A Multi-Omics Approach Reveals New Signatures in Obese Allergic Asthmatic Children
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Subjects
2.2. Determination of Clinical, Biochemical and Anthropometric Parameters
2.3. Inflammatory Biomarkers Determination
2.4. 1H NMR Spectroscopy of Blood Plasma
2.5. Gas Chromatography-Mass Spectrometry (GC-MS) Based SCFA Analysis
2.6. Fecal D and L lactate and Intestinal Microbiota Analysis
2.7. Statistical Analysis
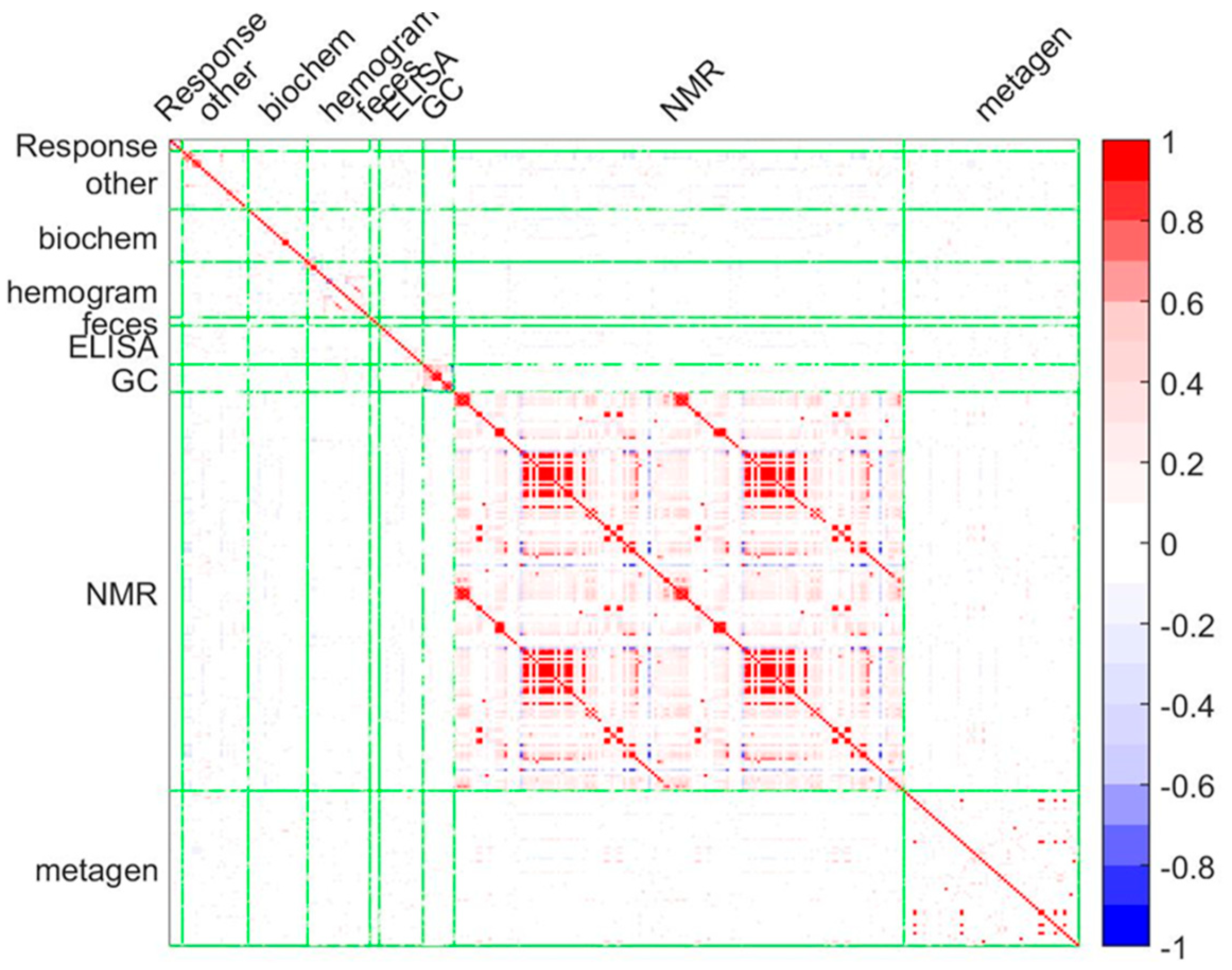
2.8. The Multi-Omics Approach
3. Results
3.1. General Characteristics of the Participants
3.2. 1H-NMR Metabolites and Intestinal Microbiota Composition in the Asthmatic Cohort
3.3. Systems Biology Characterization of the Asthmatic Cohort
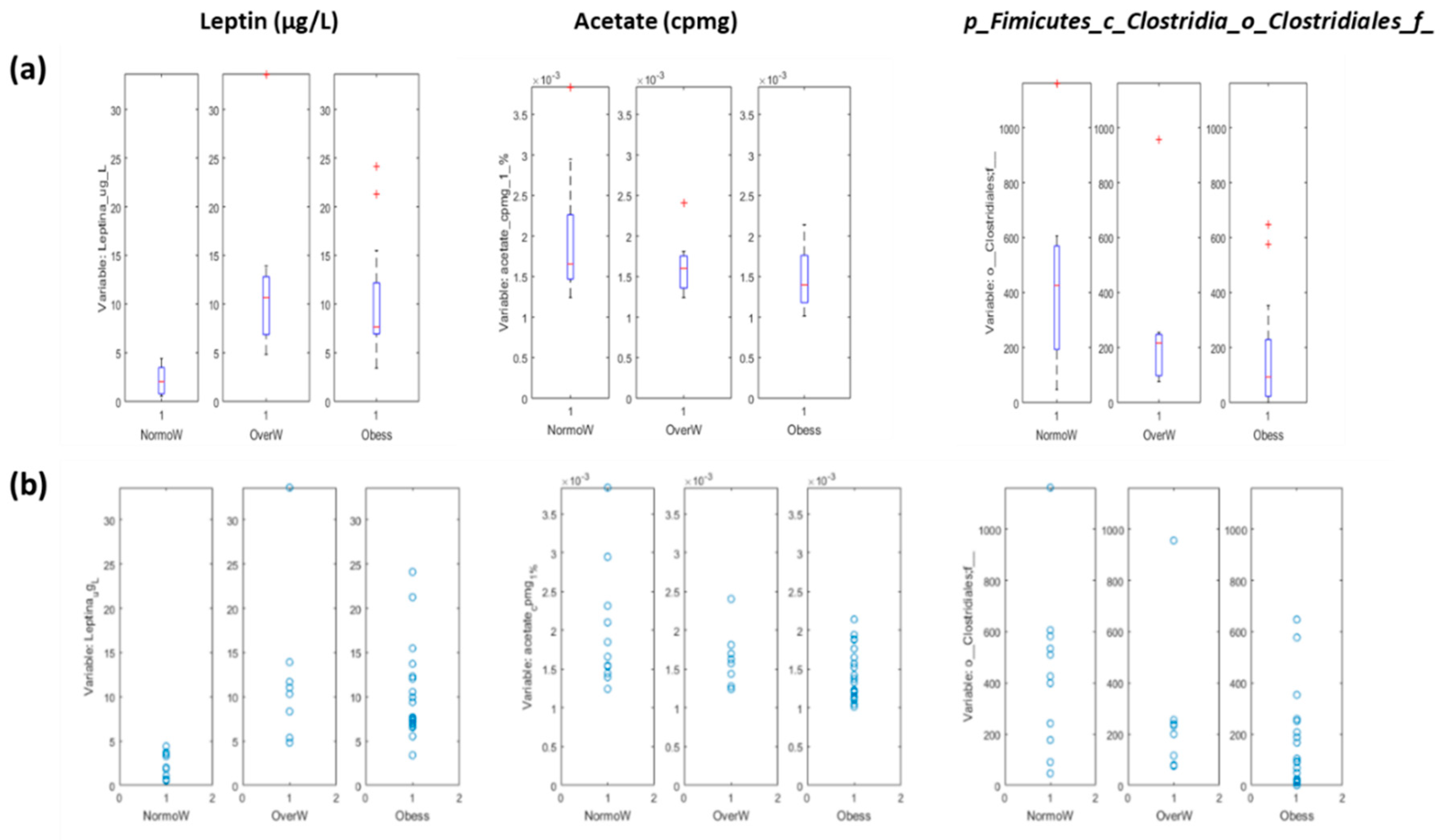
3.3.1. Systems Biology Characterization of the Asthmatic Cohort According to the Obese Status
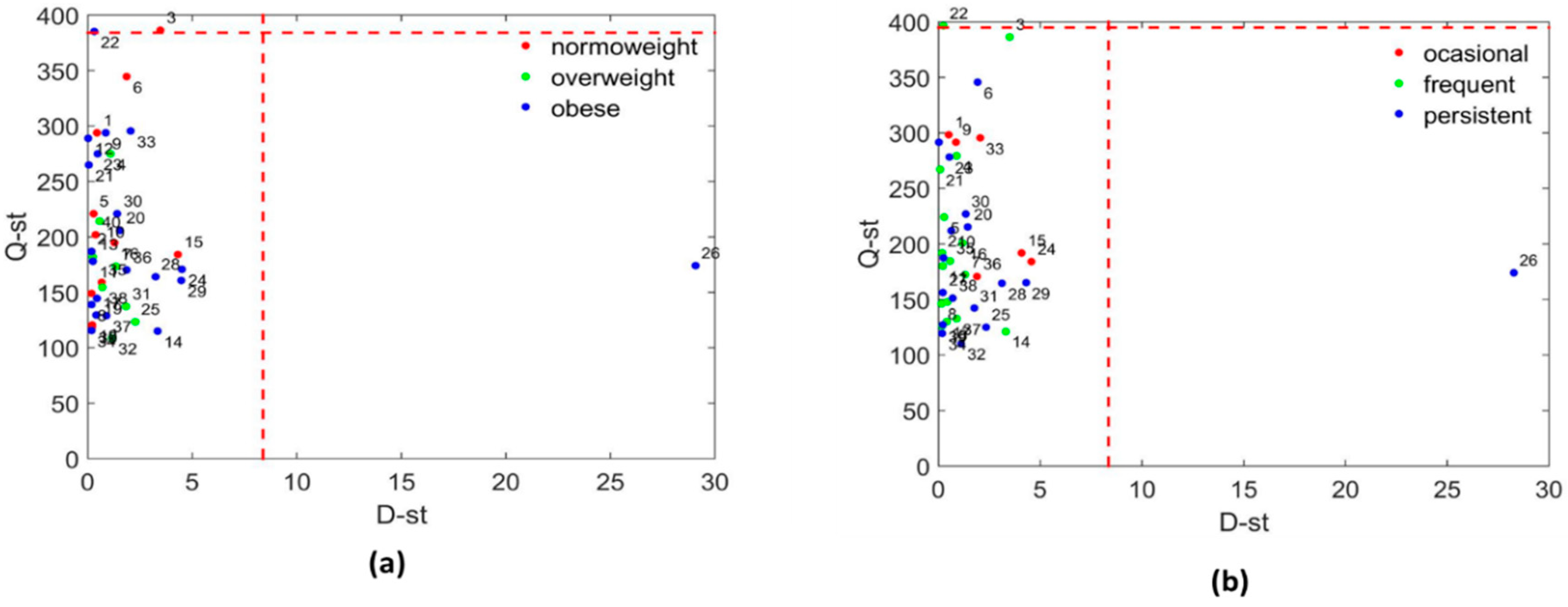
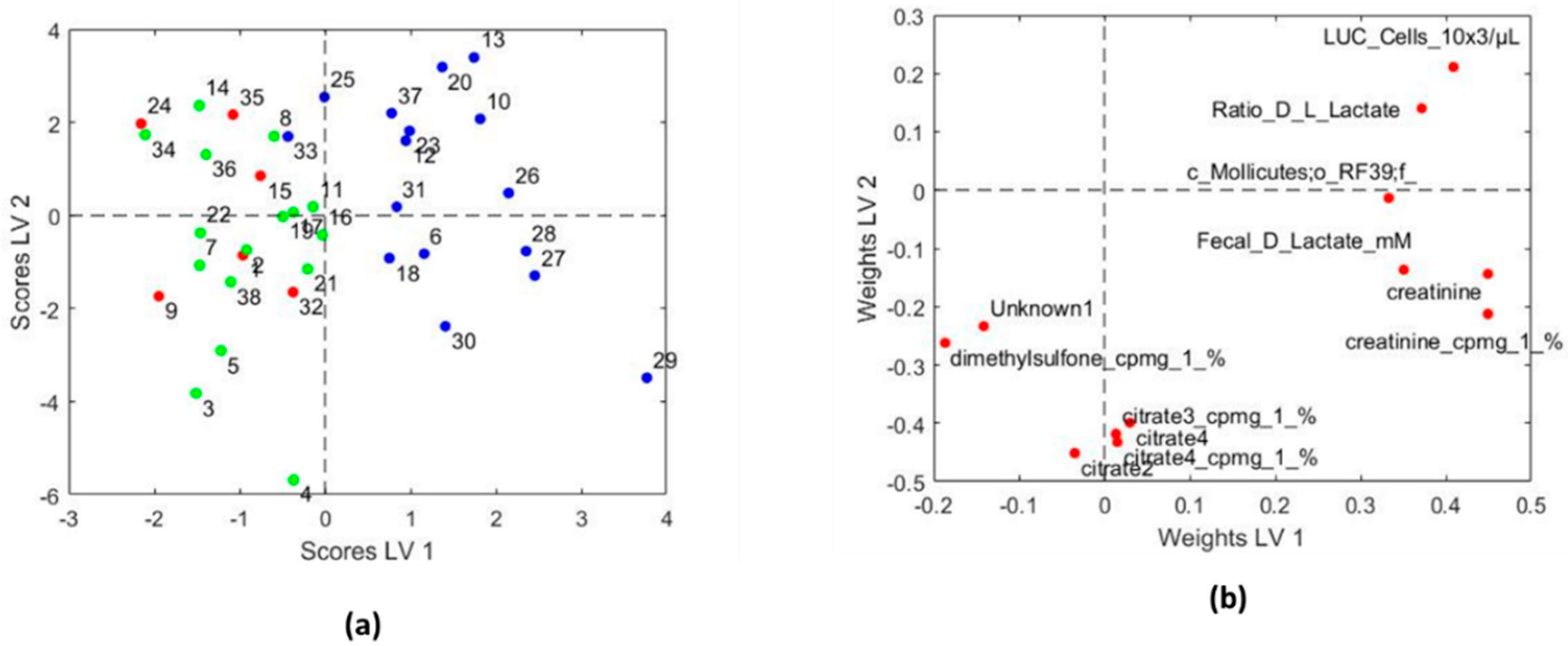
3.3.2. Systems Biology Characterization of the Asthmatic Cohort According to the Asthma Severity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lang, J.E. Obesity and childhood asthma. Curr. Opin. Pulm. Med. 2019, 25, 34–43. [Google Scholar] [CrossRef]
- Eder, W.; Ege, M.J.; Von Mutius, E. The asthma epidemic. N. Engl. J. Med. 2006, 355, 2226–2235. [Google Scholar] [CrossRef]
- Chung, K.F.; Wenzel, S.E.; Brozek, J.L.; Bush, A.; Castro, M.; Sterk, P.J.; Adcock, I.M.; Bateman, E.D.; Bel, E.H.; Bleecker, E.R.; et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur. Respir. J. 2014, 43, 343–373. [Google Scholar] [CrossRef]
- Kuruvilla, M.E.; Lee, F.E.H.; Lee, G.B. Understanding asthma phenotypes, endotypes and mechanisms of disease. Clin. Rev. Allergy Immunol. 2019, 56, 219–233. [Google Scholar] [CrossRef]
- Wenzel, S.E. Asthma phenotypes: The evolution from clinical to molecular approaches. Nat. Med. 2012, 18, 716. [Google Scholar] [CrossRef]
- Sidevela, O.; Dixon, A.E. The many faces of asthma in obesity. J. Cell. Biochem. 2014, 115, 421–426. [Google Scholar] [CrossRef]
- Dixon, A.E.; Pounter, M.E. Mechanisms of asthma in obesity pleiotropic aspects of obesity produce distinct asthma phenotypes. Am. J. Respir. Cell Moll. Biol. 2016, 54, 601–608. [Google Scholar] [CrossRef]
