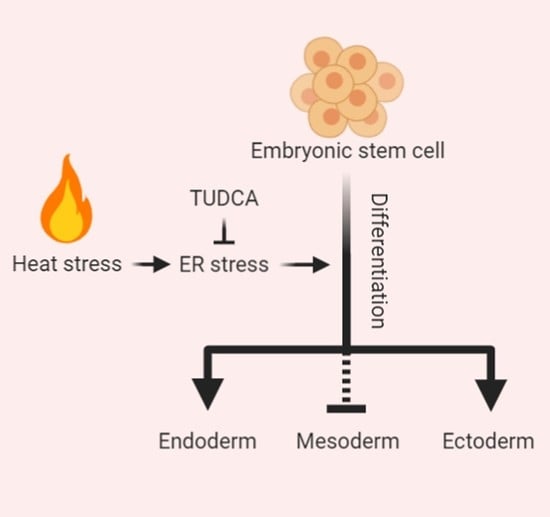
Hyperthermia Disturbs and Delays Spontaneous Differentiation of Human Embryoid Bodies
Abstract
:1. Introduction
2. Experimental Section
2.1. Culture and Maintenance of hESCs
2.2. EB Generation to Induce Spontaneous In Vitro Differentiation
2.3. Hyperthermia and Chemical Chaperone Treatment
2.4. RNA Isolation and cDNA Synthesis
2.5. Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
2.6. RT2 Profiler PCR Array and TaqMan hPSC Scorecard Assay
2.7. Statistical Analysis
3. Results
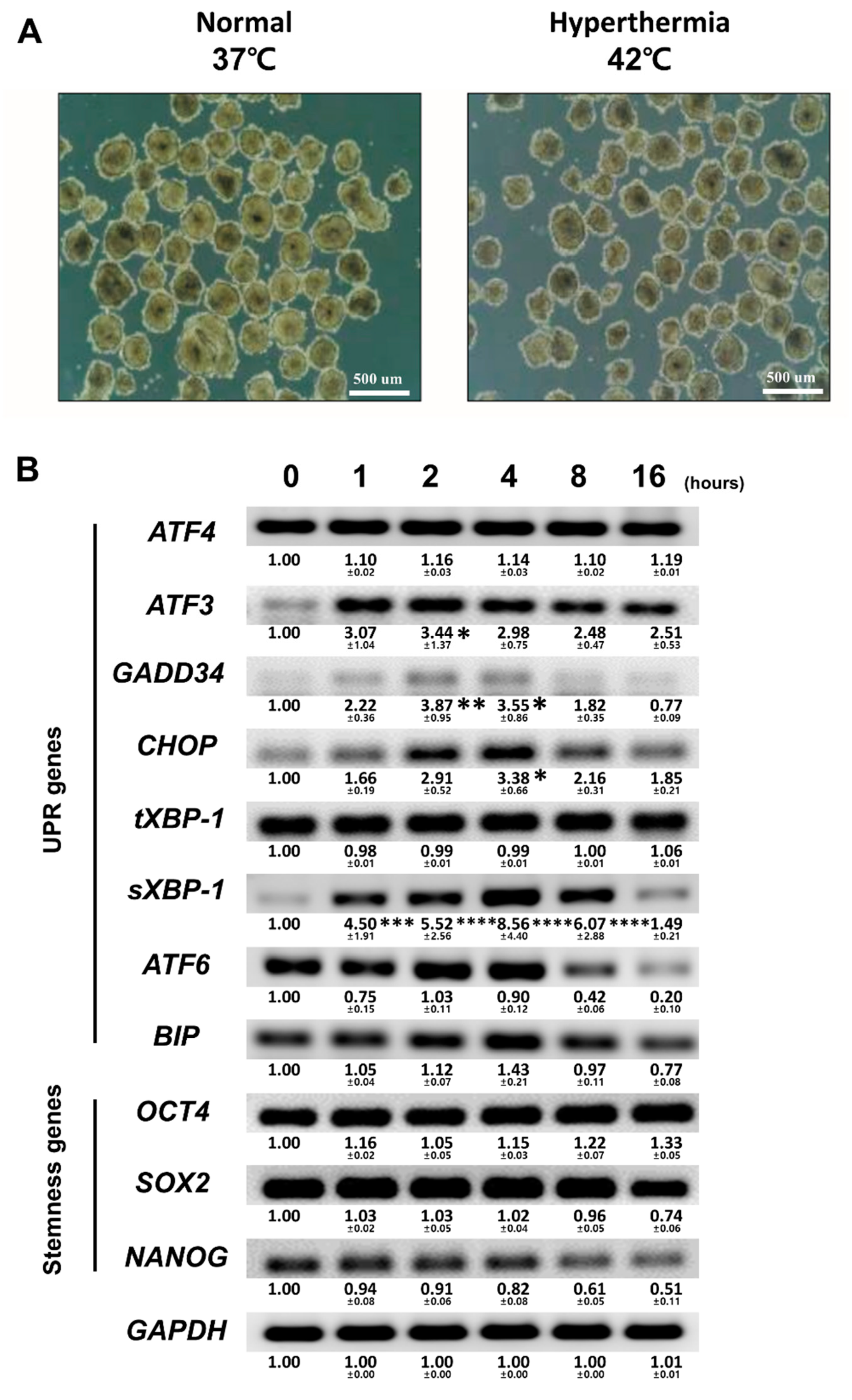
3.1. Optimizing the Conditions of Hyperthermia
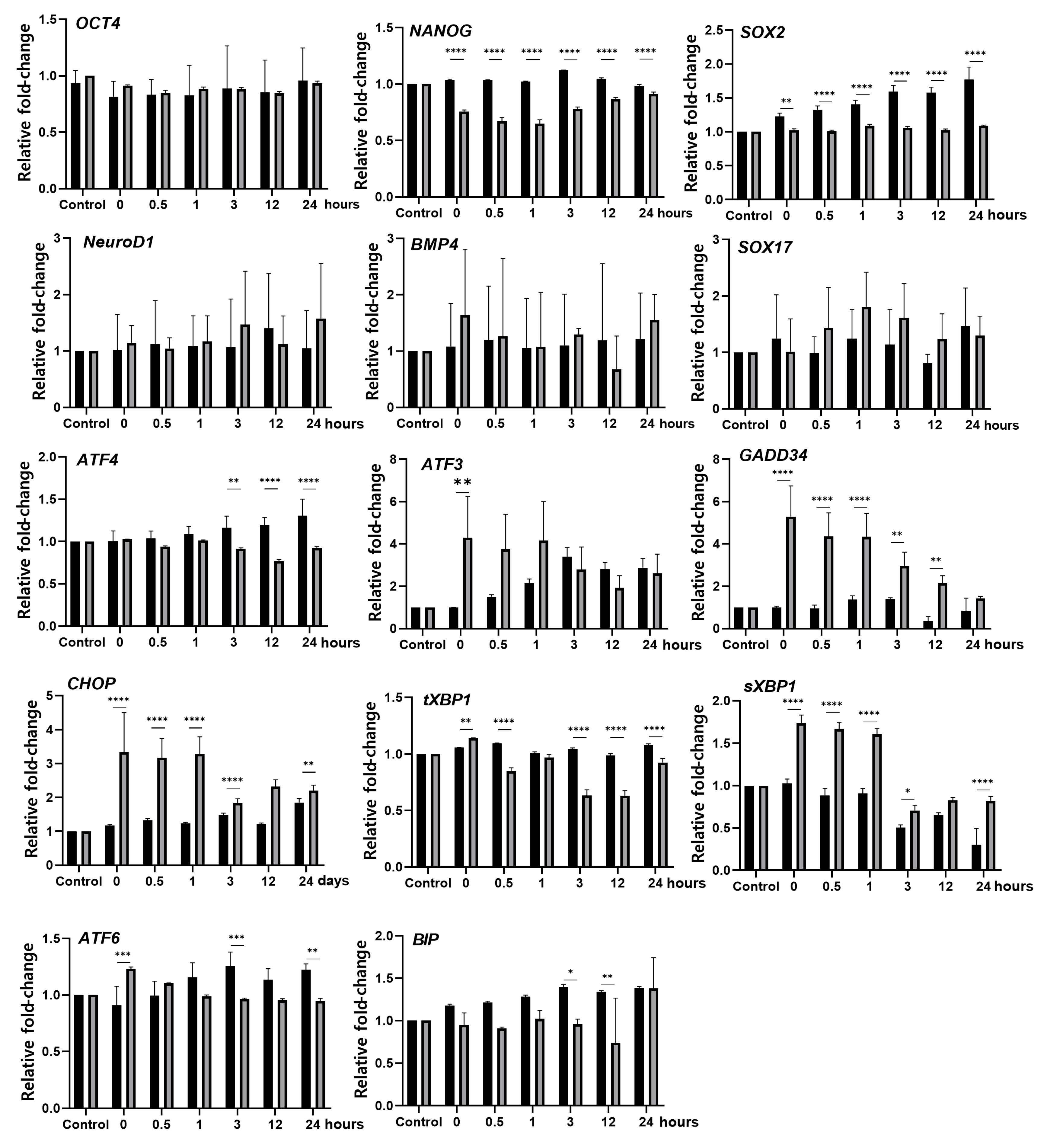
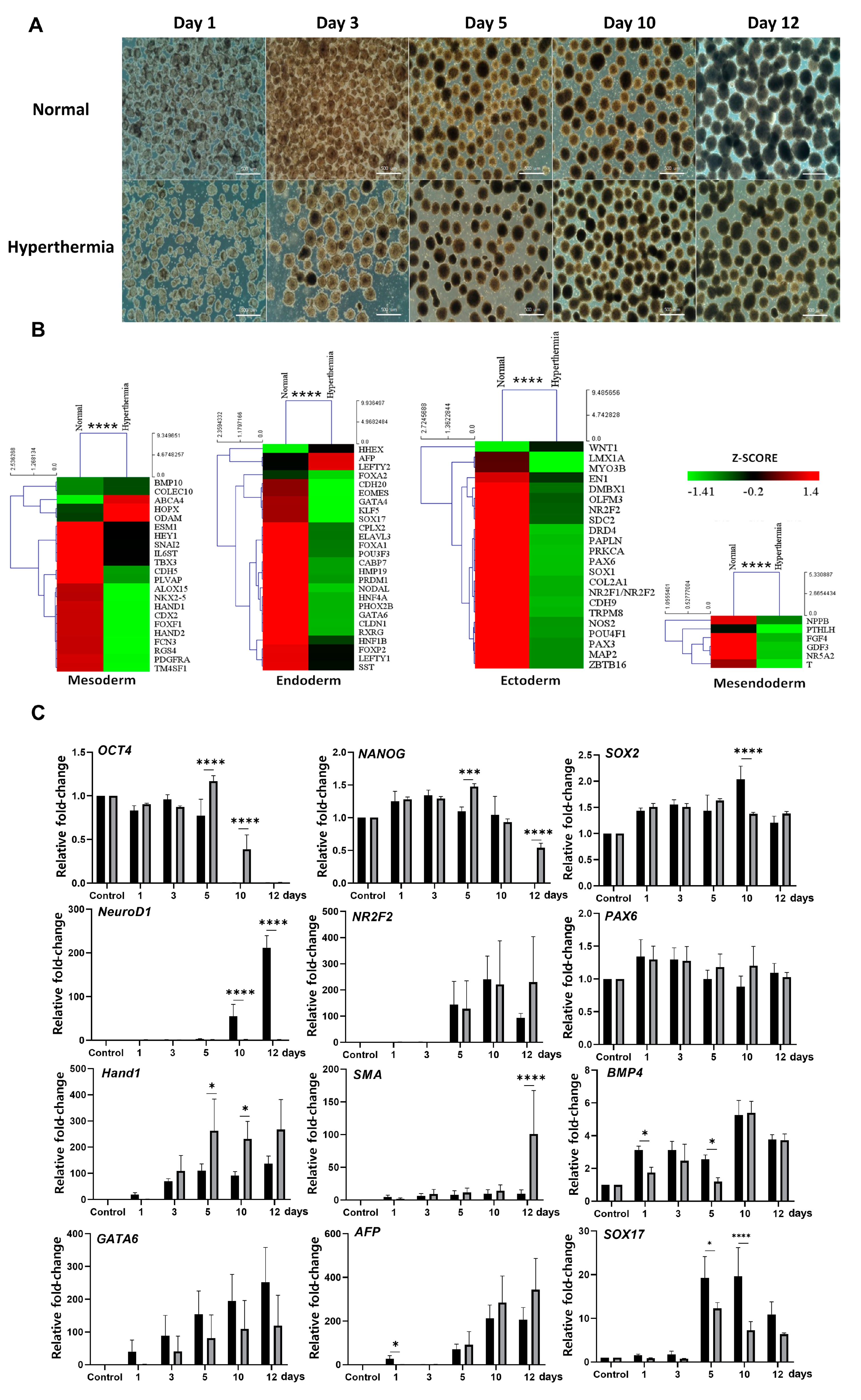
3.2. Hyperthermia Disturbs Spontaneously Differentiating hEBs through UPR Induction
3.3. TUDCA Suppresses the Expression of UPR-Related Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ziskin, M.C.; Morrissey, J. Thermal Thresholds for Teratogenicity, Reproduction, and Development. Int. J. Hyperth. 2011, 27, 374–387. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Guo, Y.; Guettouche, T.; Smith, D.F.; Voellmy, R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 1998, 94, 471–480. [Google Scholar] [CrossRef] [Green Version]
- Anckar, J.; Sistonen, L. Regulation of HSF1 function in the heat stress response: Implications in aging and disease. Annu. Rev. Biochem. 2011, 80, 1089–1115. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Guettouche, T.; Fenna, M.; Boellmann, F.; Pratt, W.B.; Toft, D.O.; Smith, D.F.; Voellmy, R. Evidence for a mechanism of repression of heat shock factor 1 transcriptional activity by a multichaperone complex. J. Biol. Chem. 2001, 276, 