The Role of Soluble Uric Acid in Modulating Autophagy Flux and Inflammasome Activation during Bacterial Infection in Macrophages
Abstract
1. Introduction
2. Experimental Section
2.1. Reagents
2.2. Bacterial Culture
2.3. Uric Acid Preparation
2.4. Macrophage Induction Assay
2.5. Macrophage Infection with Live Bacteria
2.6. Autophagy Flux Measurement
2.7. Confocal Microscopy Imaging and Image Analysis
2.8. Human and Murine IL-1β Detection and Quantification
2.9. Nitric Oxide Release Quantification
2.10. Statistical Analysis
3. Results
3.1. Soluble Uric Acid Modulates Autophagy Flux Induction in Macrophages during Bacterial Infection In Vitro
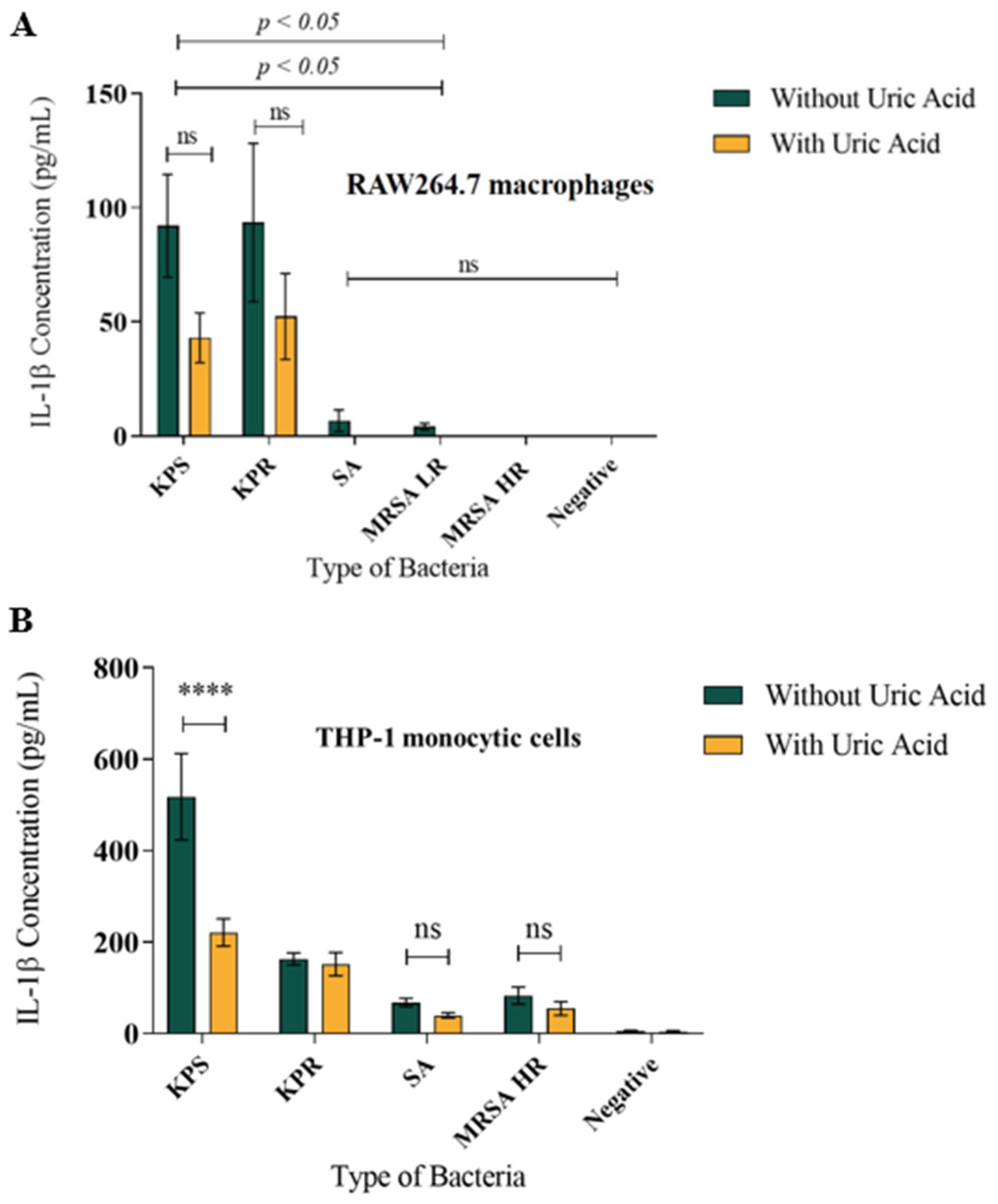
3.2. Soluble Uric Acid Decreases IL-1β Production in Murine Macrophages and Human Monocytes during Bacterial Infection In Vitro
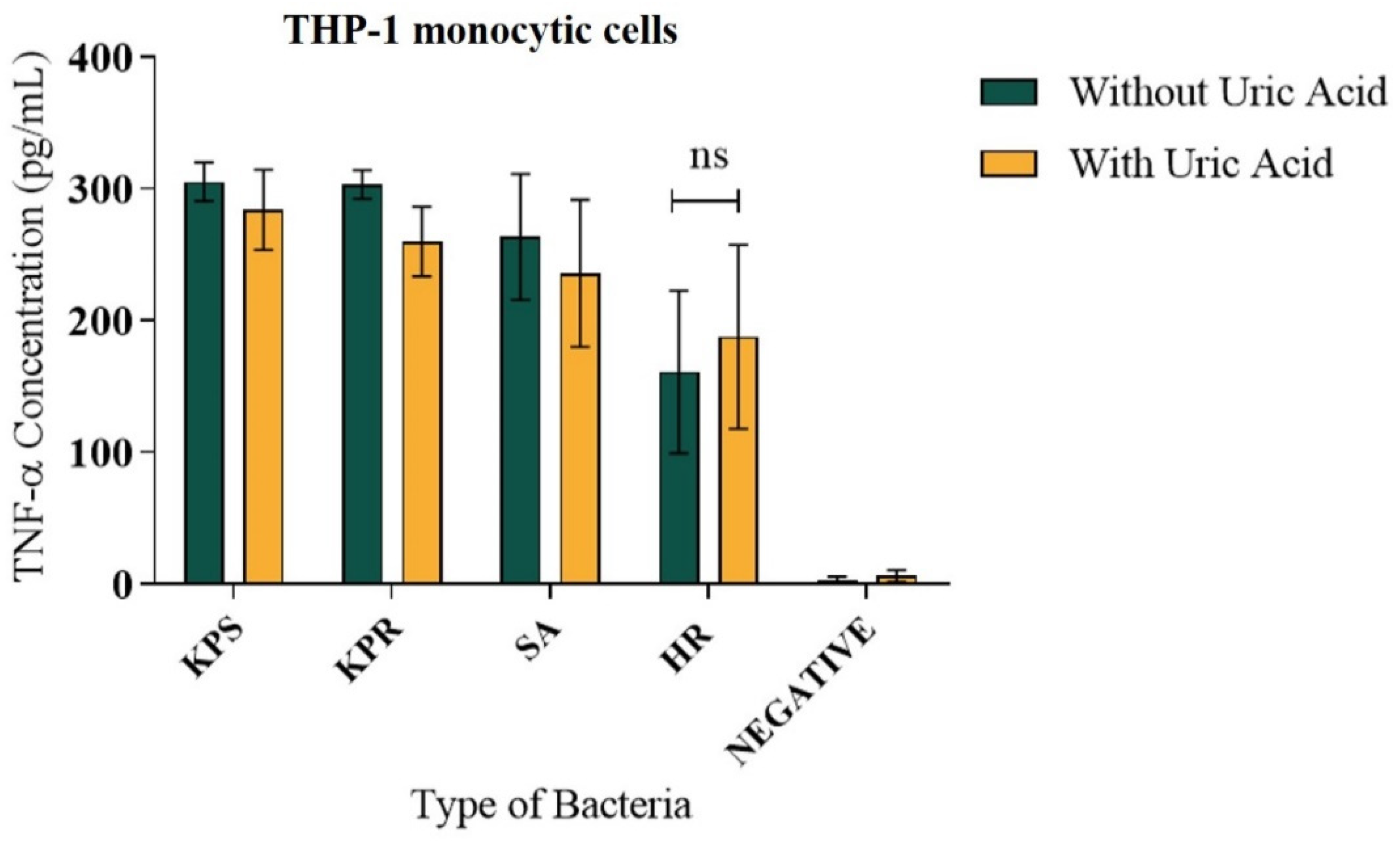
3.3. Soluble Uric Acid Does Not Affect TNF-α Release from Human Monocytes during Bacterial Infection
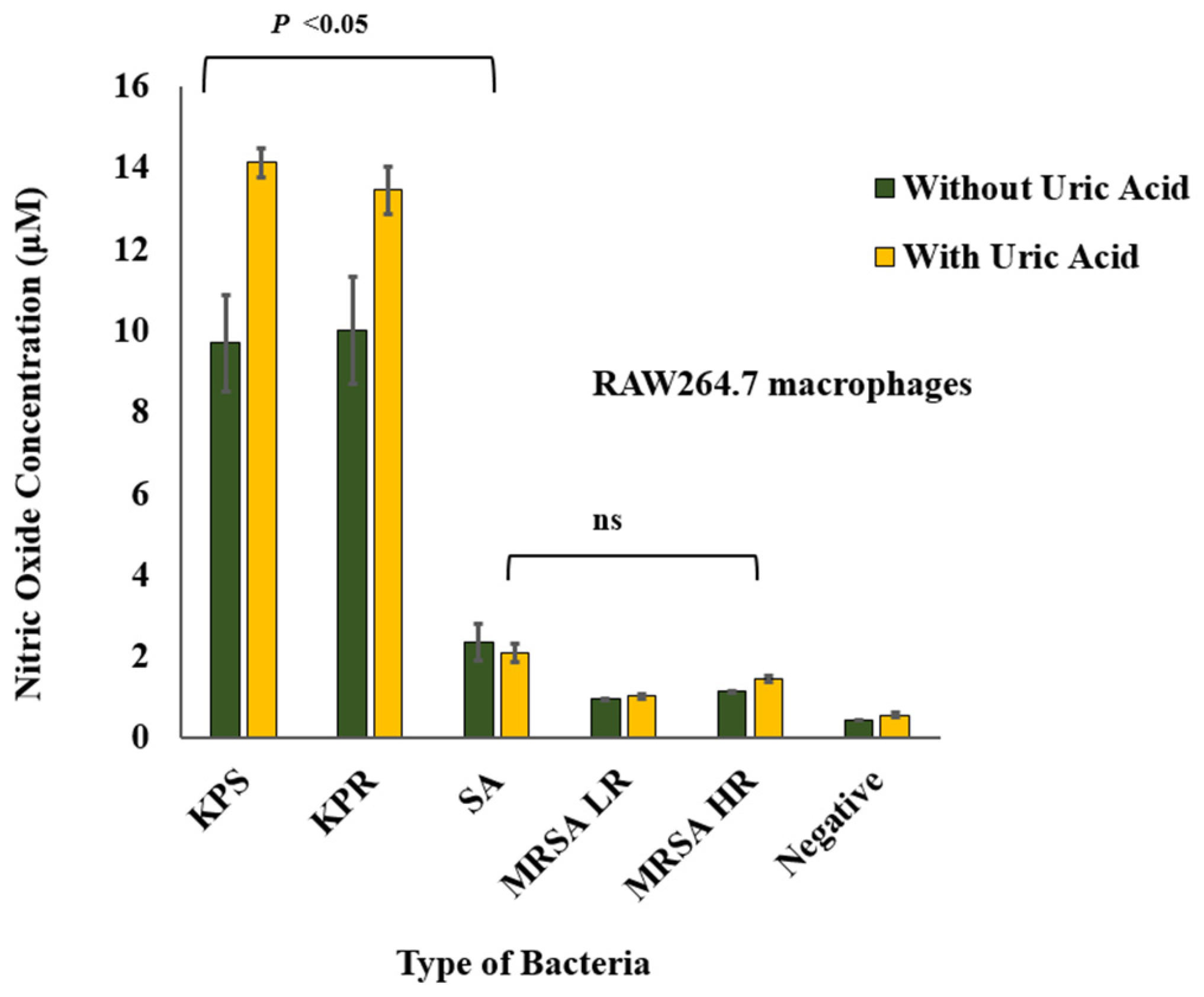
3.4. Soluble Uric Acid Enhances Nitric Oxide Production in Murine Macrophages during Bacterial Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Johnson, R.J.; Bakris, G.L.; Borghi, C.; Chonchol, M.B.; Feldman, D.; Lanaspa, M.A.; Merriman, T.R.; Moe, O.W.; Mount, D.B.; Lozada, L.G.S. Hyperuricemia, acute and chronic kidney disease, hypertension, and cardiovascular disease: Report of a scientific workshop organized by the National Kidney Foundation. Am. J. Kidney Dis. 2018, 71, 851–865. [Google Scholar] [CrossRef] [PubMed]
- Bursill, D.; Taylor, W.J.; Terkeltaub, R.; Kuwabara, M.; Merriman, T.R.; Grainger, R.; Pineda, C.; Louthrenoo, W.; Edwards, N.L.; Andres, M.; et al. Gout, Hyperuricemia, and Crystal-Associated Disease Network Consensus Statement Regarding Labels and Definitions for Disease Elements in Gout. Arthritis Care Res. (Hoboken) 2019, 7171, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.J.; Lanaspa, M.A.; Gaucher, E.A. Uric acid: A danger signal from the RNA world that may have a role in the epidemic of obesity, metabolic syndrome, and cardiorenal disease: Evolutionary considerations. Semin. Nephrol. 2011, 31, 394–399. [Google Scholar] [CrossRef] [PubMed]