- Contreras, Z.A.; Chen, Z.; Roumeliotaki, T.; Annesi-Maesano, I.; Baïz, N.; von Berg, A.; Bergström, A.; Crozier, S.; Duijts, L.; Ekström, S.; et al. Does early-onset asthma increase childhood obesity risk? A pooled analysis of 16 European cohorts. Eur. Respir. J. 2018, 52, 1800504. [Google Scholar] [CrossRef]
- Rastogi, D.; Fraser, S.; Oh, J.; Huber, A.M.; Schulman, Y.; Bhagtani, R.H.; Khan, Z.S.; Tesfa, L.; Hall, C.B.; Macian, F. Inflammation, metabolic dysregulation, and pulmonary function among obese urban adolescents with asthma. Am. J. Respir. Crit. Care Med. 2015, 191, 149–160. [Google Scholar] [CrossRef]
- Olza, J.; Gil-Campos, M.; Leis, R.; Bueno, G.; Aguilera, C.M.; Valle, M.; Cañete, R.; Tojo, R.; Moreno, L.A.; Gil, A. Presence of the metabolic syndrome in obese children at prepubertal age. Ann. Nutr. Metab. 2011, 58, 343–350. [Google Scholar] [CrossRef]
- Forno, E.; Han, Y.Y.; Muzumdar, R.H.; Celedon, J.C. Insulin resistance, metabolic syndrome, and lung function in US adolescents with and without asthma. J. Allergy Clin. Immunol. 2015, 136, 304–311. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Rampelli, S.; Guenther, K.; Turroni, S.; Wolters, M.; Veidebaum, T.; Kourides, Y.; Molnár, D.; Lissner, L.; Benitez-Paez, A.; Sanz, Y.; et al. Pre-obese children’s dysbiotic gut microbiome and unhealthy diets may predict the development of obesity. Commun. Biol. 2018, 1, 222. [Google Scholar] [CrossRef]
- Stokholm, J.; Blaser, M.J.; Thorsen, J.; Rasmussen, M.A.; Waage, J.; Vinding, R.K.; Schoos, A.M.M.; Kunøe, A.; Fink, N.R.; Chawes, B.L.; et al. Maturation of gut microbiome and risk of asthma in childhood. Nat. Commun. 2018, 9, 141. [Google Scholar] [CrossRef]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–168. [Google Scholar] [CrossRef]
- Thorburn, A.N.; McKenzie, C.I.; Shen, S.J.; Stanley, D.; Macia, L.; Mason, L.J.; Roberts, L.K.; Wong, C.H.; Shim, R.; Robert, R.; et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 2015, 6, 7320. [Google Scholar] [CrossRef]
- Boulet, L.P.; Franssen, E. Influence of obesity response to fluticasone with or whitout salmeterol in moderate asthma. Respir. Med. 2007, 101, 2240–2247. [Google Scholar] [CrossRef]
