The Prevalence of Diamine Oxidase Polymorphisms and Their Association with Histamine Intolerance Symptomatology in the Mexican Population
Abstract
1. Introduction
- (1)
- diamine oxidase (DAO), due to oxidative deamination in the case of extracellular histamines, and
- (2)
2. Materials and Methods
2.1. Study Design and Participants
2.2. Population and Biological Samples
2.3. Diamine Oxidase SNP Genotyping Analysis
2.4. Statistical Analysis
3. Results
3.1. Study Population
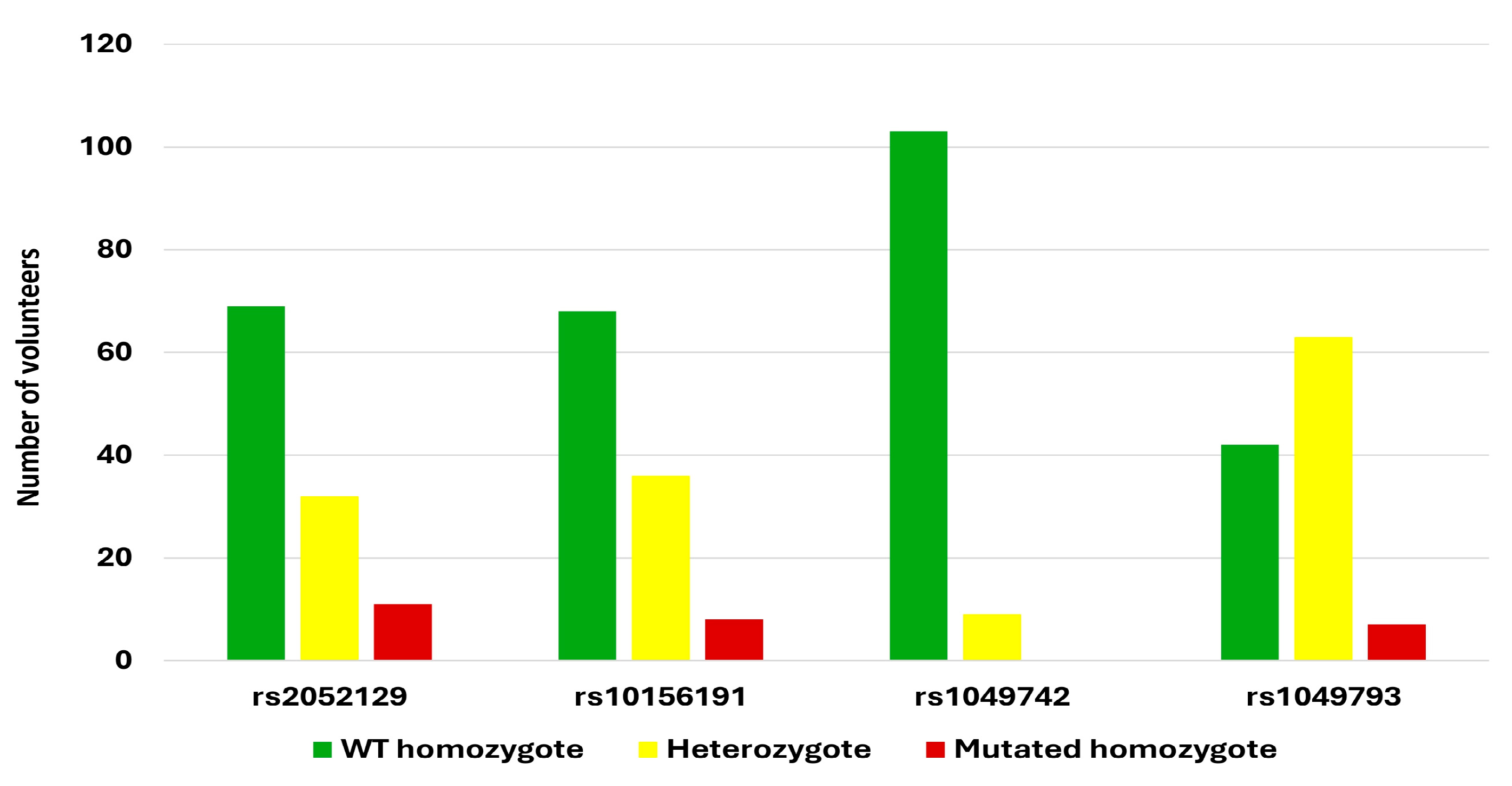
3.2. DAO SNP Genotyping
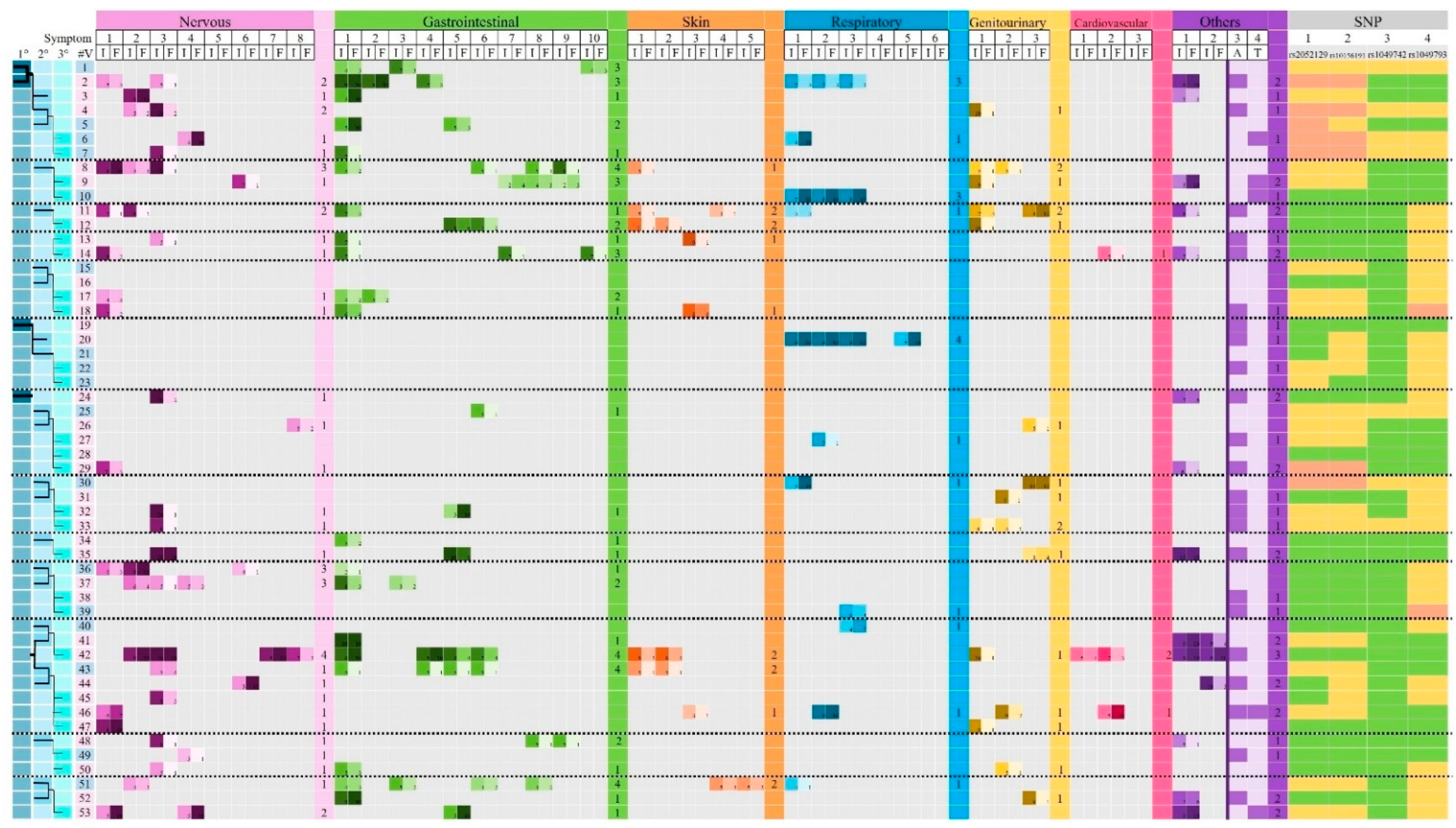
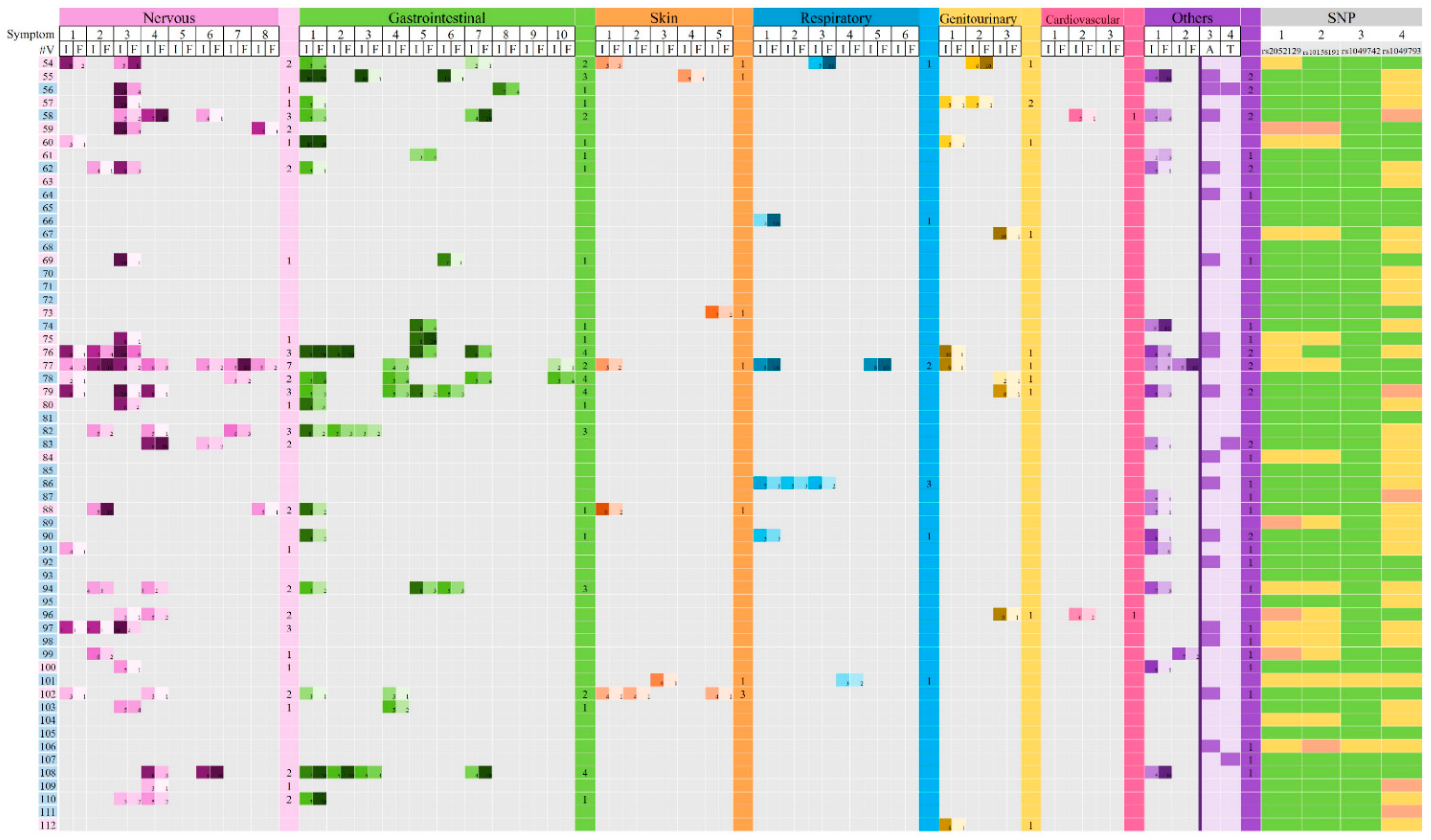
3.3. Association Between DAO SNPs and Clinical Manifestations Related to HIT
3.4. Statistical Analysis
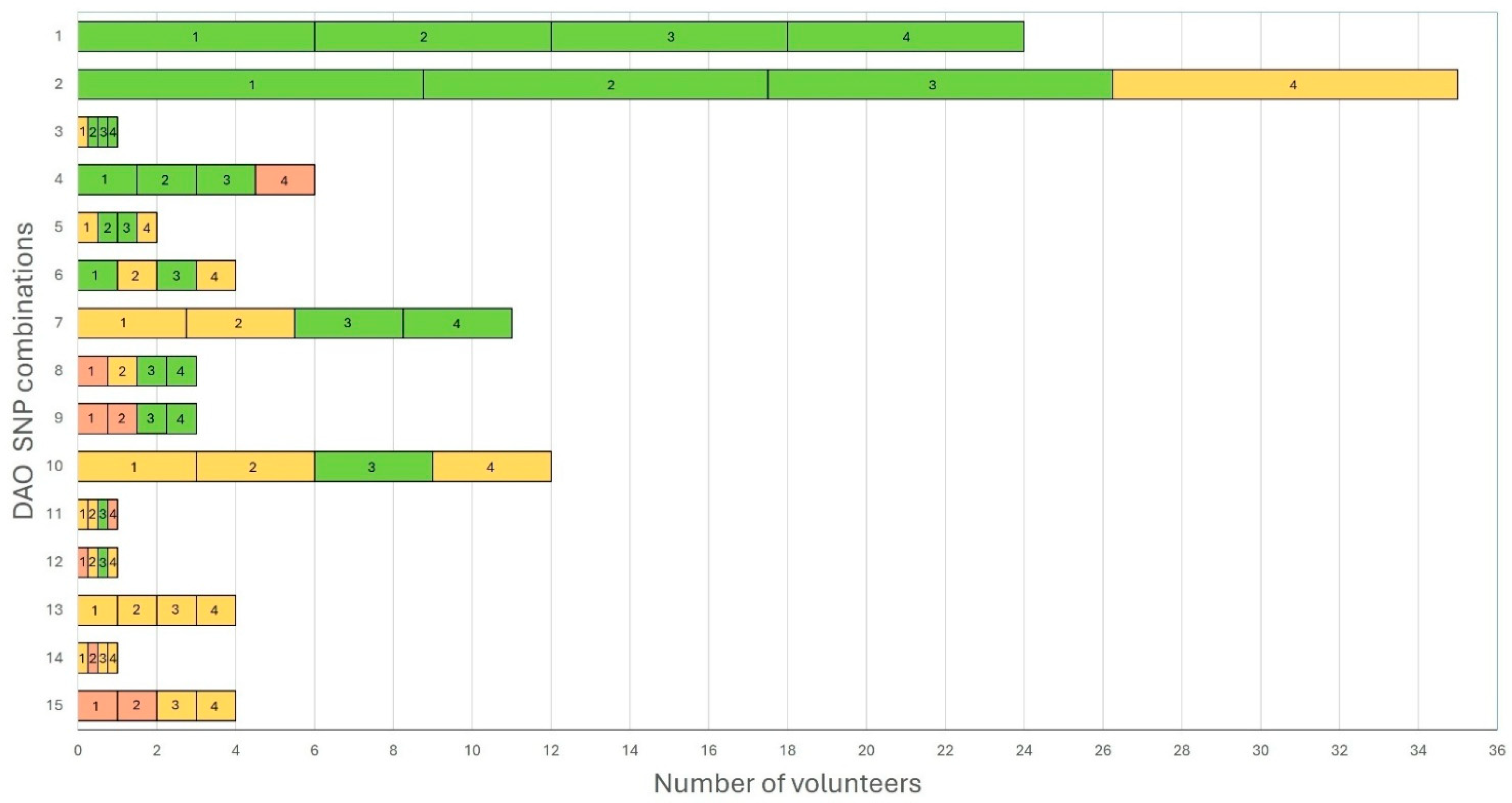
3.5. DAO Deficiency Genetic Risk Score
3.6. Comparison Between Family Groups Versus Individual Volunteers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Comas-Basté, O.; Sánchez-Pérez, S.; Veciana-Nogués, M.T.; Latorre-Moratalla, M.; Vidal-Carou, M.d.C. Histamine Intolerance: The Current State of the Art. Biomolecules 2020, 10, 1181. [Google Scholar] [CrossRef]
- Smolinska, S.; Winiarska, E.; Globinska, A.; Jutel, M. Histamine: A Mediator of Intestinal Disorders—A Review. Metabolites 2022, 12, 895. [Google Scholar] [CrossRef] [PubMed]
- Okutan, G.; Casares, E.R.; Alcalde, T.P.; Niño, G.M.S.; Penadés, B.F.; Lora, A.T.; Estríngana, L.T.; Oliva, S.L.; Martín, I.S.M. Prevalence of Genetic Diamine Oxidase (DAO) Deficiency in Female Patients with Fibromyalgia in Spain. Biomedicines 2023, 11, 660. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Ou, Y.; Cai, W.; Zheng, Y.; Li, H.; Mao, Y.; Zhou, S.; Tu, J. Advances in the Clinical Application of Histamine and Diamine Oxidase (DAO) Activity: A Review. Catalysts 2023, 13, 48. [Google Scholar] [CrossRef]
