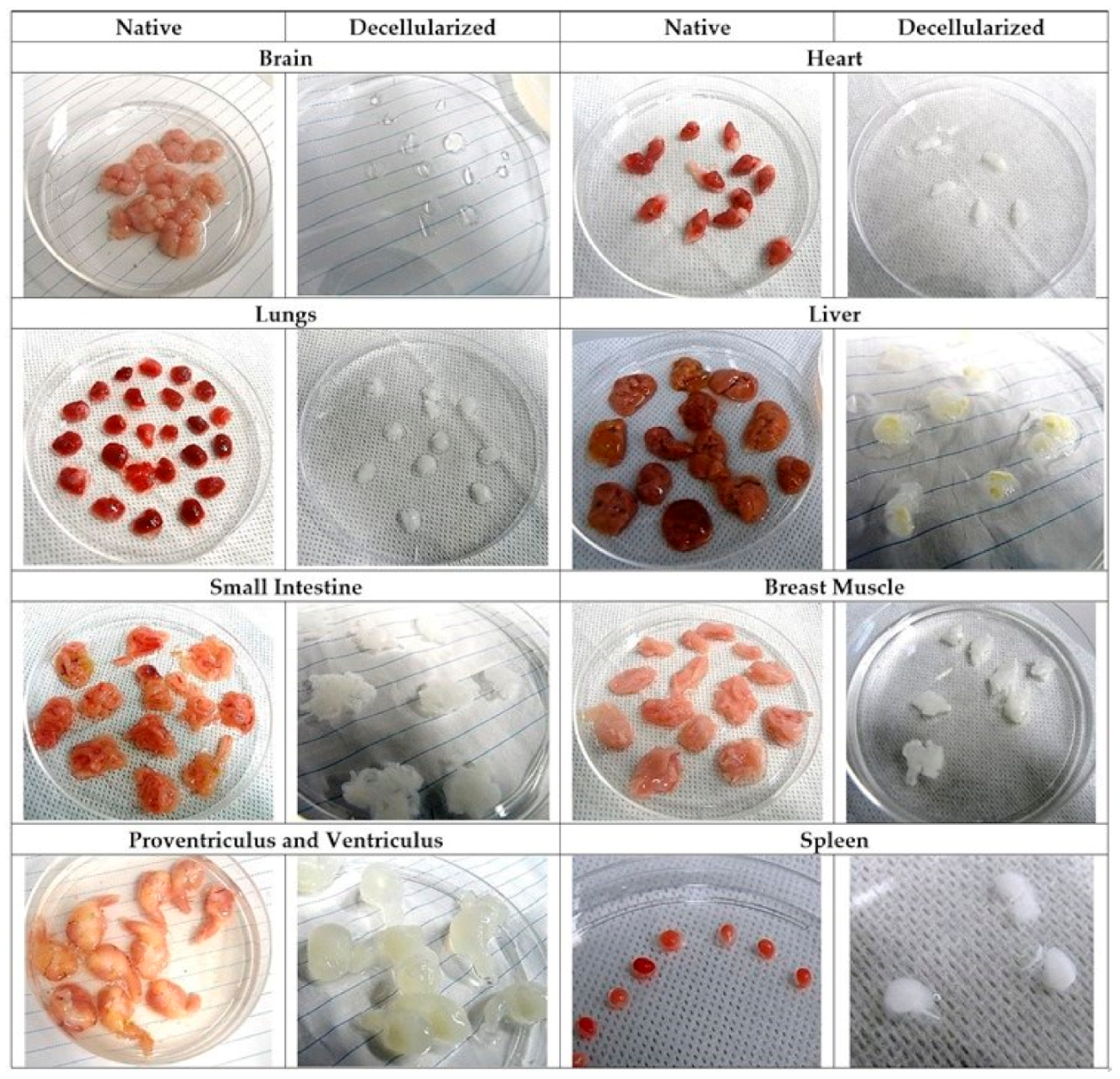
Appendix A.3. Development and Feasibility Testing of 3D Tissue Engineering Model of TNBC Micrometastases to the Liver
Appendix A.3.1. Native Structure of Chick Embryo Liver Tissue and the Effect of Decellularization
Appendix A.3.2. Effect of iWO-CL on CE Livers



Appendix A.3.3. Structural and Morphometrical Analysis of 3D TECs


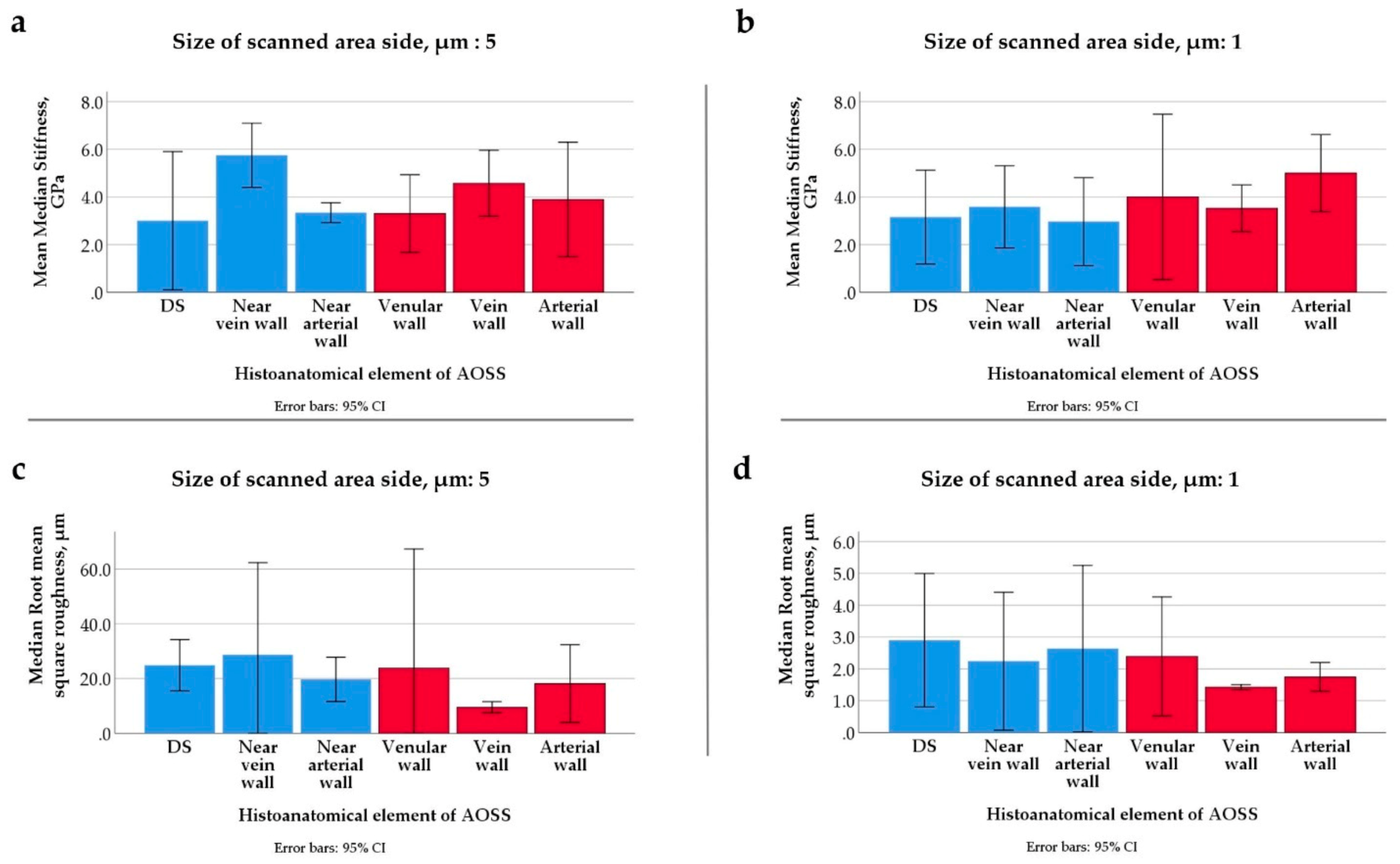

| Sampling Time Point | Compartment | ||
|---|---|---|---|
| Parenchymal | Stromal | Mixed | |
| Week 1 | 5.1 (±6.9) | 16.3 (±15.2) | - |
| Week 2 | 6.8 (±6.5) | 17.3 (±9.4) | 5.5 (±3.4) |
| Week 3 | 10.0 (±3.0) | 37.7 (±21.0) | 10.6 (±7.8) |
| Week 4 | 1.8 (±1.4) | 1.2 (±NA) | 5.6 (±NA) |
| Parameters | 2D | 3D | ||
|---|---|---|---|---|
| Estimated Value | CI95% | Estimated Value | CI95% | |
| C0 | 0.03181 | −0.2501, 0.3137 | 0.006906 | −0.01293, 0.02675 |
| Cmax | 0.8891 | 0.6392, 1.139 | 0.1524 | 0.1107, 0.1941 |
| d | 0.6325 | −0.7616, 2.027 | 0.2826 | −0.005646, 0.5708 |
| Characteristics of Fitting | 2D | 3D |
|---|---|---|
| SSE | 0.01974 | 0.0002222 |
| R-squared | 0.9634 | 0.9864 |
| Adjusted R-squared | 0.9267 | 0.9729 |
| RMSE | 0.09935 | 0.01054 |

Appendix A.3.4. Angiogenic Assay on Chick Embryo CAMs
Appendix A.3.4.1. Egg Preparation and Grafting Procedure for Angiogenic Analysis
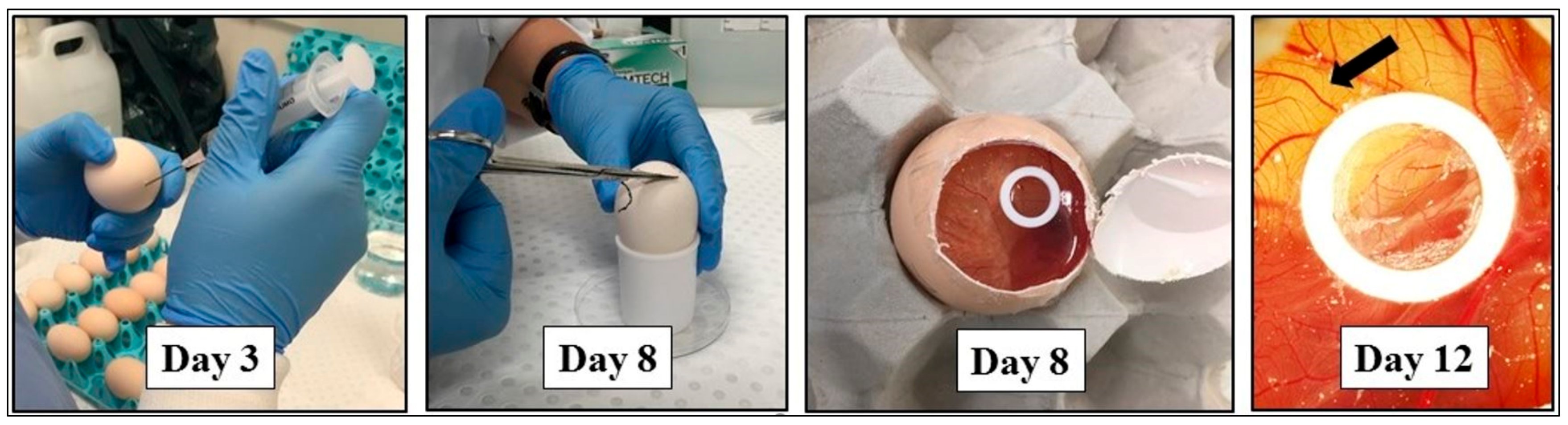
Appendix A.3.4.2. Imaging of CAM In Vivo
Appendix A.3.4.3. Quantification of Angiogenesis
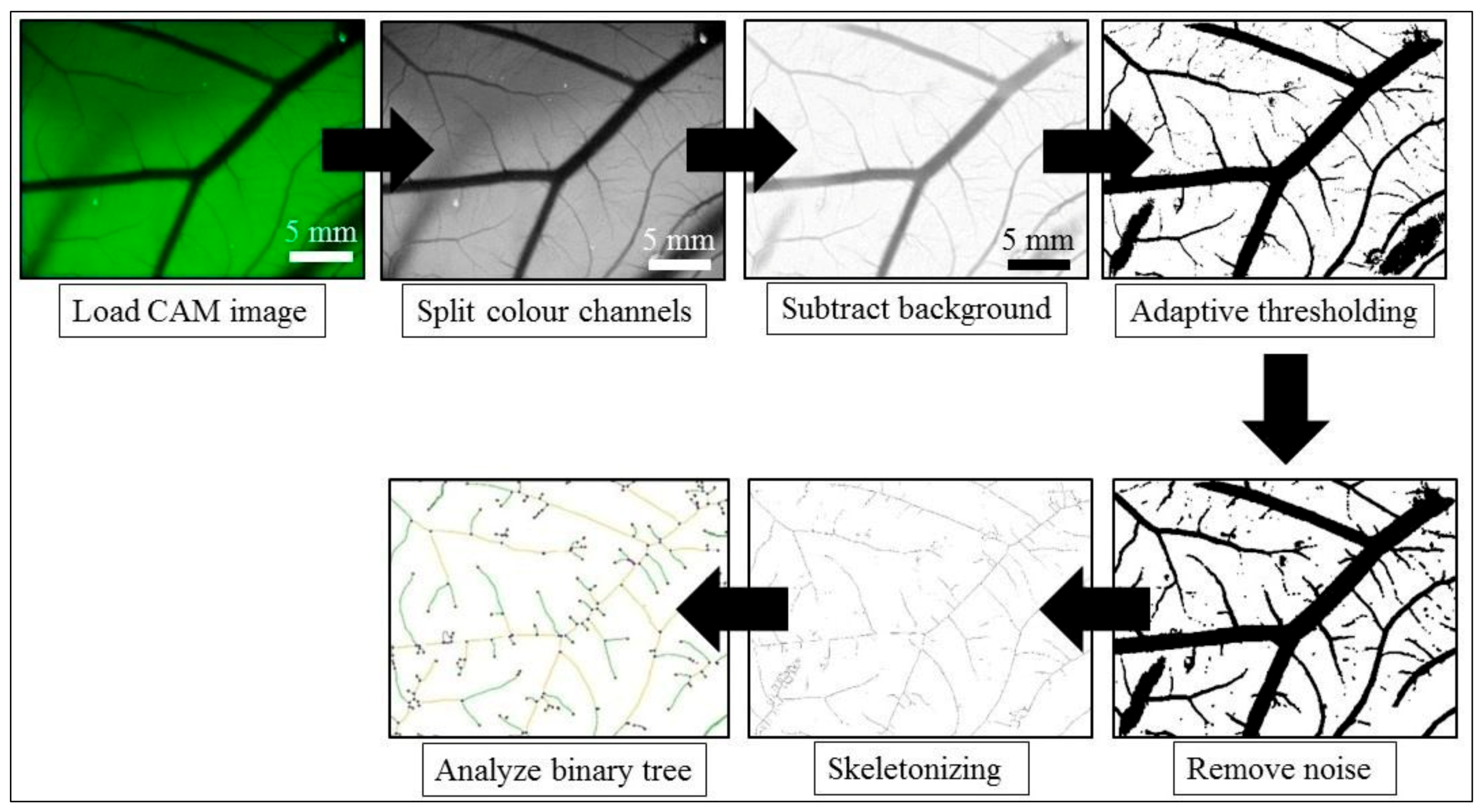


Appendix A.3.5. 3D TECs of TNBC Micrometastases to the Liver as Drug and Nanomedicine Testing Platform
Appendix A.3.5.1. Characterization of Nanoparticles

| Sample | Temperature, °C | pH | Polydispersity Index ((Dw/Dm)2) | Zeta-Potential, mV |
|---|---|---|---|---|
| Pure AMS-6 in PBS | 25 | 7.2 | 0.544 | −10.9 |
| Pure AMS-6 in CCM | 25 | 7.4 | 0.685 | −8.3 |
| AMS-6-Dox in PBS | 25 | 7.2 | 1.000 | −11.2 |
| AMS-6-Dox in CCM | 25 | 7.4 | 1.000 | −8.8 |
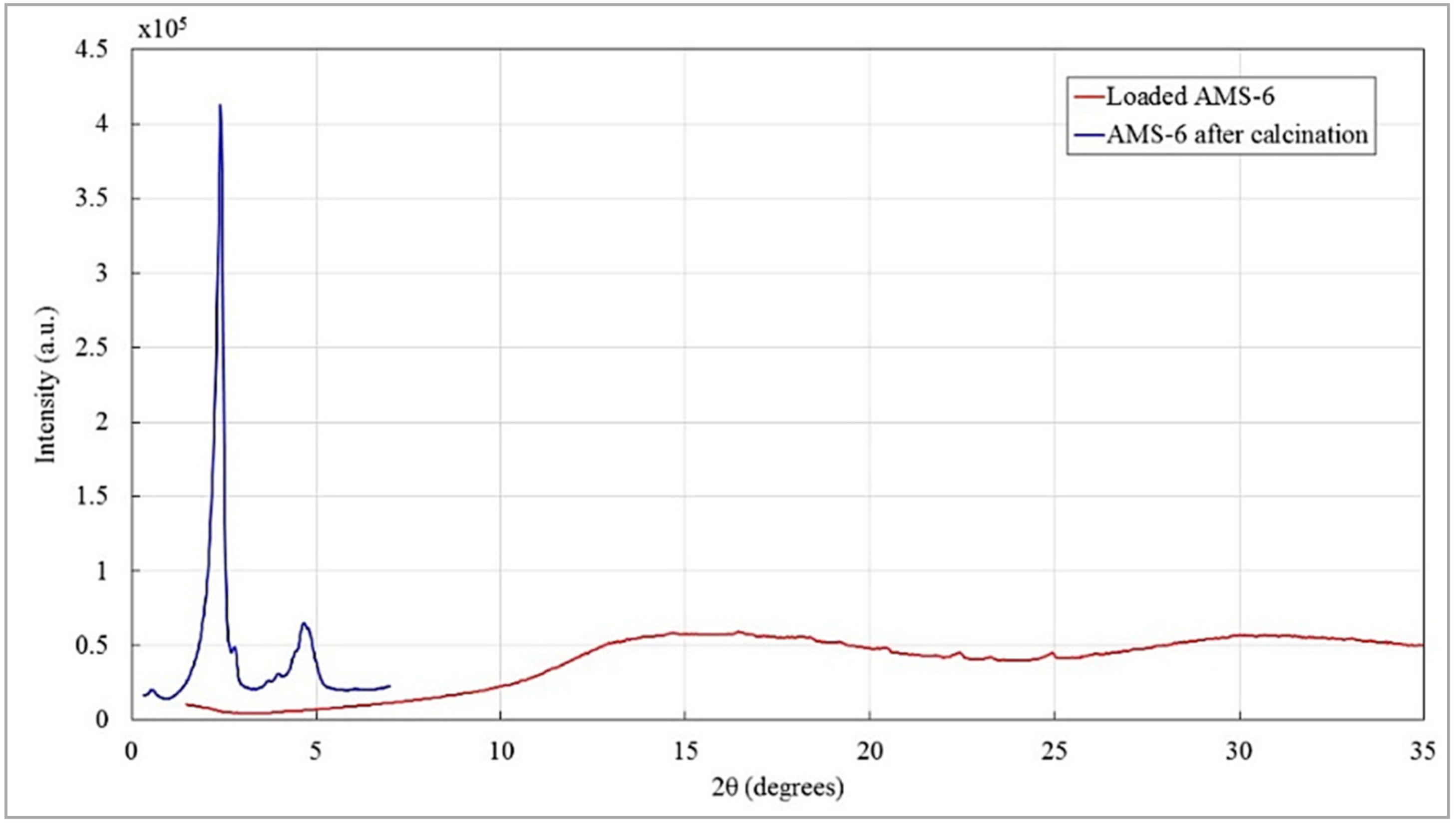



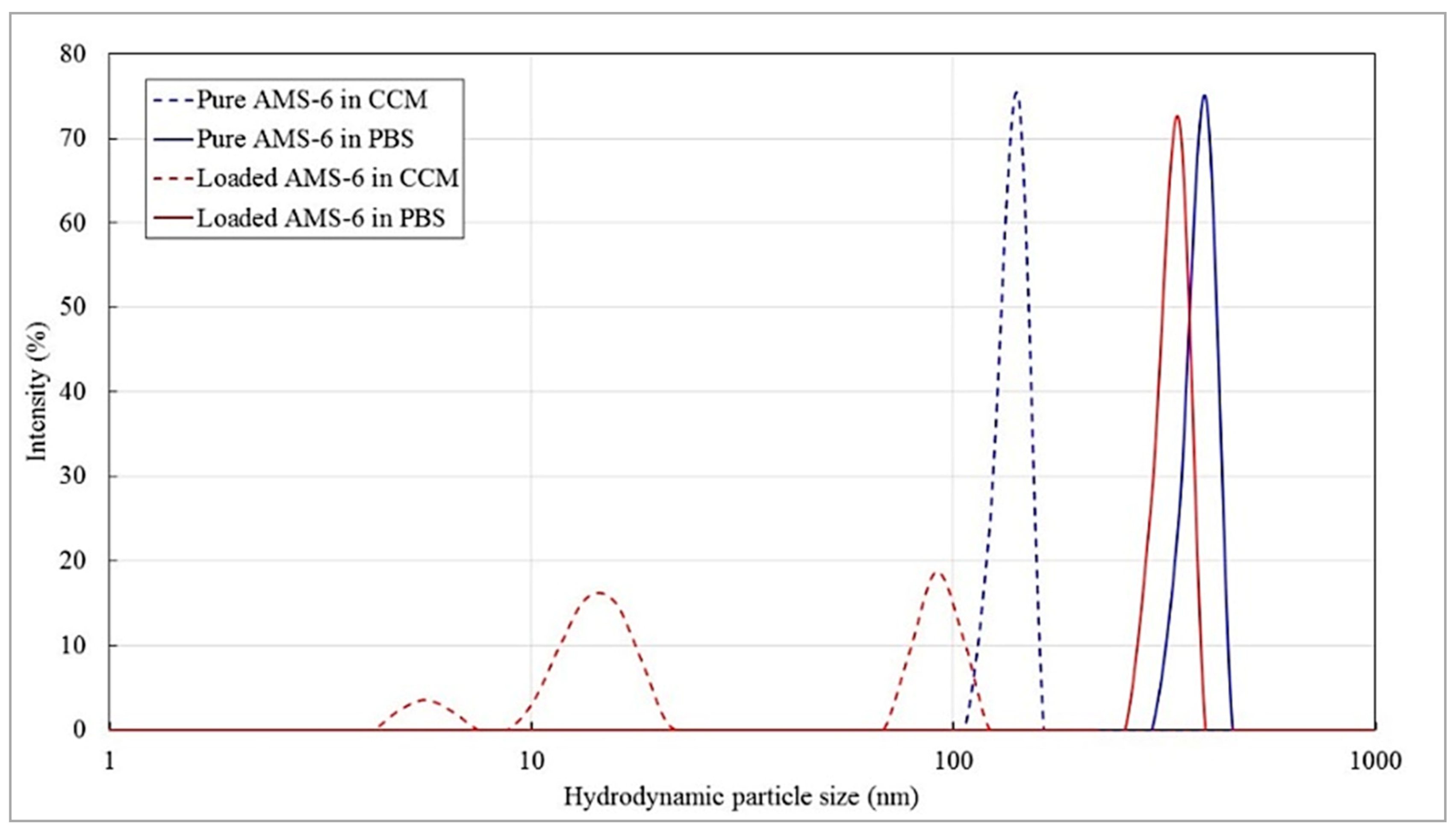
Appendix A.3.5.2. Effect of Free and Nanoformulated Doxorubicin on MDA-MB-231 Cells in 2D and 3D (TECs) Cultures
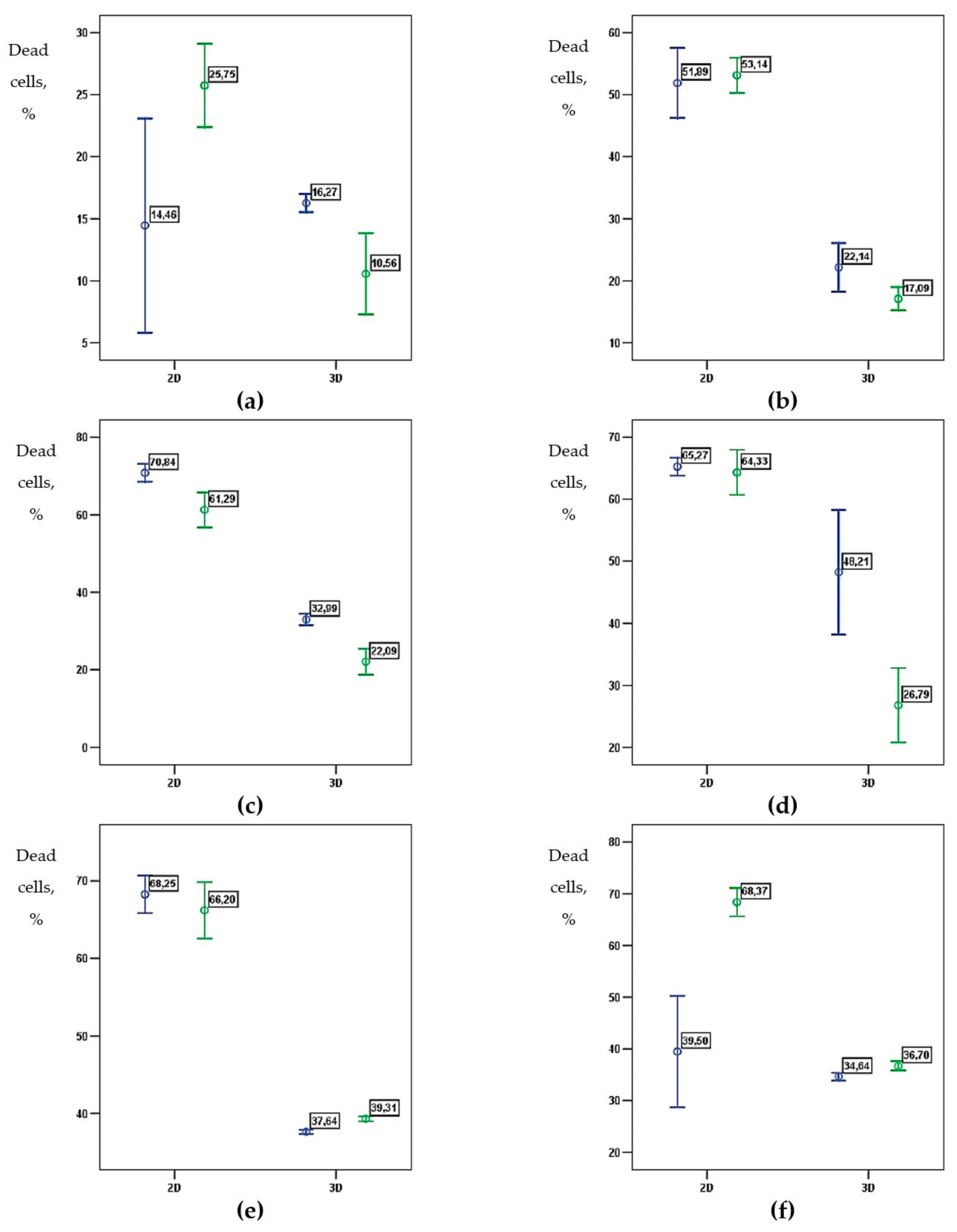
| 2D | 3D | |||
|---|---|---|---|---|
| Estimated Value | Standard Error | Estimated Value | Standard Error | |
| Maximum effect, % | 64.85 | 2.83 | 34.34 | 2.71 |
| EC50, µg/mL | 0.27 | 0.09 | 2.40 | 0.41 |
| Hill coefficient | 3.20 | 1.27 | 0.68 | 0.39 |
| IC50, µg/mL | 0.43 | 0.12 | >10 | |
| 2D | 3D | |||
|---|---|---|---|---|
| Estimated Value | Standard Error | Estimated Value | Standard Error | |