- Mukadam, S.; Zacharias, J.; Henao, M.P.; Kraschnewski, J.; Ishmael, F. Differential effects of obesity on eosinophilic vs. non-eosinophilic asthma subtypes. J. Asthma 2017, 28, 1–6. [Google Scholar] [CrossRef]
- Michalovich, D.; Rodriguez-Perez, N.; Smolinska, S.; Pirozynski, M.; Mayhew, D.; Uddin, S.; Van Horn, S.; Sokolowska, M.; Altunbulakli, C.; Eljaszewicz, A.; et al. Obesity and disease severity magnify disturbed microbiome-immune interactions in asthma patients. Nat. Commun. 2019, 10, 5711. [Google Scholar] [CrossRef]
- Plaza Moral, V.; Alonso Mostaza, S. Comité Ejecutivo de GEMA. GEMA (4.0). Spanish guideline on the management of asthma. J. Investig. Allergol. Immnunol. 2016, 26, 1–92. [Google Scholar] [CrossRef]
- Cole, T.J.; Bellizi, M.C.; Flegal, K.M.; Dietz, W.H. Establising a standard definition for child overweight and obesity worldwide: International survey. BMJ 2000, 320, 1240–1243. [Google Scholar] [CrossRef]
- Anguita-Ruiz, A.; Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Ruperez, A.I.; Leis, R.; Bueno, G.; Gil-Campos, M.; Vazquez-Cobela, R.; Cañete, R.; Moreno, L.A.; et al. X chromosome genetic data in a Spanish children cohort, dataset description and analysis pipeline. Sci. Data 2019, 6, 130. [Google Scholar] [CrossRef]
- Gardner, M.D. Standarization of spirometry: A summary of recommendations from the American Thoracic Society. The 1987 update. Ann. Intern. Med. 1988, 108, 217–220. [Google Scholar] [CrossRef]
- Sharek, P.J.; Mayer, M.L.; Loewy, L.; Robinson, T.N.; Shames, R.S.; Umetsu, D.T.; Bergman, D.A. Agreement among measures of asthma status: A prospective study of low-income children with moderate to severe asthma. Pediatrics 2002, 110, 797–804. [Google Scholar] [CrossRef]
- Dweik, R.A.; Boggs, P.B.; Erzurum, S.C.; Irvin, C.G.; Leigh, M.W.; Lundberg, J.O.; Olin, A.C.; Plummer, A.L.; Taylor, D.R.; American Thoracic Society Committee on Interpretation of Exhaled Nitric Oxide Levels (FENO) for Clinical Applications. An official ATS clinical practice guideline: Interpretation of exhaled nitric oxide levels (FENO) for clinical applications. Am. J. Respir. Crit. Care Med. 2011, 184, 602–615. [Google Scholar] [CrossRef]
- Mccrindle, B.W. Assessment and management of hyperthension in children and adolescents. Nat. Rev. Cardio 2010, 7, 155–163. [Google Scholar] [CrossRef]
