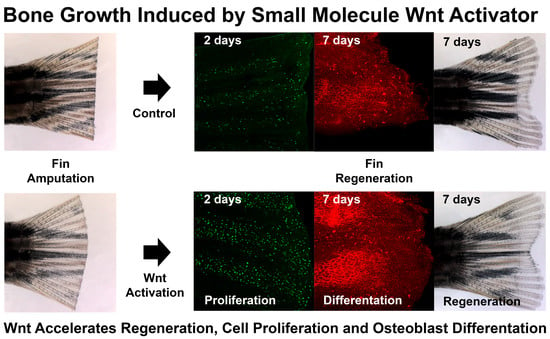
The Glycogen Synthase Kinase-3β Inhibitor LSN 2105786 Promotes Zebrafish Fin Regeneration
Abstract
1. Introduction
2. Experimental Section
2.1. Zebrafish Husbandry
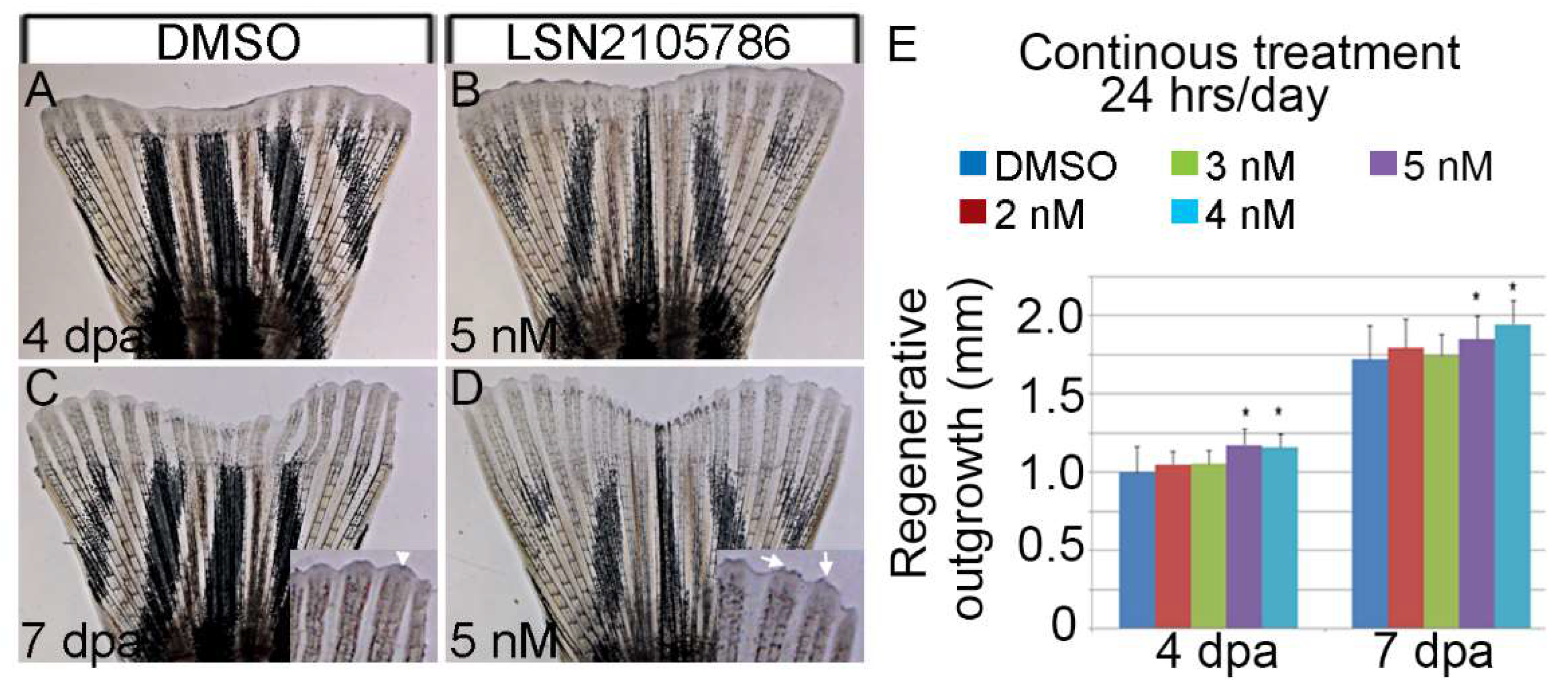
2.2. Adult Fin Amputation Assay
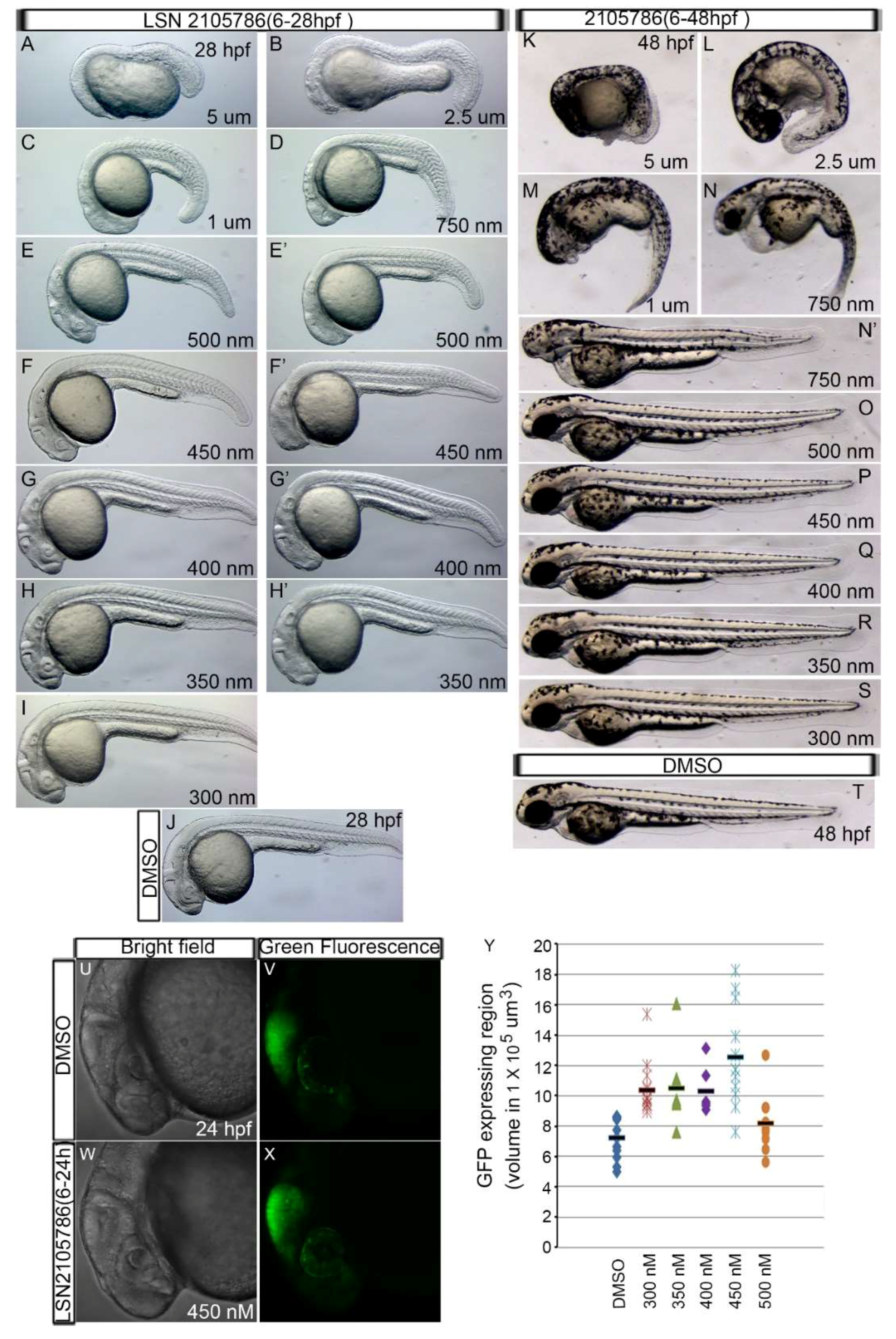
2.3. Embryo Treatment with GSK3β Inhibitor
2.4. Whole Mount in situ Hybridization
2.5. Whole Mount Immunostaining and Image Analysis
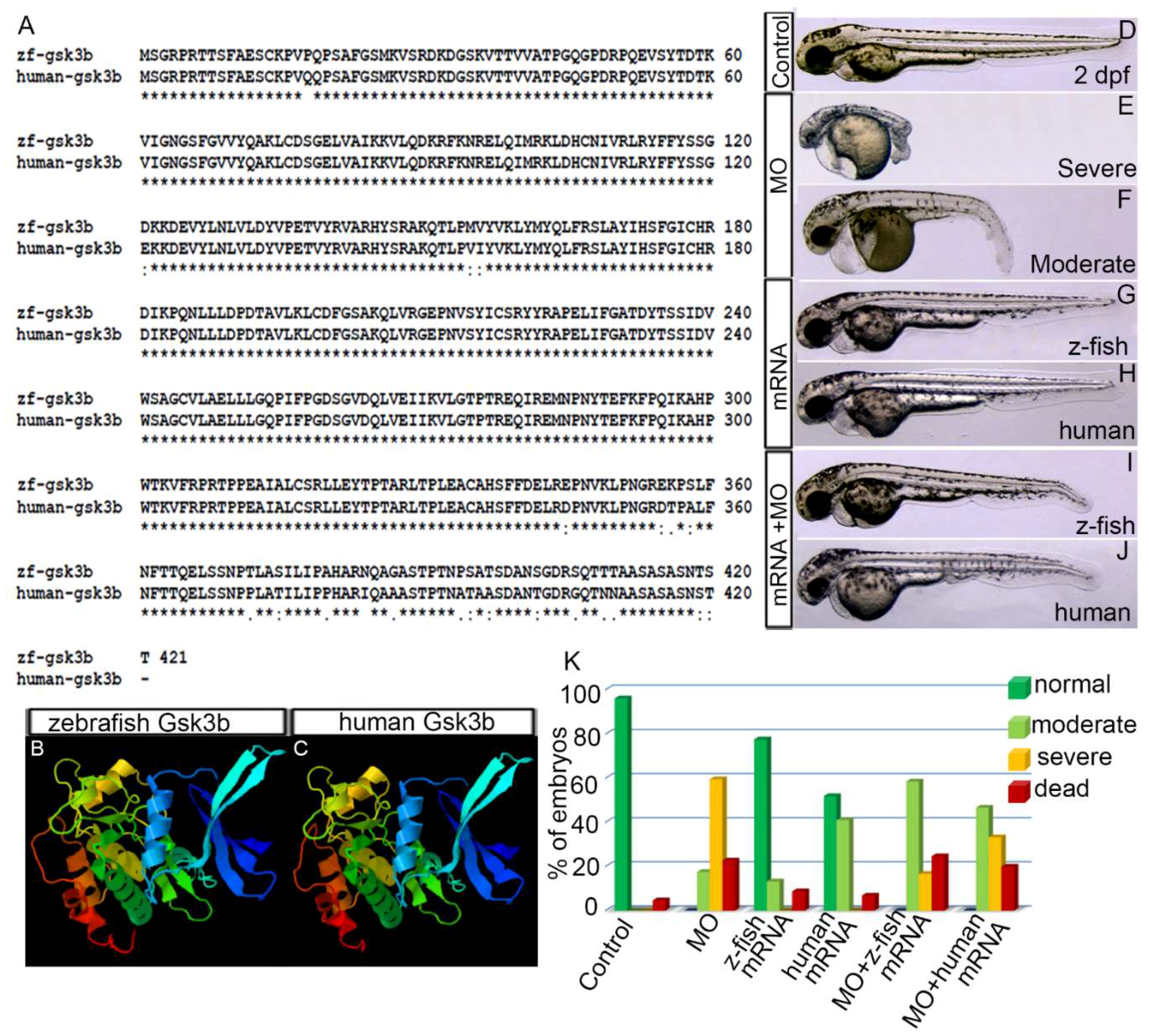
2.6. DNA Sequence Alignment and 3D Protein Structure Prediction
2.7. Knockdown of gsk3β by Morpholino and Injection of Zebrafish gsk3β or Human GSK3β mRNA
2.8. Statistical Analysis
3. Results
3.1. Human GSK3β is Structurally and Functionally Equivalent to Zebrafish Gsk3β
3.2. LSN 2105786 Treatment Upregulates Wnt Signaling in Zebrafish and Produces Eyeless Embryos
3.3. LSN 2105786 Continuous Treatment after Fin Amputation Accelerates Fin Regeneration
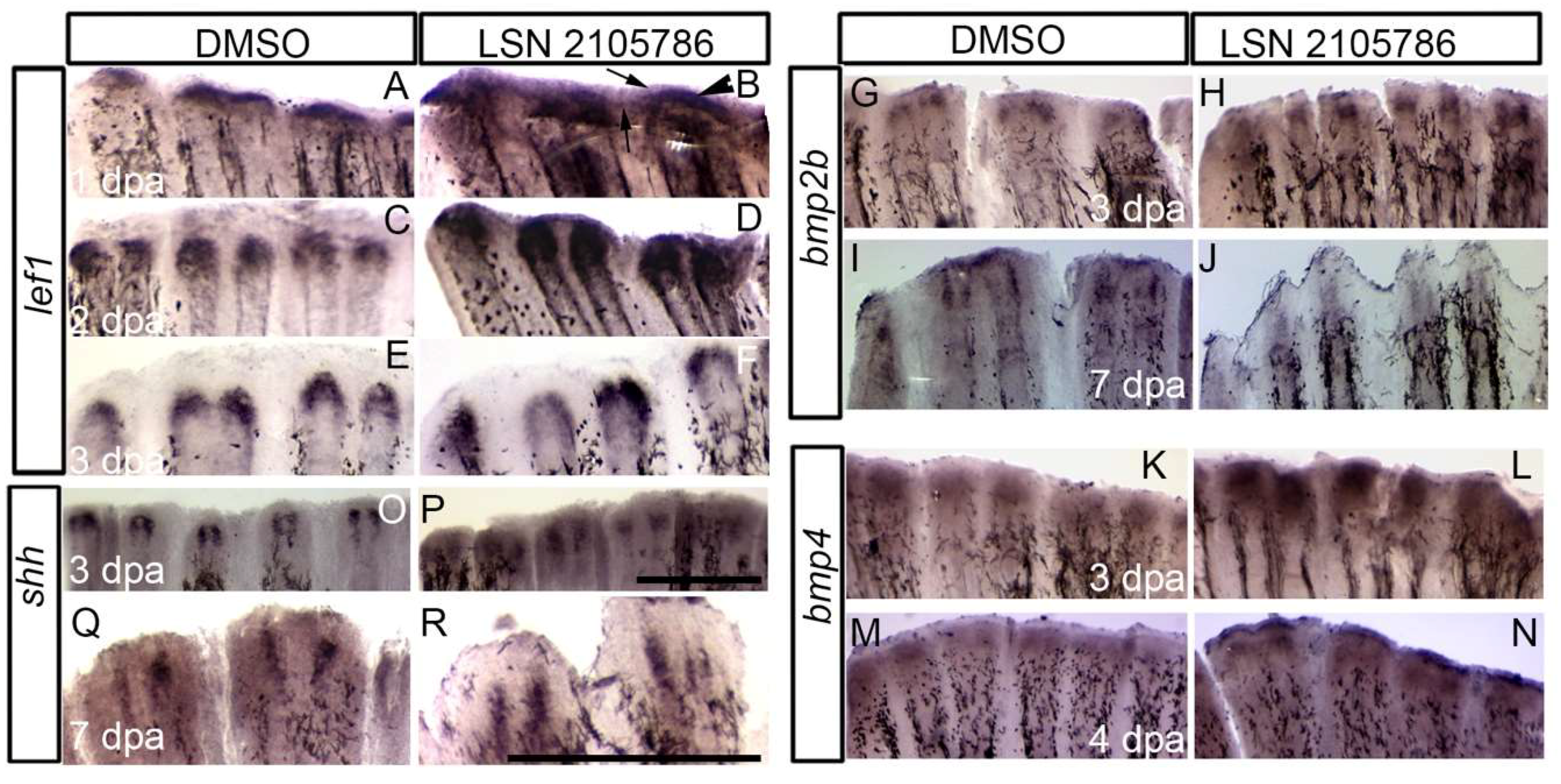
3.4. LSN 2105786 Treatment Increased Expression of Wnt Targets and Downstream Genes in Regenerating Fin Tissue
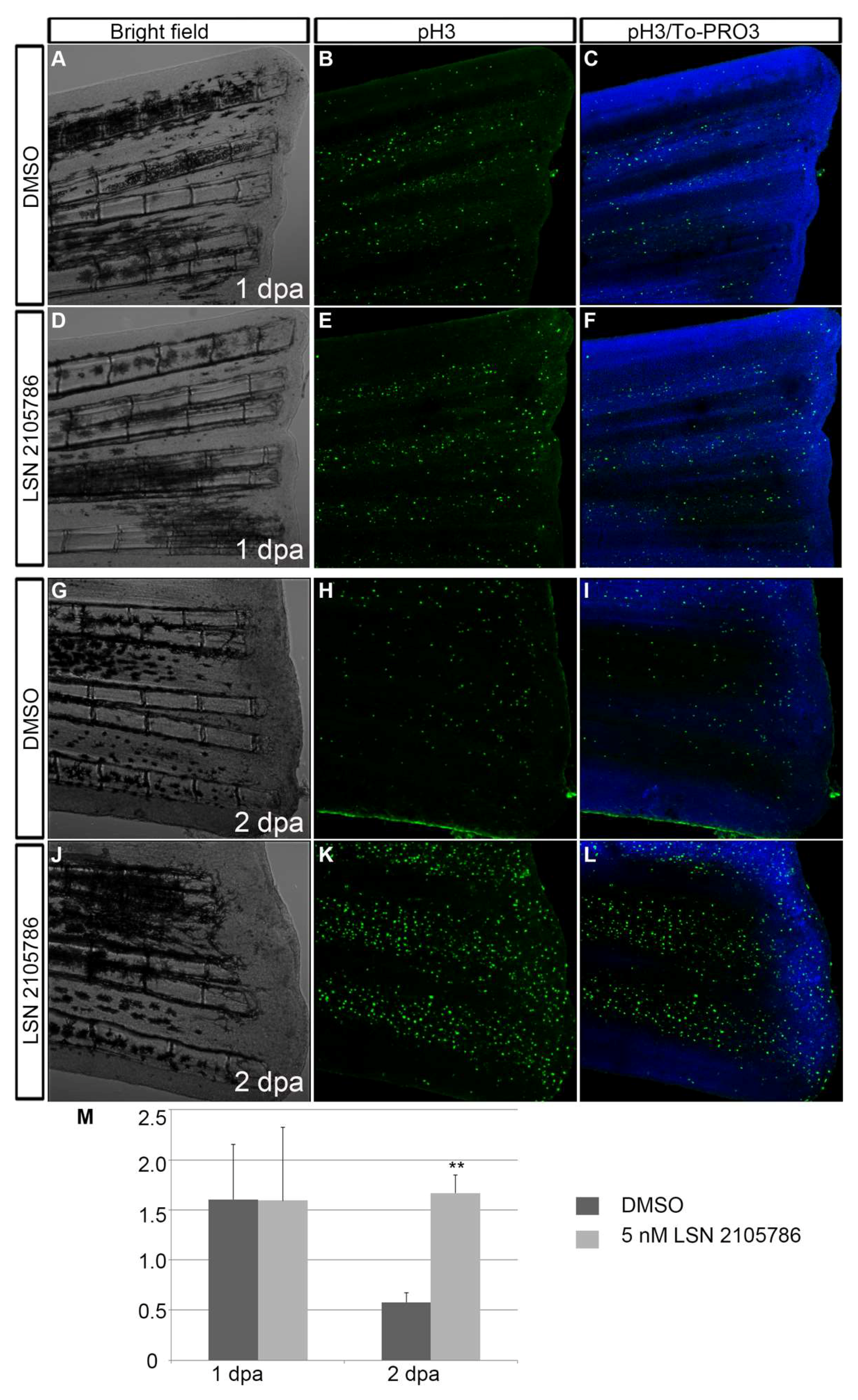
3.5. LSN 2105786 Increases Cell Proliferation in Regenerating Fins
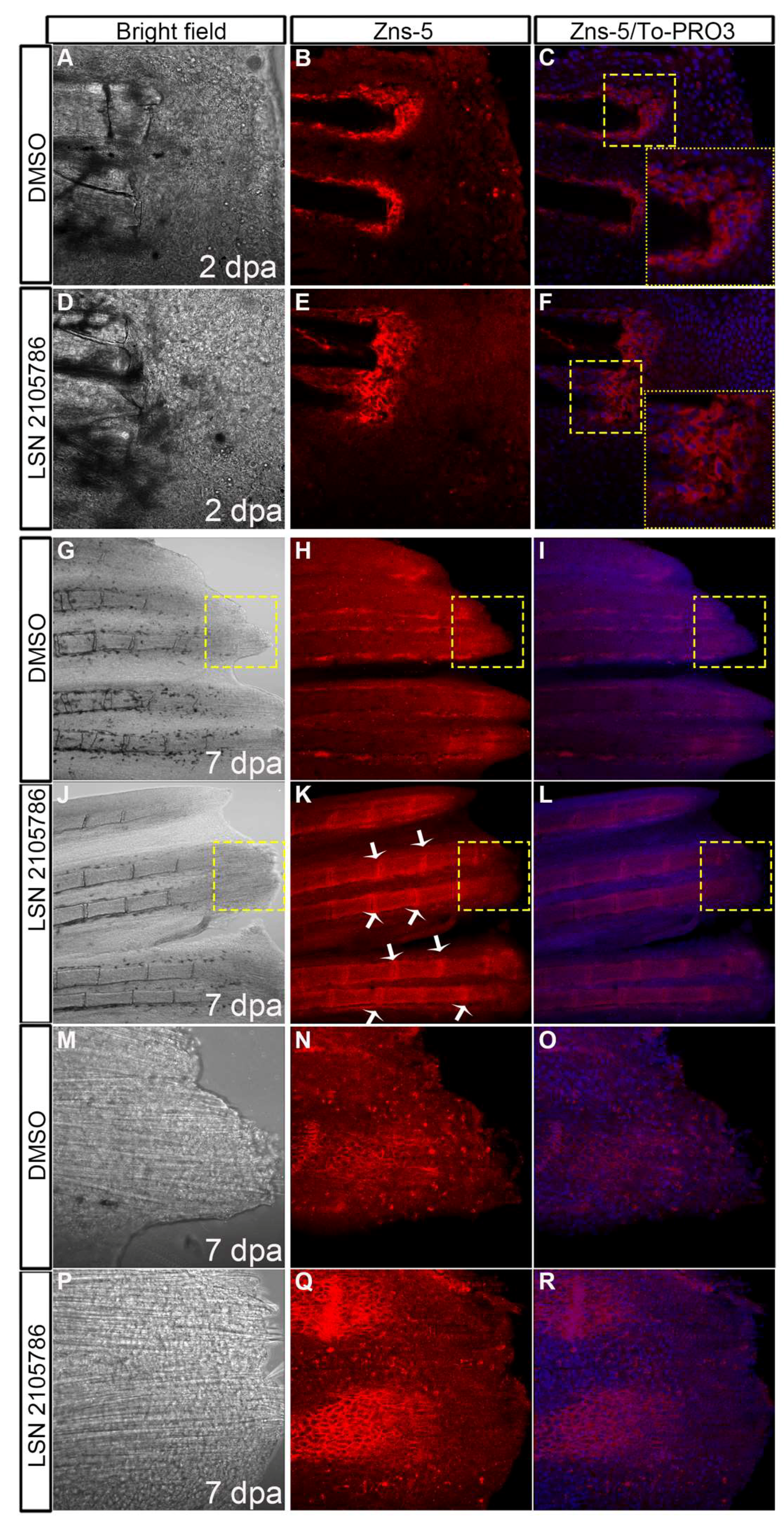
3.6. LSN 2105786 Treatment Increases Osteoblast Differentiation