| Maximum effect, % | 63.06 | 1.97 | 40.53 | 5.47 |
| EC50, µg/mL | 0.26 | 0.06 | 2.38 | 0.86 |
| Hill coefficient | 2.81 | 0.80 | 0.34 | 0.18 |
| IC50, µg/mL | 0.46 | 0.10 | >10 | |
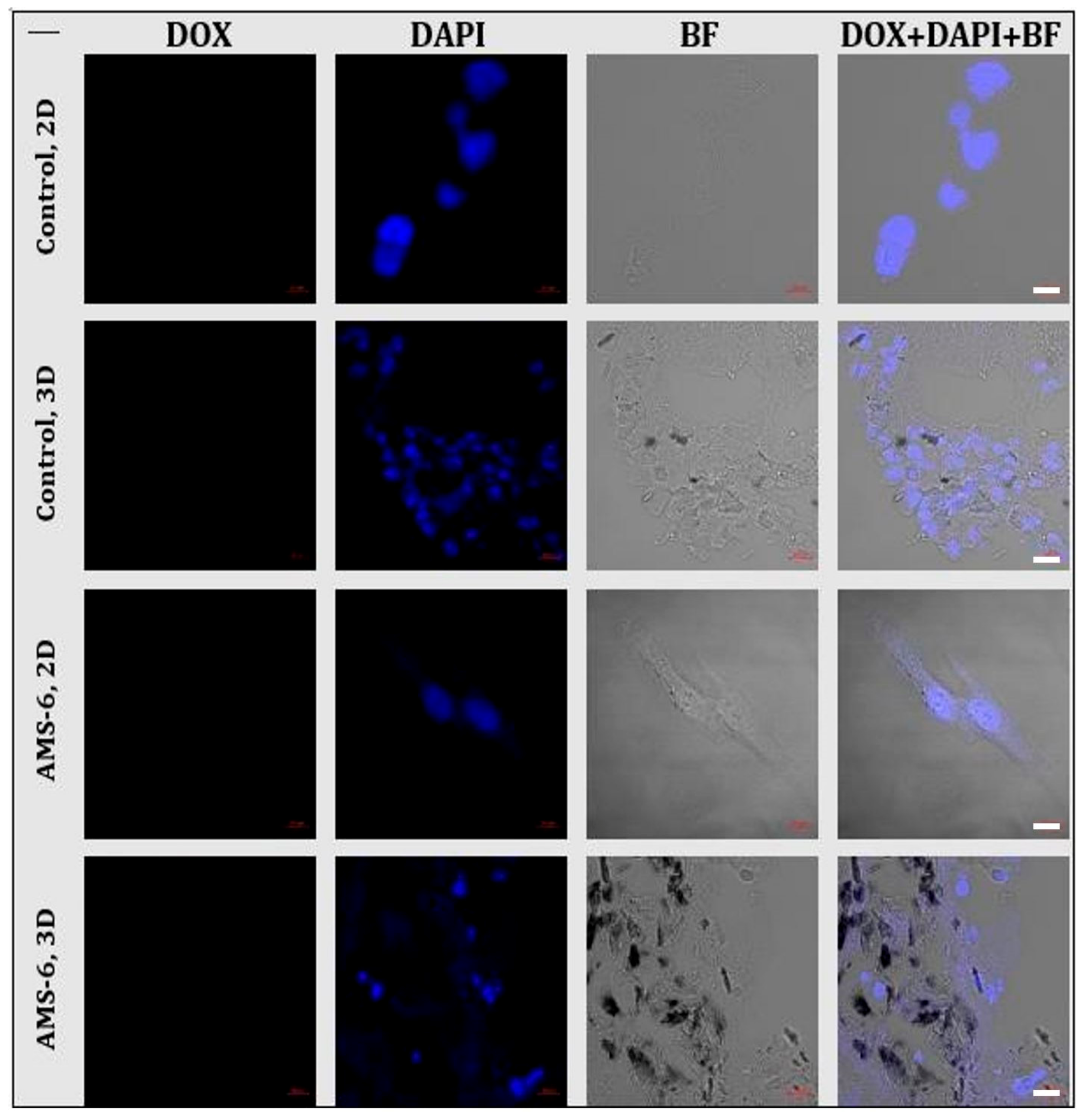

AAppendix A.4. Future Development of the AOSS-Based Models

References
- Valastyan, S.; Weinberg, R.A. Tumor metastasis: Molecular insights and evolving paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Dillekas, H.; Rogers, M.S.; Straume, O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019, 8, 5574–5576. [Google Scholar] [CrossRef]
- Riggio, A.I.; Varley, K.E.; Welm, A.L. The lingering mysteries of metastatic recurrence in breast cancer. Br. J. Cancer 2021, 124, 13–26. [Google Scholar] [CrossRef]
- Brierley, J.D.; Gospodarowicz, M.K.; Wittekind, C. (Eds.) TNM Classification of Malignant Tumours, 8th ed.; Wiley: Oxford, UK, 2017; p. 272. [Google Scholar]
- Baeriswyl, V.; Christofori, G. The angiogenic switch in carcinogenesis. Semin. Cancer Biol. 2009, 19, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.X.; Bos, P.D.; Massague, J. Metastasis: From dissemination to organ-specific colonization. Nat. Rev. Cancer 2009, 9, 274–284. [Google Scholar] [CrossRef]
- Hanahan, D.; Folkman, J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell 1996, 86, 353–364. [Google Scholar] [CrossRef]
- Holmgren, L.; O’Reilly, M.S.; Folkman, J. Dormancy of micrometastases: Balanced proliferation and apoptosis in the presence of angiogenesis suppression. Nat. Med. 1995, 1, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Hoffmann, A.D.; Liu, H.; Liu, X. Organotropism: New insights into molecular mechanisms of breast cancer metastasis. NPJ Precis. Oncol. 2018, 2, 4. [Google Scholar] [CrossRef]
- Nan, X.; Wang, J.; Liu, H.N.; Wong, S.T.C.; Zhao, H. Epithelial-Mesenchymal Plasticity in Organotropism Metastasis and Tumor Immune Escape. J. Clin. Med. 2019, 8, 747. [Google Scholar] [CrossRef]
- Mathot, L.; Stenninger, J. Behavior of seeds and soil in the mechanism of metastasis: A deeper understanding. Cancer Sci. 2012, 103, 626–631. [Google Scholar] [CrossRef]
- Chambers, A.F.; Groom, A.C.; MacDonald, I.C. Metastasis: Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2002, 2, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Nicolson, G.L. Organ specificity of tumor metastasis: Role of preferential adhesion, invasion and growth of malignant cells at specific secondary sites. Cancer Metastasis Rev. 1988, 7, 143–188. [Google Scholar] [CrossRef]