45791–45799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Lim, Y.; Lee, D.; Elvira, R.; Lee, J.M.; Lee, M.R.; Han, J. Modulation of Protein Synthesis by eIF2alpha Phosphorylation Protects Cell from Heat Stress-Mediated Apoptosis. Cells 2018, 7, 254. [Google Scholar] [CrossRef] [Green Version]
- Gagnon, D.; Dorman, L.E.; Jay, O.; Hardcastle, S.; Kenny, G.P. Core temperature differences between males and females during intermittent exercise: Physical considerations. Eur. J. Appl. Physiol. 2009, 105, 453–461. [Google Scholar] [CrossRef]
- Browning, R.C.; Baker, E.A.; Herron, J.A.; Kram, R. Effects of obesity and sex on the energetic cost and preferred speed of walking. J. Appl. Physiol. (Bethesda Md. 1985) 2006, 100, 390–398. [Google Scholar] [CrossRef] [Green Version]
- Wyndham, C.H.; McPherson, R.K.; Munro, A. Reactions to Heat of Aborigines and Caucasians. J. Appl. Physiol. 1964, 19, 1055–1058. [Google Scholar] [CrossRef]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef]
- Bertolotti, A.; Zhang, Y.; Hendershot, L.M.; Harding, H.P.; Ron, D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2000, 2, 326–332. [Google Scholar] [CrossRef]
- Edwards, M.J.; Shiota, K.; Smith, M.S.; Walsh, D.A. Hyperthermia and birth defects. Reprod. Toxicol. (Elmsford N.Y.) 1995, 9, 411–425. [Google Scholar] [CrossRef]
- Wang, L.; Deng, Y.; Duan, D.; Sun, S.; Ge, L.; Zhuo, Y.; Yuan, T.; Wu, P.; Wang, H.; Lu, M.; et al. Hyperthermia influences fate determination of neural stem cells with lncRNAs alterations in the early differentiation. PLoS ONE 2017, 12, e0171359. [Google Scholar] [CrossRef] [PubMed]
