Intrinsic Type 1 Interferon (IFN1) Profile of Uncultured Human Bone Marrow CD45lowCD271+ Multipotential Stromal Cells (BM-MSCs): The Impact of Donor Age, Culture Expansion and IFNα and IFNβ Stimulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Cells
2.2. Sample Processing and Cell Sorting
2.3. Quantitative Polymerase Chain Reaction (qPCR)
2.4. BM-MSC Culture and IFN1 Stimulation
2.5. Intracellular ROS Measurements
2.6. Statistics
3. Results
3.1. IFN1 Profile and Reduced Proliferation of Cultured BM-MSCs upon IFN1 Stimulation
3.2. IFN1 Profile of Uncultured CD45lowCD271+ BM-MSCs in Comparison to Cultured and IFN1 Stimulated BM-MSCs
3.3. Intrinsic IFN1 Profile of Uncultured CD45lowCD271+ BM-MSCs in Comparison to Control CD45+CD271− BM-HLCs
3.4. Age-Related Changes in IFN1 Profile of CD45lowCD271+ BM-MSCs Compared to CD45+CD271− BM-HLCs
3.5. Differences in IFN1 Profile of CD45lowCD271+ BM-MSCs and CD45+CD271− BM-HLCs in Patients with Osteoarthritis (OA)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| IFN1 | Type 1 interferon |
| IFNα | Interferon alpha |
| IFNβ | Interferon beta |
| MSCs | Mesenchymal stromal cells |
| BM | Bone marrow |
| HSCs | Hematopoietic stem cells |
| KL | Klotho |
| SIRT6 | Sirtuin 6 |
| ISGs | Interferon stimulated genes |
| DNA | Deoxyribonucleic acid |
| SSEA4 | Stage specific embryonic antigen 4 |
| cGAS/STING | Cyclic GMP-AMP synthase/Stimulator of interferon genes |
| IFNAR1 | Interferon alpha receptor 1 |
| IFNAR2 | Interferon alpha receptor 2 |
| JAK | Janus kinase |
| STAT1 | Signal transducer and activator of transcription1 |
| Mx1 | MX dynamine like GTPase 1 |
| HERC5 | HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| IFTM1 | Interferon induced trans membrane protein 1 |
| IFITM3 | Interferon induced trans membrane protein 3 |
| IRF7 | Interferon regulatory factor 7 |
| RPT4 | Receptor transporter protein 4 |
| ISG20 | Inteferon stimulated gene 20 |
| BID | BH3 interacting domain |
| MTCH2 | Mitochondrial homologue carrier 2 |
| ROS | Reactive oxygen species |
| OA | Osteoarthritis |
| OAS3 | 2′-5′-oligoadenylate synthetase 3 |
| SASP | Senescence associated secretory phenotype |
| IL6 | Interleukin 6 |
| IL8 | Interleukin 8 |
| USP18 | Ubiquitin specific peptidase 18 |
| FABP4 | Fatty acid binding protein 4 |
| PPARγ | Peroxisome proliferator activator receptor gamma |
| RUNX2 | Runt related transcription factor 2 |
| SPARC | Secreted protein acid and cysteine rich/Osteonectic |
| SOX9 | SRY-Box Transcription Factor 9 |
| ACAN | Aggrecan |
| IL7 | Interleukin 7 |
| CXCL12 | C-X-C chemokine 12 |
| TNFSF11 | Tumour necrosis factor super family 11 |
| Ly6E | Lymphocyte antigen 6E |
| EIF2AK2 | Eukaryotic translation initiation factor 2-alpha kinase 2 |
| TMEM173 | Transmembrane protein 173/STING |
| IRF3 | Interferon regulatory factor 3 |
| CCND2 | CyclinD2 |
| IRF9 | Interferon regulatory factor 9 |
| IFI35 | Interferon induced 35 |
| RNF213 | Ring finger protein 213 |
| DDR | DNA-damage response |
| 3D | Three dimensional |
| SERPING1 | Serpin family G member1 |
| EDTA | Ethylene diamine tetra acetic acid |
| BMA | Bone marrow aspirate |
| DMSO | Dimethylsulfoxide |
| qPCR | quantitative Polymerised chain reaction |
| IFC | Integrated fluid circuits |
| DPBS | Dulbeccos phosphate buffer saline |
References
- Kehler, D.S. Age-related disease burden as a measure of population ageing. Lancet Public Health 2019, e123–e124. [Google Scholar] [CrossRef]
- World Health Organisation, Global Burden of Disease. 2004. Available online: http://www.who.int/healthinfo/global_burden_disease/GBD_report_2004update_full.pdf (accessed on 10 July 2020).
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- World Health Organisation, Fact Sheet Musculoskeletal Conditions. 2019. Available online: http://www.who.int/news-room/fact-sheets/detail/musculoskeletal-conditions (accessed on 10 July 2020).
