Potential Impact of Microbial Dysbiosis and Tryptophan Metabolites in Advanced Stages of Colorectal Cancer
Abstract
1. Introduction
2. Materials and Methods
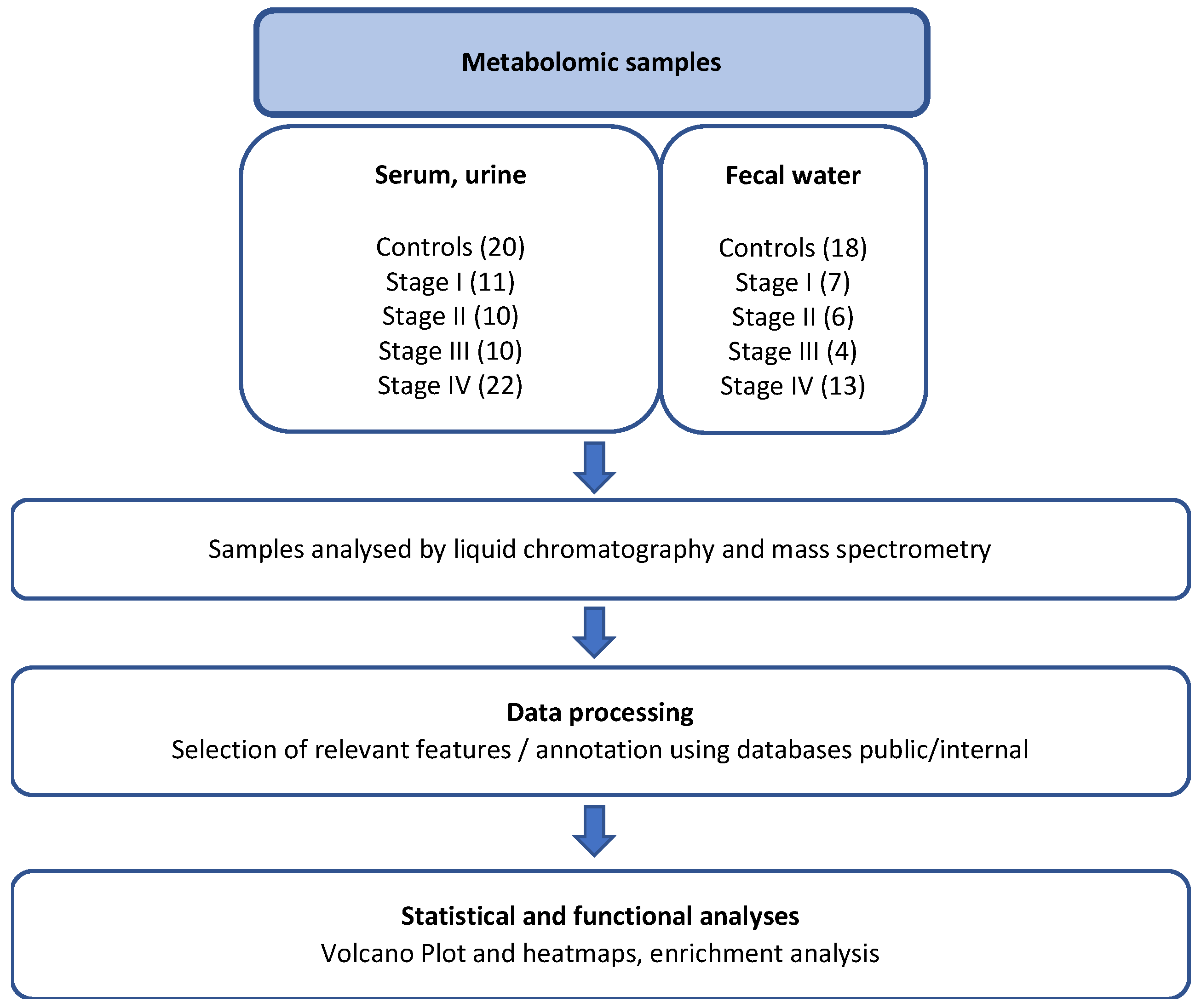
2.1. Study Patient and Sample Collections
2.2. Metabolomics Analyses
2.3. Data Processing and Bioinformatic Analyses of Metabolites
2.4. Microbiota Analysis
2.5. Quantification of Inflammatory Biomarkers and Statistical Analysis
3. Results
3.1. Characteristics of Individuals
3.2. Metabolomic Profiling in CRC Patients
3.3. In-Depth Analysis of Amino Acid Metabolism in Stage IV CRC Patients
3.4. Biomarkers of Inflammation
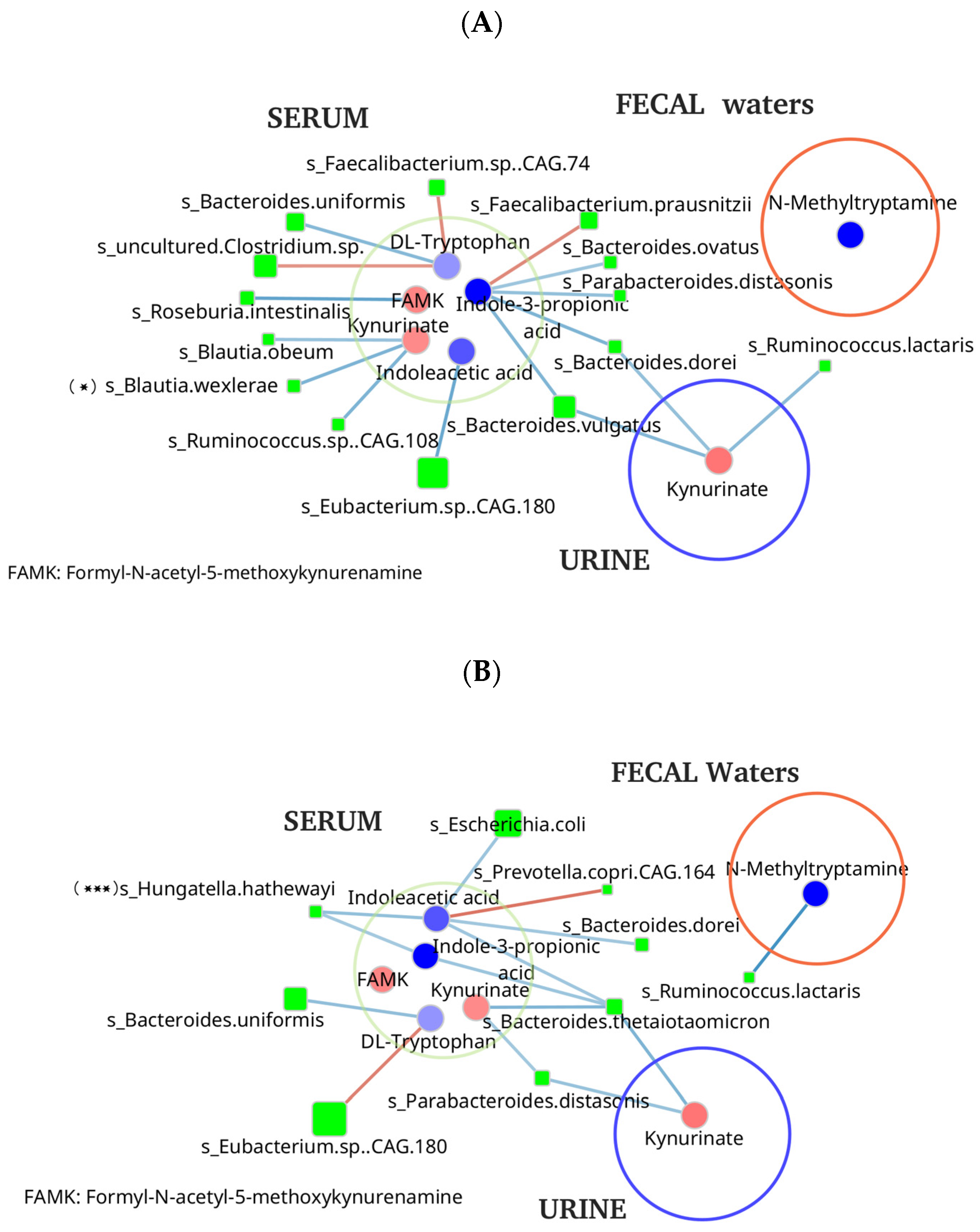
3.5. Microbial Metabolites and Stool Microbiome Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRC | Colorectal cancer |
| IL | Interleukin |
| Trp | Tryptophan |
| Kyn | Kynurenin |
| CRP | C-Reactive Protein |
| TNF | Tumor Nuclear Factor |
| IDO1 | Indoleamine 2,3 dioxygenase 1 |
| AhR | Aryl hydrocarbon Receptor |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Stoffel, E.M.; Murphy, C.C. Epidemiology and Mechanisms of the Increasing Incidence of Colon and Rectal Cancers in Young Adults. Gastroenterology 2020, 158, 341–353. [Google Scholar] [CrossRef]
- Flom, E.; Schultz, K.S.; Pantel, H.J.; Leeds, I.L. The Predictors of Complete Pathologic Response in Rectal Cancer during the Total Neoadjuvant Therapy Era: A Systematic Review. Cancers 2023, 15, 5853. [Google Scholar] [CrossRef]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, J.; Wang, L. Role and Mechanism of Gut Microbiota in Human Disease. Front. Cell Infect. Microbiol. 2021, 11, 625913. [Google Scholar] [CrossRef]
- Sobhani, I.; Rotkopf, H.; Khazaie, K. Bacteria-Related Changes in Host DNA Methylation and the Risk for CRC. Gut Microbes 2020, 12, 1800898. [Google Scholar] [CrossRef]
- O’Keefe, S.J.D.; Li, J.V.; Lahti, L.; Ou, J.; Carbonero, F.; Mohammed, K.; Posma, J.M.; Kinross, J.; Wahl, E.; Ruder, E.; et al. Fat, Fibre and Cancer Risk in African Americans and Rural Africans. Nat. Commun. 2015, 6, 6342. [Google Scholar] [CrossRef]
- De Oliveira Alves, N.; Dalmasso, N.; Nikitina, D.; Vaysse, A.; Ruez, R.; Ledoux, L.; Pedron, T.; Bergsten, E.; Boulard, O.; Autier, L.; et al. The Colibactin-Producing Escherichia Coli Alters the Tumor Microenvironment to Immunosuppressive Lipid Overload Facilitating Colorectal Cancer Progression and Chemoresistance. Gut Microbes 2024, 16, 2320291. [Google Scholar] [CrossRef] [PubMed]