- Hrubisko, M.; Danis, R.; Huorka, M.; Wawruch, M. Histamine Intolerance—The More We Know the Less We Know. A Review. Nutrients 2021, 13, 2228. [Google Scholar] [CrossRef]
- Meza-Velázquez, R.; López-Márquez, F.; Espinosa-Padilla, S.; Rivera-Guillen, M.; Ávila-Hernández, J.; Rosales-González, M. Association of diamine oxidase and histamine N-methyltransferase polymorphisms with presence of migraine in a group of Mexican mothers of children with allergies. Neurologia 2017, 32, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Arih, K.; Đorđević, N.; Košnik, M.; Rijavec, M. Evaluation of Serum Diamine Oxidase as a Diagnostic Test for Histamine Intolerance. Nutrients 2023, 15, 4246. [Google Scholar] [CrossRef]
- Schnedl, W.J.; Enko, D. Histamine Intolerance Originates in the Gut. Nutrients 2021, 13, 1262. [Google Scholar] [CrossRef]
- Izquierdo-Casas, J.; Comas-Basté, O.; Latorre-Moratalla, M.L.; Lorente-Gascón, M.; Duelo, A.; Soler-Singla, L.; Vidal-Carou, M.C. Diamine oxidase (DAO) supplement reduces headache in episodic migraine patients with DAO deficiency: A randomized double-blind trial. Clin. Nutr. 2019, 38, 152–158. [Google Scholar] [CrossRef]
- Schnedl, W.J.; Lackner, S.; Enko, D.; Schenk, M.; Holasek, S.J.; Mangge, H. Evaluation of symptoms and symptom combinations in histamine intolerance. Intest. Res. 2019, 17, 427–433. [Google Scholar] [CrossRef]
- Maintz, L.; Novak, N. Histamine and histamine intolerance. Am. J. Clin. Nutr. 2007, 85, 1185–1196. [Google Scholar] [CrossRef] [PubMed]
- DAO Deficit International Society. August 2023. Available online: https://www.deficitdao.org/en/.