- Florencio-Silva, R.; Sasso, S.G.R.; Sasso-Cerri, I.; Simões, M.J.; Cerri, P.S. Biology of Bone Tissue: Structure, Function, and Factors That Influence Bone Cells. BioMed. Res. Int. 2015, 1–17. [Google Scholar] [CrossRef]
- Kovtonyuk, L.V.; Fritsch, K.; Feng, X.; Manz, M.G.; Takizawa, H. Inflamm-Aging of Hematopoiesis, Hematopoietic Stem Cells, and the Bone Marrow Microenvironment. Front. Immunol. 2016, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Garagnani, P.; Vitale, G.; Capri, M.; Salvioli, S. Inflammaging and ‘Garb-aging’. Trends Endocrinol. Metab. 2017, 28, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Pang, W.W.; Schrier, S.L.; Weisman, I.L. Age-associated changes in human hematopoietic stem cells and Causes and Mechanisms of Hematopoietic Stem Cell Aging. Semin. Hematol. 2017, 54, 39–42. [Google Scholar] [CrossRef]
- Becerikli, M.; Jaurich, H.; Schira, J.; Schulte, M.; Döbele, C.; Wallner, C.; Abraham, S.; Wagner, J.M.; Dadras, M.; Kneser, U.; et al. Age-dependent alterations in osteoblast and osteoclast activity in human cancellous bone. J. Cell Mol. Med. 2017, 21, 2773–2781. [Google Scholar] [CrossRef]
- Ganani, D.; Crippa, S.; Volpe, L.D.; Rossella, V.; Conti, A.; Lettera, E.; Rivis, S.; Ometti, M.; Fraschini, G.; Bernardo, M.E.; et al. An early-senescence state in aged mesenchymal stromal cells contributes to hematopoietic stem and progenitor cell clonogenic impairment through the activation of a pro-inflammatory program. Aging Cell 2019, 18, 1–21. [Google Scholar] [CrossRef]
- Farr, J.N.; Khosla, S. Cellular senescence in bone. Bone 2019, 121, 121–133. [Google Scholar] [CrossRef]
- Pontikoglou, C.; Deschaseaux, F.; Sensebé, L.; Papadaki, H.A. Bone Marrow Mesenchymal Stem Cells: Biological Properties and Their Role in Hematopoiesis and Hematopoietic Stem Cell Transplantation. Stem Cell Rev. Rep. 2011, 7, 569–589. [Google Scholar] [CrossRef]
- Kawamura, H.; Nakatsuka, R.; Matsuoka, Y.; Sumide, K.; Fujioka, T.; Asano, H.; Iida, H.; Sonoda, Y. TGF-β Signaling Accelerates Senescence of Human Bone-Derived CD271 and SSEA-4 Double-Positive Mesenchymal Stromal Cells. Stem Cell Rep. 2018, 10, 920–932. [Google Scholar] [CrossRef] [PubMed]
- Josephson, A.M.; Bradaschia-Correa, V.; Lee, S.; Leclerc, K.; Patel, K.S.; Lopez, E.M.; Litwa, H.P.; Neibart, S.S.; Kadiyala, M.; Wong, M.Z.; et al. Age-related inflammation triggers skeletal stem/progenitor cell dysfunction. Proc. Natl. Acad. Sci. USA 2019, 14, 6995–7004. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, P.; El-Jawhari, J.J.; Burska, A.N.; Ponchel, F.; Giannoudis, P.V.; Jones, E.A. The Analysis of In Vivo Aging in Human Bone Marrow Mesenchymal Stromal Cells Using Colony-Forming Unit-Fibroblast Assay and the CD45lowCD271+ Phenotype. Stem Cells Int. 2019, 2019, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Katlinskaya, Y.V.; Carbone, C.J.; Zhao, B.; Katlinski, K.V.; Zheng, H.; Guha, M.; Li, N.; Chen, Q.; Yang, T.; et al. DNA damage-induced type I interferon promotes senescence and inhibits stem cell function. Cell Rep. 2015, 11, 785–797. [Google Scholar] [CrossRef]
- Li, T.; Chen, Z.J. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 2018, 215, 1287–1299. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Thi, V.L.D.; Huang, Y.; Billerbeck, E.; Saha, D.; Hoffmann, H.-H.; Wang, Y.; Silva, L.A.V.; Sarbanes, S.; Sun, T.; et al. Intrinsic immunity shapes viral resistance of stem cells. Cell 2018, 172, 423–438. [Google Scholar] [CrossRef]