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baron, R.; Kneissel, M. WNT signaling in bone homeostasis and disease: From human mutations to treatments. Nat. Med. 2013, 19, 179–192. [Google Scholar] [CrossRef]
- Moon, R.T.; Bowerman, B.; Boutros, M.; Perrimon, N. The promise and perils of Wnt signaling through beta-catenin. Science 2002, 296, 1644–1646. [Google Scholar] [CrossRef] [PubMed]
- Agholme, F.; Aspenberg, P. Wnt signaling and orthopedics, an overview. Acta Orthopaed. 2011, 82, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Westendorf, J.J.; Kahler, R.A.; Schroeder, T.M. Wnt signaling in osteoblasts and bone diseases. Gene 2004, 341, 19–39. [Google Scholar] [CrossRef]
- Gong, Y.; Slee, R.B.; Fukai, N.; Rawadi, G.; Roman-Roman, S.; Reginato, A.M.; Wang, H.; Cundy, T.; Glorieux, F.H.; Lev, D.; et al. Osteoporosis-Pseudoglioma Syndrome Collaborative, G. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 2001, 107, 513–523. [Google Scholar] [CrossRef]
- Boyden, L.M.; Mao, J.; Belsky, J.; Mitzner, L.; Farhi, A.; Mitnick, M.A.; Wu, D.; Insogna, K.; Lifton, R.P. High bone density due to a mutation in LDL-receptor-related protein 5. N. Engl. J. Med. 2002, 346, 1513–1521. [Google Scholar] [CrossRef]
- Holmen, S.L.; Zylstra, C.R.; Mukherjee, A.; Sigler, R.E.; Faugere, M.C.; Bouxsein, M.L.; Deng, L.; Clemens, T.L.; Williams, B.O. Essential role of beta-catenin in postnatal bone acquisition. J. Biol. Chem. 2005, 280, 21162–21168. [Google Scholar] [CrossRef]