- Edwards, M.J. Review: Hyperthermia and fever during pregnancy. Birth Defects Res. Part A Clin. Mol. Teratol. 2006, 76, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Chambers, C.D. Risks of hyperthermia associated with hot tub or spa use by pregnant women. Birth Defects Res. Part A Clin. Mol. Teratol. 2006, 76, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.R.; Kim, J.S.; Kim, K.S. miR-124a is important for migratory cell fate transition during gastrulation of human embryonic stem cells. Stem Cells (Dayt. Ohio) 2010, 28, 1550–1559. [Google Scholar] [CrossRef]
- Elvira, R.; Cha, S.J.; Noh, G.M.; Kim, K.; Han, J. Perk-Mediated Eif2α Phosphorylation Contributes to the Protection of Dopaminergic Neurons from Chronic Heat Stress in Drosophila. Int. J. Mol. Sci. 2020, 21, 845. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.L.; Huang, C.C.; Chi, M.S.; Chiang, H.C.; Wang, Y.S.; Hsia, C.C.; Andocs, G.; Wang, H.E.; Chi, K.H. In vitro comparison of conventional hyperthermia and modulated electro-hyperthermia. Oncotarget 2016, 7, 84082–84092. [Google Scholar] [CrossRef] [Green Version]
- Boone, D.R.; Micci, M.A.; Taglialatela, I.G.; Hellmich, J.L.; Weisz, H.A.; Bi, M.; Prough, D.S.; DeWitt, D.S.; Hellmich, H.L. Pathway-focused PCR array profiling of enriched populations of laser capture microdissected hippocampal cells after traumatic brain injury. PLoS ONE 2015, 10, e0127287. [Google Scholar] [CrossRef]
- Fergus, J.; Quintanilla, R.; Lakshmipathy, U. Characterizing Pluripotent Stem Cells Using the TaqMan(R) hPSC Scorecard(TM) Panel. Methods Mol. Biol. (Clifton N.J.) 2016, 1307, 25–37. [Google Scholar] [CrossRef]