- Kreienkamp, R.; Graziano, S.; Coll-Bonfill, N.; Bedia-Diaz, G.; Cybulla, E.; Vindigni, A.; Dorsett, D.; Kubben, N.; Batista, L.F.Z.; Gonzalo, S. A Cell-Intrinsic Interferon-like Response Links Replication Stress to Cellular Aging Caused by Progerin. Cell Rep. 2018, 22. [Google Scholar] [CrossRef]
- James, S.; Fox, J.; Afsari, F.; Lee, J.; Clough, S.; Knight, C.; Ashmore, J.; Ashton, P.; Preham, O.; Hoogduijn, M.; et al. Multiparameter Analysis of Human Bone Marrow Stromal Cells Identifies Distinct Immunomodulatory and Differentiation-Competent Subtypes. Stem Cell Rep. 2015, 4, 1004–1015. [Google Scholar] [CrossRef]
- Banchereau, J.; Pacual, V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity 2006, 25, 383–392. [Google Scholar] [CrossRef]
- Gu, Z.; Jiang, J.; Tan, W.; Xia, Y.; Cao, H.; Meng, Y.; Da, Z.; Liu, H.; Chen, C. p53/p21 Pathway involved in mediating cellular senescence of bone marrow-derived mesenchymal stem cells from systemic lupus erythematosus patients. Clin. Dev. Immunol. 2013, 2013, 1–13. [Google Scholar] [CrossRef]
- Gao, L.; Bird, A.K.; Meednu, N.; Dauenhauer, K.; Liesveld, J.; Anolik, J.; Looney, R.J. Bone Marrow-Derived Mesenchymal Stem Cells From Patients With Systemic Lupus Erythematosus Have a Senescence-Associated Secretory Phenotype Mediated by a Mitochondrial Antiviral Signaling Protein-Interferon-β Feedback Loop. Arthritis Rheumatol. 2017, 69, 1623–1635. [Google Scholar] [CrossRef]
- Churchman, S.M.; Ponchel, F.; Boxall, S.A.; Cuthbert, R.; Kouroupis, D.; Roshdy, T.; Giannoudis, P.V.; Emery, P.; McGonagle, D.; Jones, E.A. Transcriptional Profile of Native CD271+ Multipotential Stromal Cells Evidence for Multiple Fates, With Prominent Osteogenic and Wnt Pathway Signaling Activity. Arthritis Rheum. 2012, 64, 2632–2643. [Google Scholar] [CrossRef]
- Ghazanfari, R.; Zacharaki, D.; Li, H.; Lim, H.C.; Soneji, S.; Scheding, S. Human Primary Bone Marrow Mesenchymal Stromal Cells and Their in Vitro Progenies Display Distinct Transcriptional Profile Signatures. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Weber, A.; Chan, P.M.B.; Wen, C. Do immune cells lead the way in subchondral bone disturbance on osteoarthritis? Prog. Biophys. Mol. Biol. 2019, 148, 21–31. [Google Scholar] [CrossRef]
- Ni, Z.; Zhao, X.; Dai, X.; Zhao, L.; Xia, J. The role of interferon regulatory factor 5 in macrophage inflammation during osteoarthritis. Inflammation 2019, 42, 1821–1829. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.; English, A.; Churchman, S.M.; Kouroupis, D.; Boxall, S.A.; Kinsey, S.; Giannoudis, P.V.; Emery, P.; McGonagle, D. Large-scale extraction and characterization of CD271+ multipotential stromal cells from trabecular bone in health and osteoarthritis: Implications for bone regeneration strategies based on uncultured or minimally cultured multipotential stromal cells published correction appears in Arthritis Rheum. Arthritis Rheum. 2010, 62, 1944–1954. [Google Scholar] [CrossRef]
- Ilas, D.C.; Churchman, S.M.; Baboolal, T.G.; Giannoudis, P.V.; Aderintho, J.; McGonagle, D.; Jones, E. The simultaneous analysis of mesenchymal stem cells and early osteocytes accumulation in osteoarthritic femoral head sclerotic bone. Rheumatology 2019, 58, 1777–1783. [Google Scholar] [CrossRef] [PubMed]
- El-Jawhari, J.J.; Ganguly, P.; Churchman, S.M.; Jones, E.; Giannnoudis, P.V. The biological fitness of bone progenitor cells in reamer/irrigator/aspirator waste. J. Bone Joint Surg. Am. 2019, 101, 2111–2119. [Google Scholar] [CrossRef] [PubMed]