- Spencer, G.J.; Utting, J.C.; Etheridge, S.L.; Arnett, T.R.; Genever, P.G. Wnt signalling in osteoblasts regulates expression of the receptor activator of NFkappaB ligand and inhibits osteoclastogenesis in vitro. J. Cell Sci. 2006, 119, 1283–1296. [Google Scholar] [CrossRef]
- Hu, H.; Hilton, M.J.; Tu, X.; Yu, K.; Ornitz, D.M.; Long, F. Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development 2005, 132, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Hill, T.P.; Spater, D.; Taketo, M.M.; Birchmeier, W.; Hartmann, C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev. Cell 2005, 8, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Day, T.F.; Guo, X.; Garrett-Beal, L.; Yang, Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev. Cell 2005, 8, 739–750. [Google Scholar] [CrossRef]
- Bennett, C.N.; Ouyang, H.; Ma, Y.L.; Zeng, Q.; Gerin, I.; Sousa, K.M.; Lane, T.F.; Krishnan, V.; Hankenson, K.D.; MacDougald, O.A. Wnt10b increases postnatal bone formation by enhancing osteoblast differentiation. J. Bone Min. Res. 2007, 22, 1924–1932. [Google Scholar] [CrossRef]
- Zhong, N.; Gersch, R.P.; Hadjiargyrou, M. Wnt signaling activation during bone regeneration and the role of Dishevelled in chondrocyte proliferation and differentiation. Bone 2006, 39, 5–16. [Google Scholar] [CrossRef]