- Blasco-Fontecilla, H.; Bella-Fernández, M.; Wang, P.; Martin-Moratinos, M.; Li, C. Prevalence and Clinical Picture of Diamine Oxidase Gene Variants in Children and Adolescents with Attention Deficit Hyperactivity Disorder: A Pilot Study. J. Clin. Med. 2024, 13, 1659. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nazar, W.; Plata-Nazar, K.; Sznurkowska, K.; Szlagatys-Sidorkiewicz, A. Histamine Intolerance in Children: A Narrative Review. Nutrients 2021, 13, 1486. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, X.; Jin, H.; Chen, L.; Ji, J.; Zhang, Z. Histamine Intolerance—A Kind of Pseudoallergic Reaction. Biomolecules 2022, 12, 454. [Google Scholar] [CrossRef]
- Maintz, L.; Yu, C.-F.; Rodríguez, E.; Baurecht, H.; Bieber, T.; Illig, T.; Weidinger, S.; Novak, N. Association of single nucleotide polymorphisms in the diamine oxidase gene with diamine oxidase serum activities. Allergy 2011, 66, 893–902. [Google Scholar] [CrossRef]
- Karki, R.; Pandya, D.; Elston, R.C.; Ferlini, C. Defining “mutation” and “polymorphism” in the era of personal genomics. BMC Med Genom. 2015, 8, 37. [Google Scholar] [CrossRef]
- Shastry, B.S. SNPs: Impact on Gene Function and Phenotype. Methods Mol. Biol. 2009, 578, 3–22. [Google Scholar] [CrossRef]
- Ramírez-Vargas, S. Polimorfismos Genéticos de la Enzima Diamino Oxidasa en Población Mexicana; Estudio Descriptivo. Bachelor’s Thesis, Instituto Politécnico Nacional, Mexico City, Mexico, 2025. [Google Scholar]
- Mejía-Ramírez, V. Estandarización y Optimización de una Prueba Genética para la Determinación de Cuatro Polimorfismos de un solo Nucleótido del gen de la Diamino Oxidasa, Asociados con Predisposición Genética a Migraña. Bachelor’s Thesis, Universidad Nacional Autónoma de México, Mexico City, Mexico, 2025. [Google Scholar]
- GraphPad Software. GraphPad Prism, Version 10.0.0; Software for Windows: Boston, MA, USA, 2023. [Google Scholar]
