Genome Profiling of H3k4me3 Histone Modification in Human Adipose Tissue during Obesity and Insulin Resistance
Abstract
:1. Introduction
2. Methods
2.1. Sample Collection
2.2. Chromatin Immunoprecipitation Sequencing (ChIP-Seq)
2.3. RNA Sequencing (RNA-Seq)
2.4. ChIP-Seq Bioinformatic Processing and Analysis
2.5. RNA-Seq Bioinformatic Processing and Analysis
3. Results
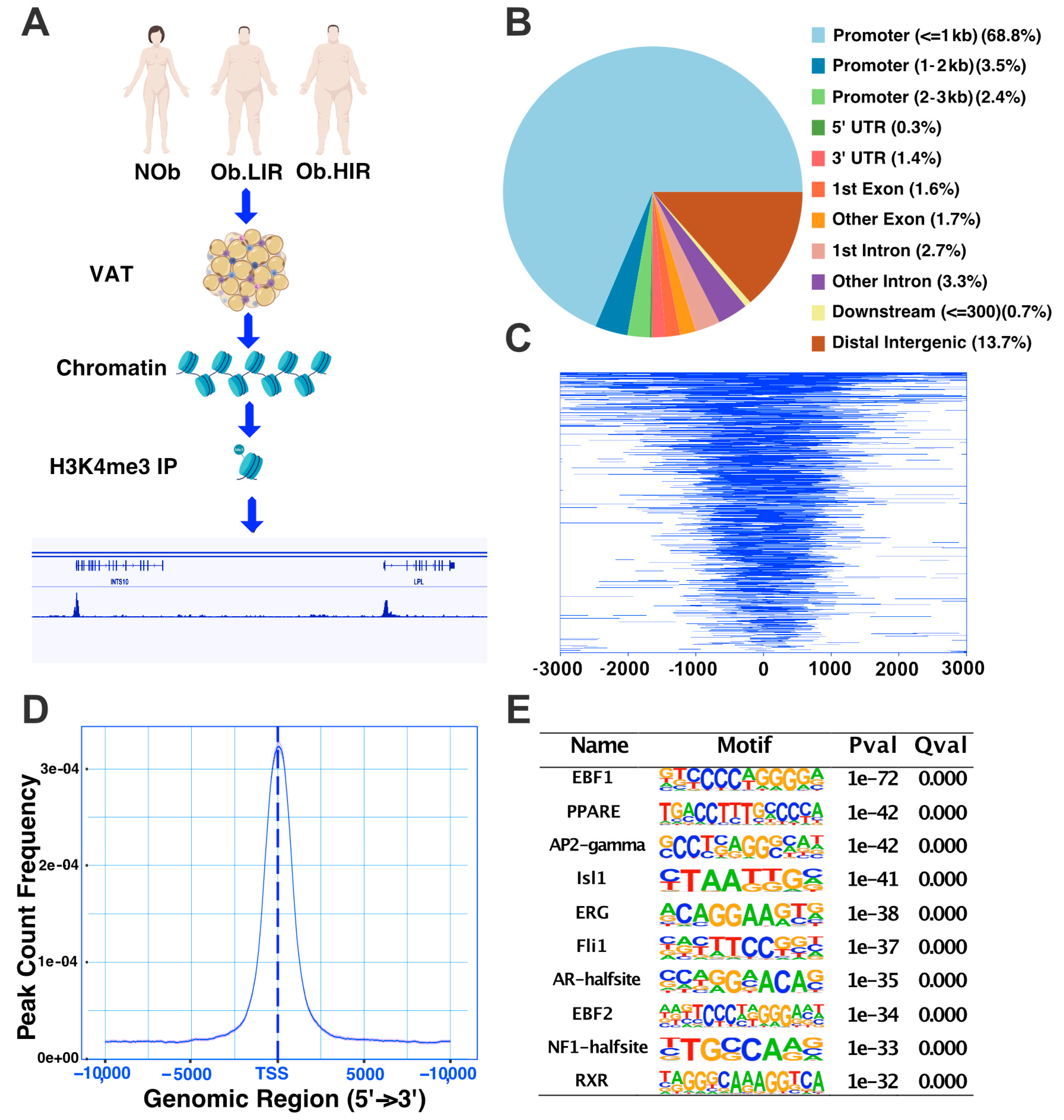
3.1. A Whole Picture of H3K4me3 Profile in Human Adipose Tissue
3.2. H3K4me3 Mark Is Associated with Gene Expression in Human Adipose Tissue
3.3. Analysis of H3K4me3 Enrichment in AT during Obesity and IR
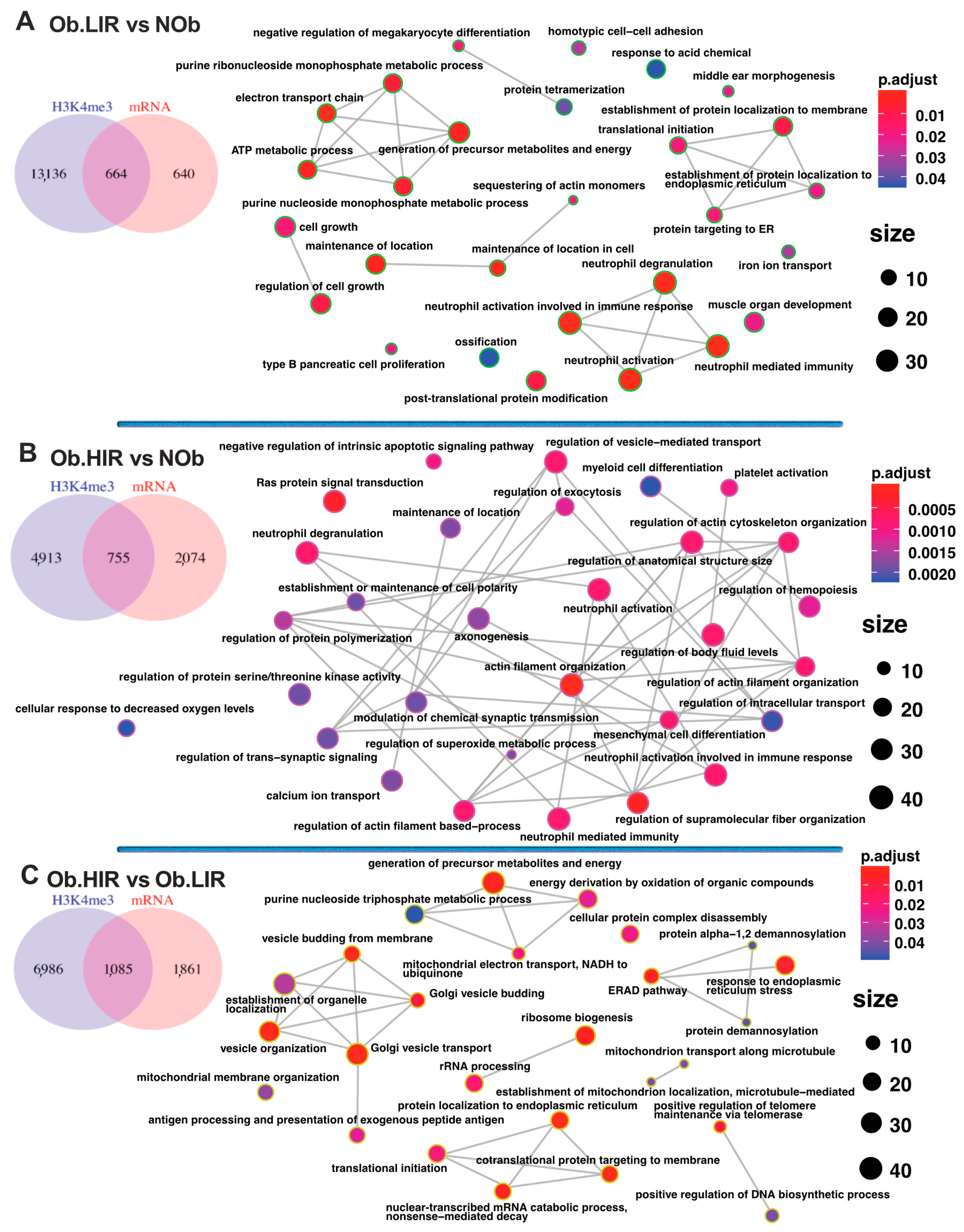
3.4. Differential Analysis of AT H3K4me3 Profile in Obesity and IR
3.5. Changes in H3K4me3 Enrichment Are Related to Transcriptional Changes
3.6. Top 10 Differentially Expressed and H3K4me3 Enriched Genes and Their Relationship with Metabolic Variables
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prins, J.B. Adipose tissue as an endocrine organ. Best Pract. Res. Clin. Endocrinol. Metab. 2002, 16, 639–651. [Google Scholar] [CrossRef]
- Schoettl, T.; Fischer, I.P.; Ussar, S. Heterogeneity of adipose tissue in development and metabolic function. J. Exp. Biol. 2018, 221, jeb162958. [Google Scholar] [CrossRef] [Green Version]
- Kahn, C.R.; Wang, G.; Lee, K.Y. Altered adipose tissue and adipocyte function in the pathogenesis of metabolic syndrome. J. Clin. Investig. 2019, 129, 3990–4000. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Dannenberg, A.J. The obese adipose tissue microenvironment in cancer development and progression Daniela. Nat. Rev. Endocrinol. 2019, 15, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Longo, M.; Zatterale, F.; Naderi, J.; Parrillo, L.; Formisano, P.; Raciti, G.A.; Beguinot, F.; Miele, C. Adipose Tissue Dysfunction as Determinant of Obesity-Associated Metabolic Complications. Int. J. Mol. Sci. 2019, 20, 2358. [Google Scholar] [CrossRef] [Green Version]