- Monsonis Centelles, S.; Hoefsloot, H.C.; Khakimov, B.; Ebrahimi, P.; Lind, M.V.; Kristensen, M.; De Roo, N.; Jacobs, D.M.; Van Duynhoven, J.; Cannet, C.; et al. Toward reliable lipoprotein particle predictions from NMR spectra of human blood: An interlaboratory ring test. Anal. Chem. 2017, 89, 8004–8012. [Google Scholar] [CrossRef]
- Khakimov, B.; Mobaraki, N.; Trimigno, A.; Aru, V.; Engelsen, S.B. Signature Mapping (SigMA): An efficient approach for processing complex human urine 1H NMR metabolomics data. Anal. Chim. Acta 2020, 1108, 142–151. [Google Scholar] [CrossRef]
- Weise, M.; Khakimov, B.; Nielsen, S.; Sørensen, H.; Van den Berg, F.; Nielsen, D.S. CoMiniGut- a small volume in vitro colon model for the screening of gut microbial fermentation processes. PeerJ 2018, 6, e4268. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glockner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 7, e1. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal Component Analysis; Springer Inc.: Secaucus, NJ, USA, 2002. [Google Scholar]
- Jackson, J.E. A User’s Guide to Principal Components; Wiley-Interscience: Hoboken, NJ, USA, 2003. [Google Scholar]
- Kourti, T.; MacGregor, J.F. Multivariate SPC methods for process and product monitoring. J. Qual. Technol. 1996, 4, 409–428. [Google Scholar] [CrossRef]
- Baker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Lê Cao, K.A.; Rossouw, D.; Robert-Granié, C.; Besse, P. A sparse PLS for variable selection when integrating omics data. Stat. Appl. Genet. Mol. Biol. 2008, 7. [Google Scholar] [CrossRef]
- Camacho, J. Missing-data theory in the context of exploratory data analysis. Chemom. Intell. Lab. Syst. 2010, 1, 8–18. [Google Scholar] [CrossRef]
- Han, Y.Y.; Forno, E.; Celedon, J.C. Adiposity, fractional exhaled nitric oxide, and asthma symptoms. Am. J. Respir. Crit. Care Med. 2014, 190, 32–39. [Google Scholar] [CrossRef]
- Rastogi, D.; Canfield, S.; Andrade, A.; Isasi, C.R.; Hall, C.B.; Rubinstein, A.; Arens, R. Obesity-associated asthma in children: A distinct entity. Chest 2012, 141, 895–905. [Google Scholar] [CrossRef]
- Mbakwa, C.A.; Hermes, G.D.A.; Penders, J.; Savelkoul, P.H.M.; Thijs, C.; Dagnelie, P.C.; Mommers, M.; Zoetendal, E.G.; Smidt, H.; Arts, I.C.W. Gut microbiota and body weight in school-aged children: The KOALA birth cohort study. Obesity 2018, 26, 1767–1776. [Google Scholar] [CrossRef]
