The Hippo–YAP Signaling as Guardian in the Pool of Intestinal Stem Cells
Abstract
1. Introduction
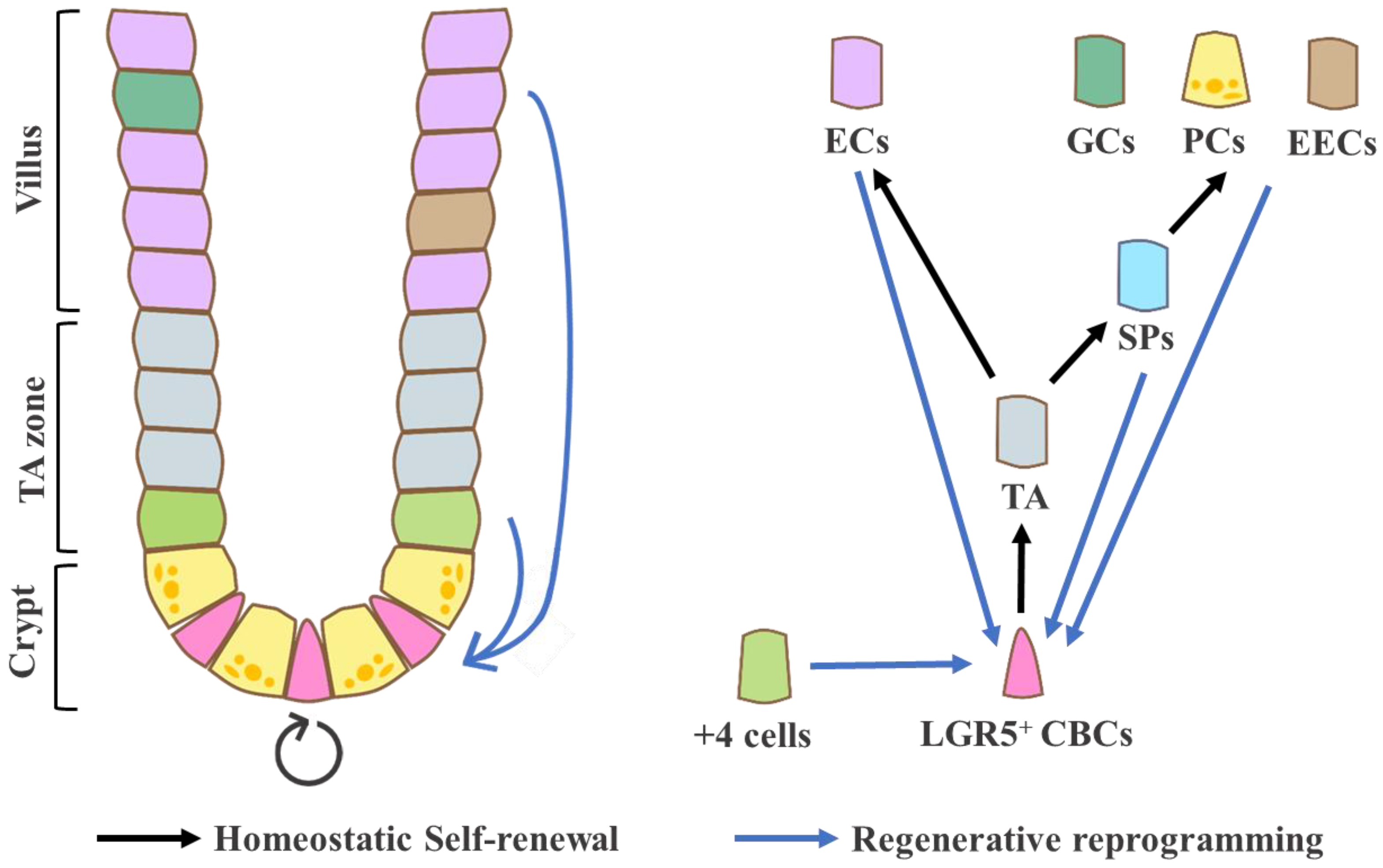
2. The Characteristics of Steady-State and Injury-Associated ISCs
2.1. Crypt Base Columnar Cells (CBCs): Homeostatic ISCs
2.2. Label-Retaining Cells (LRCs) or +4 Cells: Quiescent, Reserve ISCs?
2.3. Damage-Induced ISCs: Reverse- or Revival Stem Cells (RSCs) and Reprogramming Cells
3. The Hippo Pathway
3.1. Core Components of the Hippo Pathway
3.2. Upstream and Cross-Talk Signaling of the Hippo Pathway
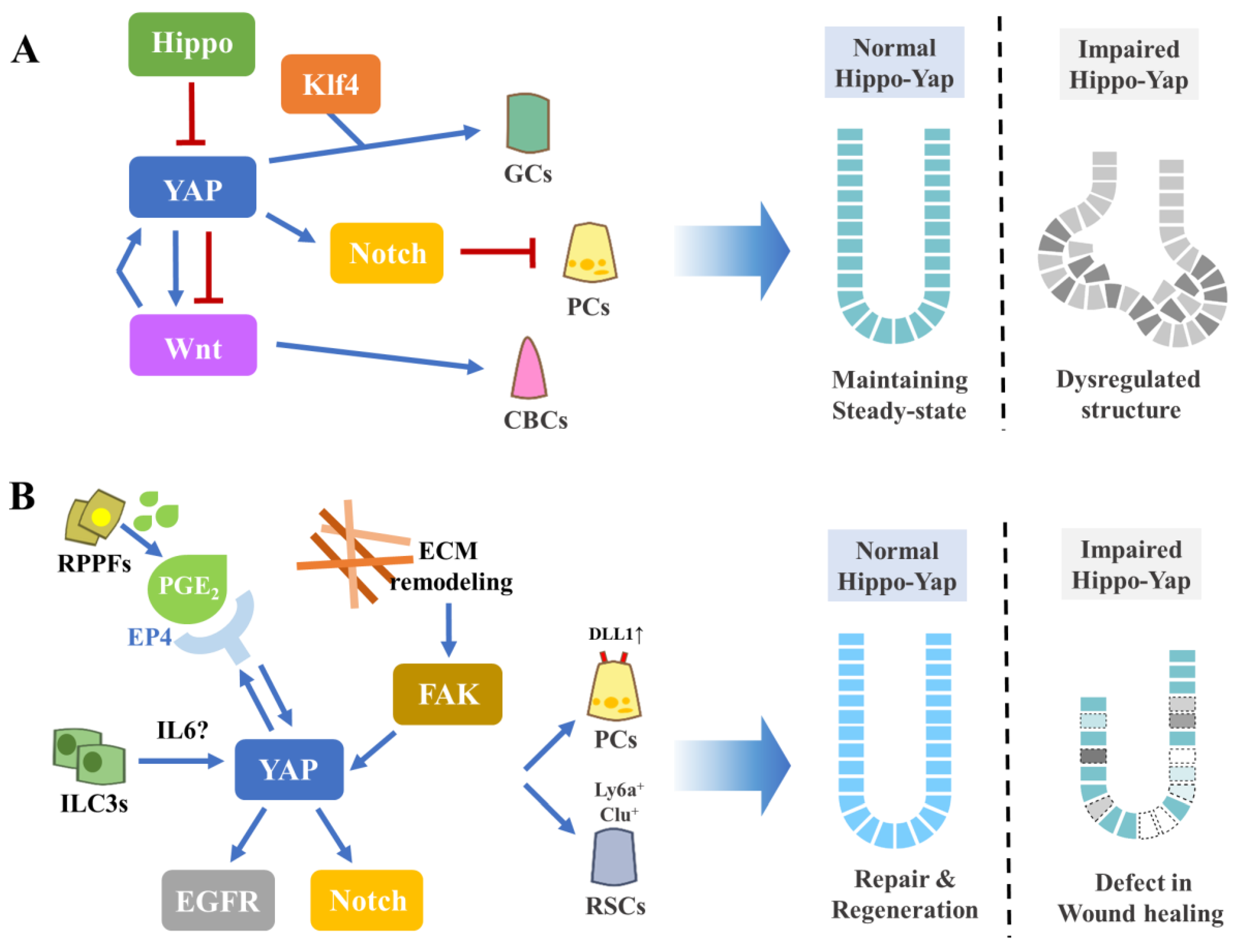
4. The Contribution of Hippo–YAP Signaling Pathway in the Intestine
4.1. Intestinal Homeostasis
4.2. Intestinal Repair and Regeneration
4.3. Intestinal Tumorigenesis
5. Summary and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Gehart, H.; Clevers, H. Tales from the crypt: New insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 19–34. [Google Scholar] [CrossRef]
- Beumer, J.; Clevers, H. Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell Biol. 2020, 1–15. [Google Scholar] [CrossRef]
- Sato, T.; Van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; Born, M.V.D.; Barker, N.; Shroyer, N.F.; Van De Wetering, M.; Clevers, H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nat. Cell Biol. 2011, 469, 415–418. [Google Scholar] [CrossRef]
- Bevins, C.L.; Salzman, N.H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Genet. 2011, 9, 356–368. [Google Scholar] [CrossRef]
- Allaire, J.M.; Crowley, S.M.; Law, H.T.; Chang, S.-Y.; Ko, H.-J.; Vallance, B.A. The Intestinal Epithelium: Central Coordinator of Mucosal Immunity. Trends Immunol. 2018, 39, 677–696. [Google Scholar] [CrossRef]
- Barbara, P.D.S.; Brink, G.R.V.D.; Roberts, D.J. Development and differentiation of the intestinal epithelium. Cell. Mol. Life Sci. 2003, 60, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-H.; Saadatpour, A.; Guo, G.; Saxena, M.; Cavazza, A.; Desai, N.; Jadhav, U.; Jiang, L.; Rivera, M.N.; Orkin, S.H.; et al. Single-Cell Transcript Profiles Reveal Multilineage Priming in Early Progenitors Derived from Lgr5 + Intestinal Stem Cells. Cell Rep. 2016, 16, 2053–2060. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; Van Es, J.H.; Kuipers, J.; Kujala, P.; Born, M.V.D.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nat. Cell Biol. 2007, 449, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Heath, J.P. Epithelial Cell Migration in the Intestine. Cell Biol. Int. 1996, 20, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Schepers, A.G.; Vries, R.; Born, M.V.D.; Van De Wetering, M.; Clevers, H. Lgr5 intestinal stem cells have high telomerase activity and randomly segregate their chromosomes. EMBO J. 2011, 30, 1104–1109. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, E.M.F.; de Sauvage, F.J. Cellular Plasticity in Intestinal Homeostasis and Disease. Cell Stem Cell 2019, 24, 54–64. [Google Scholar] [CrossRef]
- Haber, A.L.; Biton, M.; Rogel, N.; Herbst, R.H.; Shekhar, K.; Smillie, C.; Burgin, G.; DeLorey, T.M.; Howitt, M.R.; Katz, Y.; et al. A single-cell survey of the small intestinal epithelium. Nat. Cell Biol. 2017, 551, 333–339. [Google Scholar] [CrossRef]
- Grün, D.; Muraro, M.J.; Boisset, J.-C.; Wiebrands, K.; Lyubimova, A.; Dharmadhikari, G.; Born, M.V.D.; Van Es, J.; Jansen, E.; Clevers, H.; et al. De Novo Prediction of Stem Cell Identity using Single-Cell Transcriptome Data. Cell Stem Cell 2016, 19, 266–277. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; Van De Wetering, M.; Barker, N.; Stange, D.E.; Van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nat. Cell Biol. 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef]
- Santos, A.J.; Lo, Y.-H.; Mah, A.T.; Kuo, C.J. The Intestinal Stem Cell Niche: Homeostasis and Adaptations. Trends Cell Biol. 2018, 28, 1062–1078. [Google Scholar] [CrossRef]
- Cheng, H.; Leblond, C.P. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine V. Unitarian theory of the origin of the four epithelial cell types. Am. J. Anat. 1974, 141, 537–561. [Google Scholar] [CrossRef]
- De Lau, W.; Peng, W.C.; Gros, P.; Clevers, H. The R-spondin/Lgr5/Rnf43 module: Regulator of Wnt signal strength. Genes Dev. 2014, 28, 305–316. [Google Scholar] [CrossRef]
- Yan, K.S.; Janda, C.Y.; Chang, J.; Zheng, G.X.Y.; Larkin, K.A.; Luca, V.C.; Chia, L.A.; Mah, A.T.; Han, A.; Terry, J.M.; et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5+ intestinal stem-cell self-renewal. Nat. Cell Biol. 2017, 545, 238–242. [Google Scholar] [CrossRef]
- Snippert, H.J.; Van Der Flier, L.G.; Sato, T.; Van Es, J.H.; Born, M.V.D.; Kroon-Veenboer, C.; Barker, N.; Klein, A.M.; Van Rheenen, J.; Simons, B.D.; et al. Intestinal Crypt Homeostasis Results from Neutral Competition between Symmetrically Dividing Lgr5 Stem Cells. Cell 2010, 143, 134–144. [Google Scholar] [CrossRef]
- Lopez-Garcia, C.; Klein, A.M.; Simons, B.D.; Winton, D.J. Intestinal Stem Cell Replacement Follows a Pattern of Neutral Drift. Science 2010, 330, 822–825. [Google Scholar] [CrossRef]