- Purcell, M.; Kruger, A.; Tainsky, M.A. Gene expression profiling of replicative and induced senescence. Cell Cycle 2014, 15, 3927–3937. [Google Scholar] [CrossRef]
- Oreffo, R.O.; Romberg, S.; Virdi, A.S.; Joyner, C.J.; Berven, S.; Triffitt, J.T. Effects of interferon alpha on human osteoprogenitor cell growth and differentiation in vitro. J. Cell. Biochem. 1999, 74, 372–385. [Google Scholar] [CrossRef]
- Hatzfeld, A.; Eid, P.; Peiffer, I.; Li, M.L.; Barbet, R.; Oostendorp, R.A.J.; Haydont, V.; Monier, M.-N.; Milon, L.; Fortunel, N.; et al. A Sub-Population of High Proliferative Potential-Quiescent Human Mesenchymal Stem Cells Is Under the Reversible Control of Interferon Alpha/Beta. Leukemia 2007, 21, 714–724. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated gene: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- El-Sherbiny, Y.; El-Jawhari, J.J.; Moseley, T.A.; McGonagle, D.; Jones, E. T Cell Immunomodulation by Clinically Used Allogeneic Human Cancellous Bone Fragments: A Potential Novel Immunotherapy Tool. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, F.; Borger, D.K.; Wei, Q.Z.; Pinho, S.; Maryanovich, M.; Zahalka, A.H.; Suzuki, M.; Cruz, C.D.; Wang, Z.C.; Xu, C.L.; et al. Engineering a haematopoietic stem cell niche by revitalizing mesenchymal stromal cells. Nat. Cell Biol. 2019, 21, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Tadogan, A.; Kumar, S.; Allies, G.; Bausinger, J.; Beckel, F.; Hofemeister, H.; Mulaw, M.; Madan, V.; Scharffetter-Kochanek, K.; Feuring-Buske, M.; et al. DNA Damage-Induced HSPC Malfunction Depends on ROS Accumulation Downstream of IFN-1 Signaling and Bid Mobilization. Cell Stem Cell 2016, 19, 752–767. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, K. Molecular Profiles of Cell-to-Cell Variation in the Regenerative Potential of Mesenchymal Stromal Cells. Stem Cells Int. 2019, 2019, 1–14. [Google Scholar] [CrossRef]
- Flint, S.; Jovanovic, V.; Teo, B.W.; Mak, A.; Thumboo, J.; McKinney, E.F.; Lee, J.C.; MacAry, P.; Kemeny, D.M.; Jayne, D.; et al. Leucocyte subset-specific type 1 interferon signatures in SLE and other immune-mediated diseases. RMD 2016, 2, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 1–15. [Google Scholar] [CrossRef]
- Liu, F.; Wu, S.; Ren, H.; Gu, J. Klotho supresses RIG-1 mediated senescence associated inflammation. Nat. Cell Biol. 2011, 13, 254–262. [Google Scholar] [CrossRef]
- Cordeirogomes, A.; Hara, T.; Lim, V.Y.; Herndler-Brandstetter, D.; Nevius, E.; Sugiyama, T.; Tani-Ichi, S.; Schlenner, S.; Richie, E.; Rodewald, H.-R.; et al. Hematopoietic Stem Cell Niches Produce Lineage Instructive Signals to Control Multipotent Progenitor Differentiation. Immunity 2016, 45, 1219–1231. [Google Scholar] [CrossRef]
- Naismith, E.N.; Pangrazzi, L. The impact of oxidative stress, inflammation, and senescence on the maintenance of immunological memory in the bone marrow in old age. Biosci. Rep. 2019, 39, 1–12. [Google Scholar] [CrossRef]
- Sugiyama, T.; Kohara, H.; Noda, M.; Nagasawa, T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 2006, 25, 977–988. [Google Scholar] [CrossRef]