- van den Berghe, G.; Vincent, M.-F.; Marie, S. Disorders of purine and pyrimidine metabolism. In Inborn Metabolic Diseases; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Loeb, J.N. The influence of temperature on the solubility of monosodium urate. Arthritis Rheumatol. 1972, 15, 189–192. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, W.R.; Khalaf, A. Nucleation of monosodium urate crystals. Ann. Rheum. Dis. 1975, 34, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Ames, B.N.; Cathcart, R.; Schwiers, E.; Hochstein, P. Uric acid provides an antioxidant defense in humans against oxidant-and radical-caused aging and cancer: A hypothesis. Proc. Natl. Acad. Sci. USA 1981, 78, 6858–6862. [Google Scholar] [CrossRef] [PubMed]
- Hernán, A.; Douglas, G.; Pablo, C.; Hugo, V.; Rosemarie, M.; Lorena, G.; Guillermo, D.-A.; Clara, Q.; Mario, C.; Sergio, L. Uric acid: A molecule with paradoxical actions in heart failure. Rev. Med. Chile 2011, 139, 505–515. [Google Scholar]
- Desideri, G.; Castaldo, G.; Lombardi, A.; Mussap, M.; Testa, A.; Pontremoli, R.; Punzi, L.; Borghi, C. Is it time to revise the normal range of serum uric acid levels? Eur. Rev. Med. Pharm. Sci. 2014, 18, 1295–1306. [Google Scholar]
- Martinon, F.; Petrilli, V.; Mayor, A.; Tardivel, A.; Tschopp, J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 2006, 440, 237–241. [Google Scholar] [CrossRef]
- Bursill, D.; Taylor, W.J.; Terkeltaub, R.; Abhishek, A.; So, A.K.; Vargas-Santos, A.B.; Gaffo, A.L.; Rosenthal, A.; Tausche, A.K.; Reginato, A.; et al. Gout, Hyperuricaemia and Crystal-Associated Disease Network (G-CAN) consensus statement regarding labels and definitions of disease states of gout. Ann. Rheum. Dis. 2019, 78, 1592–1600. [Google Scholar] [CrossRef]