- Hua, X.; Goedert, J.J.; Pu, A.; Yu, G.; Shi, J. Allergy associations with the adult fecal microbiota: Analysis of the American gut project. EBioMedicine 2016, 3, 172–179. [Google Scholar] [CrossRef]
- Dalile, B.; Van oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota-gut-brain communications. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef]
- Vanker, N.; Ipp, H. Large unstained cells: A potentially valuable parameter in the assessment of immune activation levels in HIV infection. Acta Haematol. 2014, 131, 208–212. [Google Scholar] [CrossRef]
- Bolton, C.F. Sepsis and the systemic inflammatory response syndrome: Neuromuscular manifestations. Crit. Care Med. 1996, 24, 1408–1416. [Google Scholar] [CrossRef]
- Saude, E.J.; Skappak, C.D.; Regush, S.; Cook, K.; Ben-Zvi, A.; Becker, A.; Moqbel, R.; Sykes, B.D.; Rowe, B.H.; Adamko, D.J. Metabolomic profiling of asthma: Diagnostic utility of urine nuclear magnetic resonance spectroscopy. J. Allergy Clin. Immunol. 2011, 127, 757–764. [Google Scholar] [CrossRef]
- Chang, C.; Guo, Z.-G.; He, B.; Yao, W.-Z. Metabolomic alterations in the sera of Chinese patients with mild persistent asthma: A GC-MS-based metabolomics analysis. Acta Pharmacol. Sin. 2015, 36, 1356–1366. [Google Scholar] [CrossRef]
- Zhang, D.; Li, S.; Wang, N.; Tan, H.Y.; Zhang, Z.; Feng, Y. The cross-talk between gut microbiota and lungs in common lung disease. Front. Microbiol. 2020, 11, 301–315. [Google Scholar] [CrossRef]
- Mayeur, C.; Gratadoux, J.J.; Bridonneau, C.; Chegdani, F.; Larroque, B.; Kapel, N.; Corcos, O.; Thomas, M.; Joly, F. Faecal D/L lactate ratio is a metabolic signature of microbiota imbalance in patients with short bowel syndrome. PLoS ONE 2013, 8, e54335. [Google Scholar] [CrossRef]
- He, X.; Slupsky, C.M. Metabolic fingerprint of dimethyl sulfone (DMSO2) in microbial-mammalian co-metabolism. J. Proteome Res. 2014, 13, 5281–5292. [Google Scholar] [CrossRef] [PubMed]
| Normal-Weight | Overweight | Obese | p Value | |
|---|---|---|---|---|
| N(F/M) | 13 (3/10) | 8 (2/6) | 25 (7/18) | - |
| Age (year) | 8.5 ± 3.0 | 9.6 ± 2.0 | 8.4 ± 2.0 | 0.4 |
| Anthropometric characteristics | ||||
| BMI | 16.1 ± 1.3 | 22.6 ± 1.9 | 24.3 ± 2.6 | 0.001 a,b,c |