- Castellano-Castillo, D.; Ramos-Molina, B.; Cardona, F.; Queipo-Ortuño, M.I. Epigenetic regulation of white adipose tissue in the onset of obesity and metabolic diseases. Obes. Rev. 2020, 21, e13054. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, G.; Heard, E. Advances in epigenetics link genetics to the environment and disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef] [Green Version]
- Seo, J.B.; Riopel, M.; Cabrales, P.; Huh, J.Y.; Bandyopadhyay, G.K.; Andreyev, A.Y.; Murphy, A.N.; Beeman, S.C.; Smith, G.; Klein, S.; et al. Knockdown of ANT2 reduces adipocyte hypoxia and improves insulin resistance in obesity. Nat. Metab. 2019, 1, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.J.; Skrtic, S.; Katsogiannos, P.; Abrahamsson, N.; Sidibeh, C.O.; Dahgam, S.; Månsson, M.; Risérus, U.; Kullberg, J.; Eriksson, J.W. Impaired adipose tissue lipid storage, but not altered lipolysis, contributes to elevated levels of NEFA in type 2 diabetes. Degree of hyperglycemia and adiposity are important factors. Metabolism 2016, 65, 1768–1780. [Google Scholar] [CrossRef]
- Sharma, M.; Boytard, L.; Hadi, T.; Koelwyn, G.; Simon, R.; Ouimet, M.; Seifert, L.; Spiro, W.; Yan, B.; Hutchison, S.; et al. Enhanced glycolysis and HIF-1α activation in adipose tissue macrophages sustains local and systemic interleukin-1β production in obesity. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Castellano-Castillo, D.; Denechaud, P.-D.; Moreno-Indias, I.; Tinahones, F.J.; Fajas, L.; Queipo-Ortuño, M.I.; Cardona, F. Chromatin immunoprecipitation improvements for the processing of small frozen pieces of adipose tissue. PLoS ONE 2018, 13, e0192314. [Google Scholar] [CrossRef] [Green Version]
- Castellano-Castillo, D.; Denechaud, P.-D.; Fajas, L.; Moreno-Indias, I.; Oliva-Olivera, W.; Tinahones, F.J.; Queipo-Ortuño, M.I.; Cardona, F. Human adipose tissue H3K4me3 histone mark in adipogenic, lipid metabolism and inflammatory genes is positively associated with BMI and HOMA-IR. PLoS ONE 2019, 14, e0215083. [Google Scholar] [CrossRef] [Green Version]
- Dai, Z.; Mentch, S.J.; Gao, X.; Nichenametla, S.; Locasale, J.W. Methionine metabolism influences genomic architecture and gene expression through H3K4me3 peak width. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Lefterova, M.I.; Haakonsson, A.K.; Lazar, M.A.; Mandrup, S. PPAR g and the global map of adipogenesis and beyond. Trends Endocrinol. Metab. 2014, 25, 293–302. [Google Scholar] [CrossRef] [Green Version]
