Pathogens and Their Effect on Exosome Biogenesis and Composition
Abstract
1. Introduction
2. Types of Extracellular Vesicles
2.1. Exosomes
2.2. Microvesicles
2.3. Apoptotic Bodies
3. Exosome Biogenesis and Composition
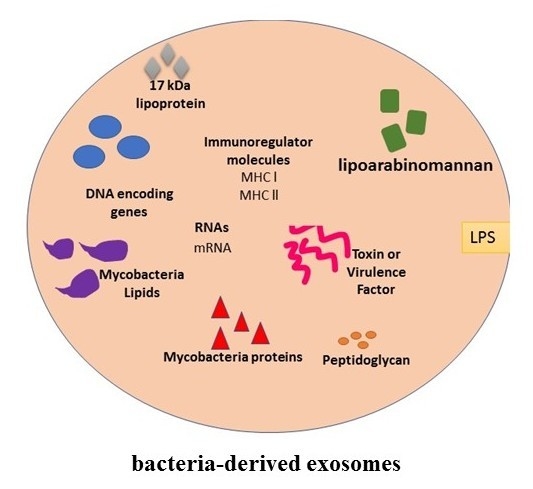
4. Exosomes and Bacterial Infections
5. Exosomes and Leishmaniasis Infections
6. Exosomes and Trypanosoma cruzi Infections
7. Conclusions
Funding
Conflicts of Interest
References
- Rilla, K.; Mustonen, A.M.; Arasu, U.T.; Harkonen, K.; Matilainen, J.; Nieminen, P. Extracellular vesicles are integral and functional components of the extracellular matrix. Matrix Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Schorey, J.S.; Cheng, Y.; Singh, P.P.; Smith, V.L. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 2015, 16, 24–43. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.P.-K.; Breakefield, X.O. Role of exosomes/microvesicles in the nervous system and use in emerging therapies. Front. Physiol. 2012, 3, 228. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.J.; Maghsoudi, T.; Wang, T. Exosomes mediate the intercellular communication after myocardial infarction. Int. J. Med. Sci. 2016, 13, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Andre, F.; Schartz, N.E.; Movassagh, M.; Flament, C.; Pautier, P.; Morice, P.; Pomel, C.; Lhomme, C.; Escudier, B.; Le Chevalier, T.; et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet 2002, 360, 295–305. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in exosome isolation techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Zabeo, D.; Cvjetkovic, A.; Lasser, C.; Schorb, M.; Lotvall, J.; Hoog, J.L. Exosomes purified from a single cell type have diverse morphology. J. Extracell. Vesicles 2017, 6, 1329476. [Google Scholar] [CrossRef] [PubMed]
- Momen-Heravi, F.; Balaj, L.; Alian, S.; Mantel, P.Y.; Halleck, A.E.; Trachtenberg, A.J.; Soria, C.E.; Oquin, S.; Bonebreak, C.M.; Saracoglu, E.; et al. Current methods for the isolation of extracellular vesicles. Biol. Chem. 2013, 394, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Moldovan, L.; Batte, K.; Wang, Y.; Wisler, J.; Piper, M. Analyzing the circulating micrornas in exosomes/extracellular vesicles from serum or plasma by qrt-pcr. Methods Mol. Biol. 2013, 1024, 129–145. [Google Scholar] [PubMed]
- Bhatnagar, S.; Schorey, J.S. Exosomes released from infected macrophages contain mycobacterium avium glycopeptidolipids and are proinflammatory. J. Biol. Chem. 2007, 282, 25779–25789. [Google Scholar] [CrossRef] [PubMed]
- Honegger, A.; Schilling, D.; Sultmann, H.; Hoppe-Seyler, K.; Hoppe-Seyler, F. Identification of e6/e7-dependent micrornas in hpv-positive cancer cells. Methods Mol. Biol. 2018, 1699, 119–134. [Google Scholar] [PubMed]
- Shenoda, B.B.; Ajit, S.K. Modulation of immune responses by exosomes derived from antigen-presenting cells. Clin. Med. Insights Pathol. 2016, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, H.M.; Fooladi, A.A.; Nourani, M.R.; Ghanezadeh, F. The role of exosomes in infectious diseases. Inflamm. Allergy Drug Targets 2013, 12, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Cestari, I.; Ansa-Addo, E.; Deolindo, P.; Inal, J.M.; Ramirez, M.I. Trypanosoma cruzi immune evasion mediated by host cell-derived microvesicles. J. Immunol. 2012, 188, 1942–1952. [Google Scholar] [CrossRef] [PubMed]
- Giri, P.K.; Schorey, J.S. Exosomes derived from m. Bovis bcg infected macrophages activate antigen-specific cd4+ and cd8+ t cells in vitro and in vivo. PLoS ONE 2008, 3, e2461. [Google Scholar] [CrossRef] [PubMed]