- Dalbeth, N.; Merriman, T.R.; Stamp, L.K. Gout. Lancet 2016, 388, 2039–2052. [Google Scholar] [CrossRef]
- Yip, K.; Cohen, R.E.; Pillinger, M.H. Asymptomatic hyperuricemia: Is it really asymptomatic? Curr. Opin. Rheumatol. 2020, 32, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Rock, K.L.; Kataoka, H.; Lai, J.-J. Uric acid as a danger signal in gout and its comorbidities. Nat. Rev. Rheumatol. 2013, 9, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Sollberger, G.; Strittmatter, G.E.; Garstkiewicz, M.; Sand, J.; Beer, H.-D. Caspase-1: The inflammasome and beyond. Innate Immun. 2014, 20, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Ravanan, P.; Srikumar, I.F.; Talwar, P. Autophagy: The spotlight for cellular stress responses. Life Sci. 2017, 188, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Thurston, T.L.; Wandel, M.P.; von Muhlinen, N.; Foeglein, Á.; Randow, F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 2012, 482, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Brumell, J.H. Bacteria–autophagy interplay: A battle for survival. Nat. Rev. Microbiol. 2014, 12, 101. [Google Scholar] [CrossRef]
- Zughaier, S.M.; Kandler, J.L.; Balthazar, J.T.; Shafer, W.M. Phosphoethanolamine Modification of Neisseria gonorrhoeae Lipid A Reduces Autophagy Flux in Macrophages. PLoS ONE 2015, 10, e0144347. [Google Scholar] [CrossRef]
- Netea-Maier, R.T.; Plantinga, T.S.; van de Veerdonk, F.L.; Smit, J.W.; Netea, M.G. Modulation of inflammation by autophagy: Consequences for human disease. Autophagy 2016, 12, 245–260. [Google Scholar] [CrossRef]
- Harris, J.; Hartman, M.; Roche, C.; Zeng, S.G.; O’Shea, A.; Sharp, F.A.; Lambe, E.M.; Creagh, E.M.; Golenbock, D.T.; Tschopp, J. Autophagy controls IL-1β secretion by targeting pro-IL-1β for degradation. J. Biol. Chem. 2011, 286, 9587–9597. [Google Scholar] [CrossRef]
- Shi, C.-S.; Shenderov, K.; Huang, N.-N.; Kabat, J.; Abu-Asab, M.; Fitzgerald, K.A.; Sher, A.; Kehrl, J.H. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat. Immunol. 2012, 13, 255. [Google Scholar] [CrossRef] [PubMed]