- Roh, H.C.; Kumari, M.; Taleb, S.; Tenen, D.; Jacobs, C.; Lyubetskaya, A.; Tsai, L.T.-Y.; Rosen, E.D. Adipocytes fail to maintain cellular identity during obesity due to reduced PPARγ activity and elevated TGFβ-SMAD signaling. Mol. Metab. 2020, 42, 101086. [Google Scholar] [CrossRef]
- Jimenez, M.A.; AÅkerblad, P.; Sigvardsson, M.; Rosen, E.D. Critical Role for Ebf1 and Ebf2 in the Adipogenic Transcriptional Cascade. Mol. Cell. Biol. 2007, 27, 743–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, H.; Mejhert, N.; Fretz, J.; Arner, E.; Lorente-Cebrián, S.; Ehrlund, A.; Dahlman-Wright, K.; Gong, X.; Strömblad, S.; Douagi, I.; et al. Early B Cell Factor 1 Regulates Adipocyte Morphology and Lipolysis in White Adipose Tissue. Cell Metab. 2014, 19, 981–992. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Zhao, J.; Meng, H.; Zhang, X. Adipose Tissue-Resident Immune Cells in Obesity and Type 2 Diabetes. Front. Immunol. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Marcelin, G.; Silveira, A.L.M.; Martins, L.B.; Ferreira, A.V.; Clément, K. Deciphering the cellular interplays underlying obesity-induced adipose tissue fibrosis. J. Clin. Investig. 2019, 129, 4032–4040. [Google Scholar] [CrossRef]
- Jokinen, R.; Pirnes-Karhu, S.; Pietiläinen, K.; Pirinen, E. Adipose tissue NAD+-homeostasis, sirtuins and poly(ADP-ribose) polymerases -important players in mitochondrial metabolism and metabolic health. Redox Biol. 2017, 12, 246–263. [Google Scholar] [CrossRef]
- Sato, F.; Maeda, N.; Yamada, T.; Namazui, H.; Fukuda, S.; Natsukawa, T.; Nagao, H.; Murai, J.; Masuda, S.; Tanaka, Y.; et al. Association of Epicardial, Visceral, and Subcutaneous Fat with Cardiometabolic Diseases. Circ. J. 2018, 82, 502–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusminski, C.M.; Scherer, P.E. Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol. Metab. 2012, 23, 435–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulven, S.M.; Dalen, K.T.; Gustafsson, J.