- Abrami, L.; Brandi, L.; Moayeri, M.; Brown, M.J.; Krantz, B.A.; Leppla, S.H.; van der Goot, F.G. Hijacking multivesicular bodies enables long-term and exosome-mediated long-distance action of anthrax toxin. Cell Rep. 2013, 5, 986–996. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.L.; Nakayasu, E.S.; Joffe, L.S.; Guimaraes, A.J.; Sobreira, T.J.; Nosanchuk, J.D.; Cordero, R.J.; Frases, S.; Casadevall, A.; Almeida, I.C.; et al. Characterization of yeast extracellular vesicles: Evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS ONE 2010, 5, e11113. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Lee, E.J.; Lee, J.H.; Jun, S.H.; Choi, C.W.; Kim, S.I.; Kang, S.S.; Hyun, S. Klebsiella pneumoniae secretes outer membrane vesicles that induce the innate immune response. FEMS Microbiol. Lett. 2012, 331, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Aline, F.; Bout, D.; Amigorena, S.; Roingeard, P.; Dimier-Poisson, I. Toxoplasma gondii antigen-pulsed-dendritic cell-derived exosomes induce a protective immune response against t. Gondii infection. Infect. Immun. 2004, 72, 4127–4137. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiu, F.; Mou, Z.; Xue, Z.; Du, H.; Zhou, C.; Li, Y.; Shi, Y.; He, S.; Zhou, H. Exosomes derived from toxoplasma gondii stimulate an inflammatory response through jnk signaling pathway. Nanomedicine 2018, 13, 1157–1168. [Google Scholar] [CrossRef] [PubMed]
- Martin-Jaular, L.; Nakayasu, E.S.; Ferrer, M.; Almeida, I.C.; Del Portillo, H.A. Exosomes from plasmodium yoelii-infected reticulocytes protect mice from lethal infections. PLoS ONE 2011, 6, e26588. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; El Andaloussi, S.; Wood, M.J. Exosomes and microvesicles: Extracellular vesicles for genetic information transfer and gene therapy. Hum. Mol. Genet. 2012, 21, R125–R134. [Google Scholar] [CrossRef] [PubMed]
- Rackov, G.; Garcia-Romero, N.; Esteban-Rubio, S.; Carrion-Navarro, J.; Belda-Iniesta, C.; Ayuso-Sacido, A. Vesicle-mediated control of cell function: The role of extracellular matrix and microenvironment. Front. Physiol. 2018, 9, 651. [Google Scholar] [CrossRef] [PubMed]
- Stahl, P.D.; Raposo, G. Exosomes and extracellular vesicles: The path forward. Essays Biochem. 2018, 62, 119–124. [Google Scholar] [CrossRef] [PubMed]
- D’Souza-Schorey, C.; Schorey, J.S. Regulation and mechanisms of extracellular vesicle biogenesis and secretion. Essays Biochem. 2018, 62, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Orekhov, A.N.; Bobryshev, Y.V. Cardiac extracellular vesicles in normal and infarcted heart. Int. J. Mol. Sci. 2016, 17, 63. [Google Scholar] [CrossRef] [PubMed]
- Caruso, S.; Poon, I.K.H. Apoptotic cell-derived extracellular vesicles: More than just debris. Front. Immunol. 2018, 9, 1486. [Google Scholar] [CrossRef] [PubMed]
- Corrado, C.; Raimondo, S.; Chiesi, A.; Ciccia, F.; De Leo, G.; Alessandro, R. Exosomes as intercellular signaling organelles involved in health and disease: Basic science and clinical applications. Int. J. Mol. Sci. 2013, 14, 5338–5366. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Cheruvanky, A.; Hu, X.; Matsumoto, T.; Hiramatsu, N.; Cho, M.E.; Berger, A.; Leelahavanichkul, A.; Doi, K.; Chawla, L.S.; et al. Urinary exosomal transcription factors, a new class of biomarkers for renal disease. Kidney Int. 2008, 74, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Eldh, M.; Ekstrom, K.; Valadi, H.; Sjostrand, M.; Olsson, B.; Jernas, M.; Lotvall, J. Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle rna. PLoS ONE 2010, 5, e15353. [Google Scholar] [CrossRef] [PubMed]
