Comparative Analysis of the Transcriptome, Proteome, and miRNA Profile of Kupffer Cells and Monocytes
Abstract
:1. Introduction
2. Results
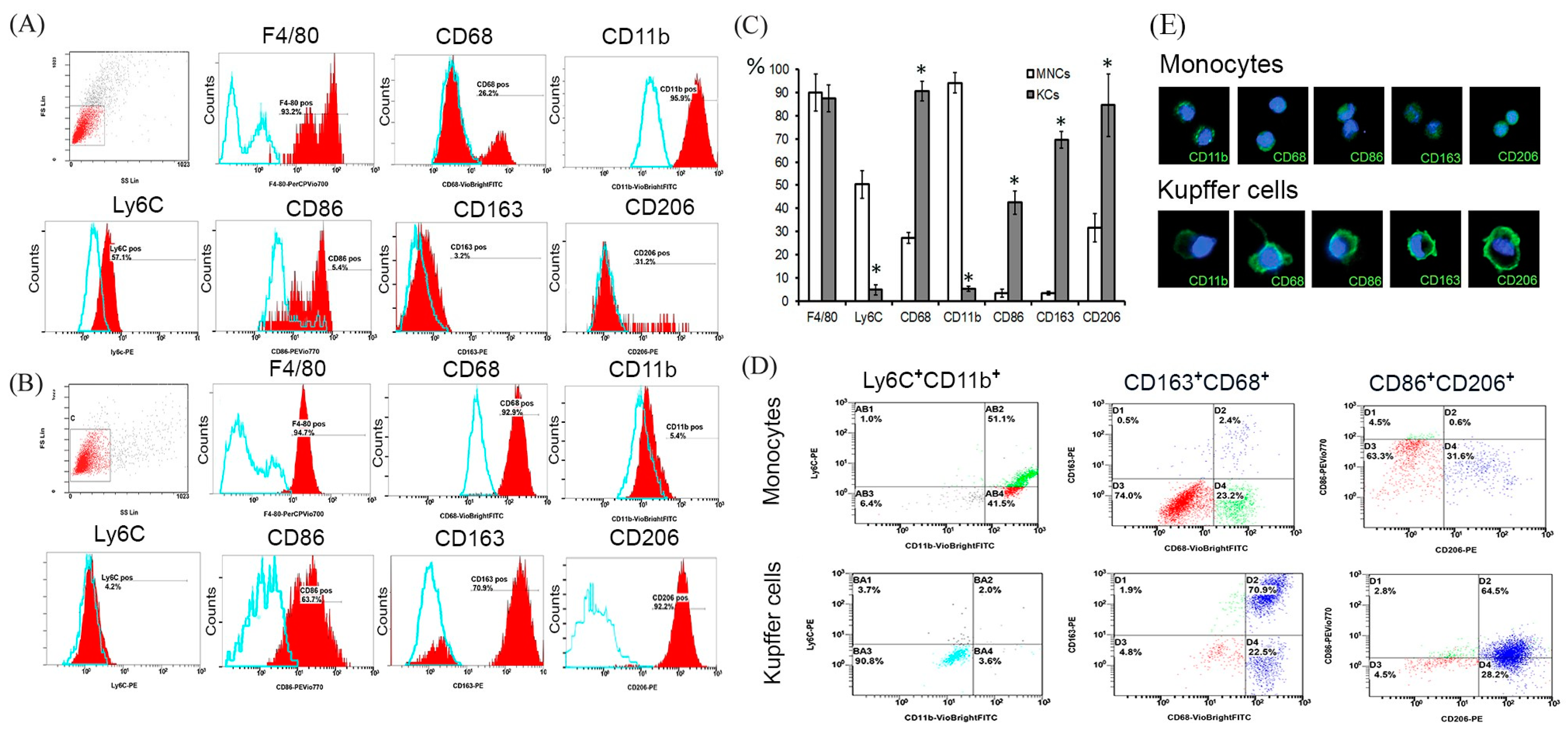
2.1. Comparative Immunophenotyping of KUPFFER Cells and Monocytes
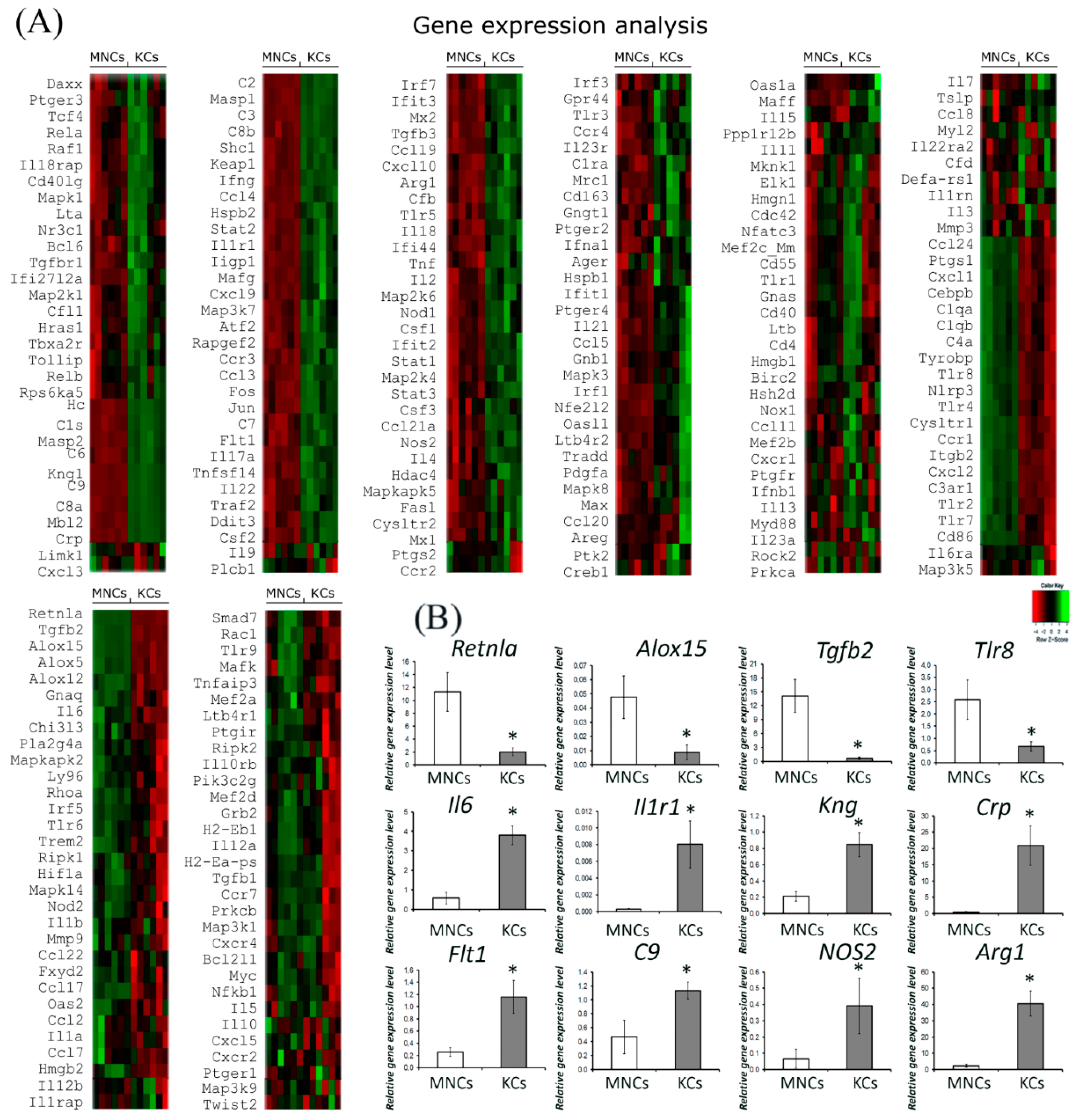
2.2. Comparative Gene Expression Profiling by Nanostring
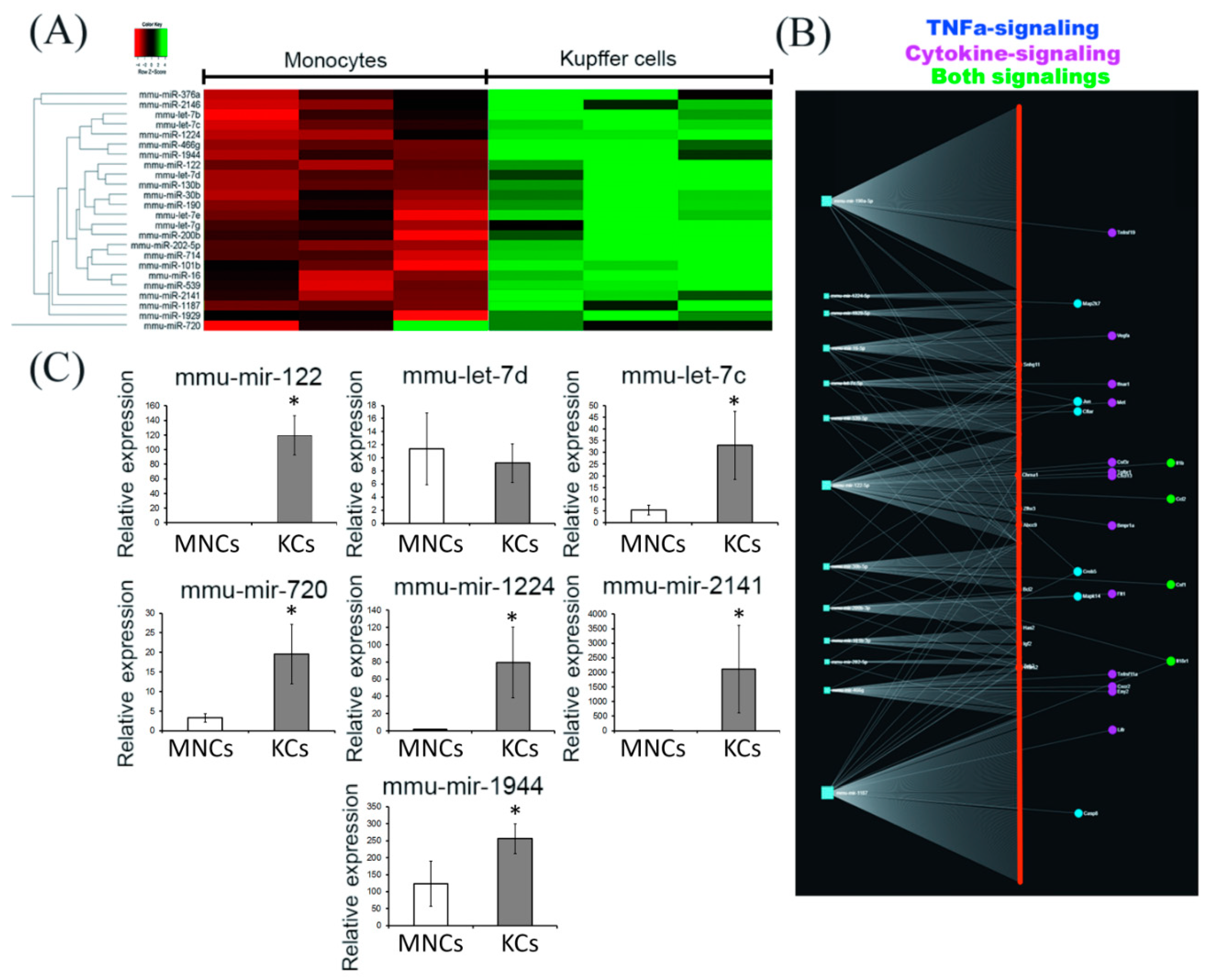
2.3. Comparative miRNA Expression Profiling by Nanostring
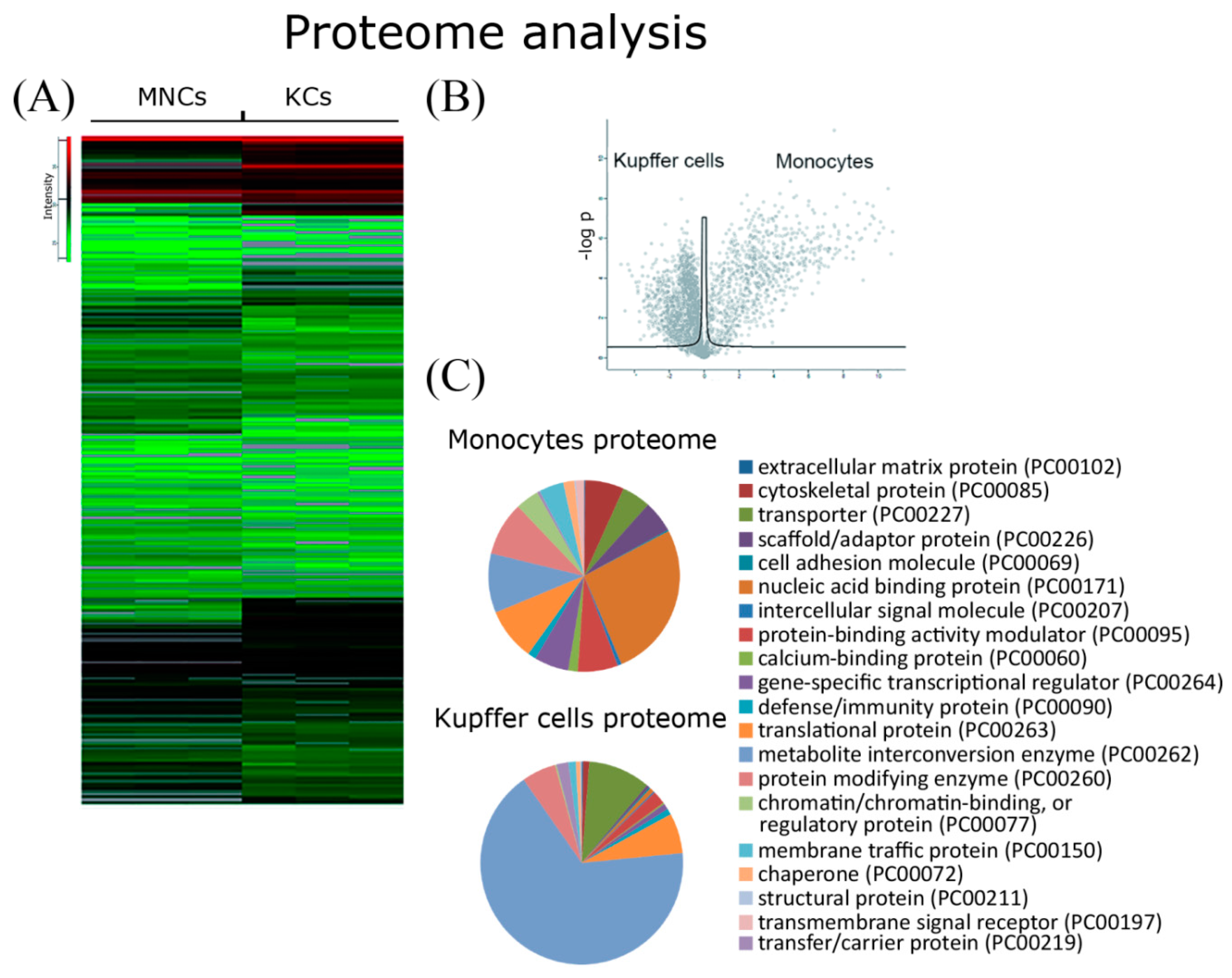
2.4. Comparative Proteome Analysis
2.5. Western-Blot Analysis of Protein Representation
3. Discussion
4. Materials and Methods
4.1. Isolation of Kupffer Cells
4.2. Isolation of Monocytes
4.3. Flow Cytometry Assay
4.4. Immunocytochemistry Assay
4.5. Nanostring Gene Expression Assay and Target Analysis
4.6. Real-Time Polymerase Chain Reaction (RT PCR) Gene Expression Assay
4.7. Proteome Analysis
4.8. Western-Blot Analysis of Protein Representation
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability Statement
References
- Theret, M.; Mounier, R.; Rossi, F. The origins and non-canonical functions of macrophages in development and regeneration. Development 2019, 146, dev156000. [Google Scholar] [CrossRef] [Green Version]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage Activation and Polarization: Nomenclature and Experimental Guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orecchioni, M.; Ghosheh, Y.; Pramod, A.B.; Ley, K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS–) vs. Alternatively Activated Macrophages. Front. Immunol. 2019, 10, 1084. [Google Scholar] [CrossRef]
- Hoeffel, G.; Ginhoux, F. Fetal monocytes and the origins of tissue-resident macrophages. Cell. Immunol. 2018, 330, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Perdiguero, E.G.; Geissmann, F. The development and maintenance of resident macrophages. Nat. Immunol. 