Structure-Activity Relationship of Hydroxycinnamic Acid Derivatives for Cooperating with Carnosic Acid and Calcitriol in Acute Myeloid Leukemia Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture and Enumeration
2.3. Annexin V/7-Aminoactinomycin D Assay
2.4. Determination of Cell Differentiation
2.5. Cytosolic Calcium Assay
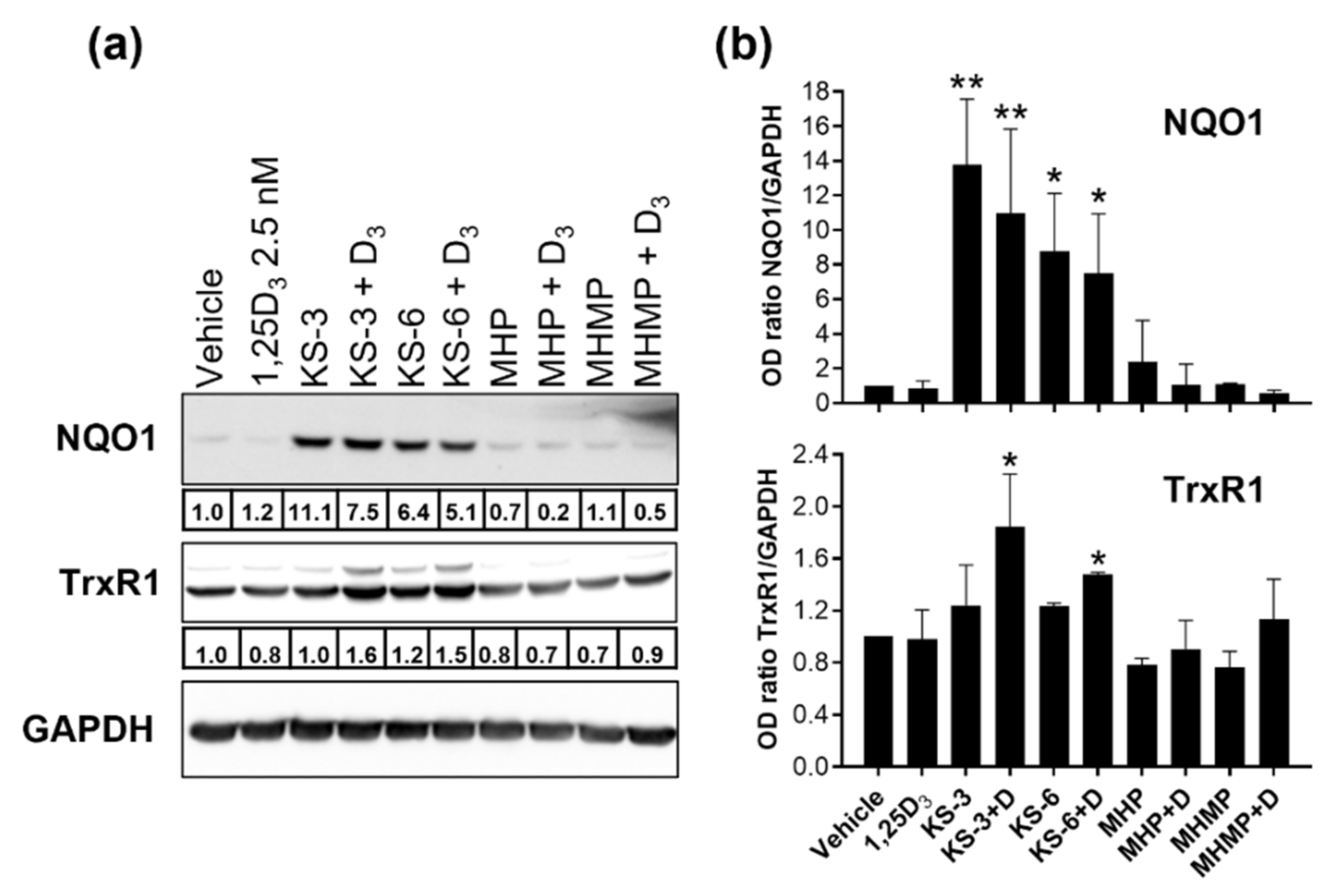
2.6. Western Blot Analysis
2.7. Molecular Modeling of the Formation of CA Anion-HCAD Complexes
2.8. Statistical Analysis
3. Results
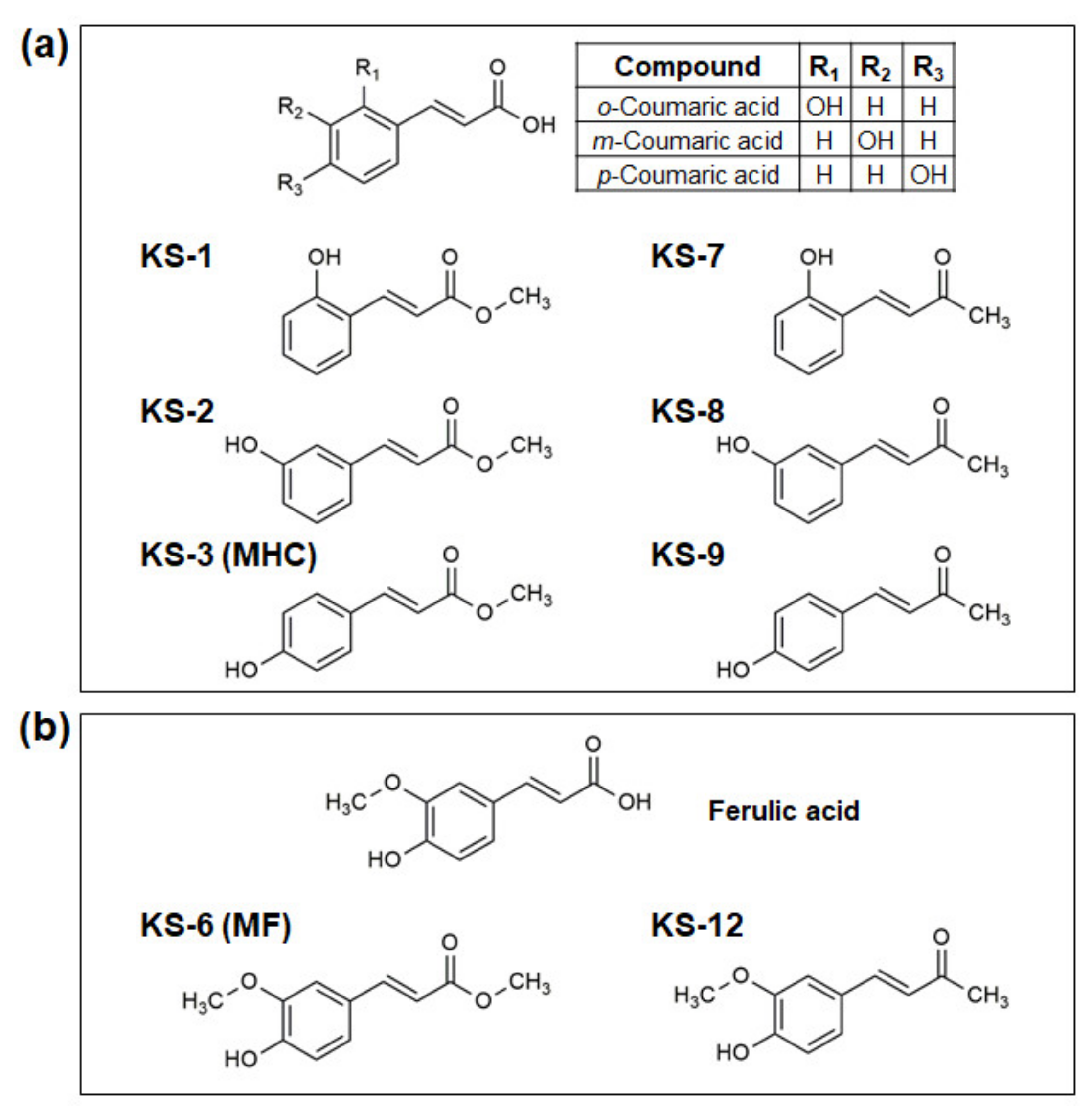
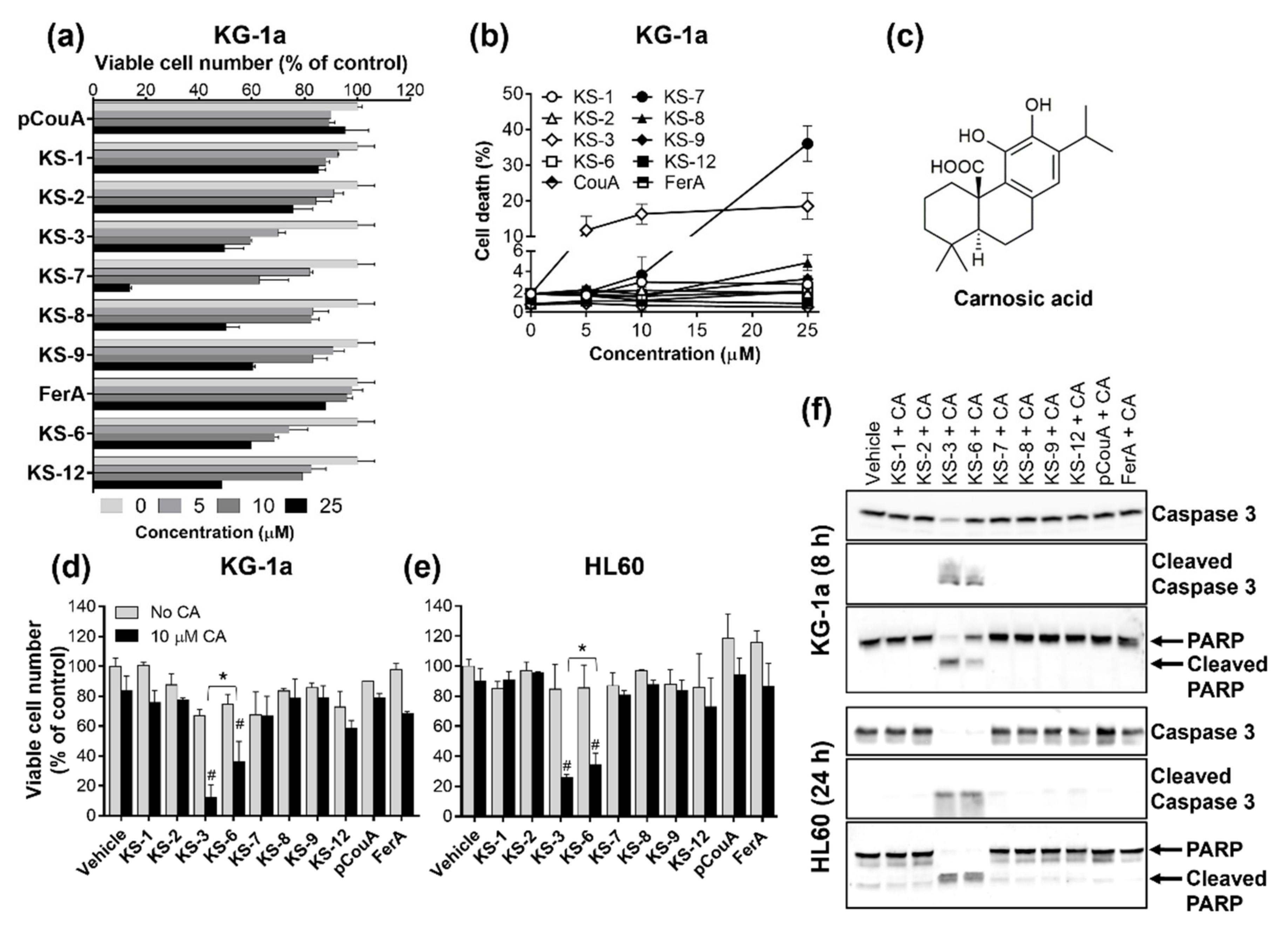
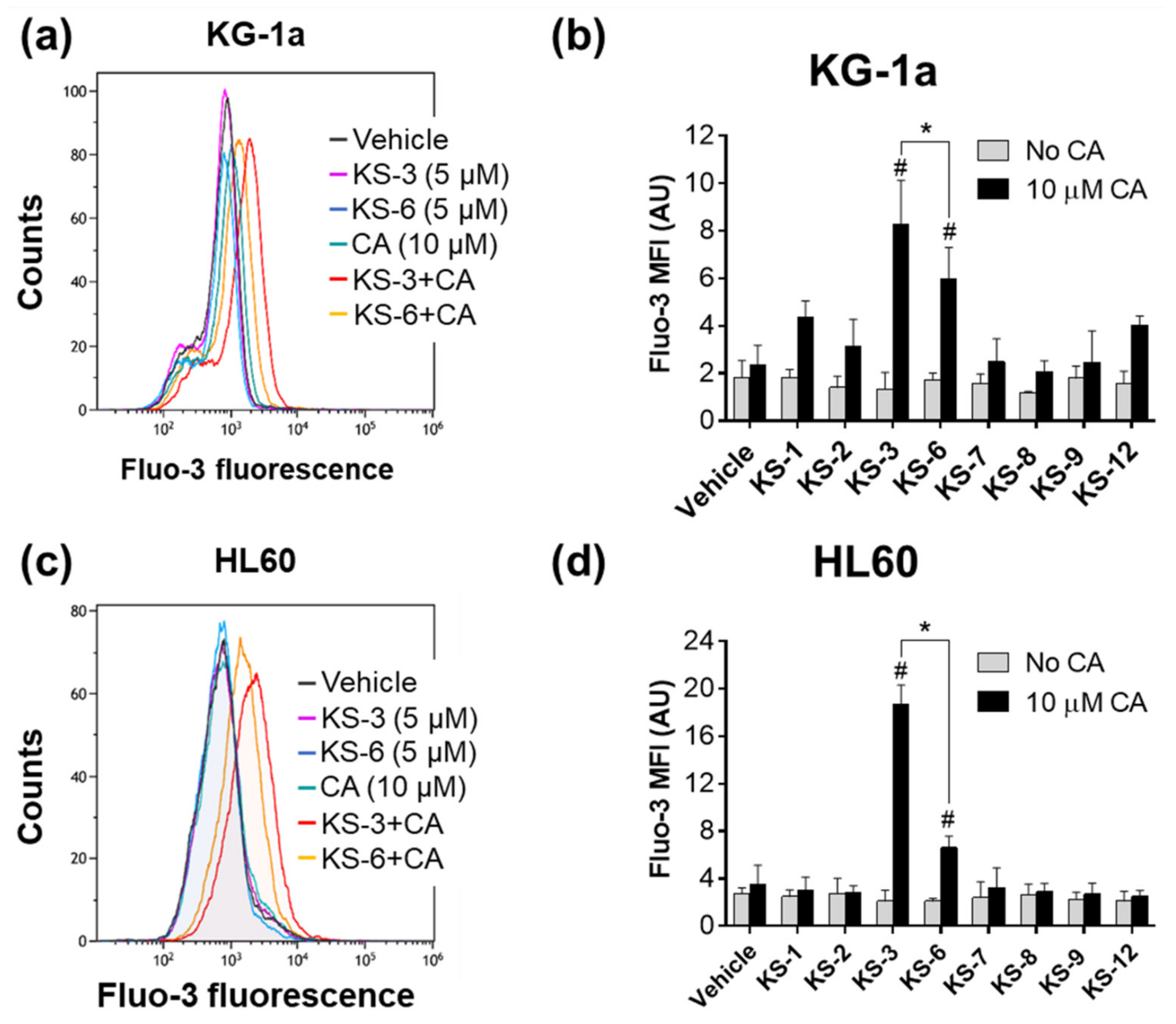
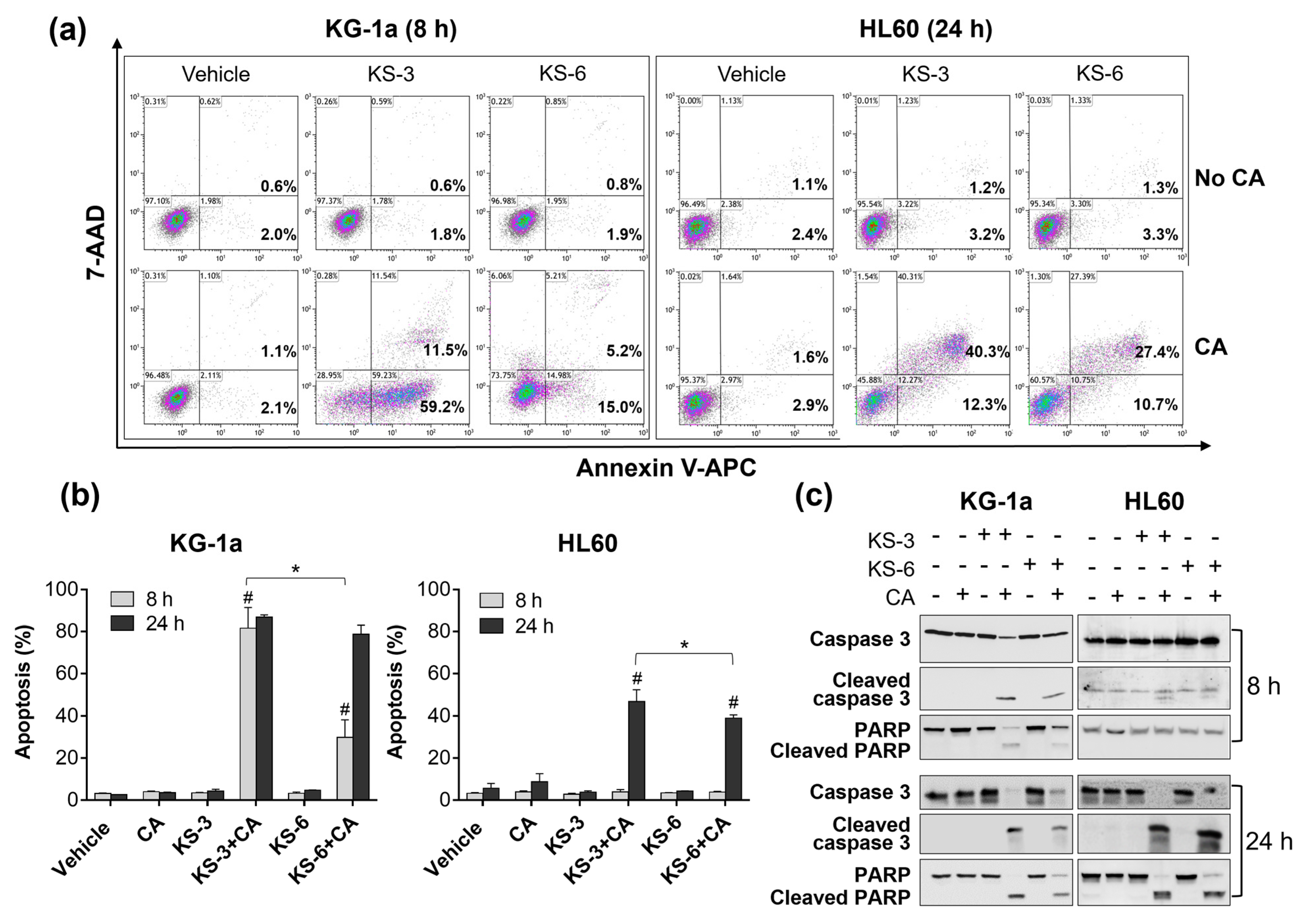
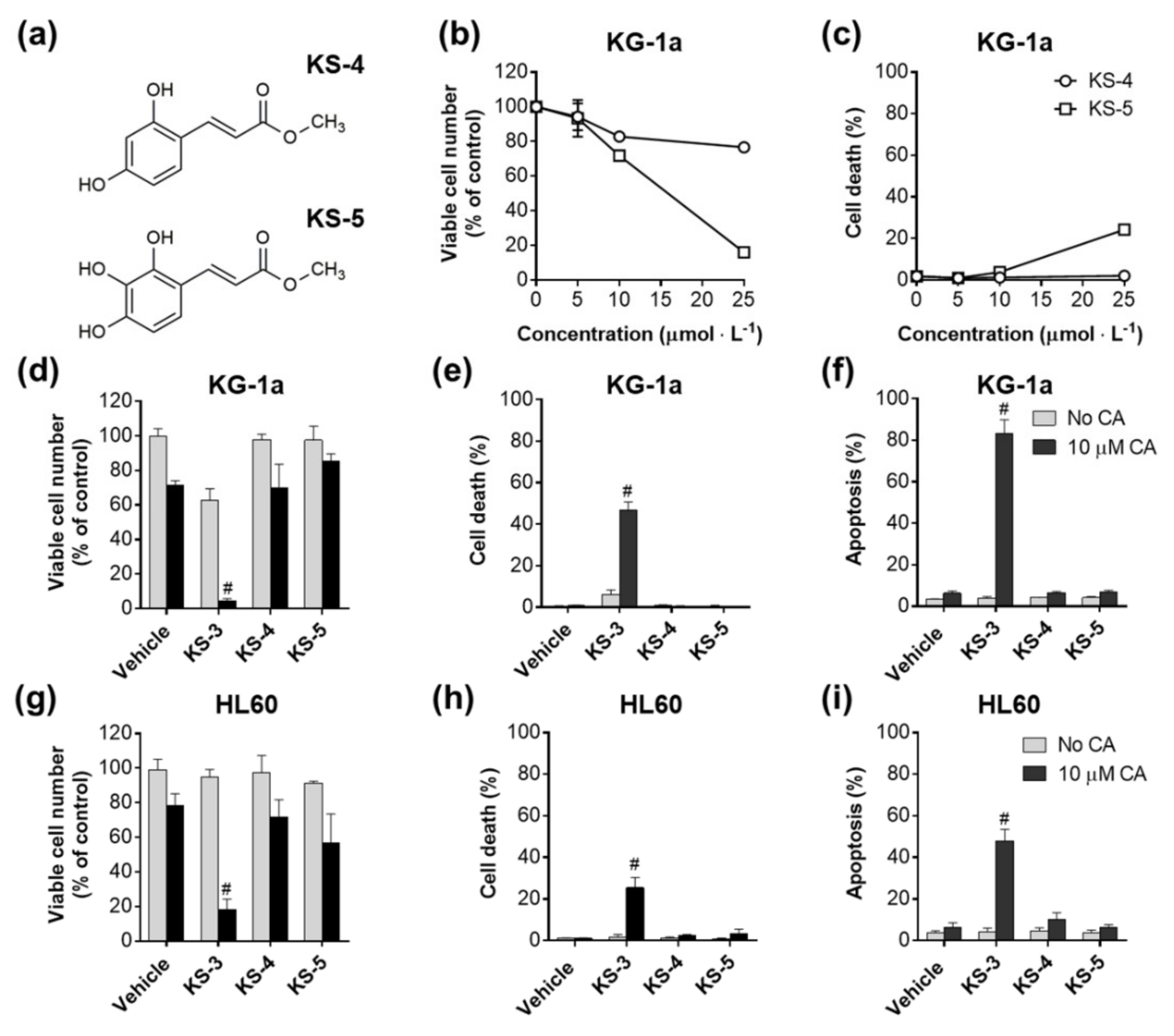
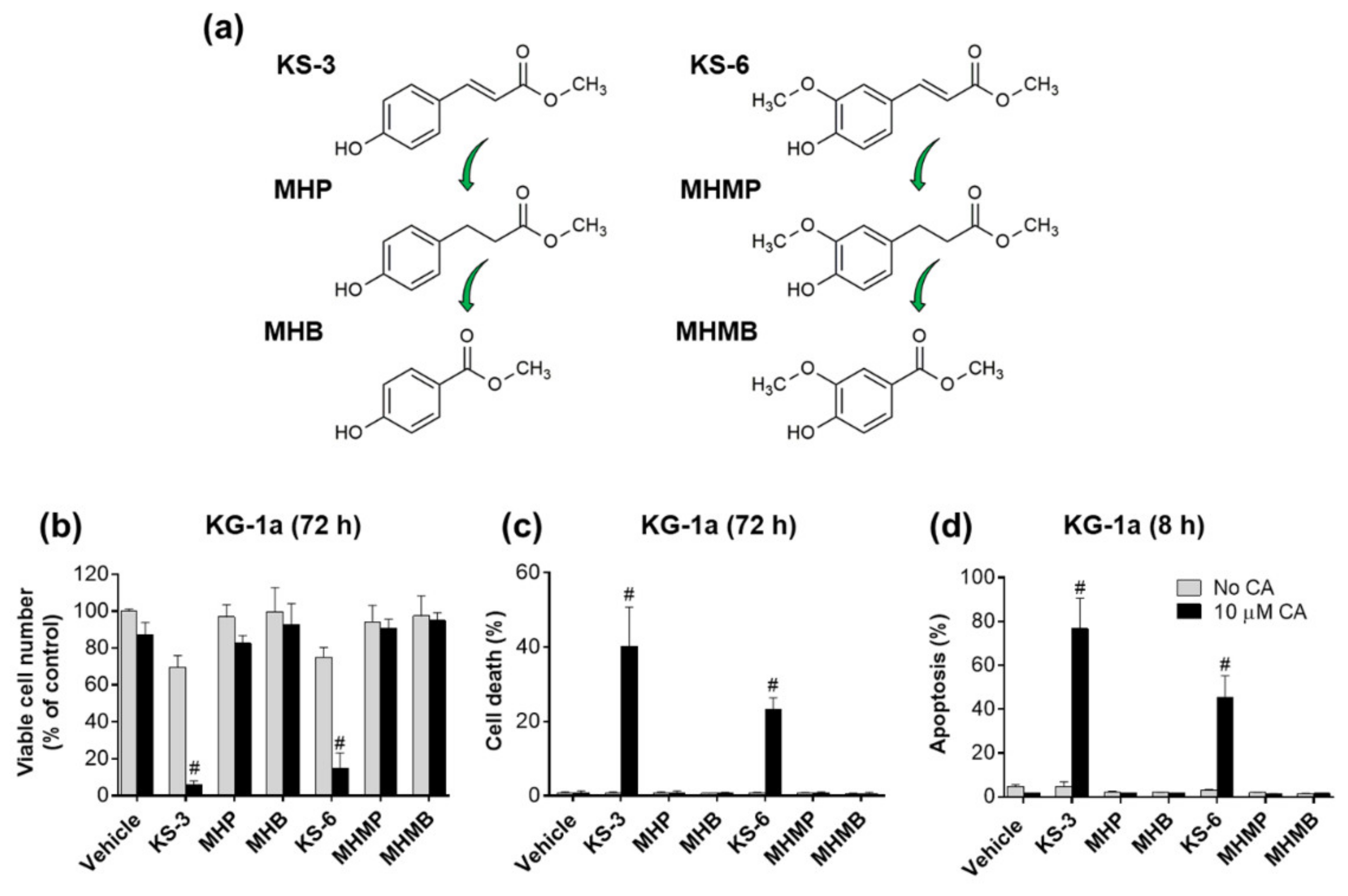
3.1. Essential Structural Features of Hydroxycinnamic Acid Derivatives Required for the Synergy with Carnosic Acid in Inducing Cytotoxicity to AML Cells
3.2. Molecular Modeling of the Formation of HCAD … CA Anion Complexes
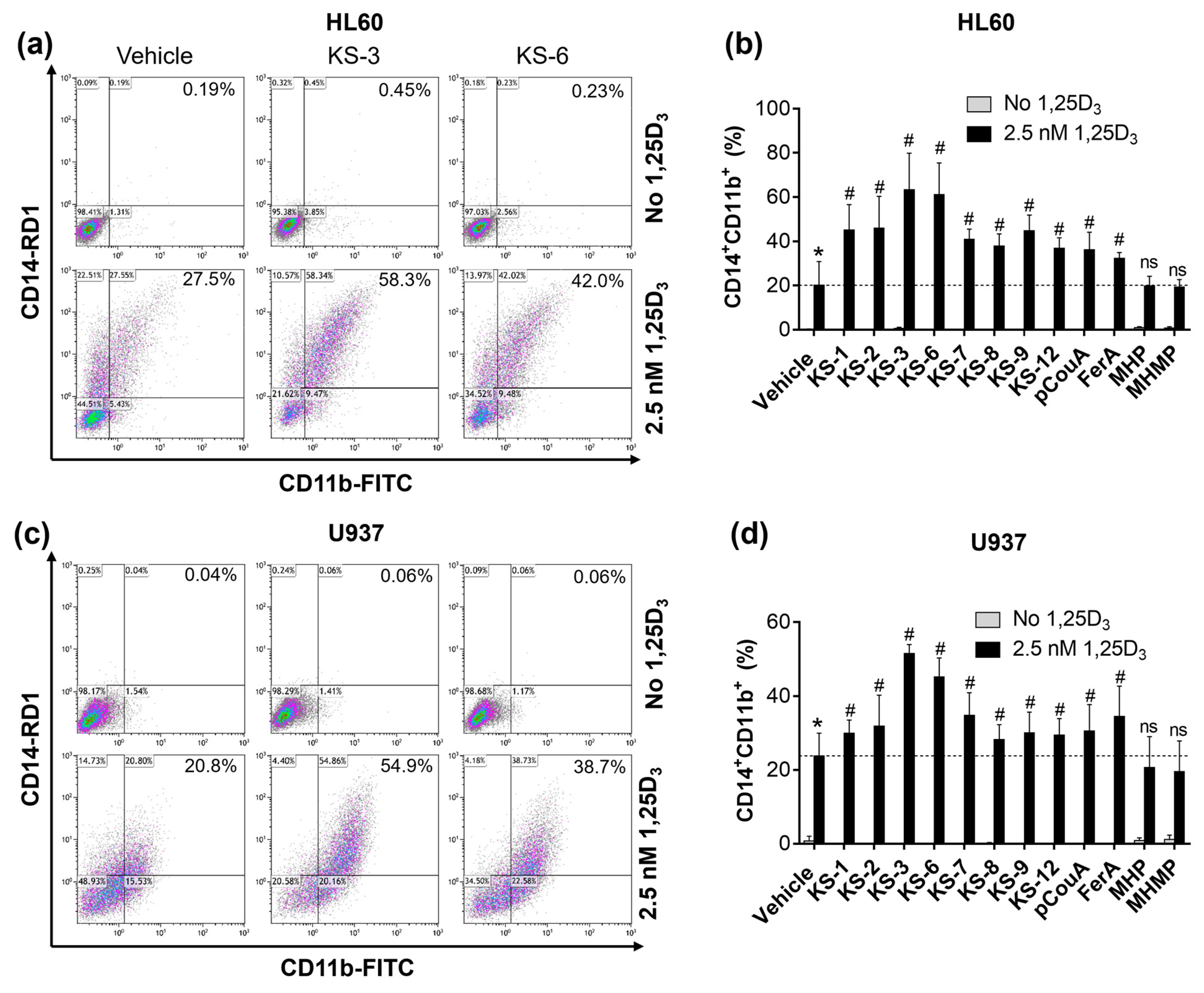
3.3. Hydroxycinnamic Acid Derivatives Can Enhance the Differentiation of AML Cells Induced by Calcitriol
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Mateen, S.; Raina, K.; Agarwal, R. Chemopreventive and anti-cancer efficacy of silibinin against growth and progression of lung cancer. Nutr. Cancer 2013, 65 (Suppl. 1), 3–11. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Sethi, G.; Um, J.Y.; Shanmugam, M.K.; Arfuso, F.; Kumar, A.P.; Bishayee, A.; Ahn, K.S. The Role of Resveratrol in Cancer Therapy. Int. J. Mol. Sci. 2017, 18, 2589. [Google Scholar] [CrossRef] [PubMed]
- Almatroodi, S.A.; Almatroudi, A.; Khan, A.A.; Alhumaydhi, F.A.; Alsahli, M.A.; Rahmani, A.H. Potential Therapeutic Targets of Epigallocatechin Gallate (EGCG), the Most Abundant Catechin in Green Tea, and Its Role in the Therapy of Various Types of Cancer. Molecules 2020, 25, 3146. [Google Scholar] [CrossRef]
- Giordano, A.; Tommonaro, G. Curcumin and Cancer. Nutrients 2019, 11, 2376. [Google Scholar] [CrossRef]
- Espinoza, J.L.; Kurokawa, Y.; Takami, A. Rationale for assessing the therapeutic potential of resveratrol in hematological malignancies. Blood Rev. 2019, 33, 43–52. [Google Scholar] [CrossRef]