- Zackular, J.P.; Rogers, M.A.M.; Ruffin, M.T.; Schloss, P.D. The Human Gut Microbiome as a Screening Tool for Colorectal Cancer. Cancer Prev. Res. 2014, 7, 1112–1121. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Du, M.; Schoen, R.E.; Hoffmeister, M.; Newcomb, P.A.; Berndt, S.I.; Caan, B.; Campbell, P.T.; Chan, A.T.; Chang-Claude, J.; et al. Determining Risk of Colorectal Cancer and Starting Age of Screening Based on Lifestyle, Environmental, and Genetic Factors. Gastroenterology 2018, 154, 2152–2164.e19. [Google Scholar] [CrossRef]
- Schmitt, M.; Greten, F.R. The Inflammatory Pathogenesis of Colorectal Cancer. Nat. Rev. Immunol. 2021, 21, 653–667. [Google Scholar] [CrossRef] [PubMed]
- Dalal, N.; Jalandra, R.; Bayal, N.; Yadav, A.K.; Harshulika, A.; Sharma, M.; Makharia, G.K.; Kumar, P.; Singh, R.; Solanki, P.R.; et al. Gut Microbiota-Derived Metabolites in CRC Progression and Causation. J. Cancer Res. Clin. Oncol. 2021, 147, 3141–3155. [Google Scholar] [CrossRef]
- Wu, J.; Wu, M.; Wu, Q. Identification of Potential Metabolite Markers for Colon Cancer and Rectal Cancer Using Serum Metabolomics. J. Clin. Lab. Anal. 2020, 34, e23333. [Google Scholar] [CrossRef]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R.; et al. Meta-Analysis of Fecal Metagenomes Reveals Global Microbial Signatures That Are Specific for Colorectal Cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, I.; Bergsten, E.; Couffin, S.; Amiot, A.; Nebbad, B.; Barau, C.; de’Angelis, N.; Rabot, S.; Canoui-Poitrine, F.; Mestivier, D.; et al. Colorectal Cancer-Associated Microbiota contributes to Oncogenic Epigenetic Signatures. Proc. Natl. Acad. Sci. USA 2019, 116, 24285–24295. [Google Scholar] [CrossRef]
- Sobhani, I.; Tap, J.; Roudot-Thoraval, F.; Roperch, J.P.; Letulle, S.; Langella, P.; Corthier, G.; Tran Van Nhieu, J.; Furet, J.P. Microbial Dysbiosis in Colorectal Cancer CRC) Patients. PLoS ONE 2011, 6, e16393. [Google Scholar] [CrossRef]
- Sadeghi, M.; Mestivier, D.; Carbonnelle, E.; Benamouzig, R.; Khazaie, K.; Sobhani, I. Loss of Symbiotic and Increase of Virulent Bacteria through Microbial Networks in Lynch Syndrome Colon Carcinogenesis. Front. Oncol. 2024, 13, 1313735. [Google Scholar] [CrossRef] [PubMed]