- Braga, T.T.; Forni, M.F.; Correa-Costa, M.; Ramos, R.N.; Barbuto, J.A.; Branco, P.; Castoldi, A.; Hiyane, M.I.; Davanso, M.R.; Latz, E. Soluble uric acid activates the NLRP3 inflammasome. Sci. Rep. 2017, 7, 39884. [Google Scholar] [CrossRef] [PubMed]
- Gasse, P.; Riteau, N.; Charron, S.; Girre, S.; Fick, L.; Pétrilli, V.; Tschopp, J.R.; Lagente, V.; Quesniaux, V.F.; Ryffel, B. Uric acid is a danger signal activating NALP3 inflammasome in lung injury inflammation and fibrosis. Am. J. Respir. Crit. Care Med. 2009, 179, 903–913. [Google Scholar] [CrossRef] [PubMed]
- Zughaier, S.M.; Ryley, H.C.; Jackson, S.K. Lipopolysaccharide (LPS) from Burkholderia cepacia is more active than LPS from Pseudomonas aeruginosa and Stenotrophomonas maltophilia in stimulating tumor necrosis factor alpha from human monocytes. Infect. Immun. 1999, 67, 1505–1507. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Zughaier, S.M.; Zimmer, S.M.; Datta, A.; Carlson, R.W.; Stephens, D.S. Differential induction of the toll-like receptor 4-MyD88-dependent and -independent signaling pathways by endotoxins. Infect. Immun. 2005, 73, 2940–2950. [Google Scholar] [CrossRef]
- Zughaier, S.M.; Tzeng, Y.L.; Zimmer, S.M.; Datta, A.; Carlson, R.W.; Stephens, D.S. Neisseria meningitidis lipooligosaccharide structure-dependent activation of the macrophage CD14/Toll-like receptor 4 pathway. Infect. Immun. 2004, 72, 371–380. [Google Scholar] [CrossRef]
- Tan, P.K.; Farrar, J.E.; Gaucher, E.A.; Miner, J.N. Coevolution of URAT1 and uricase during primate evolution: Implications for serum urate homeostasis and gout. Mol. Biol. Evol. 2016, 33, 2193–2200. [Google Scholar] [CrossRef]
- Mestre, M.B.; Colombo, M.I. Staphylococcus aureus promotes autophagy by decreasing intracellular cAMP levels. Autophagy 2012, 8, 1865–1867. [Google Scholar] [CrossRef]