- Obenauf, A.C.; Massague, J. Surviving at a Distance: Organ-Specific Metastasis. Trends Cancer 2015, 1, 76–91. [Google Scholar] [CrossRef]
- Wei, S.; Siegal, G.P. Surviving at a distant site: The organotropism of metastatic breast cancer. Semin. Diagn. Pathol. 2018, 35, 108–111. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabieres, C.; Riethdorf, S. Cancer micrometastases. Nat. Rev. Clin. Oncol. 2009, 6, 339–351. [Google Scholar] [CrossRef]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Qutaish, M.; Han, Z.; Schur, R.M.; Liu, Y.; Wilson, D.L.; Lu, Z.R. MRI detection of breast cancer micrometastases with a fibronectin-targeting contrast agent. Nat. Commun. 2015, 6, 7984. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhu, X.; Cui, K.; Mancuso, J.; Federley, R.; Fisscher, K.; Teng, G.; Mittal, V.; Gao, D.; Zhao, H.; et al. In Vivo Visualization and Characterization of Epithelial-Mesenchymal Transition in Breast Tumors. Cancer Res. 2016, 76, 2094–2104. [Google Scholar] [CrossRef]
- Massague, J.; Obenauf, A.C. Metastatic colonization by circulating tumour cells. Nature 2016, 529, 298–306. [Google Scholar] [CrossRef]
- Rafaeva, M.; Erler, J.T. Framing cancer progression: Influence of the organ- and tumour-specific matrisome. FEBS J. 2020, 287, 1454–1477. [Google Scholar] [CrossRef] [PubMed]
- Cox, T.R. The matrix in cancer. Nat. Rev. Cancer 2021, 21, 217–238. [Google Scholar] [CrossRef]
- Gkretsi, V.; Stylianopoulos, T. Cell Adhesion and Matrix Stiffness: Coordinating Cancer Cell Invasion and Metastasis. Front. Oncol. 2018, 8, 145. [Google Scholar] [CrossRef]
- Pathak, A.; Kumar, S. Independent regulation of tumor cell migration by matrix stiffness and confinement. Proc. Natl. Acad. Sci. USA 2012, 109, 10334–10339. [Google Scholar] [CrossRef] [PubMed]
- Guzman, A.; Ziperstein, M.J.; Kaufman, L.J. The effect of fibrillar matrix architecture on tumor cell invasion of physically challenging environments. Biomaterials 2014, 35, 6954–6963. [Google Scholar] [CrossRef]
- Choudhury, D.; Tun, H.W.; Wang, T.; Naing, M.W. Organ-Derived Decellularized Extracellular Matrix: A Game Changer for Bioink Manufacturing? Trends Biotechnol. 2018, 36, 787–805. [Google Scholar] [CrossRef]
- Naba, A.; Clauser, K.R.; Hoersch, S.; Liu, H.; Carr, S.A.; Hynes, R.O. The matrisome: In silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell. Proteom. 2012, 11, M111.014647. [Google Scholar] [CrossRef]
- Guimarães, C.F.; Gasperini, L.; Marques, A.P.; Reis, R.L. The stiffness of living tissues and its implications for tissue engineering. Nat. Rev. Mater. 2020, 5, 351–370. [Google Scholar] [CrossRef]