- Giacomoni, F.; Le Corguillé, G.; Monsoor, M.; Landi, M.; Pericard, P.; Pétéra, M.; Duperier, C.; Tremblay-Franco, M.; Martin, J.F.; Jacob, D.; et al. Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinformatics 2015, 31, 1493–1495. [Google Scholar] [CrossRef]
- Boudah, S.; Olivier, M.F.; Aros-Calt, S.; Oliveira, L.; Fenaille, F.; Tabet, J.C.; Junot, C. Annotation of the Human Serum Metabo lome by Coupling Three Liquid Chromatography Methods to High-Resolution Mass Spectrometry. J. Chromatogr. B Analyt Technol. Biomed. Life Sci. 2014, 966, 34–47. [Google Scholar] [CrossRef]
- Wieczorek, S.; Combes, F.; Lazar, C.; Giai Gianetto, Q.; Gatto, L.; Dorffer, A.; Hesse, A.-M.; Couté, Y.; Ferro, M.; Bruley, C.; et al. DAPAR & ProStaR: Software to Perform Statistical Analyses in Quantitative Discovery Proteomics. Bioinformatics 2017, 33, 135–136. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Giai Gianetto, Q.; Combes, F.; Ramus, C.; Bruley, C.; Couté, Y.; Burger, T. Calibration Plot for Proteomics: A Graphical Tool to Visually Check the Assumptions Underlying FDR Control in Quantitative Experiments. Proteomics 2016, 16, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Pounds, S.; Cheng, C. Robust Estimation of the False Discovery Rate. Bioinformatics 2006, 22, 1979–1987. [Google Scholar] [CrossRef]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A Web Server for Metabolomic Data Analysis and Interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef]
- Zeller, G.; Tap, J.; Voigt, A.Y.; Sunagawa, S.; Kultima, J.R.; Costea, P.I.; Amiot, A.; Böhm, J.; Brunetti, F.; Habermann, N.; et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014, 10, 766. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN analysis of metagenomic data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Rea, I.M.; Gibson, D.S.; McGilligan, V.; McNerlan, S.E.; Alexander, H.D.; Ross, O.A. Age and Age-Related Diseases: Role of Inflammation Triggers and Cytokines. Front. Immunol. 2018, 9, 586. [Google Scholar] [CrossRef]
- Nishiumi, S.; Kobayashi, T.; Ikeda, A.; Yoshie, T.; Kibi, M.; Izumi, Y.; Okuno, T.; Hayashi, N.; Kawano, S.; Takenawa, T.; et al. A Novel Serum Metabolomics-Based Diagnostic Approach for Colorectal Cancer. PLoS ONE 2012, 7, e40459. [Google Scholar] [CrossRef]