-Å.; Nebb, H.I. LXR is crucial in lipid metabolism. Prostaglandins Leukot. Essent. Fat. Acids 2005, 73, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, R.; Duncan, W.C. The SLIT–ROBO pathway: A regulator of cell function with implications for the reproductive system. Reproduction 2010, 139, 697–704. [Google Scholar] [CrossRef] [Green Version]
- Svensson, K.J.; Long, J.Z.; Jedrychowski, M.P.; Cohen, P.; Lo, J.C.; Serag, S.; Kir, S.; Shinoda, K.; Tartaglia, J.A.; Rao, R.R.; et al. A Secreted Slit2 Fragment Regulates Adipose Tissue Thermogenesis and Metabolic Function. Cell Metab. 2016, 23, 454–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega-Prieto, P.; Postic, C. Carbohydrate Sensing Through the Transcription Factor ChREBP. Front. Genet. 2019, 10, 472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, M.S.; White, A.; Perry, R.J.; Camporez, J.-P.; Hidalgo, J.; Shulman, G.I.; Davis, R.J. Regulation of adipose tissue inflammation by interleukin 6. Proc. Natl. Acad. Sci. USA 2020, 117, 2751–2760. [Google Scholar] [CrossRef] [Green Version]
- Smith, U.; Kahn, B.B. Adipose tissue regulates insulin sensitivity: Role of adipogenesis, de novo lipogenesis and novel lipids. J. Intern. Med. 2016, 280, 465–475. [Google Scholar] [CrossRef] [Green Version]
- Cereijo, R.; Gavaldà-Navarro, A.; Cairó, M.; López, T.P.Q.; Villarroya, J.; Morón-Ros, S.; Sánchez-Infantes, D.; Peyrou, M.; Iglesias, R.; Mampel, T.; et al. CXCL14, a Brown Adipokine that Mediates Brown-Fat-to-Macrophage Communication in Thermogenic Adaptation. Cell Metab. 2018, 28, 750–763.e6. [Google Scholar] [CrossRef] [Green Version]
- Wei, S.; Zhang, M.; Zheng, Y.; Yan, P. ZBTB16 Overexpression Enhances White Adipogenesis and Induces Brown-Like Adipocyte Formation of Bovine White Intramuscular Preadipocytes. Cell. Physiol. Biochem. 2018, 48, 2528–2538. [Google Scholar] [CrossRef]