- Tsankov, A.M.; Akopian, V.; Pop, R.; Chetty, S.; Gifford, C.A.; Daheron, L.; Tsankova, N.M.; Meissner, A. A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat. Biotechnol. 2015, 33, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.M.; Lee, J.H.; Yun, S.P.; Han, Y.S.; Yun, C.W.; Lee, H.J.; Noh, H.; Lee, S.J.; Han, H.J.; Lee, S.H. Tauroursodeoxycholic acid reduces ER stress by regulating of Akt-dependent cellular prion protein. Sci. Rep. 2016, 6, 39838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, T.; Lee, J.E.; Oqani, R.K.; Kim, S.Y.; Cho, E.S.; Jeong, Y.D.; Baek, J.J.; Jin, D.I. Delayed blastocyst formation or an extra day culture increases apoptosis in pig blastocysts. Anim. Reprod. Sci. 2017, 185, 128–139. [Google Scholar] [CrossRef]
- Asamoah, B.A.-O.; Kjellstrom, T.; Östergren, P.O. Is ambient heat exposure levels associated with miscarriage or stillbirths in hot regions? A cross-sectional study using survey data from the Ghana Maternal Health Survey 2007. Int. J. Biometeorol. 2018, 62, 319–330. [Google Scholar] [CrossRef] [Green Version]
- Auger, N.; Fraser Wd Fau-Sauve, R.; Sauve R Fau-Bilodeau-Bertrand, M.; Bilodeau-Bertrand M Fau-Kosatsky, T.; Kosatsky, T. Risk of Congenital Heart Defects after Ambient Heat Exposure Early in Pregnancy. Environ. health Perspect. 2017, 125, 8–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, T.; Lee, J.E.; Kang, J.W.; Shin, H.Y.; Lee, J.B.; Jin, D.I. Endoplasmic Reticulum (ER) Stress and Unfolded Protein Response (UPR) in Mammalian Oocyte Maturation and Preimplantation Embryo Development. Int. J. Mol. Sci. 2019, 20, 409. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Primers | Sequences | Length (bp) | Annealing Temperature (°C) |
|---|---|---|---|
| GAPDH | F: 5’- GGT GTG AAC CAT GAG AAG TAT GA-3’ | 123 | 60 |
| R: 5 - GAG TCC TTC CAC GAT ACC AAA G-3’ | |||