- Boyce, B.F.; Xing, L. Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch. Biochem. Biophys. 2008, 473, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.; Tasdogan, A.; Fehling, H.J. The IFN-1 > BID > ROS pathway: Linking DNA damage with HSPC malfunction. Cell Cycle 2017, 16, 819–820. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, B.C.; Sarhan, J.; Poltorak, A. Host-intrinsic interferon status in infection and immunity. Trend Mol. Med. 2019, 24, 658–668. [Google Scholar] [CrossRef]
- Jazwinski, S.M.; Kim, S. Examination of the Dimensions of Biological Age. Front. Genet. 2019, 10, 1–7. [Google Scholar] [CrossRef]
- Doss, F.; Menard, J.; Hauschild, M.; Kreutzer, H.-J.; Mittlmeier, T.; Müller-Steinhardt, M.; Müller, B. Elevated IL-6 levels in the synovial fluid of osteoarthritis patients stem from plasma cells. Scand. J. Rheumatol. 2007, 36, 136–139. [Google Scholar] [CrossRef]
- Honke, N.; Shaabani, N.; Zhang, D.-E.; Hardt, C.; Lang, K.S. Multiple functions of USP18. Cell Death Dis. 2016, 7, e2444. [Google Scholar] [CrossRef]
- Li, Y.; Ma, M.X.; Qin, B.; Lin, L.-T.; Richardson, C.D.; Feld, J.; McGilvray, I.D.; Chen, L. The Ubiquitin-Specific Protease 18 Promotes Hepatitis C Virus Production by Increasing Viral Infectivity. Mediat. Inflamm. 2019, 3124745. [Google Scholar] [CrossRef]
- Schreiber, G. The molecular basis for differential type I interferon signaling. J. Biol. Chem. 2017, 292, 7285–7294. [Google Scholar] [CrossRef] [PubMed]
- Woeckel, V.J.; Eijken, M.; van de Peppel, J.; Chiba, H.; van der Eerden, B.C.J.; van Leeuwen, J.P.T.M. IFNβ impairs extracellular matrix formation leading to inhibition of mineralization by effects in the early stage of human osteoblast differentiation. J. Cell Physiol. 2012, 227, 2668–2676. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Ferrer, S.; Michurina, T.V.; Ferraro, F.; Mazloom, A.R.; MacArthur, B.D.; Lira, S.A.; Scadden, D.T.; Ma’ayan, A.; Enikolopov, G.N.; Frenette, P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 2010, 466, 829–834. [Google Scholar] [CrossRef]
- Hiroshi, T.; Kojiro, S.; Akinori, T.; Tadatsugu, T. Interplay between interferon and other cytokine systems in bone metabolism. Immunol. Rev. 2005, 208, 181–193. [Google Scholar] [CrossRef]
- Takeuchi, T.; Shimakawa, G.; Tamura, M.; Yokosawa, H.; Arata, Y. ISG15 Regulates RANKL-induced Osteoclastogenic Differentiation of RAW264 Cells. Biol. Pharm Bull. 2015, 38, 452–486. [Google Scholar] [CrossRef]
- Van Roon, J.A.G.; Lafeber, F.P.J.G. Role of interleukin-7 in degenerative and inflammatory joint diseases. Arthritis Res. Ther. 2008, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhi, X.; Wang, J.; Su, J. RANKL Signaling in Bone Marrow Mesenchymal Stem Cells Negatively Regulates Osteoblastic Bone Formation. Bone Res. 2018, 6, 34. [Google Scholar] [CrossRef]
- Tormin, A.; Li, Q.; Brune, J.C.; Walsh, S.; Schütz, B.; Ehinger, M.; Ditzel, N.; Kassem, M.; Scheding, S. CD146 expression on primary nonhematopoietic bone marrow stem cells is correlated with in situ localization. Blood 2011, 117, 5067–5077. [Google Scholar] [CrossRef]
- Souliotis, V.L.; Vlachogiannis, N.I.; Pappa, M.; Argyriou, A.; Ntouros, P.A.; Sfikakis, P.P. DNA Damage Response and Oxidative Stress in Systemic Autoimmunity. Int. J. Mol. Sci. 2020, 21, 55. [Google Scholar] [CrossRef]
- Harichandan, A.; Buhring, H.-J. Prospective isolation of human MSCs. Best Pract. Res. Clin. Haematol. 2011, 24, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Mo, M.; Wang, S.; Zhou, Y.; Li, H.; Wu, Y. Mesenchymal stem cell subpopulations: Phenotype, property and therapeutic potential. Cell. Mol. Life Sci. 2016, 73, 3311–3321. [Google Scholar] [CrossRef]