- Schmeichel, K.L.; Bissell, M.J. Modeling tissue-specific signaling and organ function in three dimensions. J. Cell Sci. 2003, 116, 2377–2388. [Google Scholar] [CrossRef]
- Sala, M.; Ros, M.; Saltel, F. A Complex and Evolutive Character: Two Face Aspects of ECM in Tumor Progression. Front. Oncol. 2020, 10, 1620. [Google Scholar] [CrossRef] [PubMed]
- Faulk, D.M.; Johnson, S.A.; Zhang, L.; Badylak, S.F. Role of the extracellular matrix in whole organ engineering. J. Cell. Physiol. 2014, 229, 984–989. [Google Scholar] [CrossRef]
- Khanna, C.; Hunter, K. Modeling metastasis in vivo. Carcinogenesis 2005, 26, 513–523. [Google Scholar] [CrossRef]
- Fidler, I.J. Models for spontaneous metastasis. Cancer Res. 2006, 66, 9787. [Google Scholar] [CrossRef][Green Version]
- Henriquez, N.V.; van Overveld, P.G.; Que, I.; Buijs, J.T.; Bachelier, R.; Kaijzel, E.L.; Lowik, C.W.; Clezardin, P.; van der Pluijm, G. Advances in optical imaging and novel model systems for cancer metastasis research. Clin. Exp. Metastasis 2007, 24, 699–705. [Google Scholar] [CrossRef]
- Khabir, Z.; Guller, A.E.; Rozova, V.S.; Liang, L.; Lai, Y.J.; Goldys, E.M.; Hu, H.; Vickery, K.; Zvyagin, A.V. Tracing upconversion nanoparticle penetration in human skin. Colloids Surf. B Biointerfaces 2019, 184, 110480. [Google Scholar] [CrossRef]
- Guller, A.E.; Grebenyuk, P.N.; Shekhter, A.B.; Zvyagin, A.V.; Deyev, S.M. Bioreactor-Based Tumor Tissue Engineering. Acta Nat. 2016, 8, 44–58. [Google Scholar] [CrossRef]
- Burdett, E.; Kasper, F.K.; Mikos, A.G.; Ludwig, J.A. Engineering tumors: A tissue engineering perspective in cancer biology. Tissue Eng. Part B Rev. 2010, 16, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Rijal, G.; Li, W. 3D scaffolds in breast cancer research. Biomaterials 2016, 81, 135–156. [Google Scholar] [CrossRef]
- Badylak, S.F. Decellularized allogeneic and xenogeneic tissue as a bioscaffold for regenerative medicine: Factors that influence the host response. Ann. Biomed. Eng. 2014, 42, 1517–1527. [Google Scholar] [CrossRef] [PubMed]
- Crapo, P.M.; Gilbert, T.W.; Badylak, S.F. An overview of tissue and whole organ decellularization processes. Biomaterials 2011, 32, 3233–3243. [Google Scholar] [CrossRef]
- Keane, T.J.; Swinehart, I.T.; Badylak, S.F. Methods of tissue decellularization used for preparation of biologic scaffolds and in vivo relevance. Methods 2015, 84, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Parmaksiz, M.; Dogan, A.; Odabas, S.; Elcin, A.E.; Elcin, Y.M. Clinical applications of decellularized extracellular matrices for tissue engineering and regenerative medicine. Biomed. Mater. 2016, 11, 022003. [Google Scholar] [CrossRef]