- Ritsma, L.; Ellenbroek, S.I.J.; Zomer, A.; Snippert, H.J.; De Sauvage, F.J.; Simons, B.D.; Clevers, H.; Van Rheenen, J. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nat. Cell Biol. 2014, 507, 362–365. [Google Scholar] [CrossRef]
- Krndija, D.; El Marjou, F.; Guirao, B.; Richon, S.; Leroy, O.; Bellaïche, Y.; Hannezo, E.; Vignjevic, D.M. Active cell migration is critical for steady-state epithelial turnover in the gut. Science 2019, 365, 705–710. [Google Scholar] [CrossRef]
- Darwich, A.S.; Aslam, U.; Ashcroft, D.M.; Rostami-Hodjegan, A. Meta-Analysis of the Turnover of Intestinal Epithelia in Preclinical Animal Species and Humans. Drug Metab. Dispos. 2014, 42, 2016–2022. [Google Scholar] [CrossRef]
- Barker, N.; Van Oudenaarden, A.; Clevers, H. Identifying the Stem Cell of the Intestinal Crypt: Strategies and Pitfalls. Cell Stem Cell 2012, 11, 452–460. [Google Scholar] [CrossRef]
- Potten, C.S.; Owen, G.; Booth, D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J. Cell Sci. 2002, 115, 2381–2388. [Google Scholar]
- Li, L.; Clevers, H. Coexistence of Quiescent and Active Adult Stem Cells in Mammals. Science 2010, 327, 542–545. [Google Scholar] [CrossRef]
- Powell, A.E.; Wang, Y.; Li, Y.; Poulin, E.J.; Means, A.L.; Washington, M.K.; Higginbotham, J.N.; Juchheim, A.; Prasad, N.; Levy, S.E.; et al. The Pan-ErbB Negative Regulator Lrig1 Is an Intestinal Stem Cell Marker that Functions as a Tumor Suppressor. Cell 2012, 149, 146–158. [Google Scholar] [CrossRef]
- Takeda, N.; Jain, R.; Leboeuf, M.R.; Wang, Q.; Lu, M.M.; Epstein, J.A. Interconversion Between Intestinal Stem Cell Populations in Distinct Niches. Science 2011, 334, 1420–1424. [Google Scholar] [CrossRef]
- Sangiorgi, E.; Capecchi, M.R. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 2008, 40, 915–920. [Google Scholar] [CrossRef]
- Wong, V.W.Y.; Stange, D.E.; Page, M.E.; Buczacki, S.J.A.; Wabik, A.; Itami, S.; Van De Wetering, M.; Poulsom, R.; Wright, N.A.; Trotter, M.W.B.; et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat. Cell Biol. 2012, 14, 401–408. [Google Scholar] [CrossRef]
- Roche, K.C.; Gracz, A.D.; Liu, X.F.; Newton, V.; Akiyama, H.; Magness, S.T. SOX9 Maintains Reserve Stem Cells and Preserves Radioresistance in Mouse Small Intestine. Gastroenterology 2015, 149, 1553–1563.e10. [Google Scholar] [CrossRef]
- Montgomery, R.K.; Carlone, D.L.; Richmond, C.A.; Farilla, L.; Kranendonk, M.E.G.; Henderson, D.E.; Baffour-Awuah, N.Y.; Ambruzs, D.M.; Fogli, L.K.; Algra, S.; et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl. Acad. Sci. USA 2010, 108, 179–184. [Google Scholar] [CrossRef]
- Yan, K.S.; Chia, L.A.; Li, X.; Ootani, A.; Su, J.; Lee, J.Y.; Su, N.; Luo, Y.; Heilshorn, S.C.; Amieva, M.R.; et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. USA 2012, 109, 466–471. [Google Scholar] [CrossRef]
- Tian, H.; Biehs, B.; Warming, S.; Leong, K.G.; Rangell, L.; Klein, O.D.; De Sauvage, F.J. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nat. Cell Biol. 2011, 478, 255–259. [Google Scholar] [CrossRef]
- Muñoz, J.; Stange, D.E.; Schepers, A.G.; Van De Wetering, M.; Koo, B.-K.; Itzkovitz, S.; Volckmann, R.; Kung, K.S.; Koster, J.; Radulescu, S.; et al. The Lgr5 intestinal stem cell signature: Robust expression of proposed quiescent ‘+4′ cell markers. EMBO J. 2012, 31, 3079–3091. [Google Scholar] [CrossRef]
- Buczacki, S.J.A.; Zecchini, H.I.; Nicholson, A.M.; Russell, R.; Vermeulen, L.; Kemp, R.; Winton, D. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nat. Cell Biol. 2013, 495, 65–69. [Google Scholar] [CrossRef]
- Yan, K.S.; Gevaert, O.; Zheng, G.X.; Anchang, B.; Probert, C.S.; Larkin, K.A.; Davies, P.S.; Cheng, Z.-F.; Kaddis, J.S.; Jonathan, E.; et al. Intestinal Enteroendocrine Lineage Cells Possess Homeostatic and Injury-Inducible Stem Cell Activity. Cell Stem Cell 2017, 21, 78–90.e6. [Google Scholar] [CrossRef]
- Van Es, J.H.; Sato, T.; Van De Wetering, M.; Lyubimova, A.; Nee, A.N.Y.; Gregorieff, A.; Sasaki, N.; Zeinstra, L.M.; Born, M.V.D.; Korving, J.; et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 2012, 14, 1099–1104. [Google Scholar] [CrossRef]
- VanDussen, K.L.; Carulli, A.J.; Keeley, T.M.; Patel, S.R.; Puthoff, B.J.; Magness, S.T.; Tran, I.T.; Maillard, I.; Siebel, C.; Kolterud, Å.; et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development 2012, 139, 488–497. [Google Scholar] [CrossRef]