| Hip circumference (cm) | 69.4 ± 7.3 | 83.7 ± 7.9 | 82.4 ± 10.2 | 0.003 a,b |
| BMIzscore | −0.5 ± 0.7 | 1.7 ± 0.2 | 2.7 ± 0.7 | 0.001 a,b,c |
| Waist to hip ratio | 0.8 ± 0.0 | 0.8 ± 0.1 | 0.9 ± 0.1 | 0.3 |
| Systolic blood pressure (mmHg) | 100 ± 11.5 | 108.7 ± 15 | 109.2 ± 12.9 | 0.2 |
| Diastolic blood pressure (mmHg) | 56.3 ± 5.3 | 59.2 ± 9.7 | 64.6 ± 12.5 | 0.1 |
| Asthmatic related clinical parameters | ||||
| GEMA classification (occasional/frequent/persistent) | 2/7/3 | 0/2/6 | 5/10/10 | - |
| IgE higher than 3.5 | 13 | 7 | 21 | - |
| FEV1 (%) | 93.3 ± 14.3 | 100.9 ± 8.6 | 99.8 ± 12.7 | 0.7 |
| FVC (%) | 96.1 ± 13.0 | 96.3 ± 6.3 | 99.0 ± 11.7 | 0.7 |
| FEF25-75 (%) | 80.9 ± 23.0 | 91.6 ± 22.3 | 86.4 ± 23.3 | 0.7 |
| FEV1/FVC ratio | 93.5 ± 14.0 | 97.4 ± 12.0 | 92.2 ± 12.1 | 0.7 |
| Biochemical and hematological parameters | ||||
| Glucose (mg/dL) | 84.2 ± 9.7 | 88.6 ± 7.1 | 85.2 ± 9.5 | 0.05 |
| Insulin (μU/mL) | 6.4 ± 3.7 | 11.3 ± 2.5 | 12.7 ± 18.4 | 0.028 b |
| HOMA index | 1.1 ± 0.6 | 2.25 ± 0.3 | 2.9 ± 5.5 | 0.011 b |
| Triacylglycerols (mg/dL) | 53.7 ± 13.7 | 78.7 ± 31.0 | 76.5 ± 36 | 0.1 |
| HDL (mg/dL) | 55.6 ± 10.4 | 56.7 ± 14.2 | 52.2 ± 10.0 | 0.6 |
| LDL (mg/dL) | 80.6 ± 18.3 | 99.2 ± 10.0 | 96.5 ± 23.5 | 0.2 |
| Total cholesterol (mg/dL) | 148.4 ± 20.0 | 164.7 ± 23.4 | 159.8 ± 29 | 0.4 |
| C reactive protein (μU/mL) | 5.0 ± 7.5 | 2.4 ± 2.3 | 4.9 ± 4.9 | 0.6 |
| Transferrin (mg/dL) | 302.1 ± 64.3 | 287.6 ± 26.5 | 287.8 ± 32.2 | 1.0 |
| GPT | 16.5 ± 4.4 | 19.6 ± 8.1 | 25.1 ± 11.7 | 0.038 a |
| γ-GT | 14.6 ± 2.7 | 17.7 ± 3.3 | 17.1 ± 4.4 | 0.1 |
| TSH (μU/mL) | 2.3 ± 0.8 | 2.4 ± 0.8 | 2.4 ± 1.2 | 1.0 |
| Uric acid (mg/dL) | 4.1 ± 0.7 | 3.7 ± 0.8 | 4.7 ± 1.0 | 0.018 c |
| Sodium (mEq/L) | 136.8 ± 1.8 | 138.6 ± 1.4 | 138.2 ± 2.0 | 0.033 a |
| Potassium (mEq/L) | 4.5 ± 0.4 | 4.5 ± 0.5 | 4.4 ± 0.2 | 0.9 |
| Calcium (mg/dL) | 10.0 ± 0.2 | 10.2 ± 0.3 | 10.1 ± 0.3 | 0.1 |
| Vitamin D (ng/mL) | 24.9 ± 10.7 | 27.4 ± 8.2 | 26.1 ± 6.3 | 0.8 |
| Erythrocyte (×106/μL) | 5.1 ± 0.4 | 4.9 ± 0.3 | 5.0 ± 0.2 | 0.3 |
| Hemoglobin (g/dL) | 13.5 ± 1.0 | 13.7 ± 0.8 | 13.7 ± 0.9 | 0.8 |
| Leukocyte (×103/μL) | 7.9 ± 3.2 | 7.2 ± 2.2 | 7.9 ± 1.7 | 0.5 |
| Neutrophils (×103/μL) | 3.9 ± 1.8 | 3.4 ± 1.3 | 3.7 ± 1.2 | 0.7 |