- Frydrychowicz, M.; Kolecka-Bednarczyk, A.; Madejczyk, M.; Yasar, S.; Dworacki, G. Exosomes—Structure, biogenesis and biological role in non-small-cell lung cancer. Scand. J. Immunol. 2015, 81, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Meckes, D.G., Jr.; Raab-Traub, N. Microvesicles and viral infection. J. Virol. 2011, 85, 12844–12854. [Google Scholar] [CrossRef] [PubMed]
- Gyorgy, B.; Szabo, T.G.; Pasztoi, M.; Pal, Z.; Misjak, P.; Aradi, B.; Laszlo, V.; Pallinger, E.; Pap, E.; Kittel, A.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011, 68, 2667–2688. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm. Sin. B 2016, 6, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Charrin, S.; Jouannet, S.; Boucheix, C.; Rubinstein, E. Tetraspanins at a glance. J. Cell Sci. 2014, 127, 3641–3648. [Google Scholar] [CrossRef] [PubMed]
- Beach, A.; Zhang, H.G.; Ratajczak, M.Z.; Kakar, S.S. Exosomes: An overview of biogenesis, composition and role in ovarian cancer. J. Ovarian Res. 2014, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Perez-Hernandez, D.; Gutiérrez-Vázquez, C.; Jorge, I.; López-Martín, S.; Ursa, A.; Sánchez-Madrid, F.; Vázquez, J.; Yáñez-Mó, M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 2013, 288, 11649–11661. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Futamata, H.; Tashiro, Y. Complexities of cell-to-cell communication through membrane vesicles: Implications for selective interaction of membrane vesicles with microbial cells. Front. Microbiol. 2015, 6, 633. [Google Scholar] [CrossRef] [PubMed]
- Kruh-Garcia, N.A.; Wolfe, L.M.; Dobos, K.M. Deciphering the role of exosomes in tuberculosis. Tuberculosis 2015, 95, 26–30. [Google Scholar] [CrossRef] [PubMed]
- De Toro, J.; Herschlik, L.; Waldner, C.; Mongini, C. Emerging roles of exosomes in normal and pathological conditions: New insights for diagnosis and therapeutic applications. Front. Immunol. 2015, 6, 203. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.; Sampey, G.; Chung, M.C.; Bailey, C.; van Hoek, M.L.; Kashanchi, F.; Hakami, R.M. The carrying pigeons of the cell: Exosomes and their role in infectious diseases caused by human pathogens. Pathog. Dis. 2014, 71, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Kruh-Garcia, N.A.; Wolfe, L.M.; Chaisson, L.H.; Worodria, W.O.; Nahid, P.; Schorey, J.S.; Davis, J.L.; Dobos, K.M. Detection of mycobacterium tuberculosis peptides in the exosomes of patients with active and latent m. Tuberculosis infection using mrm-ms. PLoS ONE 2014, 9, e103811. [Google Scholar] [CrossRef] [PubMed]
- Maurin, M. Real-time pcr as a diagnostic tool for bacterial diseases. Expert Rev. Mol. Diagn. 2012, 12, 731–754. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Sobel, M.L.; Poole, K. Antibiotic inducibility of the mexxy multidrug efflux system of pseudomonas aeruginosa: Involvement of the antibiotic-inducible pa5471 gene product. J. Bacteriol. 2006, 188, 1847–1855. [Google Scholar] [CrossRef] [PubMed]
- Coakley, G.; Maizels, R.M.; Buck, A.H. Exosomes and other extracellular vesicles: The new communicators in parasite infections. Trends Parasitol. 2015, 31, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Coakley, G.; McCaskill, J.L.; Borger, J.G.; Simbari, F.; Robertson, E.; Millar, M.; Harcus, Y.; McSorley, H.J.; Maizels, R.M.; Buck, A.H. Extracellular vesicles from a helminth parasite suppress macrophage activation and constitute an effective vaccine for protective immunity. Cell Rep. 2017, 19, 1545–1557. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Leishmaniasis. Available online: http://www.who.int/leishmaniasis/en/ (accessed on 17 March 2018).