- Carulli, A.J.; Keeley, T.M.; Demitrack, E.S.; Chung, J.; Maillard, I.; Samuelson, L.C. Notch receptor regulation of intestinal stem cell homeostasis and crypt regeneration. Dev. Biol. 2015, 402, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Azofeifa, D.; Fazio, E.N.; Nattiv, R.; Good, H.J.; Wald, T.; Pest, M.A.; De Sauvage, F.J.; Klein, O.D.; Asfaha, S. Atoh1+ secretory progenitors possess renewal capacity independent of Lgr5+ cells during colonic regeneration. EMBO J. 2019, 38, e99984. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, U.; Saxena, M.; O’Neill, N.K.; Saadatpour, A.; Yuan, G.-C.; Herbert, Z.; Murata, K.; Shivdasani, R.A. Dynamic Reorganization of Chromatin Accessibility Signatures during Dedifferentiation of Secretory Precursors into Lgr5+ Intestinal Stem Cells. Cell Stem Cell 2017, 21, 65–77.e5. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.; Schewe, M.; Sacchetti, A.; Feijtel, D.; Van De Geer, W.S.; Teeuwssen, M.; Sleddens, H.F.; Joosten, R.; Van Royen, M.E.; Van De Werken, H.J.; et al. Paneth Cells Respond to Inflammation and Contribute to Tissue Regeneration by Acquiring Stem-like Features through SCF/c-Kit Signaling. Cell Rep. 2018, 24, 2312–2328.e7. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Tong, K.; Zhao, Y.; Balasubramanian, I.; Yap, G.S.; Ferraris, R.P.; Bonder, E.M.; Verzi, M.P.; Gao, N. Paneth Cell Multipotency Induced by Notch Activation following Injury. Cell Stem Cell 2018, 23, 46–59.e5. [Google Scholar] [CrossRef] [PubMed]
- Tetteh, P.W.; Basak, O.; Farin, H.F.; Wiebrands, K.; Kretzschmar, K.; Begthel, H.; Born, M.V.D.; Korving, J.; De Sauvage, F.; Van Es, J.H.; et al. Replacement of Lost Lgr5-Positive Stem Cells through Plasticity of Their Enterocyte-Lineage Daughters. Cell Stem Cell 2016, 18, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Yui, S.; Azzolin, L.; Maimets, M.; Pedersen, M.T.; Fordham, R.P.; Hansen, S.L.; Larsen, H.L.; Guiu, J.; Alves, M.R.; Rundsten, C.F.; et al. YAP/TAZ-Dependent Reprogramming of Colonic Epithelium Links ECM Remodeling to Tissue Regeneration. Cell Stem Cell 2018, 22, 35–49.e7. [Google Scholar] [CrossRef]
- Nusse, Y.M.; Savage, A.K.; Marangoni, P.; Rosendahl-Huber, A.K.M.; Landman, T.A.; De Sauvage, F.J.; Locksley, R.M.; Klein, O.D. Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nat. Cell Biol. 2018, 559, 109–113. [Google Scholar] [CrossRef]
- Ayyaz, A.; Kumar, S.; Sangiorgi, B.; Ghoshal, B.; Gosio, J.; Ouladan, S.; Fink, M.; Barutcu, S.; Trcka, D.; Shen, J.; et al. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nat. Cell Biol. 2019, 569, 121–125. [Google Scholar] [CrossRef]
- Udan, R.S.; Kango-Singh, M.; Nolo, R.; Tao, C.; Halder, G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003, 5, 914–920. [Google Scholar] [CrossRef]
- Wu, S.; Huang, J.; Dong, J.; Pan, D. hippo Encodes a Ste-20 Family Protein Kinase that Restricts Cell Proliferation and Promotes Apoptosis in Conjunction with salvador and warts. Cell 2003, 114, 445–456. [Google Scholar] [CrossRef]
- Harvey, K.F.; Pfleger, C.M.; Hariharan, I.K. The Drosophila Mst Ortholog, hippo, Restricts Growth and Cell Proliferation and Promotes Apoptosis. Cell 2003, 114, 457–467. [Google Scholar] [CrossRef]
- Pantalacci, S.; Tapon, N.; Léopold, P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 2003, 5, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Tapon, N.; Harvey, K.F.; Bell, D.W.; Wahrer, D.C.; Schiripo, T.A.; Haber, D.A.; Hariharan, I.K. salvador Promotes Both Cell Cycle Exit and Apoptosis in Drosophila and Is Mutated in Human Cancer Cell Lines. Cell 2002, 110, 467–478. [Google Scholar] [CrossRef]
- Justice, R.W.; Zilian, O.; Woods, D.F.; Noll, M.; Bryant, P.J. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995, 9, 534–546. [Google Scholar] [CrossRef]
- Lai, Z.-C.; Wei, X.; Shimizu, T.; Ramos, E.; Rohrbaugh, M.; Nikolaidis, N.; Ho, L.-L.; Li, Y. Control of Cell Proliferation and Apoptosis by Mob as Tumor Suppressor, Mats. Cell 2005, 120, 675–685. [Google Scholar] [CrossRef]
- Huang, J.; Wu, S.; Barrera, J.; Matthews, K.; Pan, D. The Hippo Signaling Pathway Coordinately Regulates Cell Proliferation and Apoptosis by Inactivating Yorkie, the Drosophila Homolog of YAP. Cell 2005, 122, 421–434. [Google Scholar] [CrossRef]
- Zhang, L.; Ren, F.; Zhang, Q.; Chen, Y.; Wang, B.; Jiang, J. The TEAD/TEF Family of Transcription Factor Scalloped Mediates Hippo Signaling in Organ Size Control. Dev. Cell 2008, 14, 377–387. [Google Scholar] [CrossRef]
- Wu, S.; Liu, Y.; Zheng, Y.; Dong, J.; Pan, D. The TEAD/TEF Family Protein Scalloped Mediates Transcriptional Output of the Hippo Growth-Regulatory Pathway. Dev. Cell 2008, 14, 388–398. [Google Scholar] [CrossRef]
- Oka, T.; Mazack, V.; Sudol, M. Mst2 and Lats Kinases Regulate Apoptotic Function of Yes Kinase-associated Protein (YAP). J. Biol. Chem. 2008, 283, 27534–27546. [Google Scholar] [CrossRef]
- Dong, J.; Feldmann, G.; Huang, J.; Wu, S.; Zhang, N.; Comerford, S.A.; Gayyed, M.F.; Anders, R.A.; Maitra, A.; Pan, D. Elucidation of a Universal Size-Control Mechanism in Drosophila and Mammals. Cell 2007, 130, 1120–1133. [Google Scholar] [CrossRef]
- Lei, Q.-Y.; Zhang, H.; Zhao, B.; Zha, Z.-Y.; Bai, F.; Pei, X.-H.; Zhao, S.; Xiong, Y.; Guan, K.-L. TAZ Promotes Cell Proliferation and Epithelial-Mesenchymal Transition and Is Inhibited by the Hippo Pathway. Mol. Cell. Biol. 2008, 28, 2426–2436. [Google Scholar] [CrossRef]
- Oh, H.; Irvine, K.D. In vivo regulation of Yorkie phosphorylation and localization. Development 2008, 135, 1081–1088. [Google Scholar] [CrossRef]
- Basu, S.; Totty, N.F.; Irwin, M.S.; Sudol, M.; Downward, J. Akt Phosphorylates the Yes-Associated Protein, YAP, to Induce Interaction with 14-3-3 and Attenuation of p73-Mediated Apoptosis. Mol. Cell 2003, 11, 11–23. [Google Scholar] [CrossRef]