- Neumann, Y.; Bruns, S.A.; Rohde, M.; Prajsnar, T.K.; Foster, S.J.; Schmitz, I. Intracellular Staphylococcus aureus eludes selective autophagy by activating a host cell kinase. Autophagy 2016, 12, 2069–2084. [Google Scholar] [CrossRef]
- Fang, L.; Wu, H.-M.; Ding, P.-S.; Liu, R.-Y. TLR2 mediates phagocytosis and autophagy through JNK signaling pathway in Staphylococcus aureus-stimulated RAW264. 7 cells. Cell. Signal. 2014, 26, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Maurer, K.; Reyes-Robles, T.; Alonzo III, F.; Durbin, J.; Torres, V.J.; Cadwell, K. Autophagy mediates tolerance to Staphylococcus aureus alpha-toxin. Cell Host Microbe 2015, 17, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Flannagan, R.S.; Heit, B.; Heinrichs, D.E. Intracellular replication of Staphylococcus aureus in mature phagolysosomes in macrophages precedes host cell death, and bacterial escape and dissemination. Cell. Microbiol. 2016, 18, 514–535. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Li, X.; Wang, W.; Ouedraogo, K.C.; Li, Y.; Gan, C.; Tan, S.; Zhou, X.; Wu, M. Atg7 deficiency impairs host defense against Klebsiella pneumoniae by impacting bacterial clearance, survival and inflammatory responses in mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014, 307, L355–L363. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, S.; Zhou, X.; Ye, Y.; Tan, S.; Zhang, S.; Li, R.; Yu, M.; Jundt, M.C.; Hidebrand, A. Lyn delivers bacteria to lysosomes for eradication through TLR2-initiated autophagy related phagocytosis. PLoS Pathog. 2016, 12, e1005363. [Google Scholar] [CrossRef] [PubMed]
- Crişan, T.O.; Cleophas, M.C.; Novakovic, B.; Erler, K.; van de Veerdonk, F.L.; Stunnenberg, H.G.; Netea, M.G.; Dinarello, C.A.; Joosten, L.A. Uric acid priming in human monocytes is driven by the AKT–PRAS40 autophagy pathway. Proc. Natl. Acad. Sci. USA 2017, 114, 5485–5490. [Google Scholar] [CrossRef]
- Xiao, J.; Zhang, X.; Fu, C.; Yang, Q.; Xie, Y.; Zhang, Z.; Ye, Z. Impaired Na+− K+-ATPase signaling in renal proximal tubule contributes to hyperuricemia-induced renal tubular injury. Exp. Mol. Med. 2018, 50, e452. [Google Scholar] [CrossRef]