- Chen, Y.; Whetstone, H.C.; Lin, A.C.; Nadesan, P.; Wei, Q.; Poon, R.; Alman, B.A. Beta-catenin signaling plays a disparate role in different phases of fracture repair: Implications for therapy to improve bone healing. PLoS Med. 2007, 4, e249. [Google Scholar] [CrossRef]
- Li, X.; Grisanti, M.; Fan, W.; Asuncion, F.J.; Tan, H.L.; Dwyer, D.; Han, C.Y.; Yu, L.; Lee, J.; Lee, E.; et al. Dickkopf-1 regulates bone formation in young growing rodents and upon traumatic injury. J. Bone Min. Res. 2011, 26, 2610–2621. [Google Scholar] [CrossRef] [PubMed]
- Ominsky, M.S.; Li, C.; Li, X.; Tan, H.L.; Lee, E.; Barrero, M.; Asuncion, F.J.; Dwyer, D.; Han, C.Y.; Vlasseros, F.; et al. Inhibition of sclerostin by monoclonal antibody enhances bone healing and improves bone density and strength of nonfractured bones. J. Bone Min. Res. 2011, 26, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Florio, M.; Gunasekaran, K.; Stolina, M.; Li, X.; Liu, L.; Tipton, B.; Salimi-Moosavi, H.; Asuncion, F.J.; Li, C.; Sun, B.; et al. A bispecific antibody targeting sclerostin and DKK-1 promotes bone mass accrual and fracture repair. Nat. Commun. 2016, 7, 11505. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, N.H.; Halladay, D.L.; Miles, R.R.; Gilbert, L.M.; Frolik, C.A.; Galvin, R.J.; Martin, T.J.; Gillespie, M.T.; Onyia, J.E. Effects of parathyroid hormone on Wnt signaling pathway in bone. J. Cell. Biochem. 2005, 95, 1178–1190. [Google Scholar] [CrossRef]