- Duelo, A.; Comas-Basté, O.; Sánchez-Pérez, S.; Veciana-Nogués, M.T.; Ruiz-Casares, E.; Vidal-Carou, M.C.; Latorre-Moratalla, M.L. Pilot Study on the Prevalence of Diamine Oxidase Gene Variants in Patients with Symptoms of Histamine Intolerance. Nutrients 2024, 16, 1142. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Phan, L.; Zhang, H.; Wang, Q.; Villamarin, R.; Hefferon, T.; Ramanathan, A.; Kattman, B. The evolution of dbSNP: 25 years of impact in genomic research. Nucleic Acids Res. 2024, 53, D925–D931. [Google Scholar] [CrossRef]
- van Odijk, J.; Weisheit, A.; Arvidsson, M.; Miron, N.; Nwaru, B.; Ekerljung, L. The Use of DAO as a Marker for Histamine Intolerance: Measurements and Determinants in a Large Random Population-Based Survey. Nutrients 2023, 15, 2887. [Google Scholar] [CrossRef]
- Marin, E.F.; Marcolin, L.C.; Melero, L.M.; Gazulla, M.T.; Porres, M.B. The Prevalence of Single Nucleotide Polymorphisms of the AOC1 Gene Associated with Diamine Oxidase (DAO) Enzyme Deficiency in Healthy Newborns: A Prospective Population-Based Cohort Study. Genes 2025, 16, 141. [Google Scholar] [CrossRef]
- Ayuso, P.; García-Martín, E.; Martínez, C.; Agúndez, J.A.G. Genetic variability of human diamine oxidase: Occurrence of three nonsynonymous polymorphisms and study of their effect on serum enzyme activity. Pharmacogenetics Genom. 2007, 17, 687–693. [Google Scholar] [CrossRef]
- Hamada, Y.; Shinohara, Y.; Yano, M.; Yamamoto, M.; Yoshio, M.; Satake, K.; Toda, A.; Hirai, M.; Usami, M. Effect of the menstrual cycle on serum diamine oxidase levels in healthy women. Clin. Biochem. 2013, 46, 99–102. [Google Scholar] [CrossRef]
- Sánchez-Pérez, S.; Comas-Basté, O.; Duelo, A.; Veciana-Nogués, M.T.; Berlanga, M.; Latorre-Moratalla, M.L.; Vidal-Carou, M.C. Intestinal Dysbiosis in Patients with Histamine Intolerance. Nutrients 2022, 14, 1774. [Google Scholar] [CrossRef]