2016, 17, 2–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautier, E.L.; Shay, T.; Miller, J.; Greter, M.; Jakubzick, C.; Ivanov, S.; Helft, J.; Chow, A.; Elpek, K.G.; Gordonov, S.; et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 2012, 13, 1118–1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishiyama, K.; Nakashima, H.; Ikarashi, M.; Kinoshita, M.; Nakashima, M.; Aosasa, S.; Seki, S.; Yamamoto, J. Mouse CD11b+Kupffer cells recruited from bone marrow accelerate liver regeneration after partial hepatectomy. PLoS ONE 2015, 10, e0136774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoeffel, G.; Chen, J.; Lavin, Y.; Low, D.; Almeida, F.F.; See, P.; Beaudin, A.E.; Lum, J.; Low, I.; Forsberg, E.C.; et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 2015, 42, 665–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginhoux, F.; Greter, M.; Leboeuf, M.; Nandi, S.; See, P.; Mehler, M.F.; Conway, S.J.; Ng, L.G.; Stanley, E.R.; Igor, M.; et al. Fate map analysis reveals that adult mice derive from primitive macrophages. Science 2010, 330, 841–845. [Google Scholar] [CrossRef] [Green Version]
- Gundra, U.M.; Girgis, N.M.; Ruckerl, D.; Jenkins, S.; Ward, L.N.; Kurtz, Z.D.; Wiens, K.E.; Tang, M.S.; Basu-Roy, U.; Mansukhani, A.; et al. Alternatively activated macrophages derived from monocytes and tissue macrophages are phenotypically and functionally distinct. Blood 2014, 123, e110–e122. [Google Scholar] [CrossRef] [Green Version]
- van de Laar, L.; Saelens, W.; De Prijck, S.; Martens, L.; Scott, C.L.; Van Isterdael, G.; Hoffmann, E.; Beyaert, R.; Saeys, Y.; Lambrecht, B.N.; et al. Yolk Sac Macrophages, Fetal Liver, and Adult Monocytes Can Colonize an Empty Niche and Develop into Functional Tissue-Resident Macrophages. Immunity 2016, 44, 755–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beattie, L.; Sawtell, A.; Mann, J.; Frame, T.C.M.; Teal, B.; de Labastida Rivera, F.; Brown, N.; Walwyn-Brown, K.; Moore, J.W.J.; MacDonald, S.; et al. Bone marrow-derived and resident liver macrophages display unique transcriptomic signatures but similar biological functions. J. Hepatol. 