- Wolock, S.L.; Krishnan, I.; Tenen, D.E.; Matkins, V.; Camacho, V.; Patel, S.; Agarwal, P.; Bhatia, R.; Tenen, D.G.; Klein, A.M.; et al. Mapping Distinct Bone Marrow Niche Populations and Their Differentiation Paths. Cell Rep. 2019, 28, 302–311. [Google Scholar] [CrossRef]
- Gough, D.J.; Messina, N.L.; Clarke, C.J.P.; Johnstone, R.W.; Levy, D.E. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity 2013, 36, 166–174. [Google Scholar] [CrossRef]
- Childs, B.; van Deursen, J. An evolving picture of cell senescence. Nature 2019, 566, 46–47. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, E.T.; Gonzalez-Nieto, D.; Ghiaur, G.; Dunn, S.K.; Ficker, A.M.; Murali, B.; Madhu, M.; Gutstein, D.E.; Fishman, G.I.; Barrio, L.C.; et al. Connexin-43 prevents hematopoietic stem cell senescence through transfer of reactive oxygen species to bone marrow stromal cells. Proc. Natl. Acad. Sci. USA 2012, 109, 9071–9076. [Google Scholar] [CrossRef] [PubMed]



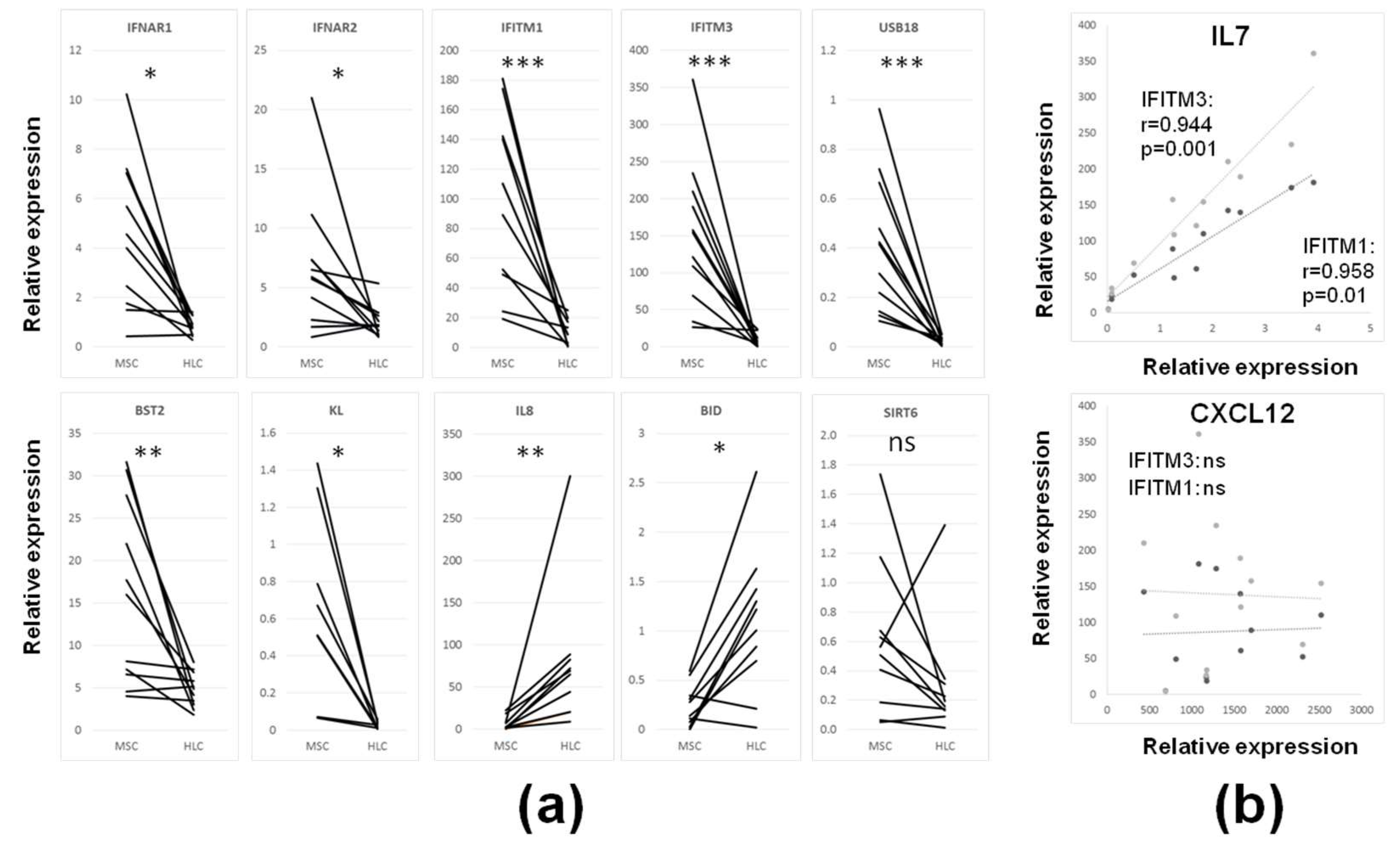

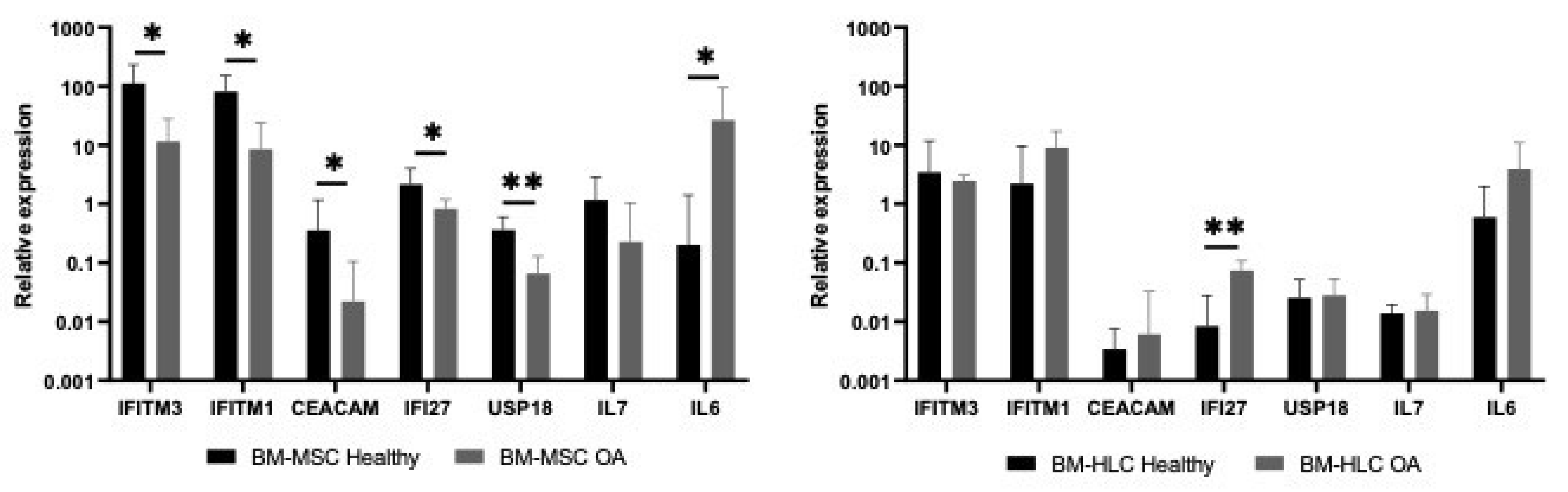
| Gene Name | Gene Id | MSC (Median) | HLC (Median) | Fold Difference | p Value |
|---|---|---|---|---|---|
| SERPING | Serine protease inhibitor, clade G (C1 inhibitor), member 1 | 22 | 0.094 | 233.3 | 0.0003 |
| IFI27 | Interferon gamma-inducible protein 27 | 1.551 | 0.01 | 155.1 | <0.0001 |
| PRDM16 | PR domain containing 16 | 0.179 | 0.002 | 105.3 | 0.042 |
| IL7 | Interleukin 7 | 1.481 | 0.016 | 93.7 | 0.0005 |
| CEACAM | Carcinoembryonic antigen-related cell adhesion molecule 1 | 0.31 | 0.007 | 44.3 | 0.0021 |
| IFITM3 | Interferon induced transmembrane protein 3 | 137.9 | 3.79 | 36.4 | <0.0001 |
| KL | Klotho | 0.51 | 0.02 | 25.5 | 0.0083 |
| USP18 | Ubiquitin specific peptidase 18 | 0.35 | 0.02 | 17.5 | <0.0001 |
| LRP1 | LDL Receptor Related Protein 1 | 48.92 | 4.03 | 12.1 | 0.0007 |
| RTP4 | Receptor (chemosensory) transporter protein 4 | 0.96 | 0.08 | 12 | 0.0036 |
| IFITM1 | Interferon induced transmembrane protein 1 | 75.05 | 6.33 | 11.9 | 0.0005 |
| LY6E | Lymphocyte antigen 6 complex, locus E | 24.58 | 2.4 | 10.2 | <0.0001 |
| SPATS2L | Spermatogenesis associated, serine-rich 2-like | 3.509 | 0.405 | 8.7 | 0.0001 |
| IFI6 | Interferon inducible alpha protein 6 | 3.285 | 0.438 | 7.5 | 0.0145 |
| IFIT5 | Interferon induced protein with tetratricopeptide repeats 5 | 0.633 | 0.11 | 5.8 | 0.0284 |
| CCND2 | CyclinD2 | 2.359 | 0.415 | 5.7 | 0.0013 |
| SOCS1 | Suppressor of cytokine signalling 3 | 2.558 | 0.461 | 5.6 | 0.0003 |
| ABCA1 | ATP Binding Cassette Subfamily A Member 1 | 1.173 | 0.24 | 4.9 | 0.0249 |
| IFNAR1 | Interferon alpha receptor 1 | 4.28 | 0.88 | 4.9 | 0.0018 |
| IFI44L | Interferon induced protein 44 like | 1.507 | 0.31 | 4.9 | 0.008 |
| OASL | 2′-5′-oligoadenylate synthetase | 1.014 | 0.22 | 4.6 | 0.0284 |
| PLSCR1 | Phospholipid scramblase 1 | 4.117 | 0.998 | 4.1 | 0.0068 |
| EIF2AK2 | Eukaryotic translation initiation factor 2-alpha kinase 2 | 3.807 | 0.956 | 4 | 0.0023 |
| OAS3 | 2′-5′-oligoadenylate synthetase 3 | 2.55 | 0.7 | 3.6 | 0.0121 |
| IFI35 | Interferon inducible alpha protein 35 | 2.68 | 0.76 | 3.5 | 0.0387 |
| NT5C3B | 5′-nucleotidase cytosolic IIIB | 0.56 | 0.159 | 3.5 | 0.0018 |
| RSAD2 | Radical S-adenosyl methionine domain containing 2 | 0.434 | 0.132 | 3.3 | 0.0169 |
| IFNAR2 | Interferon alpha receptor 2 | 5.82 | 1.84 | 3.2 | 0.0332 |