- Ferreira, L.P.; Gaspar, V.M.; Mano, J.F. Decellularized Extracellular Matrix for Bioengineering Physiomimetic 3D in Vitro Tumor Models. Trends Biotechnol. 2020, 38, 1397–1414. [Google Scholar] [CrossRef] [PubMed]
- Grey, J.F.E.; Campbell-Ritchie, A.; Everitt, N.M.; Fezovich, A.J.; Wheatley, S.P. The use of decellularised animal tissue to study disseminating cancer cells. J. Cell Sci. 2018, 132, jcs219907. [Google Scholar] [CrossRef] [PubMed]
- Shologu, N.; Szegezdi, E.; Lowery, A.; Kerin, M.; Pandit, A.; Zeugolis, D.I. Recreating complex pathophysiologies in vitro with extracellular matrix surrogates for anticancer therapeutics screening. Drug Discov. Today 2016, 21, 1521–1531. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, J.; Roy, S.; Ghosh, S. Regulation of decellularized matrix mediated immune response. Biomater. Sci. 2020, 8, 1194–1215. [Google Scholar] [CrossRef]
- Ott, H.C.; Matthiesen, T.S.; Goh, S.K.; Black, L.D.; Kren, S.M.; Netoff, T.I.; Taylor, D.A. Perfusion-decellularized matrix: Using nature’s platform to engineer a bioartificial heart. Nat. Med. 2008, 14, 213–221. [Google Scholar] [CrossRef]
- Mayorca-Guiliani, A.E.; Madsen, C.D.; Cox, T.R.; Horton, E.R.; Venning, F.A.; Erler, J.T. ISDoT: In situ decellularization of tissues for high-resolution imaging and proteomic analysis of native extracellular matrix. Nat. Med. 2017, 23, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Park, K.M.; Woo, H.M. Systemic Decellularization for Multi-organ Scaffolds in Rats. Transplant. Proc. 2012, 44, 1151–1154. [Google Scholar] [CrossRef]
- Swinehart, I.T.; Badylak, S.F. Extracellular matrix bioscaffolds in tissue remodeling and morphogenesis. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2016, 245, 351–360. [Google Scholar] [CrossRef]
- Shekhter, A.B.; Guller, A.E.; Istranov, L.P.; Istranova, E.V.; Butnaru, D.V.; Vinarov, A.Z.; Zakharkina, O.L.; Kurkov, A.V.; Kantimerov, D.F.; Antonov, E.N.; et al. Morphology of collagen matrices for tissue engineering (biocompatibility, biodegradation, tissue response). Arkh. Patol. 2015, 77, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.F.; Diogo, G.S.; Pina, S.; Oliveira, J.M.; Silva, T.H.; Reis, R.L. Collagen-based bioinks for hard tissue engineering applications: A comprehensive review. J. Mater. Sci. Mater. Med. 2019, 30, 32. [Google Scholar] [CrossRef]