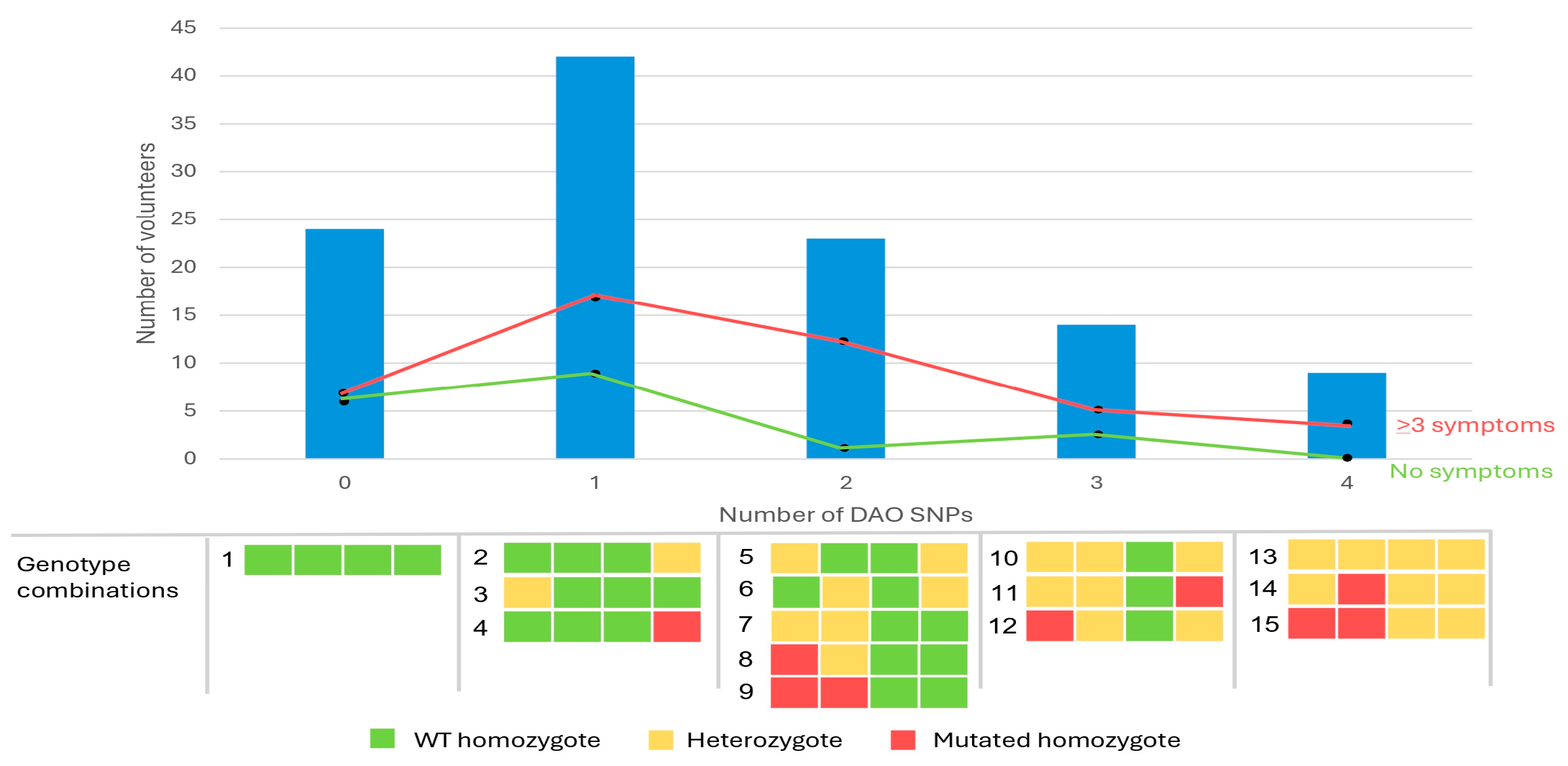
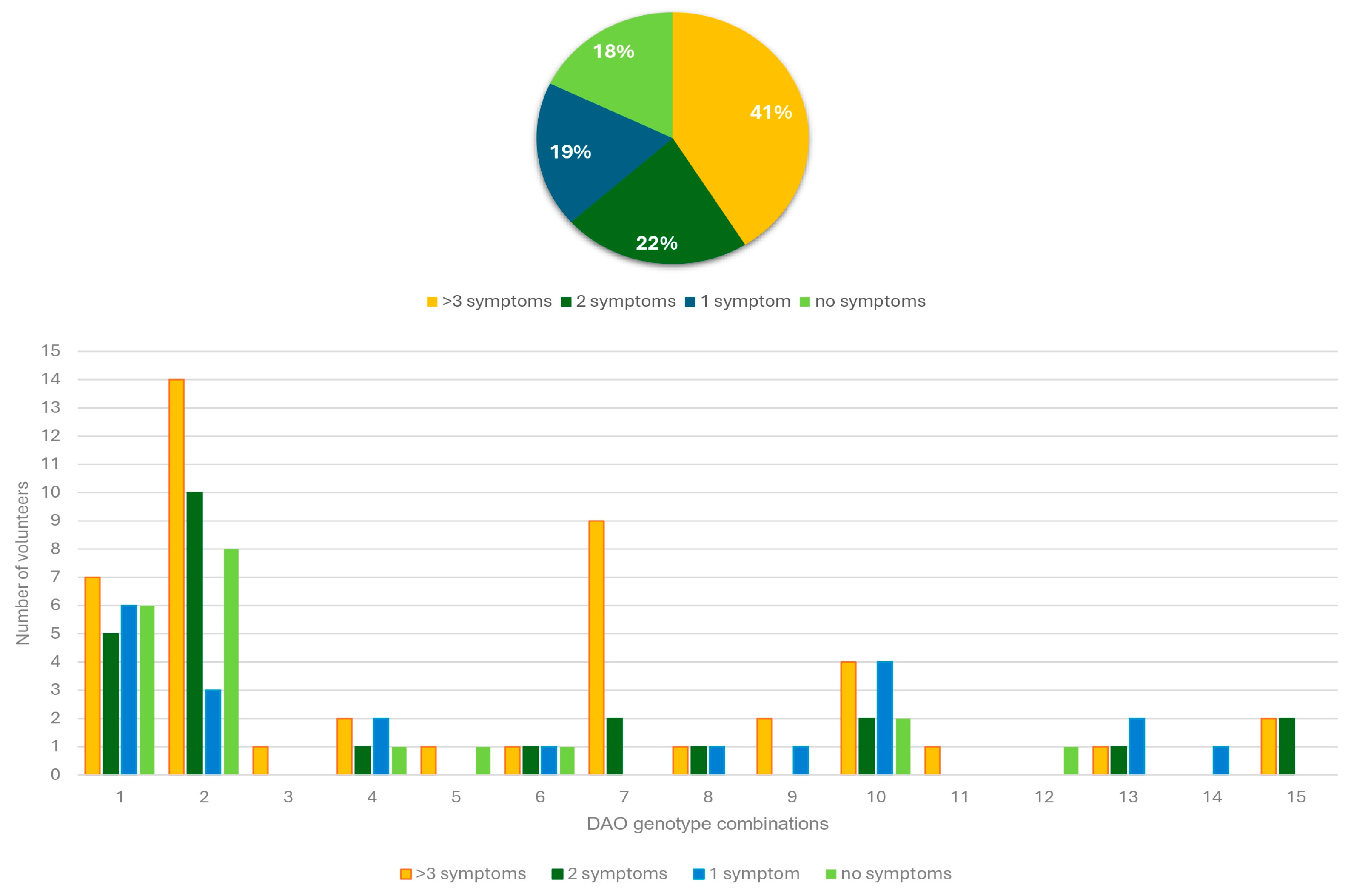

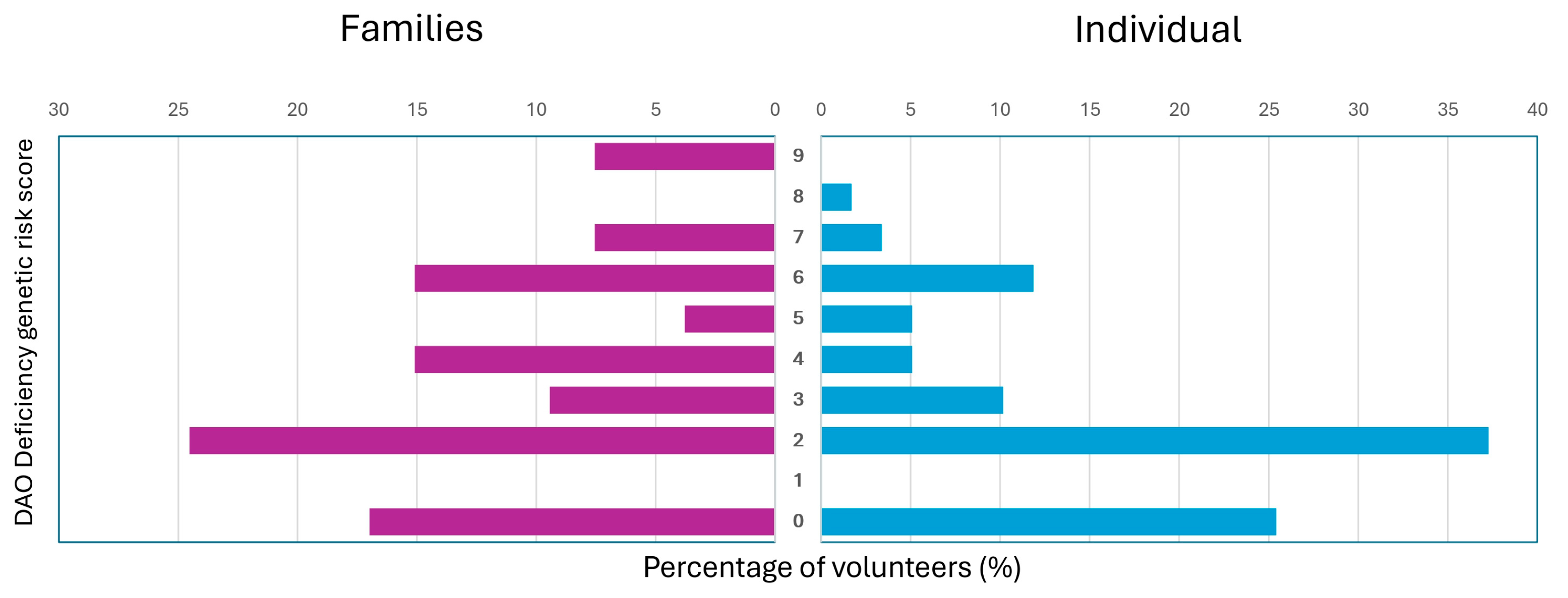
| SNP | Amino Acid Change | Polymorphism | Context Sequence [VIC/FAM] | |
|---|---|---|---|---|
| rs2052129 | N/A | G/T | Transversion substitution | CGCCCTTCTCAACCTGACTAAGTGG[G/T]CGTTAGCACTGTCCGCCCACTGCAT |
| rs10156191 | Thr16Met | C/T | Transition substitution | GTGGCTGCCATCCTGATGCTGCAGA[C/T]GGCCATGGCGGAGCCCTCCCCGGGG |
| rs1049742 | Ser332Phe | C/T | Transition substitution | TTTGCCTTCCGGCTGCGCTCCTCCT[C/T]CGGGCTGCAGGTCCTGAACGTGCAC |
| rs1049793 | His645Asp | C/G | Transversion substitution | CATCTACCACCAGAACGACCCCTGG[C/G]ACCCGCCCGTGGTCTTTGAGCAGTT |
| Regressors | Gastrointestinal System | Genitourinary System | ||
|---|---|---|---|---|
| Odds Ratio | p-Value | Odds Ratio | p-Value | |
| Age | 1.01 | 0.43 | 1.00 | 0.84 |
| Female | 3.05 | 0.01 | 37.41 | 0.01 |
| rs2052129 | 15.16 | 0.03 | 16.35 | 0.10 |
| rs10156191 | 0.09 | 0.05 | 0.07 | 0.13 |
| rs1049742 | 0.56 | 0.54 | 32.47 | 0.06 |
| rs1049793 | 1.15 | 0.77 | 0.67 | 0.63 |
| SNP | rs2052129 | rs10156191 | rs1049742 | rs1049793 |
|---|---|---|---|---|
| Global sample size | 580,272 | 452,454 | 501,300 | 114,592 |
| Reference allele | G = 0.7651 | C = 0.7299 | C = 0.9288 | C = 0.6805 |
| Altered allele | T = 0.2349 | T = 0.2701 | T = 0.0712 | G = 0.3195 |
| Latin American 1 | 8318 | 6478 | 8370 | 790 |
| Reference allele | G = 0.7795 | C = 0.6874 | C = 0.9397 | C = 0.622 |
| Altered allele | T = 0.2205 | T = 0.3126 | T = 0.0603 | G = 0.378 |
| Latin American 2 | 16,656 | 11,560 | 12,768 | 946 |
| Reference allele | G = 0.8403 | C = 0.8177 | C = 0.9656 | C = 0.614 |
| Altered allele | T = 0.1597 | T = 0.1824 | T = 0.0344 | G = 0.386 |