- Deng, X.; Li, Q.B.; Lu, Y.H.; He, N.; Jiang, J. Genetic Engineering of E. coli SE5000 and Its Potential for Ni2+ Bioremediation. Process Biochem. 2005, 40, 425–430. [Google Scholar] [CrossRef]
- Lin, Y.; Ma, C.; Bezabeh, T.; Wang, Z.; Liang, J.; Huang, Y.; Zhao, J.; Liu, X.; Ye, W.; Tang, W.; et al. 1 H NMR-Based Metabolomics Reveal Overlapping Discriminatory Metabolites and Metabolic Pathway Disturbances between Colorectal Tumor Tissues and Fecal Samples. Int. J. Cancer 2019, 145, 1679–1689. [Google Scholar] [CrossRef]
- Palego, L.; Betti, L.; Rossi, A.; Giannaccini, G. Tryptophan Biochemistry: Structural, Nutritional, Metabolic, and Medical Aspects in Humans. J. Amino Acids 2016, 2016, 8952520. [Google Scholar] [CrossRef] [PubMed]
- Ayabe, R.I.; White, M.G. Metastasis and the Microbiome: The Impact of Bacteria in Disseminated Colorectal Cancer. Front. Biosci. (Landmark Ed.) 2024, 29, 152. [Google Scholar] [CrossRef]
- Marsland, B.J. Regulating Inflammation with Microbial Metabolites. Nat. Med. 2016, 22, 581–583. [Google Scholar] [CrossRef]
- Santhanam, S.; Alvarado, D.M.; Ciorba, M.A. Therapeutic Targeting of Inflammation and Tryptophan Metabolism in Colon and Gastrointestinal Cancer. Transl. Res. 2016, 167, 67–79. [Google Scholar] [CrossRef]
- Zhang, X.; Browman, G.; Siu, W.; Basen-Engquist, K.M.; Hanash, S.M.; Hoffman, K.L.; Okhuysen, P.C.; Scheet, P.; Petrosino, J.F.; Kopetz, S.; et al. The BE GONE Trial Study Protocol: A Randomized Crossover Dietary Intervention of Dry Beans Targeting the Gut Microbiome of Overweight and Obese Patients with a History of Colorectal Polyps or Cancer. BMC Cancer 2019, 19, 1233. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tong, X.; Zou, Y.; Lin, X.; Zhao, H.; Tian, L.; Jie, Z.; Wang, Q.; Zhang, Z.; Lu, H.; et al. Mendelian Randomization Analyses Support Causal Relationships between Blood Metabolites and the Gut Microbiome. Nat. Genet. 2022, 54, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Fong, W.; Li, Q.; Ji, F.; Liang, W.; Lau, H.C.H.; Kang, X.; Liu, W.; To, K.K.-W.; Zuo, Z.; Li, X.; et al. Lactobacillus gallinarum-derived metabolites boost anti-PD1 efficacy in colorectal cancer by inhibiting regulatory T cells through modulating IDO1/Kyn/AHR axis. Gut 2023, 72, 2272–2285. [Google Scholar] [CrossRef]