- Sheng, Y.-L.; Chen, X.; Hou, X.-O.; Yuan, X.; Yuan, B.-S.; Yuan, Y.-Q.; Zhang, Q.-L.; Cao, X.; Liu, C.-F.; Luo, W.-F. Urate promotes SNCA/α-synuclein clearance via regulating mTOR-dependent macroautophagy. Exp. Neurol. 2017, 297, 138–147. [Google Scholar] [CrossRef]
- Günther, J.; Esch, K.; Poschadel, N.; Petzl, W.; Zerbe, H.; Mitterhuemer, S.; Blum, H.; Seyfert, H.-M. Comparative kinetics of Escherichia coli-and Staphylococcus aureus-specific activation of key immune pathways in mammary epithelial cells demonstrates that S. aureus elicits a delayed response dominated by interleukin-6 (IL-6) but not by IL-1A or tumor necrosis factor alpha. Infect. Immun. 2011, 79, 695–707. [Google Scholar]
- Sitia, R.; Rubartelli, A. The Unconventional Secretion of IL-1β: Handling a Dangerous Weapon to Optimize Inflammatory Responses. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 12–21. [Google Scholar]
- Takahama, M.; Akira, S.; Saitoh, T. Autophagy limits activation of the inflammasomes. Immunol. Rev. 2018, 281, 62–73. [Google Scholar] [CrossRef]
- Idriss, H.T.; Naismith, J.H. TNFα and the TNF receptor superfamily: Structure-function relationship(s). Microsc. Res. Technol. 2000, 50, 184–195. [Google Scholar] [CrossRef]
- Foster, M.W.; Hess, D.T.; Stamler, J.S. Protein S-nitrosylation in health and disease: A current perspective. Trends Mol. Med. 2009, 15, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Giordano, S.; Zhang, J. Autophagy, mitochondria and oxidative stress: Cross-talk and redox signalling. Biochem. J. 2012, 441, 523–540. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Korolchuk, V.I.; Renna, M.; Imarisio, S.; Fleming, A.; Williams, A.; Garcia-Arencibia, M.; Rose, C.; Luo, S.; Underwood, B.R. Complex inhibitory effects of nitric oxide on autophagy. Mol. Cell 2011, 43, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Li, L.; Chen, S.; Yu, Y.; Qi, M.; Tashiro, S.-I.; Onodera, S.; Ikejima, T. Silibinin induced-autophagic and apoptotic death is associated with an increase in reactive oxygen and nitrogen species in HeLa cells. Free Radic. Res. 2011, 45, 1307–1324. [Google Scholar] [CrossRef] [PubMed]