- Avtanski, D.; Poretsky, L. Phyto-polyphenols as potential inhibitors of breast cancer metastasis. Mol. Med. 2018, 24, 29. [Google Scholar] [CrossRef] [PubMed]
- Borges, G.A.; Elias, S.T.; Amorim, B.; de Lima, C.L.; Coletta, R.D.; Castilho, R.M.; Squarize, C.H.; Guerra, E.N.S. Curcumin downregulates the PI3K-AKT-mTOR pathway and inhibits growth and progression in head and neck cancer cells. Phytother. Res. 2020, 3311–3324. [Google Scholar] [CrossRef]
- Petiti, J.; Rosso, V.; Lo Iacono, M.; Panuzzo, C.; Calabrese, C.; Signorino, E.; Pironi, L.; Cartella, A.; Bracco, E.; Pergolizzi, B.; et al. Curcumin induces apoptosis in JAK2-mutated cells by the inhibition of JAK2/STAT and mTORC1 pathways. J. Cell Mol. Med. 2019, 23, 4349–4357. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, N.; Saeed, M.E.M.; Sugimoto, Y.; Klinger, A.; Fleischer, E.; Efferth, T. Putative molecular determinants mediating sensitivity or resistance towards carnosic acid tumor cell responses. Phytomedicine 2020, 77, 153271. [Google Scholar] [CrossRef]
- Steiner, M.; Priel, I.; Giat, J.; Levy, J.; Sharoni, Y.; Danilenko, M. Carnosic acid inhibits proliferation and augments differentiation of human leukemic cells induced by 1,25-dihydroxyvitamin D3 and retinoic acid. Nutr. Cancer 2001, 41, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Jimenez, A.; Garcia-Caballero, M.; Medina, M.A.; Quesada, A.R. Anti-angiogenic properties of carnosol and carnosic acid, two major dietary compounds from rosemary. Eur. J. Nutr. 2013, 52, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, H.; Yuan, B.; Yuhara, E.; Imai, M.; Furutani, R.; Fukushima, S.; Hazama, S.; Hirobe, C.; Ohyama, K.; Takagi, N.; et al. Involvement of histone H3 phosphorylation via the activation of p38 MAPK pathway and intracellular redox status in cytotoxicity of HL-60 cells induced by Vitex agnus-castus fruit extract. Int. J. Oncol. 2014, 45, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.W.; Chang, C.L.; Lee, H.C.; Chi, C.W.; Pan, J.P.; Yang, W.C. Curcumin induces the apoptosis of human monocytic leukemia THP-1 cells via the activation of JNK/ERK pathways. BMC Complement Altern. Med. 2012, 12, 22. [Google Scholar] [CrossRef]