- Wei, L.; Surma, M.; Yang, Y.; Tersey, S.; Shi, J. ROCK2 inhibition enhances the thermogenic program in white and brown fat tissue in mice. FASEB J. 2020, 34, 474–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diep, D.T.V.; Hong, K.; Khun, T.; Zheng, M.; Ul-Haq, A.; Jun, H.-S.; Kim, Y.-B.; Chun, K.-H. Anti-adipogenic effects of KD025 (SLx-2119), a ROCK2-specific inhibitor, in 3T3-L1 cells. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Senthivinayagam, S.; McIntosh, A.L.; Moon, K.C.; Atshaves, B.P. Plin2 Inhibits Cellular Glucose Uptake through Interactions with SNAP23, a SNARE Complex Protein. PLoS ONE 2013, 8, e73696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, K.; Kang, P. PLIN2 inhibits insulin-induced glucose uptake in myoblasts through the activation of the NLRP3 inflammasome. Int. J. Mol. Med. 2015, 36, 839–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hajri, T.; Hall, A.M.; Jensen, D.R.; Pietka, T.A.; Drover, V.A.; Tao, H.; Eckel, R.; Abumrad, N.A. CD36-Facilitated Fatty Acid Uptake Inhibits Leptin Production and Signaling in Adipose Tissue. Diabetes 2007, 56, 1872–1880. [Google Scholar] [CrossRef] [Green Version]
- Cai, L.; Wang, Z.; Ji, A.; Meyer, J.M.; Van Der Westhuyzen, D.R. Scavenger Receptor CD36 Expression Contributes to Adipose Tissue Inflammation and Cell Death in Diet-Induced Obesity. PLoS ONE 2012, 7, e36785. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, D.J.; Kuchibhotla, S.; Westfall, K.M.; Silverstein, R.L.; Morton, R.E.; Febbraio, M. A CD36-dependent pathway enhances macrophage and adipose tissue inflammation and impairs insulin signalling. Cardiovasc. Res. 2010, 89, 604–613. [Google Scholar] [CrossRef] [Green Version]
- Myneni, V.D.; Mousa, A.; Kaartinen, M.T. Factor XIII-A transglutaminase deficient mice show signs of metabolically healthy obesity on high fat diet. Sci. Rep. 2016, 6, 35574. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, N.; Asada, R.; Saito, A.; Kanemoto, S.; Imaizumi, K. Obesity-induced endoplasmic reticulum stress causes chronic inflammation in adipose tissue. Sci. Rep. 2012, 2, 799. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Wang, Y.; Song, L.; Zeng, L.; Yi, W.; Liu, T.; Chen, H.; Wang, M.; Ju, Z.; Cong, Y.-S. A critical role of DDRGK1 in endoplasmic reticulum homoeostasis via regulation of IRE1α stability. Nat. Commun. 2017, 8, 14186. [Google Scholar] [CrossRef] [PubMed]