- Liang, J.; Yi, P.; Wang, X.; Huang, F.; Luan, X.; Zhao, Z.; Liu, C. Acellular matrix hydrogel for repair of the temporomandibular joint disc. J. Biomed. Mater. Res. Part B Appl. Biomater. 2020, 108, 2995–3007. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O. The extracellular matrix: Not just pretty fibrils. Science 2009, 326, 1216–1219. [Google Scholar] [CrossRef]
- Hutter, H.; Vogel, B.E.; Plenefisch, J.D.; Norris, C.R.; Proenca, R.B.; Spieth, J.; Guo, C.; Mastwal, S.; Zhu, X.; Scheel, J.; et al. Conservation and novelty in the evolution of cell adhesion and extracellular matrix genes. Science 2000, 287, 989–994. [Google Scholar] [CrossRef]
- Tottey, S.; Johnson, S.A.; Crapo, P.M.; Reing, J.E.; Zhang, L.; Jiang, H.; Medberry, C.J.; Reines, B.; Badylak, S.F. The effect of source animal age upon extracellular matrix scaffold properties. Biomaterials 2011, 32, 128–136. [Google Scholar] [CrossRef]
- Badylak, S.F. Xenogeneic extracellular matrix as a scaffold for tissue reconstruction. Transpl. Immunol. 2004, 12, 367–377. [Google Scholar] [CrossRef] [PubMed]
- McNeal, W.D.; Fletcher, D.L.; Buhr, R.J. Effects of stunning and decapitation on broiler activity during bleeding, blood loss, carcass, and breast meat quality. Poult. Sci. 2003, 82, 163–168. [Google Scholar] [CrossRef]
- Guller, A.; Trusova, I.; Petersen, E.; Shekhter, A.; Kurkov, A.; Qian, Y.; Zvyagin, A. Acellular organ scaffolds for tumor tissue engineering. In Proceedings of the Micro+Nano Materials, Devices, and Systems, Sydney, New South Wales, Australia, 7–9 December 2015; p. 96684G. [Google Scholar]
- Ibidi, G.; Live/Dead Staining with FDA and PI. Application Note # 33. Available online: https://ibidi.com/img/cms/support/AN/AN33_Live_Dead_staining_with_FDA_and_PI.pdf (accessed on 27 October 2021).
- Kotova, S.L.; Timashev, P.S.; Guller, A.E.; Shekhter, A.B.; Misurkin, P.I.; Bagratashvili, V.N.; Solovieva, A.B. Collagen structure deterioration in the skin of patients with pelvic organ prolapse determined by atomic force microscopy. Microsc. Microanal. 2015, 21, 324–333. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Nowak-Sliwinska, P.; Segura, T.; Iruela-Arispe, M.L. The chicken chorioallantoic membrane model in biology, medicine and bioengineering. Angiogenesis 2014, 17, 779–804. [Google Scholar] [CrossRef]
- Steger, C. An unbiased detector of curvilinear structures. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 113–125. [Google Scholar] [CrossRef]
- Carpentier, G. Angiogenesis Analyzer for ImageJ. Available online: http://image.bio.methods.free.fr/ImageJ/?Angiogenesis-Analyzer-for-ImageJ&artpage=6-6#outil_sommaire_6 (accessed on 27 October 2021).