2016, 65, 758–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merlin, S.; Bhargava, K.K.; Ranaldo, G.; Zanolini, D.; Palestro, C.J.; Santambrogio, L.; Prat, M.; Follenzi, A.; Gupta, S. Kupffer Cell Transplantation in Mice for Elucidating Monocyte/Macrophage Biology and for Potential in Cell or Gene Therapy. Am. J. Pathol. 2016, 186, 539–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, C.L.; Zheng, F.; De Baetselier, P.; Martens, L.; Saeys, Y.; De Prijck, S.; Lippens, S.; Abels, C.; Schoonooghe, S.; Raes, G.; et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 2016, 7, 10321. [Google Scholar] [CrossRef] [PubMed]
- Gomez Perdiguero, E.; Klapproth, K.; Schulz, C.; Busch, K.; Azzoni, E.; Crozet, L.; Garner, H.; Trouillet, C.; De Bruijn, M.F.; Geissmann, F.; et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 2015, 518, 547–551. [Google Scholar] [CrossRef]
- Guilliams, M.; Scott, C.L. Does niche competition determine the origin of tissue-resident macrophages? Nat. Rev. Immunol. 2017, 17, 451–460. [Google Scholar] [CrossRef]
- You, Q.; Holt, M.; Yin, H.; Li, G.; Hu, C.J.; Ju, C. Role of hepatic resident and infiltrating macrophages in liver repair after acute injury. Biochem. Pharmacol. 2013, 86, 836–843. [Google Scholar] [CrossRef] [Green Version]
- Lokhonina, A.; Elchaninov, A.; Fatkhudinov, T.; Makarov, A.; Arutyunyan, I.; Grinberg, M.; Glinkina, V.; Surovtsev, V.; Bolshakova, G.; Goldshtein, D.; et al. Activated Macrophages of Monocytic Origin Predominantly Express Proinflammatory Cytokine Genes, Whereas Kupffer Cells Predominantly Express Anti-Inflammatory Cytokine Genes. Biomed Res. Int. 2019, 2019, 3912142. [Google Scholar] [CrossRef] [Green Version]
- Bain, C.C.; Scott, C.L.; Uronen-Hansson, H.; Gudjonsson, S.; Jansson, O.; Grip, O.; Guilliams, M.; Malissen, B.; Agace, W.W.; Mowat, A.M. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 2013, 6, 498–510. [Google Scholar] [CrossRef]
- Lokhonina, A.V.; Elchaninov, A.V.; Makarov, A.V.; Nikitina, M.P.; Goldshtein, D.V.; Paltsev, M.A.; Fatkhudinov, T.K. Comparative characteristics of the susceptibility of kupffer cells and macrophages of bone-background origin to activation factors. Mol. Meditsina 2019, 17. [Google Scholar] [CrossRef]