- Montane, X.; Kowalczyk, O.; Reig-Vano, B.; Bajek, A.; Roszkowski, K.; Tomczyk, R.; Pawliszak, W.; Giamberini, M.; Mocek-Plociniak, A.; Tylkowski, B. Current Perspectives of the Applications of Polyphenols and Flavonoids in Cancer Therapy. Molecules 2020, 25, 3342. [Google Scholar] [CrossRef]
- Niedzwiecki, A.; Roomi, M.W.; Kalinovsky, T.; Rath, M. Anticancer Efficacy of Polyphenols and Their Combinations. Nutrients 2016, 8, 552. [Google Scholar] [CrossRef]
- Maiti, P.; Plemmons, A.; Dunbar, G.L. Combination treatment of berberine and solid lipid curcumin particles increased cell death and inhibited PI3K/Akt/mTOR pathway of human cultured glioblastoma cells more effectively than did individual treatments. PloS ONE 2019, 14, e0225660. [Google Scholar] [CrossRef]
- Hu, W.H.; Chan, G.K.; Duan, R.; Wang, H.Y.; Kong, X.P.; Dong, T.T.; Tsim, K.W. Synergy of Ginkgetin and Resveratrol in Suppressing VEGF-Induced Angiogenesis: A Therapy in Treating Colorectal Cancer. Cancers (Basel) 2019, 11, 1828. [Google Scholar] [CrossRef] [PubMed]
- Bulut, G.; Atmaca, H.; Karaca, B. Trastuzumab in combination with AT-101 induces cytotoxicity and apoptosis in Her2 positive breast cancer cells. Future Oncol. 2020, 16, 4485–4495. [Google Scholar] [CrossRef]
- Srivastava, N.S.; Srivastava, R.A.K. Curcumin and quercetin synergistically inhibit cancer cell proliferation in multiple cancer cells and modulate Wnt/beta-catenin signaling and apoptotic pathways in A375 cells. Phytomedicine 2019, 52, 117–128. [Google Scholar] [CrossRef]
- Santana-Galvez, J.; Villela-Castrejon, J.; Serna-Saldivar, S.O.; Cisneros-Zevallos, L.; Jacobo-Velazquez, D.A. Synergistic Combinations of Curcumin, Sulforaphane, and Dihydrocaffeic Acid against Human Colon Cancer Cells. Int. J. Mol. Sci. 2020, 21, 3108. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chang, R.L.; Cui, X.X.; Newmark, H.L.; Conney, A.H. Synergistic effects of curcumin on all-trans retinoic acid- and 1a,25-dihydroxyvitamin D3-induced differentiation in human promyelocytic leukemia HL-60 cells. Oncol. Res. 1997, 9, 19–29. [Google Scholar] [PubMed]