| IFI44 | Interferon inducible alpha protein 44 | 3.1 | 1.01 | 3.1 | 0.0205 |
| EPSTI1 | Epithelial stromal interaction 1 | 1.59 | 0.57 | 2.8 | 0.0045 |
| IFIH1 | Interferon induced with helicase C domain 1 | 0.45 | 0.17 | 2.7 | 0.0106 |
| UBE2L6 | Ubiquitin/ISG15-conjugating enzyme E2 L6 | 11.86 | 4.5 | 2.6 | 0.0083 |
| SAMD9L | Sterile alpha motif domain containing 9-like | 1.67 | 0.642 | 2.6 | 0.0387 |
| BST2 | Bone marrow stromal antigen 2, Tetherin (CD317) | 12.06 | 4.97 | 2.4 | 0.0083 |
| IFI16 | Interferon inducible alpha protein 16 | 1.664 | 0.809 | 2.1 | 0.0068 |
| RGS1 | Regulator of G protein signalling 1 | 0.621 | 32.29 | 52 | 0.0184 |
| IL8 | Interleukin 8 | 2.919 | 67.21 | 23 | 0.0022 |
| TNF | Tumour necrosis factor | 0.17 | 3.67 | 21.6 | 0.013 |
| IRF5 | Interferon regulatory factor 5 | 0.129 | 1.285 | 10 | 0.0157 |
| HPSE | Heparanase | 0.097 | 0.732 | 7.6 | 0.0171 |
| FCGR1B | Fc fragment of IgG receptor 1B | 0.19 | 1.04 | 5.5 | 0.0031 |
| BID | BH3 interacting domain | 0.21 | 1 | 4.8 | 0.0188 |
| PRDM1 | PR domain containing 1 | 0.51 | 1.72 | 3.4 | 0.0148 |
| LAIR1 | Leukocyte associated immunoglobulin like receptor1 | 0.43 | 1.4 | 3.3 | 0.0278 |
| CASP1 | Caspase1 | 3.06 | 6.16 | 2 | 0.0205 |
| Gene ID | Gene Name | Young (Median) | Old (Median) | Fold Difference | p Value |
|---|---|---|---|---|---|
| IL7R | Interleukin7 receptor | 0.491 | 0.098 | 5.01 | 0.026 |
| PRKRA | Protein Kinase, Interferon-Inducible Double Stranded RNA Dependent Activator | 0.970 | 0.200 | 4.85 | 0.0152 |
| SCARB1 | Scavenger Receptor Class B Member 1 | 0.431 | 0.131 | 3.29 | 0.0087 |
| CXCL10 | C-X-C motif chemokine 10 | 0.580 | 0.180 | 3.22 | 0.0667 |
| RTP4 | Receptor transporter protein 4 | 0.150 | 0.050 | 3.00 | 0.0152 |
| CEACAM | Carcinoembryonic antigen-related cell adhesion molecule 1 | 0.008 | 0.003 | 2.67 | 0.0667 |
| Tp53 | Tumour protein 53 | 2.930 | 1.210 | 2.42 | 0.0043 |
| IRF9 | Interferon regulatory factor 9 | 1.520 | 0.628 | 2.42 | 0.026 |
| IRF3 | Interferon regulatory factor 3 | 1.561 | 0.660 | 2.37 | 0.0411 |
| ABCG1 | ATP Binding Cassette Subfamily G Member 1 | 0.070 | 0.030 | 2.33 | 0.0649 |
| TMEM173/STING | Transmembrane protein 173/Stimulator of interferon genes | 2.293 | 1.033 | 2.22 | 0.026 |
| IFIT5 | Interferon induced protein with tetratricopeptide repeats 5 | 0.124 | 0.058 | 2.14 | 0.0411 |
| MTCH2 | Mitochondrial carrier homolog 2 | 1.610 | 0.790 | 2.04 | 0.0152 |
| Gene ID | Gene Name | MSC (Median) | HLC (Median) | Fold Difference | p Value |
|---|---|---|---|---|---|
| PRDM16 | PR domain containing 16 | 0.11 | 0.001 | 110 | 0.055 |
| SERPING | Serine protease inhibitor, clade G (C1 inhibitor), member 1 | 8.8 | 0.086 | 102.3 | 0.0006 |
| Kl | Klotho | 0.1 | 0.002 | 50 | 0.0175 |
| LRP1 | LDL Receptor Related Protein 1 | 8 | 0.31 | 25.8 | 0.0111 |
| IL7 | Interleukin 7 | 0.22 | 0.015 | 14.7 | 0.0012 |
| IFI27 | Interferon gamma-inducible protein 27 | 0.82 | 0.07 | 11.7 | 0.0006 |
| OAS1 | 2′-5′-oligoadenylate synthetase 1 | 0.23 | 0.024 | 9.6 | 0.022 |
| CHMP5 | Charged multivesicular body protein 5 | 1.78 | 0.21 | 8.5 | 0.0728 |
| IL6 | Interleukin 6 | 26.57 | 3.93 | 6.8 | 0.0728 |
| SPATS2L | Spermatogenesis associated, serine-rich 2-like | 2.12 | 0.317 | 6.7 | 0.0012 |
| TLR4 | Toll Like Receptor 4 | 0.88 | 0.16 | 5.5 | 0.011 |
| NT5C3B | 5′-nucleotidase cytosolic IIIB | 0.37 | 0.07 | 5.3 | 0.0041 |
| CCL8 | Chemokine C-C motif ligand 8 | 0.12 | 0.023 | 5.2 | 0.0006 |