| This study/individual | 59 | 59 | 59 | 59 |
| Reference allele | G = 0.822 | C = 0.856 | C = 0.983 | C = 0.644 |
| Altered allele | T = 0.178 | T = 0.144 | T = 0.017 | G = 0.356 |
| Comparisons with Latin American 1 | p = 0.4324 | p = 0.0054 | p = 0.1628 | p = 0.7365 |
| Comparisons with Latin American 2 | p = 0.7017 | p = 0.4460 | p = 0.4639 | p = 0.6458 |
| This study/families | 53 | 53 | 53 | 53 |
| Reference allele | G = 0.689 | C = 0.670 | C = 0.934 | C = 0.670 |
| Altered allele | T = 0.311 | T = 0.330 | T = 0.066 | G = 0.330 |
| Comparisons with Latin American 1 | p = 0.1135 | p = 0.7855 | p = 0.8621 | p = 0.4847 |
| Comparisons with Latin American 2 | p = 0.0027 | p = 0.0056 | p = 0.2086 | p = 0.4144 |
| This study/total | 112 | 112 | 112 | 112 |
| Reference allele | G = 0.759 | C = 0.768 | C = 0.960 | C = 0.656 |
| Altered allele | T = 0.241 | T = 0.232 | T = 0.040 | G = 0.344 |
| Comparisons with the global sample | p = 0.879 | p = 0.364 | p = 0.199 | p = 0.578 |
| SNP | rs2052129 | rs10156191 | rs1049742 | rs1049793 |
|---|---|---|---|---|
| Global sample size | ||||
| WT allele | GG = 0.5854, 58.5% | CC = 0.5327, 53.3% | CC = 0.8627, 86.3% | CC = 0.4631, 46.3% |
| Heterozygote allele | GT = 0.3594, 35.9% | CT = 0.3942, 39.4% | CT = 0.1322, 13.2% | CG = 0.4350, 43.5% |
| Mutated allele | TT = 0.0552, 5.5% | TT = 0.0729, 7.3% | TT = 0.0051, 0.5% | GG = 0.1021, 10.2% |
| Chi-squared value (p-value) | 0.0078 (0.9295) | 0.0024 (0.9610) | 0.1115 (0.7385) | 0.0014 (0.9707) |
| Latin American 1 | ||||
| WT allele | GG = 0.6076, 60.8% | CC = 0.4725, 47.3% | CC = 0.8830, 88.3% | CC = 0.3868, 38.7% |
| Heterozygote allele | GT = 0.3437, 34.4% | CT = 0.4296, 43.0% | CT = 0.1133, 11.3% | CG = 0.4700, 47% |
| Mutated allele | TT = 0.0486, 4.9% | TT = 0.0977, 9.8% | TT = 0.0036, 0.4% | GG = 0.1429, 14.3% |