- Alkhiary, Y.M.; Gerstenfeld, L.C.; Krall, E.; Westmore, M.; Sato, M.; Mitlak, B.H.; Einhorn, T.A. Enhancement of experimental fracture-healing by systemic administration of recombinant human parathyroid hormone (PTH 1-34). J. Bone Joint Surg. 2005, 87, 731–741. [Google Scholar]
- Aspenberg, P.; Genant, H.K.; Johansson, T.; Nino, A.J.; See, K.; Krohn, K.; Garcia-Hernandez, P.A.; Recknor, C.P.; Einhorn, T.A.; Dalsky, G.P.; et al. Teriparatide for acceleration of fracture repair in humans: A prospective, randomized, double-blind study of 102 postmenopausal women with distal radial fractures. J. Bone Min. Res. 2010, 25, 404–414. [Google Scholar] [CrossRef]
- Aspenberg, P.; Johansson, T. Teriparatide improves early callus formation in distal radial fractures. Acta Orthopaed. 2010, 81, 234–236. [Google Scholar] [CrossRef] [PubMed]
- Stoick-Cooper, C.L.; Moon, R.T.; Weidinger, G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes Dev. 2007, 21, 1292–1315. [Google Scholar] [CrossRef]
- Poss, K.D.; Shen, J.; Keating, M.T. Induction of lef1 during zebrafish fin regeneration. Dev. Dyn. 2000, 219, 282–286. [Google Scholar] [CrossRef]
- Kawakami, Y.; Rodriguez Esteban, C.; Raya, M.; Kawakami, H.; Marti, M.; Dubova, I.; Izpisua Belmonte, J.C. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006, 20, 3232–3237. [Google Scholar] [CrossRef]