- Nachliely, M.; Sharony, E.; Kutner, A.; Danilenko, M. Novel analogs of 1,25-dihydroxyvitamin D2 combined with a plant polyphenol as highly efficient inducers of differentiation in human acute myeloid leukemia cells. J. Steroid Biochem. Mol. Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Danilenko, M.; Wang, X.; Studzinski, G.P. Carnosic acid and promotion of monocytic differentiation of HL60-G cells initiated by other agents. J. Natl. Cancer Inst. 2001, 93, 1224–1233. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Salman, H.; Danilenko, M.; Studzinski, G.P. Cooperation between antioxidants and 1,25-dihydroxyvitamin D3 in induction of leukemia HL60 cell differentiation through the JNK/AP-1/Egr-1 pathway. J. Cell Physiol. 2005, 204, 964–974. [Google Scholar] [CrossRef] [PubMed]
- Sharabani, H.; Izumchenko, E.; Wang, Q.; Kreinin, R.; Steiner, M.; Barvish, Z.; Kafka, M.; Sharoni, Y.; Levy, J.; Uskokovic, M.; et al. Cooperative antitumor effects of vitamin D3 derivatives and rosemary preparations in a mouse model of myeloid leukemia. Int. J. Cancer 2006, 118, 3012–3021. [Google Scholar] [CrossRef] [PubMed]
- Shabtay, A.; Sharabani, H.; Barvish, Z.; Kafka, M.; Amichay, D.; Levy, J.; Sharoni, Y.; Uskokovic, M.R.; Studzinski, G.P.; Danilenko, M. Synergistic antileukemic activity of carnosic acid-rich rosemary extract and the 19-nor Gemini vitamin D analogue in a mouse model of systemic acute myeloid leukemia. Oncology 2008, 75, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Bobilev, I.; Novik, V.; Levi, I.; Shpilberg, O.; Levy, J.; Sharoni, Y.; Studzinski, G.P.; Danilenko, M. The Nrf2 transcription factor is a positive regulator of myeloid differentiation of acute myeloid leukemia cells. Cancer Biol. Ther. 2011, 11, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gocek, E.; Novik, V.; Harrison, J.S.; Danilenko, M.; Studzinski, G.P. Inhibition of Cot1/Tlp2 oncogene in AML cells reduces ERK5 activation and up-regulates p27Kip1 concomitant with enhancement of differentiation and cell cycle arrest induced by silibinin and 1,25-dihydroxyvitamin D3. Cell Cycle 2010, 9, 4542–4551. [Google Scholar] [CrossRef] [PubMed]