- Pestana, C.R.; Oishi, J.C.; Salistre-Araújo, H.S.; Rodrigues, G.J. Inhibition of autophagy by chloroquine stimulates nitric oxide production and protects endothelial function during serum deprivation. Cell. Physiol. Biochem. 2015, 37, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Chen, S.; Chen, M.; Ma, Y.; Wang, Y.; Huang, B.; He, Z.; Zeng, Y.; Hu, Y.; Sun, S. Nitric oxide suppresses NLRP3 inflammasome activation and protects against LPS-induced septic shock. Cell Res. 2013, 23, 201–212. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Awad, D.; Al-Emadi, N.; Abu-Madi, M.; Al-Thani, A.A.; Zughaier, S.M. The Role of Soluble Uric Acid in Modulating Autophagy Flux and Inflammasome Activation during Bacterial Infection in Macrophages. Biomedicines 2020, 8, 598. https://doi.org/10.3390/biomedicines8120598
Al-Awad D, Al-Emadi N, Abu-Madi M, Al-Thani AA, Zughaier SM. The Role of Soluble Uric Acid in Modulating Autophagy Flux and Inflammasome Activation during Bacterial Infection in Macrophages. Biomedicines. 2020; 8(12):598. https://doi.org/10.3390/biomedicines8120598
Chicago/Turabian StyleAl-Awad, Duha, Nada Al-Emadi, Marawan Abu-Madi, Asmaa A. Al-Thani, and Susu M. Zughaier. 2020. "The Role of Soluble Uric Acid in Modulating Autophagy Flux and Inflammasome Activation during Bacterial Infection in Macrophages" Biomedicines 8, no. 12: 598. https://doi.org/10.3390/biomedicines8120598
APA StyleAl-Awad, D., Al-Emadi, N., Abu-Madi, M., Al-Thani, A. A., & Zughaier, S. M. (2020). The Role of Soluble Uric Acid in Modulating Autophagy Flux and Inflammasome Activation during Bacterial Infection in Macrophages. Biomedicines, 8(12), 598. https://doi.org/10.3390/biomedicines8120598