- T’Jonck, W.; Guilliams, M.; Bonnardel, J. Niche signals and transcription factors involved in tissue-resident macrophage development. Cell. Immunol. 2018, 330, 43–53. [Google Scholar] [CrossRef]
- Ziegler-Heitbrock, L. Monocyte subsets in man and other species. Cell. Immunol. 2014, 289, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Boyette, L.B.; MacEdo, C.; Hadi, K.; Elinoff, B.D.; Walters, J.T.; Ramaswami, B.; Chalasani, G.; Taboas, J.M.; Lakkis, F.G.; Metes, D.M. Phenotype, function, and differentiation potential of human monocyte subsets. PLoS ONE 2017, 12, e0176460. [Google Scholar] [CrossRef]
- Teh, Y.C.; Ding, J.L.; Ng, L.G.; Sz, C. Capturing the Fantastic Voyage of Monocytes Through Time and Space. Front. Immunol 2019, 10, 834. [Google Scholar] [CrossRef]
- Guilliams, M.; Mildner, A.; Yona, S. Developmental and Functional Heterogeneity of Monocytes. Immunity 2018, 49, 595–613. [Google Scholar] [CrossRef] [Green Version]
- Mildner, A.; Schönheit, J.; Giladi, A.; David, E.; Lara-Astiaso, D.; Lorenzo-Vivas, E.; Paul, F.; Chappell-Maor, L.; Priller, J.; Leutz, A.; et al. Genomic Characterization of Murine Monocytes Reveals C/EBPβ Transcription Factor Dependence of Ly6C− Cells. Immunity 2017, 46, 849–862.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlin, L.M.; Stamatiades, E.G.; Auffray, C.; Hanna, R.N.; Glover, L.; Vizcay-Barrena, G.; Hedrick, C.C.; Cook, H.T.; Diebold, S.; Geissmann, F. Nr4a1-dependent Ly6Clow monocytes monitor endothelial cells and orchestrate their disposal. Cell 2013, 153, 362–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabriek, B.O.; van Bruggen, R.; Deng, D.M.; Ligtenberg, A.J.M.; Nazmi, K.; Schornagel, K.; Vloet, R.P.M.; Dijkstra, C.D.; van den Berg, T.K. The macrophage scavenger receptor CD163 functions as an innate immune sensor for bacteria. Blood 2009, 113, 887–892. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Pomares, L. The mannose receptor. J. Leukoc. Biol. 2012, 92, 1177–1186. [Google Scholar] [CrossRef]