- Graves, J.D.; Gotoh, Y.; Draves, K.E.; Ambrose, D.; Han, D.K.M.; Wright, M.; Chernoff, J.; Clark, E.A.; Krebs, E.G. Caspase-mediated activation and induction of apoptosis by the mammalian Ste20-like kinase Mst1. EMBO J. 1998, 17, 2224–2234. [Google Scholar] [CrossRef]
- Kodaka, M.; Hata, Y. The mammalian Hippo pathway: Regulation and function of YAP1 and TAZ. Cell. Mol. Life Sci. 2015, 72, 285–306. [Google Scholar] [CrossRef]
- Galli, G.G.; Carrara, M.; Yuan, W.-C.; Valdes-Quezada, C.; Gurung, B.; Pepe-Mooney, B.; Zhang, T.; Geeven, G.; Gray, N.S.; De Laat, W.; et al. YAP Drives Growth by Controlling Transcriptional Pause Release from Dynamic Enhancers. Mol. Cell 2015, 60, 328–337. [Google Scholar] [CrossRef]
- Stein, C.; Bardet, A.F.; Roma, G.; Bergling, S.; Clay, I.; Ruchti, A.; Agarinis, C.; Schmelzle, T.; Bouwmeester, T.; Schuebeler, D.; et al. YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers. PLoS Genet. 2015, 11, e1005465. [Google Scholar] [CrossRef]
- Zanconato, F.; Forcato, M.; Battilana, G.; Azzolin, L.; Quaranta, E.; Bodega, B.; Rosato, A.; Bicciato, S.; Cordenonsi, M.; Piccolo, S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 2015, 17, 1218–1227. [Google Scholar] [CrossRef]
- Zhao, B.; Ye, X.; Yu, J.; Li, L.; Li, W.; Li, S.; Lin, J.D.; Wang, C.-Y.; Chinnaiyan, A.M.; Lai, Z.-C.; et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008, 22, 1962–1971. [Google Scholar] [CrossRef]
- Murakami, M.; Nakagawa, M.; Olson, E.N.; Nakagawa, O. A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc. Natl. Acad. Sci. USA 2005, 102, 18034–18039. [Google Scholar] [CrossRef]
- Strano, S.; Monti, O.; Pediconi, N.; Baccarini, A.; Fontemaggi, G.; Lapi, E.; Mantovani, F.; Damalas, A.; Citro, G.; Sacchi, A.; et al. The Transcriptional Coactivator Yes-Associated Protein Drives p73 Gene-Target Specificity in Response to DNA Damage. Mol. Cell 2005, 18, 447–459. [Google Scholar] [CrossRef]
- Wang, K.; Degerny, C.; Xu, M.; Yang, X.-J. YAP, TAZ, and Yorkie: A conserved family of signal-responsive transcriptional coregulators in animal development and human diseaseThis paper is one of a selection of papers published in this Special Issue, entitled CSBMCB’s 51st Annual Meeting–Epigenetics and Chromatin Dynamics, and has undergone the Journal’s usual peer review process. Biochem. Cell Biol. 2009, 87, 77–91. [Google Scholar] [CrossRef]
- Yagi, R.; Chen, L.-F.; Shigesada, K.; Murakami, Y.; Ito, Y. A WW domain-containing Yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999, 18, 2551–2562. [Google Scholar] [CrossRef]
- Chen, C.-L.; Gajewski, K.M.; Hamaratoglu, F.; Bossuyt, W.; Sansores-Garcia, L.; Tao, C.; Halder, G. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc. Natl. Acad. Sci. USA 2010, 107, 15810–15815. [Google Scholar] [CrossRef]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nat. Cell Biol. 2011, 474, 179–183. [Google Scholar] [CrossRef]
- Hamaratoglu, F.; Willecke, M.; Kango-Singh, M.; Nolo, R.; Hyun, E.; Tao, C.; Jafar-Nejad, H.; Halder, G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 2005, 8, 27–36. [Google Scholar] [CrossRef]
- Kim, N.-G.; Gumbiner, B.M. Adhesion to fibronectin regulates Hippo signaling via the FAK–Src–PI3K pathway. J. Cell Biol. 2015, 210, 503–515. [Google Scholar] [CrossRef]
- Kim, N.-G.; Koh, E.; Chen, X.; Gumbiner, B.M. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. USA 2011, 108, 11930–11935. [Google Scholar] [CrossRef]
- Wada, K.-I.; Itoga, K.; Okano, T.; Yonemura, S.; Sasaki, H. Hippo pathway regulation by cell morphology and stress fibers. Development 2011, 138, 3907–3914. [Google Scholar] [CrossRef]
- Zhao, B.; Li, L.; Wang, L.; Wang, C.-Y.; Yu, J.; Guan, K.-L. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012, 26, 54–68. [Google Scholar] [CrossRef]
- Hong, A.W.; Meng, Z.; Yuan, H.; Plouffe, S.W.; Moon, S.; Kim, W.; Jho, E.-H.; Guan, K.-L. Osmotic stress-induced phosphorylation by NLK at Ser128 activates YAP. EMBO Rep. 2016, 18, 72–86. [Google Scholar] [CrossRef]
- Ma, B.; Chen, Y.; Chen, L.; Cheng, H.; Mu, C.; Li, J.; Gao, R.; Zhou, C.; Cao, L.; Liu, J.; et al. Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nat. Cell Biol. 2014, 17, 95–103. [Google Scholar] [CrossRef]
- Mo, J.-S.; Meng, Z.; Kim, Y.C.; Park, H.W.; Hansen, C.G.; Kim, S.; Lim, D.-S.; Guan, K.-L. Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat. Cell Biol. 2015, 17, 500–510. [Google Scholar] [CrossRef]
- Baumgartner, R.; Poernbacher, I.; Buser, N.; Hafen, E.; Stocker, H. The WW Domain Protein Kibra Acts Upstream of Hippo in Drosophila. Dev. Cell 2010, 18, 309–316. [Google Scholar] [CrossRef]
- Genevet, A.; Wehr, M.C.; Brain, R.; Thompson, B.; Tapon, N. Kibra Is a Regulator of the Salvador/Warts/Hippo Signaling Network. Dev. Cell 2010, 18, 300–308. [Google Scholar] [CrossRef]
- Yu, J.; Zheng, Y.; Dong, J.; Klusza, S.; Deng, W.-M.; Pan, D. Kibra Functions as a Tumor Suppressor Protein that Regulates Hippo Signaling in Conjunction with Merlin and Expanded. Dev. Cell 2010, 18, 288–299. [Google Scholar] [CrossRef]