| Chi-squared value (p-value) | 0.0002 (0.9899) | 0.0001 (0.9904) | 0.0046 (0.9458) | 0.0010 (0.9744) |
| Latin American 2 | ||||
| WT allele | GG = 0.7061, 70.6% | CC = 0.6686, 66.9% | CC = 0.9324, 93.2% | CC = 0.3770, 37.7% |
| Heterozygote allele | GT = 0.2685, 26.9% | CT = 0.2983, 29.8% | CT = 0.0664, 6.6% | CG = 0.4740, 47.4% |
| Mutated allele | TT = 0.0255, 2.6% | TT = 0.0333, 3.3% | TT = 0.0012, 0.1% | GG = 0.1490, 14.9% |
| Chi-squared value (p-value) | 0.0000 (0.9965) | 0.0004 (0.9850) | 0.0006 (0.9800) | 0.0005 (0.9814) |
| This study/individual | ||||
| WT allele | GG = 0.6756, 67.6% | CC = 0.7327, 73.3% | CC = 0.9663, 96.6% | CC = 0.4147, 41.5% |
| Heterozygote allele | GT = 0.2924, 29.2% | CT = 0.2465, 24.7% | CT = 0.0334, 3.3% | CG = 0.4586, 45.9% |
| Mutated allele | TT = 0.0317, 3.2% | TT = 0.0207, 2.1% | TT = 0.0003, 0.03% | GG = 0.1267, 12.7% |
| Chi-squared value (p-value) | 0.0137 (0.9069) | 0.0562 (0.8126) | 0.0175 (0.8946) | 0.0199 (0.8877) |
| This study/families | ||||
| WT allele | GG = 0.474, 47.4% | CC = 0.4489, 44.9% | CC = 0.8724, 87.2% | CC = 0.4489, 44.9% |
| Heterozygote allele | GT = 0.428, 42.8% | CT = 0.4422, 44.2% | CT = 0.1233, 12.3% | CG = 0.4422, 44.2% |
| Mutated allele | TT = 0.097, 9.7% | TT = 0.1089, 10.9% | TT = 0.0044, 0.4% | GG = 0.1089, 10.9% |
| Chi-squared value (p-value) | 0.0077 (0.9302) | 0.0190 (0.8905) | 0.2650 (0.6067) | 0.0190 (0.8905) |
| This study/total | ||||
| WT allele | GG = 0.5761, 57.6% | CC = 0.5898, 59.0% | CC = 0.9216, 92.2% | CC = 0.4303, 43.0% |
| Heterozygote allele | GT = 0.3660, 36.6% | CT = 0.3564, 35.6% | CT = 0.0768, 7.7% | CG = 0.4513, 45.1% |
| Mutated allele | TT = 0.0581, 5.8% | TT = 0.0538, 5.4% | TT = 0.0016, 0.2% | GG = 0.1183, 11.8% |