- Italiani, P.; Boraschi, D. From Monocytes to M1/M2 Macrophages: Phenotypical vs. Functional Differentiation. Front. Immunol. 2014, 5, 514. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, M.; Uchida, T.; Sato, A.; Nakashima, M.; Nakashima, H.; Shono, S.; Habu, Y.; Miyazaki, H.; Hiroi, S.; Seki, S. Characterization of two F4/80-positive Kupffer cell subsets by their function and phenotype in mice. J. Hepatol. 2010, 53, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Blagih, J.; Jones, R.G. Polarizing macrophages through reprogramming of glucose metabolism. Cell Metab. 2012, 15, 793–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadiku, P.; Walmsley, S.R. Hypoxia and the regulation of myeloid cell metabolic imprinting: Consequences for the inflammatory response. EMBO Rep. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.; Nicolas, E.; Marks, D.; Sander, C.; Lerro, A.; Buendia, M.A.; Xu, C.; Mason, W.S.; Moloshok, T.; Bort, R.; et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004, 1, 106–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, S.-H.; Delgado, E.R.; Otero, P.A.; Teng, K.-Y.; Kutay, H.; Meehan, K.M.; Moroney, J.B.; Monga, J.K.; Hand, N.J.; Friedman, J.R.; et al. MicroRNA-122 regulates polyploidization in the murine liver. Hepatology 2016, 64, 599–615. [Google Scholar] [CrossRef] [Green Version]
- Tsai, W.C.; Hsu, S.D.; Hsu, C.S.; Lai, T.C.; Chen, S.J.; Shen, R.; Huang, Y.; Chen, H.C.; Lee, C.H.; Tsai, T.F.; et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Investig. 2012, 122, 2884–2897. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Liu, J.; Luo, Y. MicroRNAs in tumor immunity: Functional regulation in tumor-associated macrophages. J. Zhejiang Univ. Sci. B 2020, 21, 12–28. [Google Scholar] [CrossRef]
- Lu, L.; McCurdy, S.; Huang, S.; Zhu, X.; Peplowska, K.; Tiirikainen, M.; Boisvert, W.A.; Garmire, L.X. Time Series miRNA-mRNA integrated analysis reveals critical miRNAs and targets in macrophage polarization. Sci. Rep. 2016, 6, 37446. [Google Scholar] [CrossRef] [Green Version]