- Angus, L.; Moleirinho, S.; Herron, L.R.; Sinha, A.; Zhang, X.; Niestrata, M.; Dholakia, K.; Prystowsky, M.; Harvey, K.; Reynolds, P.A.; et al. Willin/FRMD6 expression activates the Hippo signaling pathway kinases in mammals and antagonizes oncogenic YAP. Oncogene 2011, 31, 238–250. [Google Scholar] [CrossRef]
- Piccolo, S.; Dupont, S.; Cordenonsi, M. The Biology of YAP/TAZ: Hippo Signaling and Beyond. Physiol. Rev. 2014, 94, 1287–1312. [Google Scholar] [CrossRef]
- Rauskolb, C.; Pan, G.; Reddy, B.V.V.G.; Oh, H.; Irvine, K.D. Zyxin Links Fat Signaling to the Hippo Pathway. PLoS Biol. 2011, 9, e1000624. [Google Scholar] [CrossRef]
- Sharma, P.; McNeill, H. Fat and Dachsous Cadherins. Prog. Mol. Biol. Transl. Sci. 2013, 116, 215–235. [Google Scholar] [PubMed]
- Bossuyt, W.; Chen, C.-L.; Chen, Q.; Sudol, M.; McNeill, H.; Pan, D.; Kopp, A.; Halder, G. An evolutionary shift in the regulation of the Hippo pathway between mice and flies. Oncogene 2013, 33, 1218–1228. [Google Scholar] [CrossRef]
- Mao, Y.; Mulvaney, J.; Zakaria, S.; Yu, T.; Morgan, K.M.; Allen, S.; Basson, M.A.; Francis-West, P.; Irvine, K.D. Characterization of a Dchs1 mutant mouse reveals requirements for Dchs1-Fat4 signaling during mammalian development. Development 2011, 138, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.W.; Lim, C.J.; Chong, Y.F.; Pobbati, A.V.; Huang, C.; Hong, W. Hippo Pathway-independent Restriction of TAZ and YAP by Angiomotin. J. Biol. Chem. 2011, 286, 7018–7026. [Google Scholar] [CrossRef] [PubMed]
- Paramasivam, M.; Sarkeshik, A.; Yates, J.R.; Fernandes, M.J.G.; Mccollum, D. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol. Biol. Cell 2011, 22, 3725–3733. [Google Scholar] [CrossRef] [PubMed]
- Oka, T.; Remue, E.; Meerschaert, K.; Vanloo, B.; Boucherie, C.; Gfeller, D.; Bader, G.D.; Sidhu, S.S.; Vandekerckhove, J.; Gettemans, J.; et al. Functional complexes between YAP2 and ZO-2 are PDZ domain-dependent, and regulate YAP2 nuclear localization and signalling. Biochem. J. 2010, 432, 461–478. [Google Scholar] [CrossRef] [PubMed]
- Remue, E.; Meerschaert, K.; Oka, T.; Boucherie, C.; Vandekerckhove, J.; Sudol, M.; Gettemans, J. TAZ interacts with zonula occludens-1 and -2 proteins in a PDZ-1 dependent manner. FEBS Lett. 2010, 584, 4175–4180. [Google Scholar] [CrossRef]
- Schlegelmilch, K.; Mohseni, M.; Kirak, O.; Pruszak, J.; Rodriguez, J.R.; Zhou, D.; Kreger, B.T.; Vasioukhin, V.; Avruch, J.; Brummelkamp, T.R.; et al. Yap1 Acts Downstream of α-Catenin to Control Epidermal Proliferation. Cell 2011, 144, 782–795. [Google Scholar] [CrossRef]
- Wang, K.-C.; Yeh, Y.-T.; Nguyen, P.; Limqueco, E.; Lopez, J.; Thorossian, S.; Guan, K.-L.; Li, Y.-S.J.; Chien, S. Flow-dependent YAP/TAZ activities regulate endothelial phenotypes and atherosclerosis. Proc. Natl. Acad. Sci. USA 2016, 113, 11525–11530. [Google Scholar] [CrossRef]
- Wang, L.; Luo, J.-Y.; Li, B.; Tian, X.Y.; Chen, L.-J.; Huang, Y.; Liu, J.; Deng, D.; Lau, C.W.; Wan, S.; et al. Integrin-YAP/TAZ-JNK cascade mediates atheroprotective effect of unidirectional shear flow. Nat. Cell Biol. 2016, 540, 579–582. [Google Scholar] [CrossRef]
- Fernández, B.G.; Gaspar, P.; Brás-Pereira, C.; Jezowska, B.; Rebelo, S.R.; Janody, F. Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development 2011, 138, 2337–2346. [Google Scholar] [CrossRef]
- Sansores-Garcia, L.; Bossuyt, W.; Wada, K.-I.; Yonemura, S.; Tao, C.; Sasaki, H.; Halder, G. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 2011, 30, 2325–2335. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kim, M.; Lee, S.; Kuninaka, S.; Saya, H.; Lee, H.; Lee, S.; Lim, D. cAMP/PKA signalling reinforces the LATS–YAP pathway to fully suppress YAP in response to actin cytoskeletal changes. EMBO J. 2013, 32, 1543–1555. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.; Yang, J.; DeRan, M.; Wu, C.; Su, A.I.; Bonamy, G.M.; Liu, J.; Peters, E.C.; Wu, X. Identification of Serum-Derived Sphingosine-1-Phosphate as a Small Molecule Regulator of YAP. Chem. Biol. 2012, 19, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.-S.; Yu, F.-X.; Gong, R.; Brown, J.H.; Guan, K.-L. Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs). Genes Dev. 2012, 26, 2138–2143. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.-X.; Zhao, B.; Panupinthu, N.; Jewell, J.L.; Lian, I.; Wang, L.H.; Zhao, J.; Yuan, H.; Tumaneng, K.; Li, H.; et al. Regulation of the Hippo-YAP Pathway by G-Protein-Coupled Receptor Signaling. Cell 2012, 150, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Nguyen, H.T.; Chen, Q.; Zhang, R.; Hagman, Z.; Voorhoeve, P.M.; Cohen, S.M. Opposing activities of the R as and H ippo pathways converge on regulation of YAP protein turnover. EMBO J. 2014, 33, 2447–2457. [Google Scholar] [CrossRef]
- Lin, L.; Sabnis, A.J.; Chan, E.; Olivas, V.; Cade, L.; Pazarentzos, E.; Asthana, S.; Neel, D.S.; Yan, J.J.; Lu, X.; et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat. Genet. 2015, 47, 250–256. [Google Scholar] [CrossRef]