- Riazati, N.; Kable, M.E.; Newman, J.W.; Adkins, Y.; Freytag, T.; Jiang, X.; Stephensen, C.B. Associations of Microbial and Indoleamine-2,3-Dioxygenase-Derived Tryptophan Metabolites with Immune Activation in Healthy Adults. Front. Immunol. 2022, 13, 917966. [Google Scholar] [CrossRef]
- Hatcher, C.; Richenberg, G.; Waterson, S.; Nguyen, L.H.; Joshi, A.D.; Carreras-Torres, R.; Moreno, V.; Chan, A.T.; Gunter, M.; Lin, Y.; et al. Application of Mendelian Randomization to Explore the Causal Role of the Human Gut Microbiome in Colorectal Cancer. Sci. Rep. 2023, 13, 5968. [Google Scholar] [CrossRef] [PubMed]
- Alexeev, E.E.; Lanis, J.M.; Kao, D.J.; Campbell, E.L.; Kelly, C.J.; Battista, K.D.; Gerich, M.E.; Jenkins, B.R.; Walk, S.T.; Kominsky, D.J.; et al. Microbiota-Derived Indole Metabolites Promote Human and Murine Intestinal Homeostasis through Regulation of Interleukin-10 Receptor. Am. J. Pathol. 2018, 188, 1183–1194. [Google Scholar] [CrossRef]
- Yan, B.; Mao, X.; Hu, S.; Wang, S.; Liu, X.; Sun, J. Spermidine protects intestinal mucosal barrier function in mice colitis via the AhR/Nrf2 and AhR/STAT3 signaling pathways. Int. Immunopharmacol. 2023, 119, 110166. [Google Scholar] [CrossRef]
- Oda, K.; Wlodawer, A. Overview of the Properties of Glutamic Peptidases That Are Present in Plant and Bacterial Pathogens and Play a Role in Celiac Disease and Cancer. Biochemistry 2023, 62, 672–694. [Google Scholar] [CrossRef]
- Damerell, V.; Klaassen-Dekker, N.; Brezina, S.; Ose, J.; Ulvik, A.; van Roekel, E.; Holowatyj, A.N.; Baierl, A.; Böhm, J.; Bours, M.J.L.; et al. Circulating tryptophan-kynurenine pathway metabolites are associated with all-cause mortality among patients with stage I–III colorectal cancer. Int. J. Cancer 2025, 156, 552–565. [Google Scholar] [CrossRef]
- Heaver, S.L.; Johnson, E.L.; Ley, R.E. Sphingolipids in Host-Microbial Interactions. Curr. Opin. Microbiol. 2018, 43, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Venkateswaran, N.; Conacci-Sorell, M. Kynurenine: An oncometabolite in colon cancer. Cell Stress 2020, 4, 24–26. [Google Scholar] [CrossRef] [PubMed]


| Characteristic | Control | CRC Patients | p Value |
|---|---|---|---|
| Individuals, n | 20 | 53 | |
| Gender (n, %) | |||
| Female | 13 (65%) | 24 (45%) | |
| Male | 7 (35%) | 29 (55%) | |
| Age (yr) | 59 | 67 | 0.005 |
| (extremes) | (35–77) | (44–87) | |
| BMI (kg) | 25.5 (20–34) | 25.3 (15–40) | 0.618 |