- Stoick-Cooper, C.L.; Weidinger, G.; Riehle, K.J.; Hubbert, C.; Major, M.B.; Fausto, N.; Moon, R.T. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 2007, 134, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, N.H.; Onyia, J.E.; Zeng, Q.; Tian, X.; Liu, M.; Halladay, D.L.; Frolik, C.A.; Engler, T.; Wei, T.; Kriauciunas, A.; et al. Orally bioavailable GSK-3alpha/beta dual inhibitor increases markers of cellular differentiation in vitro and bone mass in vivo. J. Bone Min. Res. 2006, 21, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Dorsky, R.I.; Sheldahl, L.C.; Moon, R.T. A transgenic Lef1/beta-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev. Biol. 2002, 241, 229–237. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book; Eugene, O.R., Ed.; The University of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Tsai, J.N.; Liao, P.Y.; Tsai, W.Y.; Lin, K.Y.; Chuang, C.C.; Sun, C.K.; Chang, W.C.; Tsai, H.J. Glycogen synthase kinase 3 alpha and 3 beta have distinct functions during cardiogenesis of zebrafish embryo. BMC Dev. Biol. 2007, 7, 93. [Google Scholar] [CrossRef]
- Van de Water, S.; van de Wetering, M.; Joore, J.; Esseling, J.; Bink, R.; Clevers, H.; Zivkovic, D. Ectopic Wnt signal determines the eyeless phenotype of zebrafish masterblind mutant. Development 2001, 128, 3877–3888. [Google Scholar]