- Garcia-Bennett, A.E.; Miyasaka, K.; Terasaki, O.; Che, S. Structural Solution of Mesocaged Material AMS-8. Chem. Mater. 2004, 16, 3597–3605. [Google Scholar] [CrossRef]
- Garcia-Bennett, A.E.; Terasaki, O.; Che, S.; Tatsumi, T. Structural investigations of AMS-n mesoporous materials by transmission electron microscopy. Chem. Mater. 2004, 16, 813–821. [Google Scholar] [CrossRef]
- Brunauer, S.; Emmett, P.H.; Teller, E. Adsorption of gases in multimolecular layers. J. Am. Chem. Soc. 1938, 60, 309–319. [Google Scholar] [CrossRef]
- Uygun, B.E.; Soto-Gutierrez, A.; Yagi, H.; Izamis, M.L.; Guzzardi, M.A.; Shulman, C.; Milwid, J.; Kobayashi, N.; Tilles, A.; Berthiaume, F.; et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat. Med. 2010, 16, 814–820. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Lessa, N.; Estrada, D.C.; Severson, E.B.; Lingala, S.; Zern, M.A.; Nolta, J.A.; Wu, J. Decellularized liver matrix as a carrier for the transplantation of human fetal and primary hepatocytes in mice. Liver Transpl. 2011, 17, 418–427. [Google Scholar] [CrossRef]
- Kajbafzadeh, A.M.; Javan-Farazmand, N.; Monajemzadeh, M.; Baghayee, A. Determining the optimal decellularization and sterilization protocol for preparing a tissue scaffold of a human-sized liver tissue. Tissue Eng. Part C Methods 2013, 19, 642–651. [Google Scholar] [CrossRef] [PubMed]
- Witasp, E.; Kupferschmidt, N.; Bengtsson, L.; Hultenby, K.; Smedman, C.; Paulie, S.; Garcia-Bennett, A.E.; Fadeel, B. Efficient internalization of mesoporous silica particles of different sizes by primary human macrophages without impairment of macrophage clearance of apoptotic or antibody-opsonized target cells. Toxicol. Appl. Pharm. 2009, 239, 306–319. [Google Scholar] [CrossRef]
- Tizard, M.L.; Jenkins, K.A.; Cooper, C.A.; Woodcock, M.E.; Challagulla, A.; Doran, T.J. Potential benefits of gene editing for the future of poultry farming. Transgenic Res. 2019, 28, 87–92. [Google Scholar] [CrossRef]
- Clement, S.; Guller, A.; Mahbub, S.B.; Goldys, E.M. Oxygen-Carrying Polymer Nanoconstructs for Radiodynamic Therapy of Deep Hypoxic Malignant Tumors. Biomedicines 2021, 9, 322. [Google Scholar] [CrossRef] [PubMed]
- Baiguera, S.; Macchiarini, P.; Ribatti, D. Chorioallantoic membrane for in vivo investigation of tissue-engineered construct biocompatibility. J. Biomed. Mater. Res. Part B Appl. Biomater. 2012, 100, 1425–1434. [Google Scholar] [CrossRef]
- Ribatti, D. The chick embryo chorioallantoic membrane (CAM). A multifaceted experimental model. Mech. Dev. 2016, 141, 70–77. [Google Scholar] [CrossRef]
- Lu, X.; Kang, Y. Organotropism of breast cancer metastasis. J. Mammary Gland Biol. Neoplasia 2007, 12, 153–162. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Carey, L.; Winer, E.; Viale, G.; Cameron, D.; Gianni, L. Triple-negative breast cancer: Disease entity or title of convenience? Nat. Rev. Clin. Oncol. 2010, 7, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; He, G.; Yan, S.; Chen, C.; Song, L.; Rosol, T.J.; Deng, X. Triple-negative breast cancer: Is there a treatment on the horizon? Oncotarget 2017, 8, 1913–1924. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.L.; Abu Jalboush, S.; Lo, H.W. Exosomal MicroRNAs and Organotropism in Breast Cancer Metastasis. Cancers 2020, 12, 1827. [Google Scholar] [CrossRef]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef]
- Wei, S.; Siegal, G.P. Metastatic Organotropism: An Intrinsic Property of Breast Cancer Molecular Subtypes. Adv. Anat. Pathol. 2017, 24, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Khoo, B.L.; Lee, S.C.; Kumar, P.; Tan, T.Z.; Warkiani, M.E.; Ow, S.G.; Nandi, S.; Lim, C.T.; Thiery, J.P. Short-term expansion of breast circulating cancer cells predicts response to anti-cancer therapy. Oncotarget 2015, 6, 15578–15593. [Google Scholar] [CrossRef]
- Disibio, G.; French, S.W. Metastatic patterns of cancers: Results from a large autopsy study. Arch. Pathol. Lab. Med. 2008, 132, 931–939. [Google Scholar] [CrossRef]
- Xiong, G.; Flynn, T.J.; Chen, J.; Trinkle, C.; Xu, R. Development of an ex vivo breast cancer lung colonization model utilizing a decellularized lung matrix. Integr. Biol. Quant. Biosci. Nano Macro 2015, 7, 1518–1525. [Google Scholar] [CrossRef]
- Tsilimigras, D.I.; Brodt, P.; Clavien, P.-A.; Muschel, R.J.; D’Angelica, M.I.; Endo, I.; Parks, R.W.; Doyle, M.; de Santibañes, E.; Pawlik, T.M. Liver metastases. Nat. Rev. Dis. Primers 2021, 7, 27. [Google Scholar] [CrossRef]
- Ma, R.; Feng, Y.; Lin, S.; Chen, J.; Lin, H.; Liang, X.; Zheng, H.; Cai, X. Mechanisms involved in breast cancer liver metastasis. J. Transl. Med. 2015, 13, 64. [Google Scholar] [CrossRef]
- Rosenow, F.; Ossig, R.; Thormeyer, D.; Gasmann, P.; Schlüter, K.; Brunner, G.; Haier, J.; Eble, J.A. Integrins as Antimetastatic Targets of RGD-Independent Snake Venom Components in Liver Metastasis. Neoplasia 2008, 10, 168–176. [Google Scholar] [CrossRef]
- Haier, J. An Intravital Model to Monitor Steps of Metastatic Tumor Cell Adhesion within the Hepatic Microcirculation. J. Gastrointest. Surg. 2003, 7, 507–515. [Google Scholar] [CrossRef]
- Roos, E.; Dingemans, K.P.; Van de Pavert, I.V.; Van den Bergh-Weerman, M.A. Mammary-carcinoma cells in mouse liver: Infiltration of liver tissue and interaction with Kupffer cells. Br. J. Cancer 1978, 38, 88–99. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, H.; Song, X.; Yang, Q. Metastatic heterogeneity of breast cancer: Molecular mechanism and potential therapeutic targets. Semin. Cancer Biol. 2020, 60, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Hernandez, A.; Amenta, P.S. The extracellular matrix in hepatic regeneration. FASEB J. 1995, 9, 1401–1410. [Google Scholar] [CrossRef] [PubMed]
- Tabaries, S.; Dong, Z.; Annis, M.G.; Omeroglu, A.; Pepin, F.; Ouellet, V.; Russo, C.; Hassanain, M.; Metrakos, P.; Diaz, Z.; et al. Claudin-2 is selectively enriched in and promotes the formation of breast cancer liver metastases through engagement of integrin complexes. Oncogene 2011, 30, 1318–1328. [Google Scholar] [CrossRef]
- Ishii, S.; Mizoi, T.; Kawano, K.; Cay, O.; Thomas, P.; Nachman, A.; Ford, R.; Shoji, Y.; Kruskal, J.B.; Steele, G., Jr.; et al. Implantation of human colorectal carcinoma cells in the liver studied by in vivo fluorescence videomicroscopy. Clin. Exp. Metastasis 1996, 14, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Barkan, D.; Green, J.E.; Chambers, A.F. Extracellular matrix: A gatekeeper in the transition from dormancy to metastatic growth. Eur. J. Cancer 2010, 46, 1181–1188. [Google Scholar] [CrossRef]
- Seo, B.R.; DelNero, P.; Fischbach, C. In vitro models of tumor vessels and matrix: Engineering approaches to investigate transport limitations and drug delivery in cancer. Adv. Drug Deliv. Rev. 2014, 69–70, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Faulk, D.M.; Wildemann, J.D.; Badylak, S.F. Decellularization and cell seeding of whole liver biologic scaffolds composed of extracellular matrix. J. Clin. Exp. Hepatol. 2015, 5, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Rozova, V.S.; Anwer, A.G.; Guller, A.E.; Es, H.A.; Khabir, Z.; Sokolova, A.I.; Gavrilov, M.U.; Goldys, E.M.; Warkiani, M.E.; Thiery, J.P.; et al. Machine learning reveals mesenchymal breast carcinoma cell adaptation in response to matrix stiffness. PLoS Comput. Biol. 2021, 17, e1009193. [Google Scholar] [CrossRef] [PubMed]
- McClelland, R.; Wauthier, E.; Uronis, J.; Reid, L. Gradients in the liver’s extracellular matrix chemistry from periportal to pericentral zones: Influence on human hepatic progenitors. Tissue Eng. Part A 2008, 14, 59–70. [Google Scholar] [CrossRef]
- Reid, L.M.; Fiorino, A.S.; Sigal, S.H.; Brill, S.; Holst, P.A. Extracellular matrix gradients in the space of Disse: Relevance to liver biology. Hepatology 1992, 15, 1198–1203. [Google Scholar] [CrossRef]