| Linfocyte (×103/μL) | 2.6 ± 0.9 | 2.7 ± 0.6 | 3.0 ± 0.8 | 0.2 |
| Monocyte (×103/μL) | 0.5 ± 0.2 | 0.5 ± 0.1 | 0.5 ± 0.2 | 0.9 |
| Eosinophils (×103/μL) | 0.7 ± 0.8 | 0.5 ± 0.6 | 0.5 ± 0.5 | 0.5 |
| LUC cells (×103/μL) | 0.2 ± 0.1 | 0.2 ± 0.1 | 0.1 ± 0.1 | 0.7 |
| Platelet (×103/μL) | 306.9 ± 57.9 | 291.6 ± 90.7 | 320.2 ± 66.8 | 0.6 |
| Normal-Weight | Overweight | Obese | p Value | |
|---|---|---|---|---|
| Adiponectin (mg/L) | 7.0 ± 4.3 | 6.9 ± 3.8 | 6.8 ± 4.4 | 0.9 |
| Resistin (μg/mL) | 13.8 ± 4.3 | 13.5 ± 2.8 | 13.4 ± 4.8 | 1.0 |
| Leptin (μg/L) | 2.2 ± 1.4 | 12.4 ± 9.1 | 10.0 ± 4.8 | 0.0001 a,b |
| MCP-1 (pg/mL) | 124.0 ± 35.8 | 115.6 ± 28.9 | 134.3 ± 54.2 | 0.6 |
| TNF-α (pg/mL) | 4.0 ± 1.1 | 3.7 ± 2.0 | 3.5 ± 1.0 | 0.3 |
| Chitinase-3 (μg/L) | 17.1 ± 7.6 | 19.4 ± 6.2 | 21.5 ± 8.8 | 0.4 |
| IL-10 (pg/mL) | 4.0 ± 1.5 | 5.7 ± 2.4 | 4.5 ± 1.7 | 0.3 |
| IL-13 (pg/mL) | 3.0 ± 1.8 | 3.1 ± 2.7 | 2.3 ± 2.6 | 0.4 |
| IL-4 (pg/mL) | 13.5 ± 6.3 | 39.9 ± 52.8 | 13.4 ± 6.7 | 0.2 |
| IL-5 (pg/mL) | 1.3 ± 0.4 | 1.7 ± 1.0 | 1.4 ± 0.4 | 0.7 |
| IL-6 (pg/mL) | 1.6 ± 1.3 | 1.7 ± 1.3 | 1.7 ± 2.3 | 0.8 |
| IL-8 (pg/mL) | 2.1 ± 0.6 | 1.7 ± 0.5 | 1.8 ± 0.7 | 0.5 |
| LBP (µg/mL) | 3.0 ± 1.4 | 3.8 ± 1.2 | 3.5 ± 1.8 | 0.6 |
| Asthmatic Children | Control Children | p-Value | |
|---|---|---|---|
| N(F/M) | 46 (12/34) | 104 (57/47) | - |
| Age (year) | 8.7 ± 2.3 | 8.0 ± 0.5 | 0.1 |
| BMI | 21.6 ± 4.2 | 22.5 ± 5.4 | 0.4 |
| BMIzscore | 1.6 ± 1.5 | 2.1 ± 2.0 | 0.2 |
| Systolic blood pressure (mmHg) | 107.3 ± 13.2 | 107 ± 12.5 | 0.9 |
| Diastolic blood pressure (mmHg) | 62.1 ± 11.3 | 67.9 ± 12.9 | 0.005 |
| GPT (U/L) | 22.0 ± 10.4 | 20.5 ± 8.3 | 0.8 |
| γ-GT (U/L) | 16.6 ± 4.0 | 14.9 ± 9.6 | 0.001 |
| Triacylglycerols (mg/dL) | 71.3 ± 32.2 | 64.6 ± 26.6 | 0.3 |
| HDL (mg/dL) | 53.8 ± 10.7 | 57.6 ± 15.6 | 0.3 |
| LDL (mg/dL) | 93.2 ± 21.3 | 94.6 ± 25.2 | 0.8 |
| Total cholesterol (mg/dL) | 157.9 ± 26.4 | 165.1 ± 29.5 | 0.2 |
| TSH (μU/mL) | 2.4 ± 1.0 | 2.3 ± 1.0 | 0.4 |
| Glucose (mg/dL) | 85.5 ± 9.1 | 82.5 ± 7.4 | 0.1 |
| Insulin (μU/mL) | 10.9 ± 14.4 | 7.9 ± 4.6 | 0.1 |
| HOMA index | 2.3 ± 4.3 | 1.6 ± 1.0 | 0.1 |
| Uric acid (mg/dL) | 4.4 ± 1.0 | 4.0 ± 1.0 | 0.02 |
| Hemoglobin (g/dL) | 13.7 ± 0.9 | 13.2 ± 3.1 | 0.000 |
| Leukocyte (×103/μL) | 7.8 ± 2.2 | 7.0 ± 2.0 | 0.02 |