- Pesakhov, S.; Nachliely, M.; Barvish, Z.; Aqaqe, N.; Schwartzman, B.; Voronov, E.; Sharoni, Y.; Studzinski, G.P.; Fishman, D.; Danilenko, M. Cancer-selective cytotoxic Ca2+ overload in acute myeloid leukemia cells and attenuation of disease progression in mice by synergistically acting polyphenols curcumin and carnosic acid. Oncotarget 2016, 7, 31847–31861. [Google Scholar] [CrossRef] [PubMed]
- Pesakhov, S.; Khanin, M.; Studzinski, G.P.; Danilenko, M. Distinct combinatorial effects of the plant polyphenols curcumin, carnosic acid, and silibinin on proliferation and apoptosis in acute myeloid leukemia cells. Nutr. Cancer 2010, 62, 811–824. [Google Scholar] [CrossRef] [PubMed]
- El-Seedi, H.R.; El-Said, A.M.; Khalifa, S.A.; Goransson, U.; Bohlin, L.; Borg-Karlson, A.K.; Verpoorte, R. Biosynthesis, natural sources, dietary intake, pharmacokinetic properties, and biological activities of hydroxycinnamic acids. J. Agric. Food Chem. 2012, 60, 10877–10895. [Google Scholar] [CrossRef] [PubMed]
- Abotaleb, M.; Liskova, A.; Kubatka, P.; Busselberg, D. Therapeutic Potential of Plant Phenolic Acids in the Treatment of Cancer. Biomolecules 2020, 10, 221. [Google Scholar] [CrossRef] [PubMed]
- Su, P.; Shi, Y.; Wang, J.; Shen, X.; Zhang, J. Anticancer agents derived from natural cinnamic acids. Anticancer Agents Med. Chem. 2015, 15, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Trachtenberg, A.; Muduli, S.; Sidoryk, K.; Cybulski, M.; Danilenko, M. Synergistic Cytotoxicity of Methyl 4-Hydroxycinnamate and Carnosic Acid to Acute Myeloid Leukemia Cells via Calcium-Dependent Apoptosis Induction. Front Pharmacol. 2019, 10, 507. [Google Scholar] [CrossRef] [PubMed]
- Sidoryk, K.; Jaromin, A.; Filipczak, N.; Cmoch, P.; Cybulski, M. Synthesis and Antioxidant Activity of Caffeic Acid Derivatives. Molecules 2018, 23, 2199. [Google Scholar] [CrossRef] [PubMed]