- Silverman, J.M.; Clos, J.; de’Oliveira, C.C.; Shirvani, O.; Fang, Y.; Wang, C.; Foster, L.J.; Reiner, N.E. An exosome-based secretion pathway is responsible for protein export from leishmania and communication with macrophages. J. Cell Sci. 2010, 123, 842–852. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, G.; Chaudhuri, M.; Pan, A.; Chang, K.P. Surface acid proteinase (gp63) of leishmania mexicana. A metalloenzyme capable of protecting liposome-encapsulated proteins from phagolysosomal degradation by macrophages. J. Biol. Chem. 1989, 264, 7483–7489. [Google Scholar] [PubMed]
- Silverman, J.M.; Clos, J.; Horakova, E.; Wang, A.Y.; Wiesgigl, M.; Kelly, I.; Lynn, M.A.; McMaster, W.R.; Foster, L.J.; Levings, M.K.; et al. Leishmania exosomes modulate innate and adaptive immune responses through effects on monocytes and dendritic cells. J. Immunol. 2010, 185, 5011–5022. [Google Scholar] [CrossRef] [PubMed]
- Schnitzer, J.K.; Berzel, S.; Fajardo-Moser, M.; Remer, K.A.; Moll, H. Fragments of antigen-loaded dendritic cells (DC) and DC-derived exosomes induce protective immunity against leishmania major. Vaccine 2010, 28, 5785–5793. [Google Scholar] [CrossRef] [PubMed]
- Udoko, A.N.; Johnson, C.A.; Dykan, A.; Rachakonda, G.; Villalta, F.; Mandape, S.N.; Lima, M.F.; Pratap, S.; Nde, P.N. Early regulation of profibrotic genes in primary human cardiac myocytes by trypanosoma cruzi. PLoS Negl. Trop. Dis. 2016, 10, e0003747. [Google Scholar] [CrossRef] [PubMed]
- Leslie, M. Infectious diseases. Drug developers finally take aim at a neglected disease. Science 2011, 333, 933–935. [Google Scholar] [CrossRef] [PubMed]