- Blum, N.; Begemann, G. Retinoic acid signaling controls the formation, proliferation and survival of the blastema during adult zebrafish fin regeneration. Development 2012, 139, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Poss, K.D.; Keating, M.T.; Nechiporuk, A. Tales of regeneration in zebrafish. Dev. Dyn. 2003, 226, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Murciano, C.; Fernandez, T.D.; Duran, I.; Maseda, D.; Ruiz-Sanchez, J.; Becerra, J.; Akimenko, M.A.; Mari-Beffa, M. Ray-interray interactions during fin regeneration of Danio rerio. Dev. Biol. 2002, 252, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Quint, E.; Smith, A.; Avaron, F.; Laforest, L.; Miles, J.; Gaffield, W.; Akimenko, M.A. Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc. Natl. Acad. Sci. USA 2002, 99, 8713–8718. [Google Scholar] [CrossRef] [PubMed]
- Willert, K.; Jones, K.A. Wnt signaling: Is the party in the nucleus? Genes Dev. 2006, 20, 1394–1404. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.; Sato, M.; Goldsmith, P. High-throughput in vivo screening for bone anabolic compounds with zebrafish. J. Biomol. Screen. 2005, 10, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Sarmah, S.; Marrs, J.A. Zebrafish as a Vertebrate Model System to Evaluate Effects of Environmental Toxicants on Cardiac Development and Function. Int. J. Mol. Sci. 2016, 17, 2123. [Google Scholar] [CrossRef]