- Danilenko, M.; Wang, Q.; Wang, X.; Levy, J.; Sharoni, Y.; Studzinski, G.P. Carnosic acid potentiates the antioxidant and prodifferentiation effects of 1α,25-dihydroxyvitamin D3 in leukemia cells but does not promote elevation of basal levels of intracellular calcium. Cancer Res. 2003, 63, 1325–1332. [Google Scholar] [PubMed]
- Nachliely, M.; Trachtenberg, A.; Khalfin, B.; Nalbandyan, K.; Cohen-Lahav, M.; Yasuda, K.; Sakaki, T.; Kutner, A.; Danilenko, M. Dimethyl fumarate and vitamin D derivatives cooperatively enhance VDR and Nrf2 signaling in differentiating AML cells in vitro and inhibit leukemia progression in a xenograft mouse model. J. Steroid Biochem. Mol. Biol. 2019, 188, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09, Revision D.01; Gaussian, Inc.: Wallingford, CT, USA, 2013; Available online: http://www.gaussian.com (accessed on 6 August 2021).
- Foresman, J.B.; Frisch, Æ. Exploring Chemistry with Electronic Structure Methods, 3rd ed.; Gaussian, Inc.: Wallingford, CT, USA, 2015; p. 500. [Google Scholar]
- Madan, V.; Koeffler, H.P. Differentiation therapy of myeloid leukemia: Four decades of development. Haematologica 2021, 106, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.; McKercher, S.R.; Lipton, S.A. Nrf2/ARE-mediated antioxidant actions of pro-electrophilic drugs. Free Radic. Biol. Med. 2013, 65, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Nioi, P.; McMahon, M.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: Reassessment of the ARE consensus sequence. Biochem. J. 2003, 374, 337–348. [Google Scholar] [CrossRef]
- Sakurai, A.; Nishimoto, M.; Himeno, S.; Imura, N.; Tsujimoto, M.; Kunimoto, M.; Hara, S. Transcriptional regulation of thioredoxin reductase 1 expression by cadmium in vascular endothelial cells: Role of NF-E2-related factor-2. J. Cell Physiol. 2005, 203, 529–537. [Google Scholar] [CrossRef]
- Gacche, R.N.; Jadhav, S.G. Antioxidant Activities and Cytotoxicity of Selected Coumarin Derivatives: Preliminary Results of a Structure–Activity Relationship Study Using Computational Tools. J. Exp. Clin. Med. 2012, 4, 165–169. [Google Scholar] [CrossRef]