- Villalta, F.; Dobish, M.C.; Nde, P.N.; Kleshchenko, Y.Y.; Hargrove, T.Y.; Johnson, C.A.; Waterman, M.R.; Johnston, J.N.; Lepesheva, G.I. Vni cures acute and chronic experimental chagas disease. J. Infect. Dis. 2013, 208, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Bern, C.; Kjos, S.; Yabsley, M.J.; Montgomery, S.P. Trypanosoma cruzi and chagas’ disease in the united states. Clin. Microbiol. Rev. 2011, 24, 655–681. [Google Scholar] [CrossRef] [PubMed]
- Gascon, J.; Bern, C.; Pinazo, M.J. Chagas disease in spain, the united states and other non-endemic countries. Acta Trop. 2010, 115, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Matthews, Q.L.; Farrow, A.L.; Rachakonda, G.; Gu, L.; Nde, P.; Krendelchtchikov, A.; Pratap, S.; Sakhare, S.S.; Sabbaj, S.; Lima, M.F.; et al. Epitope capsid-incorporation: New effective approach for vaccine development for chagas disease. Pathog. Immun. 2016, 1, 214–233. [Google Scholar] [CrossRef] [PubMed]
- Borges, B.C.; Uehara, I.A.; Dias, L.O.; Brígido, P.C.; da Silva, C.V.; Silva, M.J. Mechanisms of infectivity and evasion derived from microvesicles cargo produced by trypanosoma cruzi. Front. Cell. Infect. Microbiol. 2016, 6, 161. [Google Scholar] [CrossRef] [PubMed]
- Van der Heyde, H.C.; Gramaglia, I.; Combes, V.; George, T.C.; Grau, G.E. Flow cytometric analysis of microparticles. Methods Mol. Biol. 2011, 699, 337–354. [Google Scholar] [PubMed]
- Affranchino, J.L.; Ibanez, C.F.; Luquetti, A.O.; Rassi, A.; Reyes, M.B.; Macina, R.A.; Aslund, L.; Pettersson, U.; Frasch, A.C. Identification of a trypanosoma cruzi antigen that is shed during the acute phase of chagas’ disease. Mol. Biochem. Parasitol. 1989, 34, 221–228. [Google Scholar] [CrossRef]
- Goncalves, M.F.; Umezawa, E.S.; Katzin, A.M.; de Souza, W.; Alves, M.J.; Zingales, B.; Colli, W. Trypanosoma cruzi: Shedding of surface antigens as membrane vesicles. Exp. Parasitol. 1991, 72, 43–53. [Google Scholar] [CrossRef]