| Ob.LIR vs. NOb | Ob.HIR vs. NOb | Ob.HIR vs. 0b.LIR | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene | FC | p | Gene | FC | p | Gene | FC | p |
| CXCL14 | 1.75 | 0.000 | ADRA2A | 1.59 | 0.000 | CD36 | 1.62 | 0.000 |
| TERC | 2.33 | 0.000 | LCP1 | 1.79 | 0.000 | F13A1 | 0.89 | 0.004 |
| ZBTB16 | 2.09 | 0.000 | NANOS1 | 1.27 | 0.000 | DTX4 | 0.89 | 0.004 |
| EDN1 | 1.28 | 0.001 | ROCK2 | 0.77 | 0.000 | ATP8B4 | 0.97 | 0.007 |
| KLF15 | 1.42 | 0.001 | PLIN2 | 1.55 | 0.000 | FGF12 | 1.12 | 0.014 |
| GRIA1 | −1.63 | 0.000 | PICK1 | −1.10 | 0.000 | DDRGK1 | −0.90 | 0.000 |
| GRIK3 | −2.50 | 0.000 | ACOX3 | −0.77 | 0.000 | PDAP1 | −0.98 | 0.000 |
| NRXN3 | −1.82 | 0.013 | GRIK3 | −2.76 | 0.000 | RPL24 | −1.02 | 0.000 |
| CSMD2 | −2.24 | 0.015 | PITPNM3 | −1.54 | 0.000 | INCENP | −0.96 | 0.000 |
| ISL1 | −0.89 | 0.016 | EPS8L1 | −2.05 | 0.000 | CSKMT | −3.09 | 0.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castellano-Castillo, D.; Ramos-Molina, B.; Oliva-Olivera, W.; Ocaña-Wilhelmi, L.; Queipo-Ortuño, M.I.; Cardona, F. Genome Profiling of H3k4me3 Histone Modification in Human Adipose Tissue during Obesity and Insulin Resistance. Biomedicines 2021, 9, 1363. https://doi.org/10.3390/biomedicines9101363
Castellano-Castillo D, Ramos-Molina B, Oliva-Olivera W, Ocaña-Wilhelmi L, Queipo-Ortuño MI, Cardona F. Genome Profiling of H3k4me3 Histone Modification in Human Adipose Tissue during Obesity and Insulin Resistance. Biomedicines. 2021; 9(10):1363. https://doi.org/10.3390/biomedicines9101363
Chicago/Turabian StyleCastellano-Castillo, Daniel, Bruno Ramos-Molina, Wilfredo Oliva-Olivera, Luis Ocaña-Wilhelmi, María Isabel Queipo-Ortuño, and Fernando Cardona. 2021. "Genome Profiling of H3k4me3 Histone Modification in Human Adipose Tissue during Obesity and Insulin Resistance" Biomedicines 9, no. 10: 1363. https://doi.org/10.3390/biomedicines9101363
APA StyleCastellano-Castillo, D., Ramos-Molina, B., Oliva-Olivera, W., Ocaña-Wilhelmi, L., Queipo-Ortuño, M. I., & Cardona, F. (2021). Genome Profiling of H3k4me3 Histone Modification in Human Adipose Tissue during Obesity and Insulin Resistance. Biomedicines, 9(10), 1363. https://doi.org/10.3390/biomedicines9101363