| TNM staging (n, %) | 53 | ||
| I | - | 11 (21%) | |
| II | - | 10 (19%) | |
| III | - | 10 (19%) | |
| IV | - | 22 (41%) | |
| Tumor location (n, %) | Number (%) | ||
| Right colon | - | 21 (40%) | |
| Left colon | - | 21 (40%) | |
| Rectum | - | 11 (20%) | |
| Molecular markers in tumor tissues (n, %) | 2 (4%) | ||
| MSI | - | 45 (84%) | |
| WIF-1 Gene methylated | - | 22 (42%) | |
| KRAS mutated | - | 0 | |
| Braf mutated |
| Metabolites | Log2FC Cancer II/Controls | Log2FC Cancer III/Controls | Log2FC Cancer IV/Controls |
|---|---|---|---|
| Serum | |||
| Cotinine | 2.2873 | ||
| Ser-Gly-Thr/Ala-ser-ser/Thr-ser-Gly | 2.0674 | 2.2671 | 3.2037 |
| Acetaminophen | 3.2661 | ||
| Aspartylphenylalanine | 1.5630 | ||
| N-Acetyl-L-glutamic acid | 1.5179 | ||
| Eicosanoids | 1.4908 | ||
| beta-citryl-L-glutamate | 1.5133 | ||
| N8-Acetylspermidine | 1.2949 | ||
| 4-Hydroxy-2-quinolinecarboxylic acid (Kynurinate) | 2.9540 | 1.2238 | |
| Dglutamic acid | 1.1606 | ||
| Cholic acid | −2.6064 | −2.4377 | |
| 2,5-Furandicarboxylic acid | −2.2914 | −2.3831 | |
| gamma-glutamyl-Se-methylselenocysteine | −2.2428 | −2.6907 | |
| (9Z,11E)-(13S)-13-Hydroxyoctadeca-9,11-dienoic acid | −2.1660 | ||
| 13 carboxygamma tocopherol | −2.1075 | −2.1619 | −2.8619 |
| 2-OH-3(2,3,4-trimethoxyphenyl)propanoic acid | −3.1326 | ||
| 7-Methylxanthine/1-Methylxanthine/3-Methylxanthine | −2.7637 | ||
| Ethyl 2-(methyldithio)propionate | −2.4824 | ||
| Guanidinosuccinic acid | −2.2632 | ||
| 5-methyl-2furancarboxyaldehyde (5-methyl-2-furfural) | −2.1402 | ||
| Urine | |||
| Galactonic acid/Gluconic acid | 2.6300 | ||
| pcresol sulfate | 2.1906 | 1.2992 | |
| 4-Hydroxy-2-quinolinecarboxylic acid (Kynurinate) | 2.0819 | 1.4458 | |
| Sphingosine 1-phosphate | 2.7050 | ||
| Taurochenodeoxycholate 3/7-sulfate | 2.5328 | ||
| Sulfoglycolithocholate/sulfolithocholylglycine | 1.9500 | ||
| Taurolithocholic acid-3-sulfate | 1.9476 | ||
| N6-(L-1,3-Dicarboxypropyl)-L-lysine (Saccharopine) | 1.9295 | ||
| Glucosylgalactosyl hydroxylysine | 1.8288 | ||
| 5-Methoxyindoleacetate/Indolelactate | 1.7167 | ||
| L-Tartaric acid | −4.5342 | ||
| {[3-(2-hydroxy-4-methoxyphenyl)prop-2-en-1-yl]oxy}sulfonic acid ou [1-(4-methoxyphenyl)-3-oxopropan-2-yl]oxy}sulfonic acid | −3.4328 | −2.7478 | −2.9375 |
| N6-Methyladenine | −2.8901 | −1.6964 | |
| N-acetyl-L-arginine | −2.7452 | ||