- Umezawa, E.S.; Shikanai-Yasuda, M.A.; Stolf, A.M. Changes in isotype composition and antigen recognition of anti-trypanosoma cruzi antibodies from acute to chronic chagas disease. J. Clin. Lab. Anal. 1996, 10, 407–413. [Google Scholar] [CrossRef]
- Jazin, E.E.; Bontempi, E.J.; Sanchez, D.O.; Aslund, L.; Henriksson, J.; Frasch, A.C.; Pettersson, U. Trypanosoma cruzi exoantigen is a member of a 160 kda gene family. Parasitology 1995, 110, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Mack, M.; Kleinschmidt, A.; Bruhl, H.; Klier, C.; Nelson, P.J.; Cihak, J.; Plachy, J.; Stangassinger, M.; Erfle, V.; Schlondorff, D. Transfer of the chemokine receptor ccr5 between cells by membrane-derived microparticles: A mechanism for cellular human immunodeficiency virus 1 infection. Nat. Med. 2000, 6, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Mantel, P.Y.; Marti, M. The role of extracellular vesicles in plasmodium and other protozoan parasites. Cell. Microbiol. 2014, 16, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Pinho, R.T.; Vannier-Santos, M.A.; Alves, C.R.; Marino, A.P.; Castello Branco, L.R.; Lannes-Vieira, J. Effect of trypanosoma cruzi released antigens binding to non-infected cells on anti-parasite antibody recognition and expression of extracellular matrix components. Acta Trop. 2002, 83, 103–115. [Google Scholar] [CrossRef]
- Trocoli Torrecilhas, A.C.; Tonelli, R.R.; Pavanelli, W.R.; da Silva, J.S.; Schumacher, R.I.; de Souza, W.; Silva, N.C.; de Almeida Abrahamsohn, I.; Colli, W.; Manso Alves, M.J. Trypanosoma cruzi: Parasite shed vesicles increase heart parasitism and generate an intense inflammatory response. Microbes Infect. 2009, 11, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Cestari, I.; Ramirez, M.I. Inefficient complement system clearance of trypanosoma cruzi metacyclic trypomastigotes enables resistant strains to invade eukaryotic cells. PLoS ONE 2010, 5, e9721. [Google Scholar] [CrossRef] [PubMed]