| CXCL10 | C-X-C motif chemokine 10 | 0.56 | 0.11 | 5.1 | 0.038 |
| ABCA1 | ATP Binding Cassette Subfamily A Member 1 | 2.88 | 0.59 | 4.9 | 0.0728 |
| IFITM3 | Interferon induced transmembrane protein 3 | 11.6 | 2.47 | 4.7 | 0.026 |
| IFI35 | Interferon alpha-inducible protein 35 | 1.29 | 0.28 | 4.6 | 0.0728 |
| RTP4 | Receptor (chemosensory) transporter protein 4 | 0.12 | 0.03 | 4 | 0.0728 |
| PRKRA | Protein Kinase, Interferon-Inducible Double Stranded RNA Dependent Activator | 0.83 | 0.21 | 4.0 | 0.073 |
| GBP1 | Guanylate binding protein 1 | 3.3 | 0.84 | 3.9 | 0.038 |
| STING | Stimulator of interferon genes | 1.25 | 0.46 | 2.7 | 0.073 |
| PLSCR1 | Phospholipid scramblase 1 | 2.04 | 0.81 | 2.5 | 0.0262 |
| IFI44L | Interferon induced protein 44 like | 0.35 | 0.16 | 2.2 | 0.0262 |
| IFIT5 | Interferon induced protein with tetratricopeptide repeats 5 | 0.26 | 0.12 | 2.2 | 0.0006 |
| IFI16 | Interferon inducible alpha protein 16 | 1.164 | 0.56 | 2.1 | 0.0041 |
| IFNAR2 | Interferon alpha receptor 2 | 1.36 | 0.66 | 2.1 | 0.035 |
| IFNG | Interferon gamma | 0.003 | 1.29 | 430 | 0.0167 |
| RGS1 | Regulator of G protein signalling 1 | 0.18 | 21.98 | 122.1 | 0.0006 |
| LAMP3 | Lysosome associated membrane glycoprotein 3 | 0.02 | 0.48 | 24 | 0.0023 |
| FCGR1B | Fc fragment of IgG receptor IB | 0.003 | 0.065 | 21.7 | 0.0513 |
| IL7R | Interleukin 7 receptor | 0.19 | 2.89 | 15.2 | 0.0012 |
| SIGLEC1 | Sialic acid binding Ig like Lectin 1 | 0.008 | 0.08 | 10 | 0.0714 |
| PRDM1 | PR domain containing 1, with ZNF domain | 0.71 | 3.6 | 5.1 | 0.011 |
| LAIR1 | Leukocyte associated immunoglobulin like receptor1 | 0.13 | 0.61 | 4.7 | 0.0023 |
| TGFB | PR domain containing 1, with ZNF domain | 2.9 | 13.23 | 4.6 | 0.0012 |
| ISG20 | Interferon stimulated exonuclease gene 20kDa | 1.17 | 5.081 | 4.3 | 0.0006 |
| HPSE | Heparanase | 0.06 | 0.16 | 2.7 | 0.053 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganguly, P.; Burska, A.N.; Davis, C.L.M.; El-Jawhari, J.J.; Giannoudis, P.V.; Jones, E.A. Intrinsic Type 1 Interferon (IFN1) Profile of Uncultured Human Bone Marrow CD45lowCD271+ Multipotential Stromal Cells (BM-MSCs): The Impact of Donor Age, Culture Expansion and IFNα and IFNβ Stimulation. Biomedicines 2020, 8, 214. https://doi.org/10.3390/biomedicines8070214
Ganguly P, Burska AN, Davis CLM, El-Jawhari JJ, Giannoudis PV, Jones EA. Intrinsic Type 1 Interferon (IFN1) Profile of Uncultured Human Bone Marrow CD45lowCD271+ Multipotential Stromal Cells (BM-MSCs): The Impact of Donor Age, Culture Expansion and IFNα and IFNβ Stimulation. Biomedicines. 2020; 8(7):214. https://doi.org/10.3390/biomedicines8070214
Chicago/Turabian StyleGanguly, Payal, Agata N. Burska, Charlotte L.M. Davis, Jehan J. El-Jawhari, Peter V. Giannoudis, and Elena A. Jones. 2020. "Intrinsic Type 1 Interferon (IFN1) Profile of Uncultured Human Bone Marrow CD45lowCD271+ Multipotential Stromal Cells (BM-MSCs): The Impact of Donor Age, Culture Expansion and IFNα and IFNβ Stimulation" Biomedicines 8, no. 7: 214. https://doi.org/10.3390/biomedicines8070214
APA StyleGanguly, P., Burska, A. N., Davis, C. L. M., El-Jawhari, J. J., Giannoudis, P. V., & Jones, E. A. (2020). Intrinsic Type 1 Interferon (IFN1) Profile of Uncultured Human Bone Marrow CD45lowCD271+ Multipotential Stromal Cells (BM-MSCs): The Impact of Donor Age, Culture Expansion and IFNα and IFNβ Stimulation. Biomedicines, 8(7), 214. https://doi.org/10.3390/biomedicines8070214