- Geng, J.; Gates, P.B.; Kumar, A.; Guenther, S.; Garza-Garcia, A.; Kuenne, C.; Zhang, P.; Looso, M.; Brockes, J.P. Identification of the orphan gene Prod 1 in basal and other salamander families. Evodevo 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Baer, C.; Squadrito, M.L.; Laoui, D.; Thompson, D.; Hansen, S.K.; Kiialainen, A.; Hoves, S.; Ries, C.H.; Ooi, C.H.; De Palma, M. Suppression of microRNA activity amplifies IFN-γ-induced macrophage activation and promotes anti-tumour immunity. Nat. Cell Biol. 2016, 18, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Liu, G.-P.; Kong, D.; Huang, W.; Sun, Y.; Zhao, D. Downregulation of miR-1224 protects against oxidative stress-induced acute liver injury by regulating hepatocyte growth factor. J. Cell. Biochem. 2019, 120, 12369–12375. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houthuys, E.; Movahedi, K.; De Baetselier, P.; Van Ginderachter, J.A.; Brouckaert, P. A method for the isolation and purification of mouse peripheral blood monocytes. J. Immunol. Methods 2010, 359, 1–10. [Google Scholar] [CrossRef]
- Elchaninov, A.; Fatkhudinov, T.; Usman, N.; Arutyunyan, I.; Makarov, A.; Lokhonina, A.; Eremina, I.; Surovtsev, V.; Goldshtein, D.; Bolshakova, G.; et al. Multipotent stromal cells stimulate liver regeneration by influencing the macrophage polarization in rat. World J. Hepatol. 2018. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4. [Google Scholar] [CrossRef]
- TargetScanHuman 7.2. Available online: http://www.targetscan.org/vert_72/ (accessed on 2 March 2020).
- miRNet. Available online: https://www.mirnet.ca/ (accessed on 2 March 2020).
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Kovalchuk, S.I.; Jensen, O.N.; Rogowska-Wrzesinska, A. FlashPack: Fast and simple preparation of ultrahigh-performance capillary columns for LC-MS. Mol. Cell. Proteomics 2019, 18, 383–390. [Google Scholar] [CrossRef] [Green Version]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- PANTHER—Gene List Analysis. Available online: http://pantherdb.org/ (accessed on 2 March 2020).