- Bayer-Santos, E.; Aguilar-Bonavides, C.; Rodrigues, S.P.; Cordero, E.M.; Marques, A.F.; Varela-Ramirez, A.; Choi, H.; Yoshida, N.; da Silveira, J.F.; Almeida, I.C. Proteomic analysis of trypanosoma cruzi secretome: Characterization of two populations of extracellular vesicles and soluble proteins. J. Proteome Res. 2013, 12, 883–897. [Google Scholar] [CrossRef] [PubMed]
- Overbye, A.; Skotland, T.; Koehler, C.J.; Thiede, B.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget 2015, 6, 30357–30376. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.L.; Nakayasu, E.S.; Oliveira, D.L.; Nimrichter, L.; Nosanchuk, J.D.; Almeida, I.C.; Casadevall, A. Extracellular vesicles produced by cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell 2008, 7, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Kooijmans, S.A.; Vader, P.; van Dommelen, S.M.; van Solinge, W.W.; Schiffelers, R.M. Exosome mimetics: A novel class of drug delivery systems. Int. J. Nanomed. 2012, 7, 1525–1541. [Google Scholar]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mrnas and micrornas is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
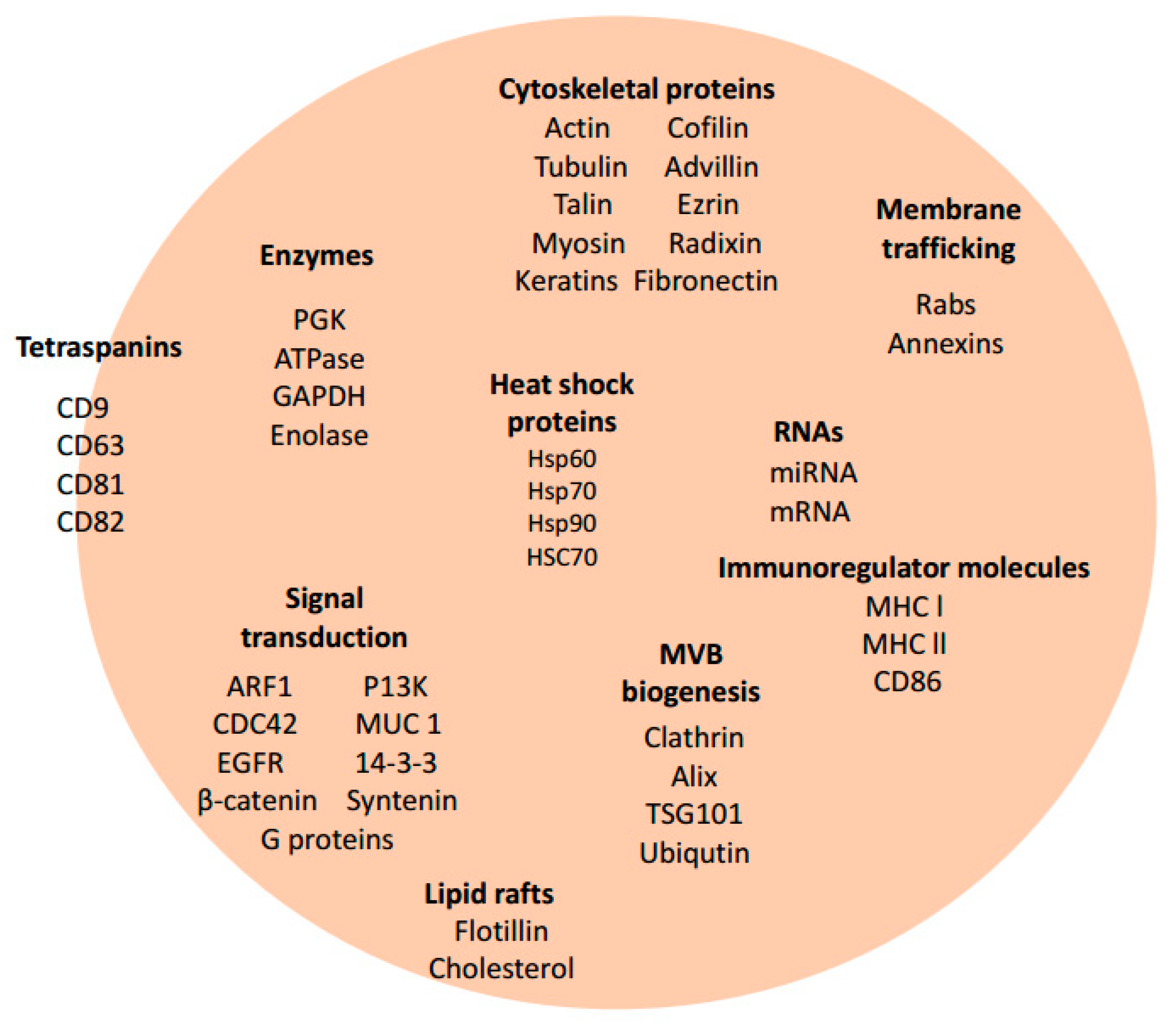
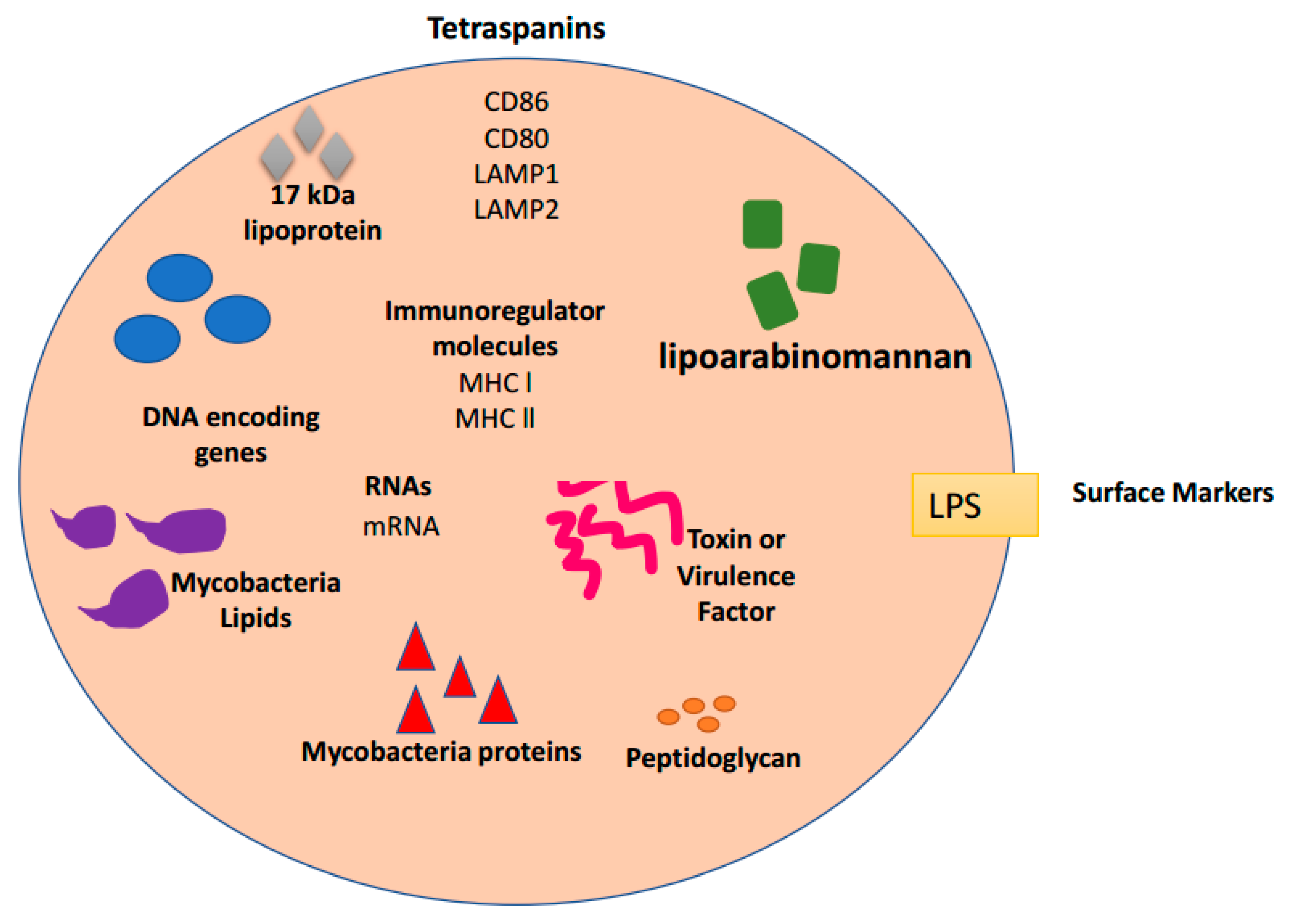
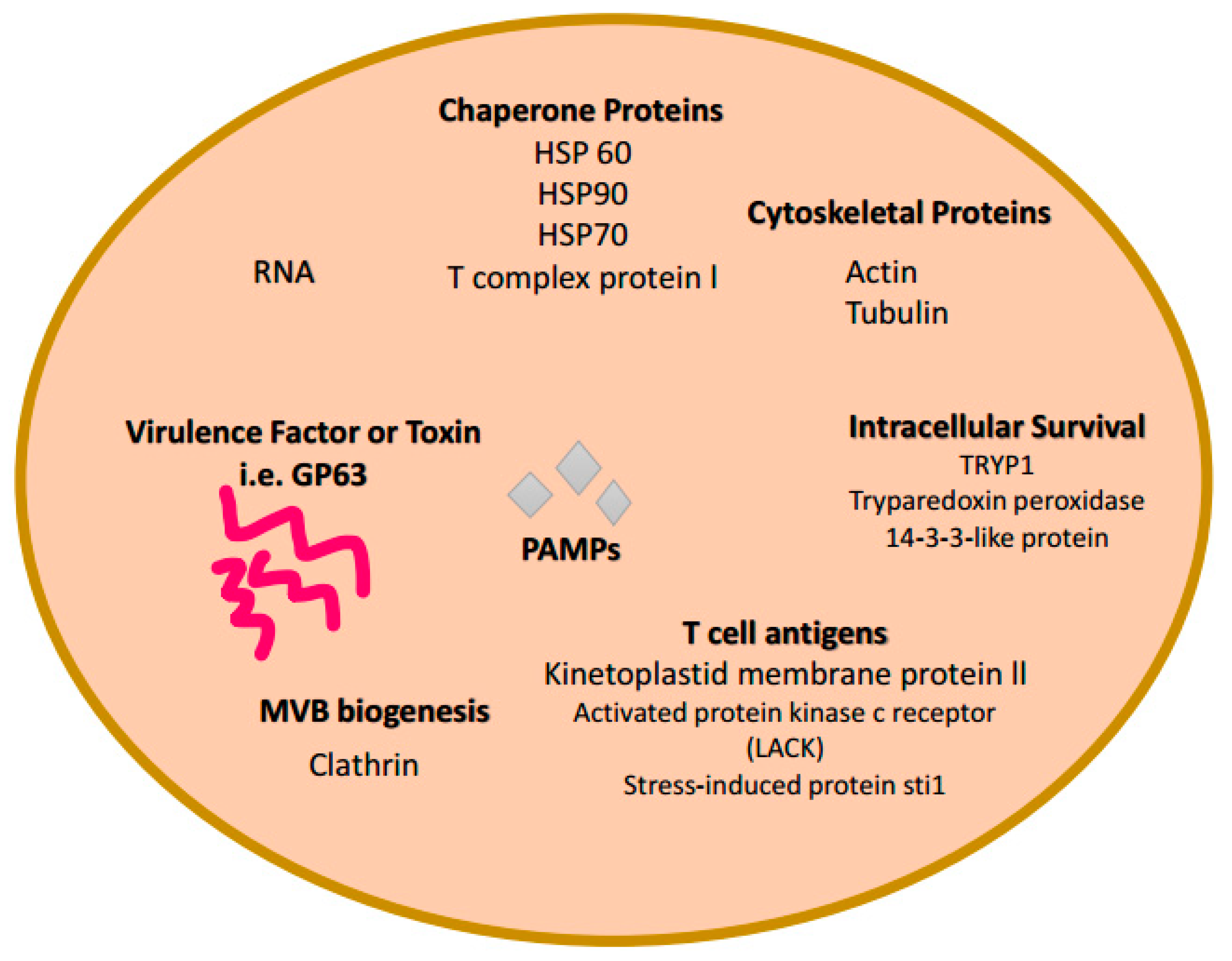
| Pathogen | Exosome Host | Role in Infection | References |
|---|---|---|---|
| Leishmania donovani | Macrophages Monoctyes (after IFN-γ treatment) Dendritic cells | • Suppression of immune system • Stimulation of IL-10, and suppression of IL-8 and TNF-α • Pro and anti-inflammatory response | [2,14] |