- Ling, W.; Lu, Q.; Lu, C.; Quan, J.; Ma, L.; Li, J.; He, D.; Liu, J.; Yang, J.; Wen, T.; et al. Effects of vascularity and differentiation of hepatocellular carcinoma on tumor and liver stiffness: In vivo and in vitro studies. Ultrasound Med. Biol. 2014, 40, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Zaman, M.H.; Trapani, L.M.; Sieminski, A.L.; Mackellar, D.; Gong, H.; Kamm, R.D.; Wells, A.; Lauffenburger, D.A.; Matsudaira, P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. USA 2006, 103, 10889–10894. [Google Scholar] [CrossRef]
- Schrader, J.; Gordon-Walker, T.T.; Aucott, R.L.; van Deemter, M.; Quaas, A.; Walsh, S.; Benten, D.; Forbes, S.J.; Wells, R.G.; Iredale, J.P. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology 2011, 53, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- Ranamukhaarachchi, S.K.; Modi, R.N.; Han, A.; Velez, D.O.; Kumar, A.; Engler, A.J.; Fraley, S.I. Macromolecular crowding tunes 3D collagen architecture and cell morphogenesis. Biomater. Sci. 2019, 7, 618–633. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Perez, M.; Voytik-Harbin, S.L.; Rickus, J.L. Extracellular Matrix Properties Regulate the Migratory Response of Glioblastoma Stem Cells in Three-Dimensional Culture. Tissue Eng. Part A 2015, 21, 2572–2582. [Google Scholar] [CrossRef]
- Viji Babu, P.K.; Rianna, C.; Mirastschijski, U.; Radmacher, M. Nano-mechanical mapping of interdependent cell and ECM mechanics by AFM force spectroscopy. Sci. Rep. 2019, 9, 12317. [Google Scholar] [CrossRef] [PubMed]
- Friedl, P.; Wolf, K. Tumour-cell invasion and migration: Diversity and escape mechanisms. Nat Rev Cancer 2003, 3, 362–374. [Google Scholar] [CrossRef]
- Liu, Y.J.; Le Berre, M.; Lautenschlaeger, F.; Maiuri, P.; Callan-Jones, A.; Heuze, M.; Takaki, T.; Voituriez, R.; Piel, M. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell 2015, 160, 659–672. [Google Scholar] [CrossRef]
- Friedl, P. Prespecification and plasticity: Shifting mechanisms of cell migration. Curr Opin Cell Biol 2004, 16, 14–23. [Google Scholar] [CrossRef]
- Grigore, A.D.; Jolly, M.K.; Jia, D.; Farach-Carson, M.C.; Levine, H. Tumor Budding: The Name is EMT. Partial EMT. Journal of Clinical Medicine 2016, 5, 51. [Google Scholar] [CrossRef]
- Holtkamp, N.; Afanasieva, A.; Elstner, A.; van Landeghem, F.K.; Konneker, M.; Kuhn, S.A.; Kettenmann, H.; von Deimling, A. Brain slice invasion model reveals genes differentially regulated in glioma invasion. Biochem. Biophys. Res. Commun. 2005, 336, 1227–1233. [Google Scholar] [CrossRef]
- Yarmenitis, S.D.; Kalogeropoulou, C.P.; Hatjikondi, O.; Ravazoula, P.; Petsas, T.; Siamblis, D.; Kalfarentzos, F. An experimental approach of the Doppler perfusion index of the liver in detecting occult hepatic metastases: Histological findings related to the hemodynamic measurements in Wistar rats. Eur. Radiol. 2000, 10, 417–424. [Google Scholar] [CrossRef]
- Alzubi, M.A.; Sohal, S.S.; Sriram, M.; Turner, T.H.; Zot, P.; Idowu, M.; Harrell, J.C. Quantitative assessment of breast cancer liver metastasis expansion with patient-derived xenografts. Clin. Exp. Metastasis 2019, 36, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.M.; Kumar, M.P.; Wheeler, S.E.; Young, C.L.; Venkataramanan, R.; Stolz, D.B.; Griffith, L.G.; Lauffenburger, D.A.; Wells, A. A Model of Dormant-Emergent Metastatic Breast Cancer Progression Enabling Exploration of Biomarker Signatures. Mol Cell Proteomics 2018, 17, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Kingston, B.R.; Syed, A.M.; Ngai, J.; Sindhwani, S.; Chan, W.C.W. Assessing micrometastases as a target for nanoparticles using 3D microscopy and machine learning. Proc. Natl. Acad. Sci. USA 2019, 116, 14937–14946. [Google Scholar] [CrossRef] [PubMed]
- Spratt, J.A.; von Fournier, D.; Spratt, J.S.; Weber, E.E. Decelerating growth and human breast cancer. Cancer 1993, 71, 2013–2019. [Google Scholar] [CrossRef]
- Tsoularis, A.; Wallace, J. Analysis of logistic growth models. Math. Biosci. 2002, 179, 21–55. [Google Scholar] [CrossRef]
- Singh, M.; Mukundan, S.; Jaramillo, M.; Oesterreich, S.; Sant, S. Three-Dimensional Breast Cancer Models Mimic Hallmarks of Size-Induced Tumor Progression. Cancer Res. 2016, 76, 3732–3743. [Google Scholar] [CrossRef]
- Schlatter, P.; Konig, M.F.; Karlsson, L.M.; Burri, P.H. Quantitative study of intussusceptive capillary growth in the chorioallantoic membrane (CAM) of the chicken embryo. Microvasc. Res. 1997, 54, 65–73. [Google Scholar] [CrossRef]
- Hillen, F.; Griffioen, A.W. Tumour vascularization: Sprouting angiogenesis and beyond. Cancer Metastasis Rev. 2007, 26, 489–502. [Google Scholar] [CrossRef]
- Fischbach, C.; Chen, R.; Matsumoto, T.; Schmelzle, T.; Brugge, J.S.; Polverini, P.J.; Mooney, D.J. Engineering tumors with 3D scaffolds. Nat. Methods 2007, 4, 855–860. [Google Scholar] [CrossRef]
- Hamburger, V.; Hamilton, H.L. A series of normal stages in the development of the chick embryo. J. Morphol. 1951, 88, 49–92. [Google Scholar] [CrossRef]
- Atluri, R.; Hedin, N.; Garcia-Bennett, A.E. Hydrothermal phase transformation of bicontinuous cubic mesoporous material AMS-6. Chem. Mater. 2008, 20, 3857–3866. [Google Scholar] [CrossRef]
- Ribatti, D.; Conconi, M.T.; Nico, B.; Baiguera, S.; Corsi, P.; Parnigotto, P.P.; Nussdorfer, G.G. Angiogenic response induced by acellular brain scaffolds grafted onto the chick embryo chorioallantoic membrane. Brain Res. 2003, 989, 9–15. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Mak, K.M.; Png, C.Y.; Lee, D.J. Type V Collagen in Health, Disease, and Fibrosis. Anat. Rec. 2016, 299, 613–629. [Google Scholar] [CrossRef]
- Blacher, S.; Devy, L.; Hlushchuk, R.; Larger, E.; Lamandé, N.; Burri, P.; Corvol, P.; Djonov, V.; Foidart, J.-M.; Noël, A. Quantification of Angiogenesis in the Chicken Chorioallantoic Membrane (Cam). Image Anal. Stereol. 2011, 23, 169–180. [Google Scholar] [CrossRef][Green Version]
- Doukas, C.N.; Maglogiannis, I.; Chatziioannou, A.A. Computer-supported angiogenesis quantification using image analysis and statistical averaging. IEEE Trans. Inf. Technol. Biomed. 2008, 12, 650–657. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guller, A.; Kuschnerus, I.; Rozova, V.; Nadort, A.; Yao, Y.; Khabir, Z.; Garcia-Bennett, A.; Liang, L.; Polikarpova, A.; Qian, Y.; et al. Chick Embryo Experimental Platform for Micrometastases Research in a 3D Tissue Engineering Model: Cancer Biology, Drug Development, and Nanotechnology Applications. Biomedicines 2021, 9, 1578. https://doi.org/10.3390/biomedicines9111578
Guller A, Kuschnerus I, Rozova V, Nadort A, Yao Y, Khabir Z, Garcia-Bennett A, Liang L, Polikarpova A, Qian Y, et al. Chick Embryo Experimental Platform for Micrometastases Research in a 3D Tissue Engineering Model: Cancer Biology, Drug Development, and Nanotechnology Applications. Biomedicines. 2021; 9(11):1578. https://doi.org/10.3390/biomedicines9111578
Chicago/Turabian StyleGuller, Anna, Inga Kuschnerus, Vlada Rozova, Annemarie Nadort, Yin Yao, Zahra Khabir, Alfonso Garcia-Bennett, Liuen (Olivia) Liang, Aleksandra Polikarpova, Yi Qian, and et al. 2021. "Chick Embryo Experimental Platform for Micrometastases Research in a 3D Tissue Engineering Model: Cancer Biology, Drug Development, and Nanotechnology Applications" Biomedicines 9, no. 11: 1578. https://doi.org/10.3390/biomedicines9111578
APA StyleGuller, A., Kuschnerus, I., Rozova, V., Nadort, A., Yao, Y., Khabir, Z., Garcia-Bennett, A., Liang, L., Polikarpova, A., Qian, Y., Goldys, E. M., & Zvyagin, A. V. (2021). Chick Embryo Experimental Platform for Micrometastases Research in a 3D Tissue Engineering Model: Cancer Biology, Drug Development, and Nanotechnology Applications. Biomedicines, 9(11), 1578. https://doi.org/10.3390/biomedicines9111578