- bioDBnet—Biological Database Network. Available online: https://biodbnet-abcc.ncifcrf.gov/db/db2db.php (accessed on 2 March 2020).
- Elchaninov, A.V.; Fatkhudinov, T.K.; Vishnyakova, P.A.; Nikitina, M.P.; Lokhonina, A.V.; Makarov, A.V.; Arutyunyan, I.V.; Kananykhina, E.Y.; Poltavets, A.S.; Butov, K.R.; et al. Molecular mechanisms of splenectomy-induced hepatocyte proliferation. PLoS ONE 2020, 15, e0233767. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef]

| Antibody | Isotypic Control | Manufacturer |
|---|---|---|
| CD11b-VioBright FITC, mouse (clone: REA592) | REA Control-VioBright FITC | Miltenyi Biotec |
| CD86-PE-Vio770, mouse | Rat IgG2b-PE-Vio770 | Miltenyi Biotec |
| Ly-6C-PE, mouse | Rat IgG2a-PE | Miltenyi Biotec |
| CD206 (MMR) Monoclonal Antibody (MR6F3), PE | Rat IgG2b kappa Isotype Control (eB149/10H5), PE | eBioscience™ |
| CD163-PE (Thermo Fisher) Monoclonal Antibody | Mouse IgG1 kappa Isotype Control, PE, eBioscience™ | eBioscience™ |
| Anti-F4/80-PerCP-Vio700, mouse | REA Control-PerCP-Vio700 | Miltenyi Biotec |
| CD68-FITC, mouse (clone: FA-11) | Rat IgG2a-FITC | Miltenyi Biotec |
| FIZZ1 (Retnla) | for | CCTTTCCTGAGATTCTGCCCC |
| rev | CGAGTAAGCACAGGCAGTTG | |
| TGFb2 | for | TCCCCTCCGAAAATGCCATC |
| rev | CCTCCGCTCTGGTTTTCACA | |
| Alox15 | for | AAGATGTAACCCACCACGTTCA |
| rev | TGCCCCGATGACACAGAAAA | |
| Tlr8 | for | TGGTCCAGCTATAGAGCACATC |
| rev | ACTGAGGGGGCATGTTTTCC | |
| Flt1 | for | ACAAGTCAAACCTGGAGCTGA |
| rev | TGCAGAGGCTTGAACGACTT | |
| IL1r1 | for | GACCCCCATATCAGCGGACC |
| rev | ACCGGATATTGCTTCCCCCG | |
| Crp | for | GACTCGTATGGCGGTGACTT |
| rev | GCTGAGTGTCCCACCAACAT | |
| Kng1 | for | GTGAAGCAGTAGTCCCAGCAA |
| rev | CCCTCTAATCATCCCTGTGGC | |
| C9 | for | CCCTTGCCATCTTTGCCTTG |
| rev | GTGGGTCATCGTCACAGTCA | |
| Il6 | for | CCACTTCACAAGTCGGAGGC |
| rev | GGAGAGCATTGGAAATTGGGGT | |
| Arg1 | for | ACGGCAGTGGCTTTAACCTT |