- Ismail, T.; Shafi, S.; Srinivas, J.; Sarkar, D.; Qurishi, Y.; Khazir, J.; Alam, M.S.; Kumar, H.M. Synthesis and tyrosinase inhibition activity of trans-stilbene derivatives. Bioorg. Chem. 2016, 64, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.P.; Hayashi, T.; Cottam, H.B.; Jin, G.; Yao, S.; Wu, C.C.; Rosenbach, M.D.; Corr, M.; Schwab, R.B.; Carson, D.A. Nrf2 responses and the therapeutic selectivity of electrophilic compounds in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2010, 107, 7479–7484. [Google Scholar] [CrossRef]
- Lee, E.J.; Kim, S.J.; Hahn, Y.I.; Yoon, H.J.; Han, B.; Kim, K.; Lee, S.; Kim, K.P.; Suh, Y.G.; Na, H.K.; et al. 15-Keto prostaglandin E2 suppresses STAT3 signaling and inhibits breast cancer cell growth and progression. Redox Biol. 2019, 23, 101175. [Google Scholar] [CrossRef] [PubMed]
- Park, J.B. Javamide-I-O-methyl ester increases p53 acetylation and induces cell death via activating caspase 3/7 in monocytic THP-1 cells. Phytomedicine 2016, 23, 1647–1652. [Google Scholar] [CrossRef] [PubMed]
- Micillo, R.; Iacomino, M.; Perfetti, M.; Panzella, L.; Koike, K.; D’Errico, G.; d’Ischia, M.; Napolitano, A. Unexpected impact of esterification on the antioxidant activity and (photo)stability of a eumelanin from 5,6-dihydroxyindole-2-carboxylic acid. Pigment Cell Melanoma Res. 2018, 31, 475–483. [Google Scholar] [CrossRef]
- Scapagnini, G.; Vasto, S.; Abraham, N.G.; Caruso, C.; Zella, D.; Fabio, G. Modulation of Nrf2/ARE pathway by food polyphenols: A nutritional neuroprotective strategy for cognitive and neurodegenerative disorders. Mol. Neurobiol. 2011, 44, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Sokoloski, J.A.; Shyam, K.; Sartorelli, A.C. Induction of the differentiation of HL-60 promyelocytic leukemia cells by curcumin in combination with low levels of vitamin D3. Oncol. Res. 1997, 9, 31–39. [Google Scholar]