| Trypanosoma cruzi | Macrophages | • Evasion of host immune response • Division of exosomes • Apoptosis resistance | [15] |
| Mycobacterium tuberculosis | Macrophages, plasma | • Pro-inflammatory response | [16] |
| Mycobacterium bovis | Macrophages | • Pro-inflammatory response | [16] |
| Mycobacterium avium | Macrophages | • Pro-inflammatory response | [11] |
| Bacillus anthracis | Retinal pigment epithelial cells | • Transfer of lethal factor to uninfected cells | [17] |
| Cryptococcus neoformans | Macrophages | • Pro-inflammatory response | [18] |
| Klebsiella pneumoniae | Epithelial cells | • Pro-inflammatory response | [19] |
| Toxoplasma gondii | TAg-pulsed-DC2.4 Macrophages | • TH1 immune response • JNK Pathway activation | [20,21] |
| Plasmodium yoelii | Reticulocytes, Plasma | • Immune response modulation • Reticulocytosis • Change of cell tropism | [2,22] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jones, L.B.; Bell, C.R.; Bibb, K.E.; Gu, L.; Coats, M.T.; Matthews, Q.L. Pathogens and Their Effect on Exosome Biogenesis and Composition. Biomedicines 2018, 6, 79. https://doi.org/10.3390/biomedicines6030079
Jones LB, Bell CR, Bibb KE, Gu L, Coats MT, Matthews QL. Pathogens and Their Effect on Exosome Biogenesis and Composition. Biomedicines. 2018; 6(3):79. https://doi.org/10.3390/biomedicines6030079
Chicago/Turabian StyleJones, Leandra B., Courtnee’ R. Bell, Kartz E. Bibb, Linlin Gu, Mamie T. Coats, and Qiana L. Matthews. 2018. "Pathogens and Their Effect on Exosome Biogenesis and Composition" Biomedicines 6, no. 3: 79. https://doi.org/10.3390/biomedicines6030079
APA StyleJones, L. B., Bell, C. R., Bibb, K. E., Gu, L., Coats, M. T., & Matthews, Q. L. (2018). Pathogens and Their Effect on Exosome Biogenesis and Composition. Biomedicines, 6(3), 79. https://doi.org/10.3390/biomedicines6030079