- Zhao, B.; Li, L.; Guan, K.-L. Hippo signaling at a glance. J. Cell Sci. 2010, 123, 4001–4006. [Google Scholar] [CrossRef]
- Huang, W.; Lv, X.; Liu, C.; Zha, Z.; Zhang, H.; Jiang, Y.; Xiong, Y.; Lei, Q.-Y.; Guan, K.-L. The N-terminal Phosphodegron Targets TAZ/WWTR1 Protein for SCFβ-TrCP-dependent Degradation in Response to Phosphatidylinositol 3-Kinase Inhibition. J. Biol. Chem. 2012, 287, 26245–26253. [Google Scholar] [CrossRef]
- Azzolin, L.; Zanconato, F.; Bresolin, S.; Forcato, M.; Basso, G.; Bicciato, S.; Cordenonsi, M.; Piccolo, S. Role of TAZ as Mediator of Wnt Signaling. Cell 2012, 151, 1443–1456. [Google Scholar] [CrossRef] [PubMed]
- Varelas, X.; Miller, B.W.; Sopko, R.; Song, S.; Gregorieff, A.; Fellouse, F.A.; Sakuma, R.; Pawson, T.; Hunziker, W.; McNeill, H.; et al. The Hippo Pathway Regulates Wnt/β-Catenin Signaling. Dev. Cell 2010, 18, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Imajo, M.; Miyatake, K.; Iimura, A.; Miyamoto, A.; Nishida, E. A molecular mechanism that links Hippo signalling to the inhibition of Wnt/β-catenin signalling. EMBO J. 2012, 31, 1109–1122. [Google Scholar] [CrossRef] [PubMed]
- Heallen, T.; Zhang, M.; Wang, J.; Bonilla-Claudio, M.; Klysik, E.; Johnson, R.L.; Martin, J.F. Hippo Pathway Inhibits Wnt Signaling to Restrain Cardiomyocyte Proliferation and Heart Size. Science 2011, 332, 458–461. [Google Scholar] [CrossRef] [PubMed]
- Rosenbluh, J.; Nijhawan, D.; Cox, A.G.; Li, X.; Neal, J.T.; Schafer, E.J.; Zack, T.I.; Wang, X.; Tsherniak, A.; Schinzel, A.C.; et al. β-Catenin-Driven Cancers Require a YAP1 Transcriptional Complex for Survival and Tumorigenesis. Cell 2012, 151, 1457–1473. [Google Scholar] [CrossRef] [PubMed]
- Camargo, F.D.; Gokhale, S.; Johnnidis, J.B.; Fu, D.; Bell, G.W.; Jaenisch, R.; Brummelkamp, T.R. YAP1 Increases Organ Size and Expands Undifferentiated Progenitor Cells. Curr. Biol. 2007, 17, 2054–2060. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Zhang, Y.; Wu, H.; Barry, E.; Yin, Y.; Lawrence, E.; Dawson, D.; Willis, J.E.; Markowitz, S.D.; Camargo, F.D.; et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc. Natl. Acad. Sci. USA 2011, 108, E1312–E1320. [Google Scholar] [CrossRef]
- Tschaharganeh, D.F.; Chen, X.; Latzko, P.; Malz, M.; Gaida, M.M.; Felix, K.; Ladu, S.; Singer, S.; Pinna, F.; Gretz, N.; et al. Yes-Associated Protein Up-regulates Jagged-1 and Activates the NOTCH Pathway in Human Hepatocellular Carcinoma. Gastroenterology 2013, 144, 1530–1542.e12. [Google Scholar] [CrossRef]
- Fernandez-L, A.; Northcott, P.A.; Dalton, J.; Fraga, C.; Ellison, D.; Angers, S.; Taylor, M.D.; Kenney, A.M. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009, 23, 2729–2741. [Google Scholar] [CrossRef]
- Song, S.; Honjo, S.; Jin, J.; Chang, S.-S.; Scott, A.W.; Chen, Q.; Kalhor, N.; Correa, A.M.; Hofstetter, W.L.; Albarracin, C.T.; et al. The Hippo Coactivator YAP1 Mediates EGFR Overexpression and Confers Chemoresistance in Esophageal Cancer. Clin. Cancer Res. 2015, 21, 2580–2590. [Google Scholar] [CrossRef]
- Adams, C.W.M.; Ayres, J.; Fagg, N.L.K.; Hicks, B.H. Lymph-vessel embolism in a case of Whipple’s disease. Histopathology 1985, 9, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Zhang, N.; Zheng, Y.; De Wilde, R.F.; Maitra, A.; Pan, D. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010, 24, 2383–2388. [Google Scholar] [CrossRef] [PubMed]
- Imajo, M.; Ebisuya, M.; Nishida, E. Dual role of YAP and TAZ in renewal of the intestinal epithelium. Nat. Cell Biol. 2015, 17, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Azzolin, L.; Panciera, T.; Soligo, S.; Enzo, E.; Bicciato, S.; Dupont, S.; Bresolin, S.; Frasson, C.; Basso, G.; Guzzardo, V.; et al. YAP/TAZ Incorporation in the β-Catenin Destruction Complex Orchestrates the Wnt Response. Cell 2014, 158, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Fallah, S.; Beaulieu, J.-F. The Hippo Pathway Effector YAP1 Regulates Intestinal Epithelial Cell Differentiation. Cells 2020, 9, 1895. [Google Scholar] [CrossRef]
- Hong, A.W.; Meng, Z.; Guan, K.-L. The Hippo pathway in intestinal regeneration and disease. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 324–337. [Google Scholar] [CrossRef]
- Llado, V.; Nakanishi, Y.; Duran, A.; Reina-Campos, M.; Shelton, P.M.; Linares, J.F.; Yajima, T.; Campos, A.; Aza-Blanc, P.; Leitges, M.; et al. Repression of Intestinal Stem Cell Function and Tumorigenesis through Direct Phosphorylation of β-Catenin and Yap by PKCζ. Cell Rep. 2015, 10, 740–754. [Google Scholar] [CrossRef]
- Barry, E.R.; Morikawa, T.; Butler, B.L.; Shrestha, K.; De La Rosa, R.; Yan, K.S.; Fuchs, C.S.; Magness, S.T.; Smits, R.; Ogino, S.; et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nat. Cell Biol. 2013, 493, 106–110. [Google Scholar] [CrossRef]
- Gjorevski, N.; Sachs, N.; Manfrin, A.; Giger, S.; Bragina, M.E.; Ordóñez-Morán, P.; Clevers, N.S.H.; Lutolf, M.P. Designer matrices for intestinal stem cell and organoid culture. Nat. Cell Biol. 2016, 539, 560–564. [Google Scholar] [CrossRef]