| Adiponectin (mg/L) | 6.9 ± 4.2 | 22.7 ± 11.0 | 0.000 |
| Resistin (mg/L) | 13.5 ± 4.3 | 13.2 ± 8.7 | 0.05 |
| Leptin (µg/L) | 8.5 ± 6.5 | 16.4 ± 14.10 | 0.005 |
| MCP-1 (pg/mL) | 128.2 ± 45.9 | 103.9 ± 61.2 | 0.001 |
| TNF-α (pg/mL) | 3.7 ± 1.2 | 3.9 ± 2.3 | 0.9 |
| IL-6 (pg/mL) | 1.7 ± 1.8 | 7.5 ± 12.5 | 0.002 |
| IL-8 (pg/mL) | 1.8 ± 0.6 | 2.4 ± 2.6 | 0.1 |
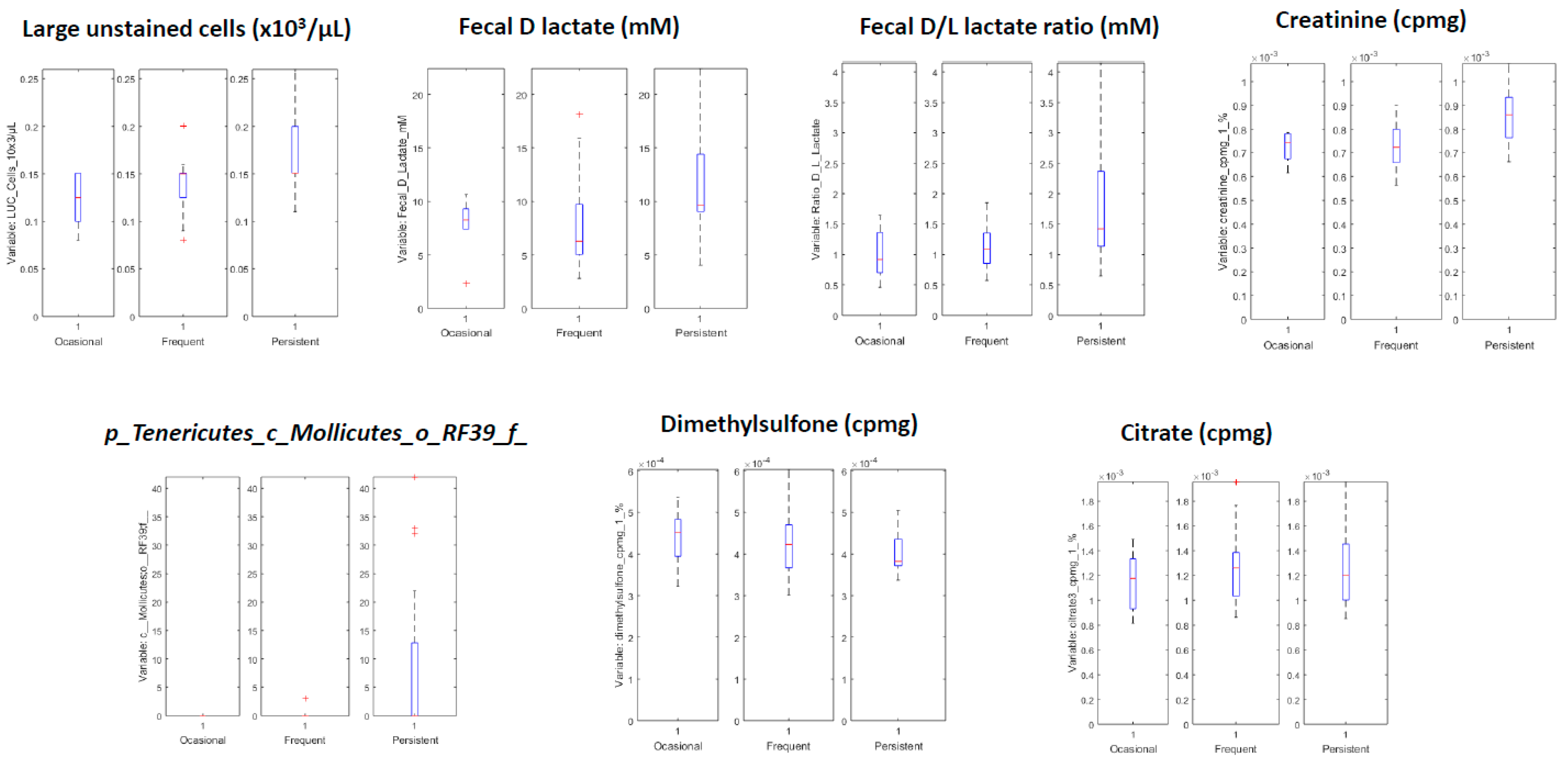
| Variables with Higher Values for Children with Persistent Asthma | Variables with Lower Values for Children with Persistent Asthma |
|---|---|
| LUC cells (×103/µL) | Citrate 3 (cpmg) |
| Fecal D lactate mM | Citrate 4 (cpmg) |
| Ratio D/L lactate | Dimethylsulfone (cpmg) |
| Creatinine (cpmg) | Citrate 2 |
| Creatinine | Citrate 4 |
| c_Mollicutes;o_RF39;f_ | Unknown NMR metabolite |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomez-Llorente, M.A.; Martínez-Cañavate, A.; Chueca, N.; Rico, M.d.l.C.; Romero, R.; Anguita-Ruiz, A.; Aguilera, C.M.; Gil-Campos, M.; Mesa, M.D.; Khakimov, B.; et al. A Multi-Omics Approach Reveals New Signatures in Obese Allergic Asthmatic Children. Biomedicines 2020, 8, 359. https://doi.org/10.3390/biomedicines8090359
Gomez-Llorente MA, Martínez-Cañavate A, Chueca N, Rico MdlC, Romero R, Anguita-Ruiz A, Aguilera CM, Gil-Campos M, Mesa MD, Khakimov B, et al. A Multi-Omics Approach Reveals New Signatures in Obese Allergic Asthmatic Children. Biomedicines. 2020; 8(9):359. https://doi.org/10.3390/biomedicines8090359
Chicago/Turabian StyleGomez-Llorente, Mª Amelia, Ana Martínez-Cañavate, Natalia Chueca, Mª de la Cruz Rico, Raquel Romero, Augusto Anguita-Ruiz, Concepción Mª Aguilera, Mercedes Gil-Campos, Maria D Mesa, Bekzod Khakimov, and et al. 2020. "A Multi-Omics Approach Reveals New Signatures in Obese Allergic Asthmatic Children" Biomedicines 8, no. 9: 359. https://doi.org/10.3390/biomedicines8090359
APA StyleGomez-Llorente, M. A., Martínez-Cañavate, A., Chueca, N., Rico, M. d. l. C., Romero, R., Anguita-Ruiz, A., Aguilera, C. M., Gil-Campos, M., Mesa, M. D., Khakimov, B., Morillo, J. A., Gil, Á., Camacho, J., & Gomez-Llorente, C. (2020). A Multi-Omics Approach Reveals New Signatures in Obese Allergic Asthmatic Children. Biomedicines, 8(9), 359. https://doi.org/10.3390/biomedicines8090359