| rev | AGGTAGTCAGTCCCTGGCTT | |
| NOS2 | for | GCTGCCAGGGTCACAACTT |
| rev | CCTCACATACTGTGGACGGG | |
| GAPDH | for | AGGCCGGTGCTGAGTATGTC |
| rev | TGCCTGCTTCACCACCTTCT |
| mmu-miR122-for | TGGAGTGTGACAATGGTGTTTG |
| mmu-let-7c-for | TGAGGTAGTAGGTTGTATGGTT |
| mmu-let-7d-for | AGAGGTAGTAGGTTGCATAGTT |
| mmu-miR-720-for | ATCTCGCTGGGGCCTCCA |
| mmu-miR-1224-for | GTGAGGACTGGGGAGGTGGAG |
| mmu-miR-1944-for | CTCTGTGCTGAATGTCAAGTTCTGATT |
| mmu-miR-2141-for | AGGAGGTGTCAGAAAAGTT |
| RNU6-for | CTCGCTTCGGCAGCACA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elchaninov, A.; Lokhonina, A.; Nikitina, M.; Vishnyakova, P.; Makarov, A.; Arutyunyan, I.; Poltavets, A.; Kananykhina, E.; Kovalchuk, S.; Karpulevich, E.; et al. Comparative Analysis of the Transcriptome, Proteome, and miRNA Profile of Kupffer Cells and Monocytes. Biomedicines 2020, 8, 627. https://doi.org/10.3390/biomedicines8120627
Elchaninov A, Lokhonina A, Nikitina M, Vishnyakova P, Makarov A, Arutyunyan I, Poltavets A, Kananykhina E, Kovalchuk S, Karpulevich E, et al. Comparative Analysis of the Transcriptome, Proteome, and miRNA Profile of Kupffer Cells and Monocytes. Biomedicines. 2020; 8(12):627. https://doi.org/10.3390/biomedicines8120627
Chicago/Turabian StyleElchaninov, Andrey, Anastasia Lokhonina, Maria Nikitina, Polina Vishnyakova, Andrey Makarov, Irina Arutyunyan, Anastasiya Poltavets, Evgenia Kananykhina, Sergey Kovalchuk, Evgeny Karpulevich, and et al. 2020. "Comparative Analysis of the Transcriptome, Proteome, and miRNA Profile of Kupffer Cells and Monocytes" Biomedicines 8, no. 12: 627. https://doi.org/10.3390/biomedicines8120627
APA StyleElchaninov, A., Lokhonina, A., Nikitina, M., Vishnyakova, P., Makarov, A., Arutyunyan, I., Poltavets, A., Kananykhina, E., Kovalchuk, S., Karpulevich, E., Bolshakova, G., Sukhikh, G., & Fatkhudinov, T. (2020). Comparative Analysis of the Transcriptome, Proteome, and miRNA Profile of Kupffer Cells and Monocytes. Biomedicines, 8(12), 627. https://doi.org/10.3390/biomedicines8120627