| OCT4 | F: 5’- TAT GGG AGC CCT CAC TTC AC-3’ | 144 | 60 |
| R: 5’- AGT TTG TGC CAG GGT TTT TG -3’ | |||
| SOX2 | F: 5′- GCT AGT CTC CAA GCG ACG AA -3′ | 144 | 60 |
| R: 5′- GCA AGA AGC CTC TCC TTG AA -3′ | |||
| NANOG | F: 5′- CAG TCT GGA CAC TGG CTG AA -3′ | 149 | 60 |
| R: 5′- CTC GCT GAT TAG GCT CCA AC -3′ | |||
| BMP4 | F: 5′- GGA GAT GGT AGT AGA GGG ATG T -3′ | 105 | 60 |
| R: 5′- CGT GTG TGT GTG GTG TAT GT -3′ | |||
| SOX17 | F: 5′- CAT GAC TCC GGT GTG AAT CTC -3′ | 99 | 60 |
| R: 5′- CAC GTC AGG ATA GTT GCA GTA ATA -3′ | |||
| ATF3 | F: 5’- CCT CTG CGC TGG AAT CAG TC-3’ | 111 | 60 |
| R: 5’- TTC TTT CTC GTC GCC TCT TTT T-3’ | |||
| ATF4 | F: 5’- ATG ACC GAA ATG AGC TTC CTG-3’ | 153 | 60 |
| R: 5’- GCT GGA GAA CCC ATG AGG T-3’ | |||
| ATF6 | F: 5’- TCC TCG GTC AGT GGA CTC TTA-3’ | 235 | 60 |
| R: 5’- CTT GGG CTG AAT TGA AGG TTT TG-3’ | |||
| CHOP | F: 5’- GGA AAC AGA GTG GTC ATT CCC-3’ | 116 | 60 |
| R: 5’- CTG CTT GAG CCG TTC ATT CTC-3’ | |||
| BIP | F: 5’- CAT CAC GCC GTC CTA TGT CG-3′ | 104 | 60 |
| R: 5′- CGT CAA AGA CCG TGT TCT CG-3′ | |||
| GADD34 | F: 5′- ATG ATG GCA TGT ATG GTG AGC-3′ | 120 | 60 |
| R: 5′- AAC CTT GCA GTG TCC TTA TCA G-3′ | |||
| sXBP-1 | F: 5′- GGT CTG CTG AGT CCG CAG CAG G-3′ | 311 | 60 |
| R: 5′- GGG CTT GGT ATA TAT GTG G-3′ | |||
| tXBP-1 | F: 5′- CCC TCC AGA ACA TCT CCC CAT-3′ | 101 | 60 |
| R: 5′- ACA TGA CTG GGT CCA AGT TGT-3′ | |||
| NEUROD1 | F: 5’- GAA CGC AGA GGA GGA CTC AC-3’ | 109 | 60 |
| R: 5’- CTT GGG CTT TTG ATC GTC AT-3’ | |||
| TBX3 | F: 5’- GAT GAG TCC TCC AGT GAA CAA G-3’ | 91 | 60 |
| R: 5’- CTT TGA GGT TCG ATG TCC CTA C-3’ | |||
| Hand1 | F: 5’- CGC CTA GCC ACC AGC TAC ATC-3’ | 106 | 60 |