- Serra, D.; Mayr, U.; Boni, A.; Lukonin, I.; Rempfler, M.; Meylan, L.C.; Stadler, M.B.; Strnad, P.; Papasaikas, P.; Vischi, D.; et al. Self-organization and symmetry breaking in intestinal organoid development. Nat. Cell Biol. 2019, 569, 66–72. [Google Scholar] [CrossRef]
- Kiesler, P.; Fuss, I.J.; Strober, W. Experimental Models of Inflammatory Bowel Diseases. Cell. Mol. Gastroenterol. Hepatol. 2015, 1, 154–170. [Google Scholar] [CrossRef] [PubMed]
- Leibowitz, B.J.; Yang, L.; Wei, L.; Buchanan, M.E.; Rachid, M.; Parise, R.A.; Beumer, J.H.; Eiseman, J.L.; Schoen, R.E.; Zhang, L.; et al. Targeting p53-dependent stem cell loss for intestinal chemoprotection. Sci. Transl. Med. 2018, 10, eaam7610. [Google Scholar] [CrossRef] [PubMed]
- Hauer-Jensen, M.; Denham, J.W.; Andreyev, H.J.N. Radiation enteropathy-pathogenesis, treatment and prevention. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Moya, I.M.; Halder, G. Hippo–YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 2019, 20, 211–226. [Google Scholar] [CrossRef]
- Gregorieff, A.; Liu, Y.; Inanlou, M.R.; Khomchuk, Y.; Wrana, J. Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer. Nat. Cell Biol. 2015, 526, 715–718. [Google Scholar] [CrossRef]
- Tao, S.; Tang, D.; Morita, Y.; Sperka, T.; Omrani, O.; Lechel, A.; Sakk, V.; Kraus, J.; Kestler, H.A.; Kühl, M.; et al. Wnt activity and basal niche position sensitize intestinal stem and progenitor cells to DNA damage. EMBO J. 2015, 34, 624–640. [Google Scholar] [CrossRef]
- Taniguchi, K.; Wu, L.-W.; Grivennikov, S.I.; De Jong, P.R.; Lian, I.; Yu, F.-X.; Wang, K.; Ho, S.B.; Boland, B.S.; Chang, J.T.; et al. A gp130–Src–YAP module links inflammation to epithelial regeneration. Nat. Cell Biol. 2015, 519, 57–62. [Google Scholar] [CrossRef]
- Deng, F.; Peng, L.; Li, Z.; Tan, G.; Liang, E.; Chen, S.; Zhao, X.; Zhi, F. YAP triggers the Wnt/β-catenin signalling pathway and promotes enterocyte self-renewal, regeneration and tumorigenesis after DSS-induced injury. Cell Death Dis. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Yu, M.; Luo, Y.; Cong, Z.; Mu, Y.; Qiu, Y.; Zhong, M. MicroRNA-590-5p Inhibits Intestinal Inflammation by Targeting YAP. J. Crohn’s Colitis 2018, 12, 993–1004. [Google Scholar] [CrossRef]
- Romera-Hernández, M.; Aparicio-Domingo, P.; Papazian, N.; Karrich, J.J.; Cornelissen, F.; Hoogenboezem, R.M.; Samsom, J.N.; Cupedo, T. Yap1-Driven Intestinal Repair Is Controlled by Group 3 Innate Lymphoid Cells. Cell Rep. 2020, 30, 37–45.e3. [Google Scholar] [CrossRef]
- Miyoshi, H.; VanDussen, K.L.; Malvin, N.P.; Ryu, S.H.; Wang, Y.; Sonnek, N.M.; Lai, C.; Stappenbeck, T. Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium. EMBO J. 2016, 36, 5–24. [Google Scholar] [CrossRef] [PubMed]
- Iwanaga, K.; Okada, M.; Murata, T.; Hori, M.; Ozaki, H. Prostaglandin E2 Promotes Wound-Induced Migration of Intestinal Subepithelial Myofibroblasts via EP2, EP3, and EP4 Prostanoid Receptor Activation. J. Pharmacol. Exp. Ther. 2011, 340, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Sheng, H. Amphiregulin Promotes Intestinal Epithelial Regeneration: Roles of Intestinal Subepithelial Myofibroblasts. Endocrinology 2010, 151, 3728–3737. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Li, J.; Tan, S.; Xu, M.; Tao, J.; Jiang, J.; Liu, H.; Wu, B. COX-1/PGE2/EP4 alleviates mucosal injury by upregulating β-arr1-mediated Akt signaling in colitis. Sci. Rep. 2017, 7, 1055. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-S.; Shin, T.; Lee, B.; Yu, K.; Seo, Y.; Lee, S.; Seo, M.; Hong, I.; Choi, S.W.; Seo, K.; et al. Human Umbilical Cord Blood Mesenchymal Stem Cells Reduce Colitis in Mice by Activating NOD2 Signaling to COX2. Gastroenterology 2013, 145, 1392–1403.e8. [Google Scholar] [CrossRef]
- Kim, H.-B.; Kim, M.; Park, Y.-S.; Park, I.; Kim, T.; Yang, S.-Y.; Cho, C.J.; Hwang, D.; Jung, J.-H.; Markowitz, S.D.; et al. Prostaglandin E2 Activates YAP and a Positive-Signaling Loop to Promote Colon Regeneration After Colitis but Also Carcinogenesis in Mice. Gastroenterology 2017, 152, 616–630. [Google Scholar] [CrossRef]
- Roulis, M.; Kaklamanos, A.; Schernthanner, M.; Bielecki, P.; Zhao, J.; Kaffe, E.; Frommelt, L.-S.; Qu, R.; Knapp, M.S.; Henriques, A.; et al. Paracrine orchestration of intestinal tumorigenesis by a mesenchymal niche. Nat. Cell Biol. 2020, 580, 524–529. [Google Scholar] [CrossRef]
- Terzić, J.; Grivennikov, S.; Karin, E.; Karin, M. Inflammation and Colon Cancer. Gastroenterology 2010, 138, 2101–2114.e5. [Google Scholar] [CrossRef]
- Miyoshi, Y.; Ando, H.; Nagase, H.; Nishisho, I.; Horii, A.; Miki, Y.; Mori, T.; Utsunomiya, J.; Baba, S.; Petersen, G. Germ-line mutations of the APC gene in 53 familial adenomatous polyposis patients. Proc. Natl. Acad. Sci. USA 1992, 89, 4452–4456. [Google Scholar] [CrossRef]
- Plouffe, S.W.; Hong, A.W.; Guan, K.-L. Disease implications of the Hippo/YAP pathway. Trends Mol. Med. 2015, 21, 212–222. [Google Scholar] [CrossRef]