| D-(+)-Neopterin | −2.6057 | −2.1259 | −2.5797 |
| N-Acetyl-L-glutamic acid | −2.5539 | −1.5816 | |
| Deoxyinosine | −2.5367 | −1.6937 | |
| L-Histidine | −2.5125 | ||
| 4-Phenoxybutyric acid/butanoic acid | −2.4893 | ||
| 4-Hydroxycoumarin/4-hydroxy-2H-chromen-2-one | −3.1658 | ||
| Fecal water | |||
| Acetylcysteine | −6.0085 | −5.4812 | |
| Hypoxanthine | −3.8516 | ||
| 13 carboxygamma tocopherol | −3.5300 | −4.8583 | |
| Citric acid | −2.3928 | ||
| N-Methyltryptamine | −5.3552 | ||
| N-(3-methylbutyl)-acetamide | −4.6979 | ||
| 2,3-dihydrobenzofuran | −3.3037 | ||
| DL-p-Hydroxyphenyllactic acid/Homovanillic acid | −3.0083 | ||
| Tyrosyl-Isoleucine | −3.0111 | ||
| 4(2-aminophenyl)-2,4 dioxobutanoic acid | −2.8709 | ||
| Valsartan | 8.8180 | ||
| Aspartylglutamate | 5.0664 | ||
| Acetyltaurine | 3.6545 | ||
| N-Acetylneuraminic acid | 3.4861 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Hulin, A.; Rifflet, A.; Castelli, F.; Giai Gianetto, Q.; Fenaille, F.; Aissat, A.; Matondo, M.; Fellahi, S.; Tournigand, C.; Junot, C.; et al. Potential Impact of Microbial Dysbiosis and Tryptophan Metabolites in Advanced Stages of Colorectal Cancer. Biomedicines 2026, 14, 26. https://doi.org/10.3390/biomedicines14010026
Hulin A, Rifflet A, Castelli F, Giai Gianetto Q, Fenaille F, Aissat A, Matondo M, Fellahi S, Tournigand C, Junot C, et al. Potential Impact of Microbial Dysbiosis and Tryptophan Metabolites in Advanced Stages of Colorectal Cancer. Biomedicines. 2026; 14(1):26. https://doi.org/10.3390/biomedicines14010026
Chicago/Turabian StyleHulin, Anne, Aline Rifflet, Florence Castelli, Quentin Giai Gianetto, François Fenaille, Abdel Aissat, Mariette Matondo, Soraya Fellahi, Christophe Tournigand, Christophe Junot, and et al. 2026. "Potential Impact of Microbial Dysbiosis and Tryptophan Metabolites in Advanced Stages of Colorectal Cancer" Biomedicines 14, no. 1: 26. https://doi.org/10.3390/biomedicines14010026
APA StyleHulin, A., Rifflet, A., Castelli, F., Giai Gianetto, Q., Fenaille, F., Aissat, A., Matondo, M., Fellahi, S., Tournigand, C., Junot, C., Sansonetti, P., Gomperts-Boneca, I., Mestivier, D., & Sobhani, I. (2026). Potential Impact of Microbial Dysbiosis and Tryptophan Metabolites in Advanced Stages of Colorectal Cancer. Biomedicines, 14(1), 26. https://doi.org/10.3390/biomedicines14010026