- Asnani, A.; Peterson, R.T. The zebrafish as a tool to identify novel therapies for human cardiovascular disease. Dis. Models Mech. 2014, 7, 763–767. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarmah, S.; Curtis, C.; Mahin, J.; Farrell, M.; Engler, T.A.; Sanchez-Felix, M.V.; Sato, M.; Ma, Y.L.; Chu, S.; Marrs, J.A. The Glycogen Synthase Kinase-3β Inhibitor LSN 2105786 Promotes Zebrafish Fin Regeneration. Biomedicines 2019, 7, 30. https://doi.org/10.3390/biomedicines7020030
Sarmah S, Curtis C, Mahin J, Farrell M, Engler TA, Sanchez-Felix MV, Sato M, Ma YL, Chu S, Marrs JA. The Glycogen Synthase Kinase-3β Inhibitor LSN 2105786 Promotes Zebrafish Fin Regeneration. Biomedicines. 2019; 7(2):30. https://doi.org/10.3390/biomedicines7020030
Chicago/Turabian StyleSarmah, Swapnalee, Courtney Curtis, Jennifer Mahin, Mark Farrell, Thomas A. Engler, Manuel V. Sanchez-Felix, Masahiko Sato, Yanfai Linda Ma, Shaoyou Chu, and James A. Marrs. 2019. "The Glycogen Synthase Kinase-3β Inhibitor LSN 2105786 Promotes Zebrafish Fin Regeneration" Biomedicines 7, no. 2: 30. https://doi.org/10.3390/biomedicines7020030
APA StyleSarmah, S., Curtis, C., Mahin, J., Farrell, M., Engler, T. A., Sanchez-Felix, M. V., Sato, M., Ma, Y. L., Chu, S., & Marrs, J. A. (2019). The Glycogen Synthase Kinase-3β Inhibitor LSN 2105786 Promotes Zebrafish Fin Regeneration. Biomedicines, 7(2), 30. https://doi.org/10.3390/biomedicines7020030