- Powell, S.M.; Zilz, N.; Beazer-Barclay, Y.; Bryan, T.M.; Hamilton, S.R.; Thibodeau, S.N.; Vogelstein, B.; Kinzler, K.W. APC mutations occur early during colorectal tumorigenesis. Nat. Cell Biol. 1992, 359, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Steinhardt, A.A.; Gayyed, M.F.; Klein, A.P.; Dong, J.; Maitra, A.; Pan, D.; Montgomery, E.A.; Anders, R.A. Expression of Yes-associated protein in common solid tumors. Hum. Pathol. 2008, 39, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shi, S.; Guo, Z.; Zhang, X.; Han, S.; Yang, A.; Wen, W.; Zhu, Q. Overexpression of YAP and TAZ Is an Independent Predictor of Prognosis in Colorectal Cancer and Related to the Proliferation and Metastasis of Colon Cancer Cells. PLoS ONE 2013, 8, e65539. [Google Scholar] [CrossRef] [PubMed]
- Yuen, H.-F.; McCrudden, C.M.; Huang, Y.-H.; Tham, J.M.; Zhang, X.; Zeng, Q.; Zhang, S.-D.; Hong, W. TAZ Expression as a Prognostic Indicator in Colorectal Cancer. PLoS ONE 2013, 8, e54211. [Google Scholar] [CrossRef]
- Touil, Y.; Igoudjil, W.; Corvaisier, M.; Dessein, A.-F.; Vandomme, J.; Monté, D.; Stechly, L.; Skrypek, N.; Langlois, C.; Grard, G.; et al. Colon Cancer Cells Escape 5FU Chemotherapy-Induced Cell Death by Entering Stemness and Quiescence Associated with the c-Yes/YAP Axis. Clin. Cancer Res. 2014, 20, 837–846. [Google Scholar] [CrossRef]
- Lu, X.; Pan, J.; Li, S.; Chi, P.; Xu, Z.; Huang, Y. Lentivirus-mediated RNA interference targeting WWTR1 in human colorectal cancer cells inhibits cell proliferation in vitro and tumor growth in vivo. Oncol. Rep. 2012, 28, 179–185. [Google Scholar] [CrossRef][Green Version]
- Bienz, M.; Clevers, H. Linking Colorectal Cancer to Wnt Signaling. Cell 2000, 103, 311–320. [Google Scholar] [CrossRef]
- Brabletz, T.; Jung, A.; Dag, S.; Hlubek, F.; Kirchner, T. β-Catenin Regulates the Expression of the Matrix Metalloproteinase-7 in Human Colorectal Cancer. Am. J. Pathol. 1999, 155, 1033–1038. [Google Scholar] [CrossRef]
- Crawford, H.C.; Fingleton, B.M.; Rudolph-Owen, L.A.; Goss, K.J.H.; Rubinfeld, B.; Polakis, P.; Matrisian, L.M. The metalloproteinase matrilysin is a target of β-catenin transactivation in intestinal tumors. Oncogene 1999, 18, 2883–2891. [Google Scholar] [CrossRef]
- He, T.-C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; Da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of c-MYC as a Target of the APC Pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef]
- Tetsu, O.; McCormick, F. β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nat. Cell Biol. 1999, 398, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Maitra, A.; Anders, R.A.; Taketo, M.M.; Pan, D. β-Catenin destruction complex-independent regulation of Hippo–YAP signaling by APC in intestinal tumorigenesis. Genes Dev. 2015, 29, 1493–1506. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Irvine, K.D. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev. Biol. 2011, 350, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; He, Z.; Kong, L.; Chen, Q.; Yuan, Q.; Zhang, S.; Ye, J.; Liu, H.; Sun, X.; Geng, J.; et al. Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration. Sci. Transl. Med. 2016, 8, 352ra108. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Wang, H.; Shi, Z.; Dong, A.; Zhang, W.; Song, X.; He, F.; Wang, Y.; Zhang, Z.; Wang, W.; et al. A Peptide Mimicking VGLL4 Function Acts as a YAP Antagonist Therapy against Gastric Cancer. Cancer Cell 2014, 25, 166–180. [Google Scholar] [CrossRef]
- Liu-Chittenden, Y.; Huang, B.; Shim, J.S.; Chen, Q.; Lee, S.-J.; Anders, R.A.; Liu, J.O.; Pan, D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012, 26, 1300–1305. [Google Scholar] [CrossRef]
- Giraud, J.; Molina-Castro, S.; Seeneevassen, L.; Sifré, E.; Izotte, J.; Tiffon, C.; Staedel, C.; Boeuf, H.; Fernandez, S.; Barthelemy, P.; et al. Verteporfin targeting YAP1/TAZ-TEAD transcriptional activity inhibits the tumorigenic properties of gastric cancer stem cells. Int. J. Cancer 2020, 146, 2255–2267. [Google Scholar] [CrossRef]
- Zhou, X.; Li, W.; Wang, S.; Zhang, P.; Wang, Q.; Xiao, J.; Zhang, C.; Zheng, X.; Xu, X.; Xue, S.; et al. YAP Aggravates Inflammatory Bowel Disease by Regulating M1/M2 Macrophage Polarization and Gut Microbial Homeostasis. Cell Rep. 2019, 27, 1176–1189.e5. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, Y.; Park, S.-Y.; Kim, H.-S.; Nam, J.-S. The Hippo–YAP Signaling as Guardian in the Pool of Intestinal Stem Cells. Biomedicines 2020, 8, 560. https://doi.org/10.3390/biomedicines8120560
Seo Y, Park S-Y, Kim H-S, Nam J-S. The Hippo–YAP Signaling as Guardian in the Pool of Intestinal Stem Cells. Biomedicines. 2020; 8(12):560. https://doi.org/10.3390/biomedicines8120560
Chicago/Turabian StyleSeo, Yoojin, So-Yeon Park, Hyung-Sik Kim, and Jeong-Seok Nam. 2020. "The Hippo–YAP Signaling as Guardian in the Pool of Intestinal Stem Cells" Biomedicines 8, no. 12: 560. https://doi.org/10.3390/biomedicines8120560
APA StyleSeo, Y., Park, S.-Y., Kim, H.-S., & Nam, J.-S. (2020). The Hippo–YAP Signaling as Guardian in the Pool of Intestinal Stem Cells. Biomedicines, 8(12), 560. https://doi.org/10.3390/biomedicines8120560