| R: 5’- CGC CAT CCG CCT TCT TGA GTT-3’ | |||
| SMA | F: 5’- ACC CAC AAT GTC CCC ATC TA-3’ | 123 | 60 |
| R: 5’- GAA GGA ATA GCC ACG CTC AG-3’ | |||
| GATA6 | F: 5’- TCC ACT CGT GTC TGC TTT TG-3’ | 140 | 60 |
| R: 5’- CCC TTC CCT TCC ATC TTC TC-3’ | |||
| AFP | F: 5’- AAA TGC GTT TCT CGT TGC TT-3’ | 136 | 60 |
| R: 5’- GCC ACA GGC CAA TAG TTT GT-3’ | |||
| OFLM3 | F: 5′- ACC CAG TTC AAG GAG GAA ATA AG-3′ | 104 | 60 |
| R: 5′- CTC AGC ACT CTT TGG TGT AGT T-3′ | |||
| NR2F2 | F: 5′- GAA GAT CTC TCC CTT CAC CTT TC-3′ | 100 | 60 |
| R: 5′- GAA TCT CCT CCT CGG TGT TTA TC-3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, J.H.; Kim, H.K.; Ha, T.W.; Im, J.S.; Song, B.H.; Hong, K.S.; Oh, J.S.; Han, J.; Lee, M.R. Hyperthermia Disturbs and Delays Spontaneous Differentiation of Human Embryoid Bodies. Biomedicines 2020, 8, 176. https://doi.org/10.3390/biomedicines8060176
Kwon JH, Kim HK, Ha TW, Im JS, Song BH, Hong KS, Oh JS, Han J, Lee MR. Hyperthermia Disturbs and Delays Spontaneous Differentiation of Human Embryoid Bodies. Biomedicines. 2020; 8(6):176. https://doi.org/10.3390/biomedicines8060176
Chicago/Turabian StyleKwon, Ji Hyun, Hyun Kyu Kim, Tae Won Ha, Jeong Suk Im, Byung Hoo Song, Ki Sung Hong, Jae Sang Oh, Jaeseok Han, and Man Ryul Lee. 2020. "Hyperthermia Disturbs and Delays Spontaneous Differentiation of Human Embryoid Bodies" Biomedicines 8, no. 6: 176. https://doi.org/10.3390/biomedicines8060176
APA StyleKwon, J. H., Kim, H. K., Ha, T. W., Im, J. S., Song, B. H., Hong, K. S., Oh, J. S., Han, J., & Lee, M. R. (2020). Hyperthermia Disturbs and Delays Spontaneous Differentiation of Human Embryoid Bodies. Biomedicines, 8(6), 176. https://doi.org/10.3390/biomedicines8060176