| Chi-squared value (p-value) | 0.0247 (0.8752) | 0.0004 (0.9849) | 0.1963 (0.6578) | 0.0096 (0.9218) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguilar-Rodea, P.; Mejía-Ramírez, V.; Hernández-Munguía, R.; Ramírez-Vargas, S.; Tovar-Vivar, D.; Leyva-Hernández, J.; Nacar-Gutiérrez, J.C.; Morales-Martínez, M.; Palafox-Zaldivar, A. The Prevalence of Diamine Oxidase Polymorphisms and Their Association with Histamine Intolerance Symptomatology in the Mexican Population. Biomedicines 2025, 13, 2280. https://doi.org/10.3390/biomedicines13092280
Aguilar-Rodea P, Mejía-Ramírez V, Hernández-Munguía R, Ramírez-Vargas S, Tovar-Vivar D, Leyva-Hernández J, Nacar-Gutiérrez JC, Morales-Martínez M, Palafox-Zaldivar A. The Prevalence of Diamine Oxidase Polymorphisms and Their Association with Histamine Intolerance Symptomatology in the Mexican Population. Biomedicines. 2025; 13(9):2280. https://doi.org/10.3390/biomedicines13092280
Chicago/Turabian StyleAguilar-Rodea, Pamela, Viviana Mejía-Ramírez, Raúl Hernández-Munguía, Saúl Ramírez-Vargas, Diana Tovar-Vivar, Jaquelin Leyva-Hernández, Juan Carlos Nacar-Gutiérrez, Miriam Morales-Martínez, and Aracely Palafox-Zaldivar. 2025. "The Prevalence of Diamine Oxidase Polymorphisms and Their Association with Histamine Intolerance Symptomatology in the Mexican Population" Biomedicines 13, no. 9: 2280. https://doi.org/10.3390/biomedicines13092280
APA StyleAguilar-Rodea, P., Mejía-Ramírez, V., Hernández-Munguía, R., Ramírez-Vargas, S., Tovar-Vivar, D., Leyva-Hernández, J., Nacar-Gutiérrez, J. C., Morales-Martínez, M., & Palafox-Zaldivar, A. (2025). The Prevalence of Diamine Oxidase Polymorphisms and Their Association with Histamine Intolerance Symptomatology in the Mexican Population. Biomedicines, 13(9), 2280. https://doi.org/10.3390/biomedicines13092280