- de Freitas Silva, M.; Pruccoli, L.; Morroni, F.; Sita, G.; Seghetti, F.; Viegas, C.; Tarozzi, A. The Keap1/Nrf2-ARE Pathway as a Pharmacological Target for Chalcones. Molecules 2018, 23, 1803. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.W.; Chun, K.S.; Kim, D.H.; Kim, S.J.; Kim, S.H.; Cho, N.C.; Na, H.K.; Surh, Y.J. Curcumin induces stabilization of Nrf2 protein through Keap1 cysteine modification. Biochem. Pharmacol. 2020, 173, 113820. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, P.; Zhao, Y.; Yang, C.; Clark, A.; Leung, T.; Chen, X.; Sang, S. Synthesis, evaluation, and metabolism of novel shogaol derivatives as potent Nrf2 activators. Free Radic. Biol. Med. 2016, 95, 243–254. [Google Scholar] [CrossRef] [PubMed]
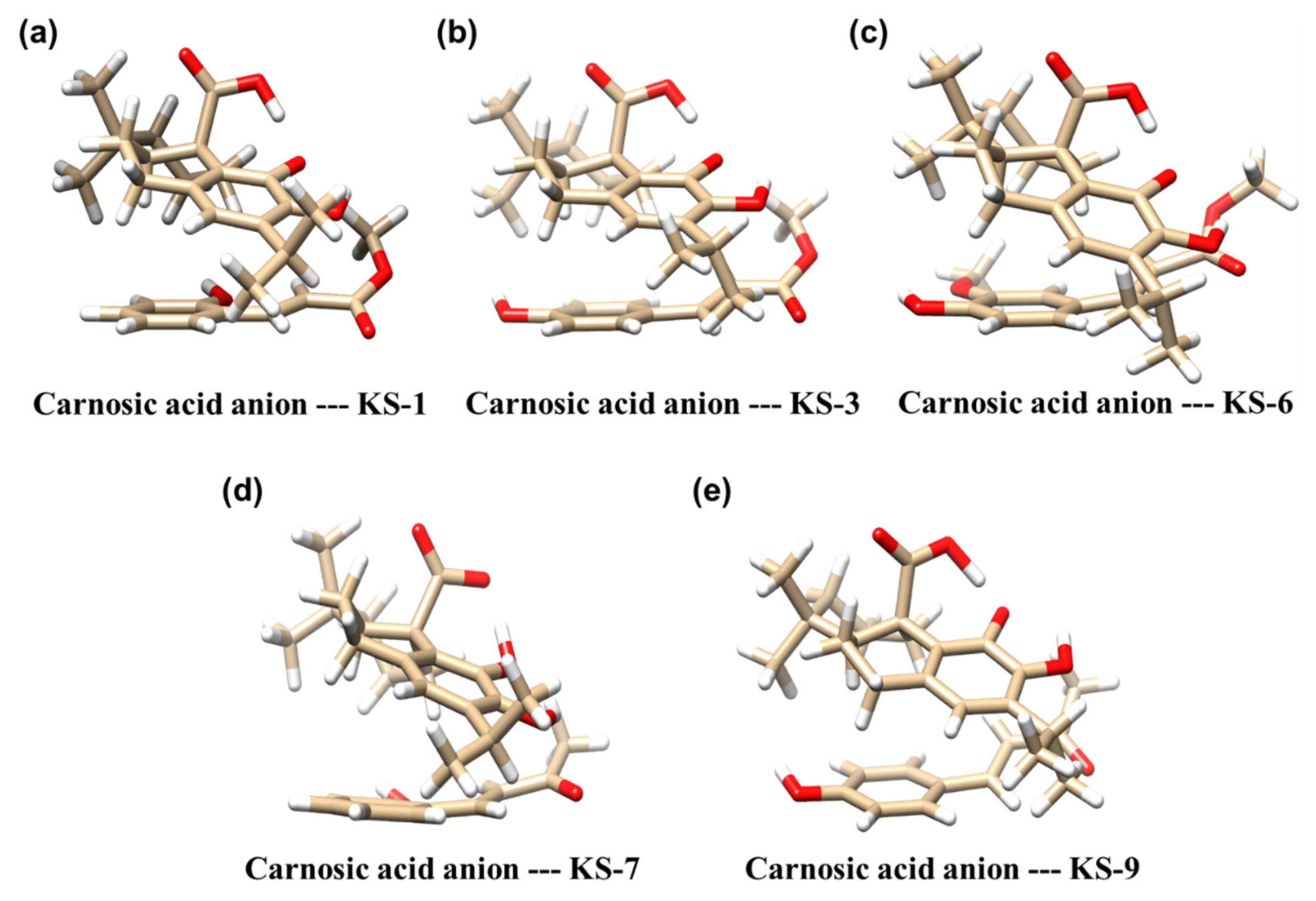
| Molecular Complex a | Binding Energy [kcal/mol] b |
|---|---|
| Carnosic acid anion—KS-1 | −17 |
| Carnosic acid anion—KS-3 | −19 |
| Carnosic acid anion—KS-6 | −16 |
| Carnosic acid anion—KS-7 | −14 |
| Carnosic acid anion—KS-9 | −16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trachtenberg, A.; Sidoryk, K.; Alreate, S.; Muduli, S.; Leś, A.; Cybulski, M.; Danilenko, M. Structure-Activity Relationship of Hydroxycinnamic Acid Derivatives for Cooperating with Carnosic Acid and Calcitriol in Acute Myeloid Leukemia Cells. Biomedicines 2021, 9, 1517. https://doi.org/10.3390/biomedicines9111517
Trachtenberg A, Sidoryk K, Alreate S, Muduli S, Leś A, Cybulski M, Danilenko M. Structure-Activity Relationship of Hydroxycinnamic Acid Derivatives for Cooperating with Carnosic Acid and Calcitriol in Acute Myeloid Leukemia Cells. Biomedicines. 2021; 9(11):1517. https://doi.org/10.3390/biomedicines9111517
Chicago/Turabian StyleTrachtenberg, Aviram, Katarzyna Sidoryk, Somaya Alreate, Suchismita Muduli, Andrzej Leś, Marcin Cybulski, and Michael Danilenko. 2021. "Structure-Activity Relationship of Hydroxycinnamic Acid Derivatives for Cooperating with Carnosic Acid and Calcitriol in Acute Myeloid Leukemia Cells" Biomedicines 9, no. 11: 1517. https://doi.org/10.3390/biomedicines9111517
APA StyleTrachtenberg, A., Sidoryk, K., Alreate, S., Muduli, S., Leś, A., Cybulski, M., & Danilenko, M. (2021). Structure-Activity Relationship of Hydroxycinnamic Acid Derivatives for Cooperating with Carnosic Acid and Calcitriol in Acute Myeloid Leukemia Cells. Biomedicines, 9(11), 1517. https://doi.org/10.3390/biomedicines9111517










