A Hitchhiker Guide to Structural Variant Calling: A Comprehensive Benchmark Through Different Sequencing Technologies
Abstract
1. Introduction
2. Materials and Methods
2.1. Benchmarking Dataset
2.2. Illumina Short-Read Sequencing
2.3. Long-Read Sequencing
2.3.1. PacBio Long-Read Sequencing
2.3.2. Oxford Nanopore Long-Read Sequencing
2.4. Performance Evaluation
3. Results
3.1. Evaluation of Different Structural Variant Calling Algorithms for Illumina Short Reads
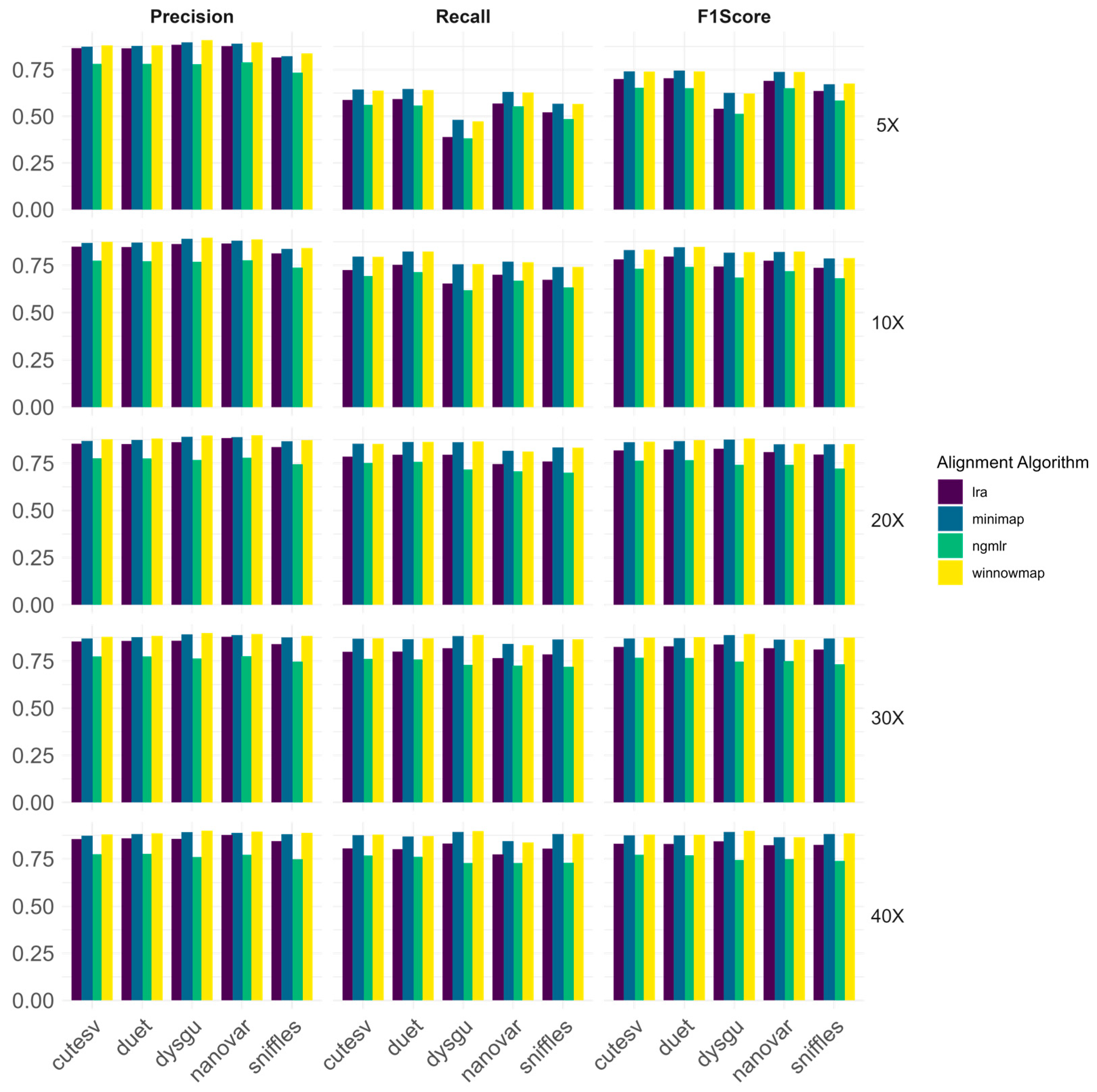
3.1.1. Evaluation of the Impact of Short-Read Alignment Algorithms on Structural Variant Calling
3.1.2. Evaluation of Deletion Calling in Low Complexity Regions for Illumina Short Reads
3.2. Evaluation of Structural Variant Calling in ONT Long Reads
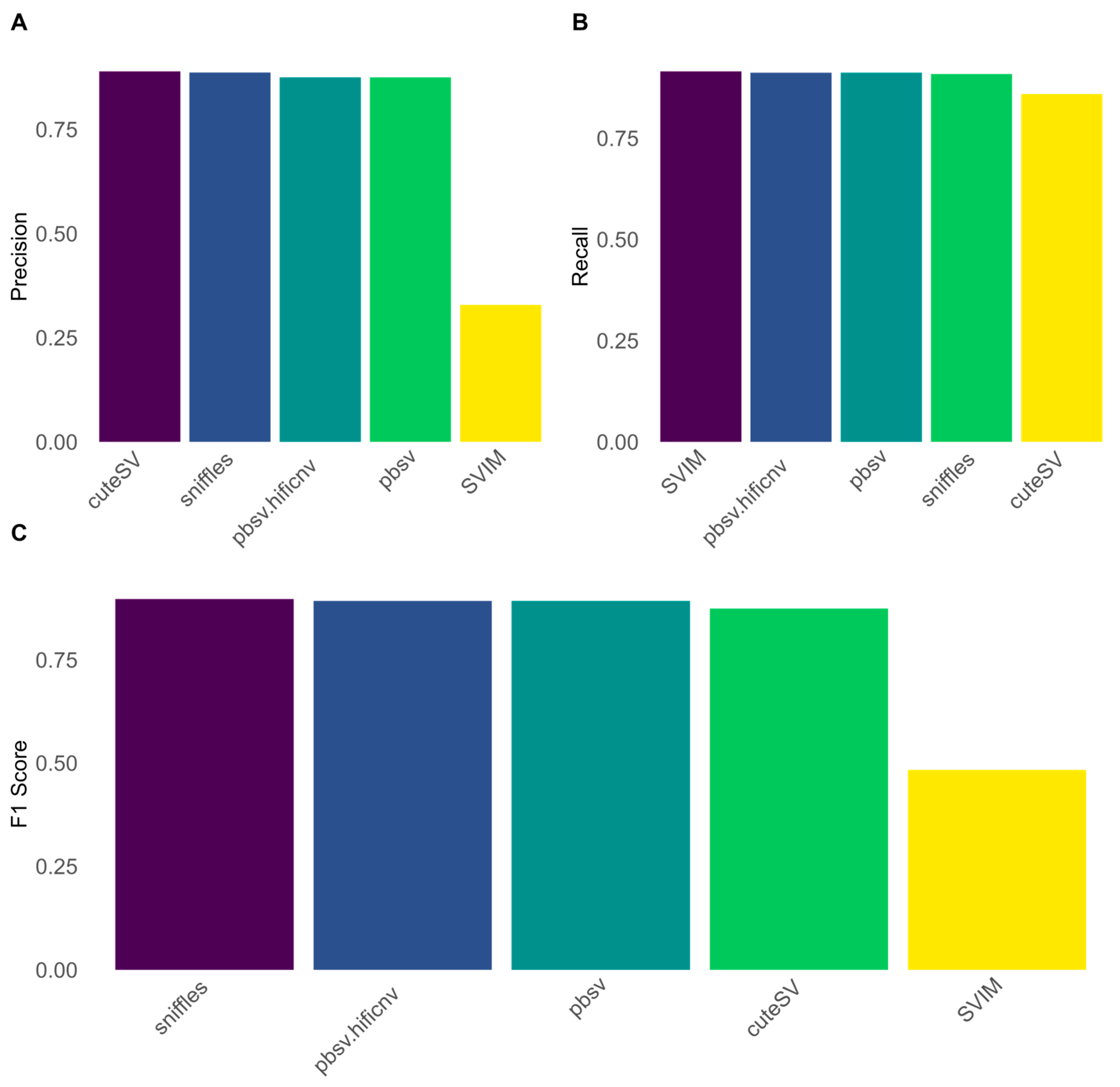
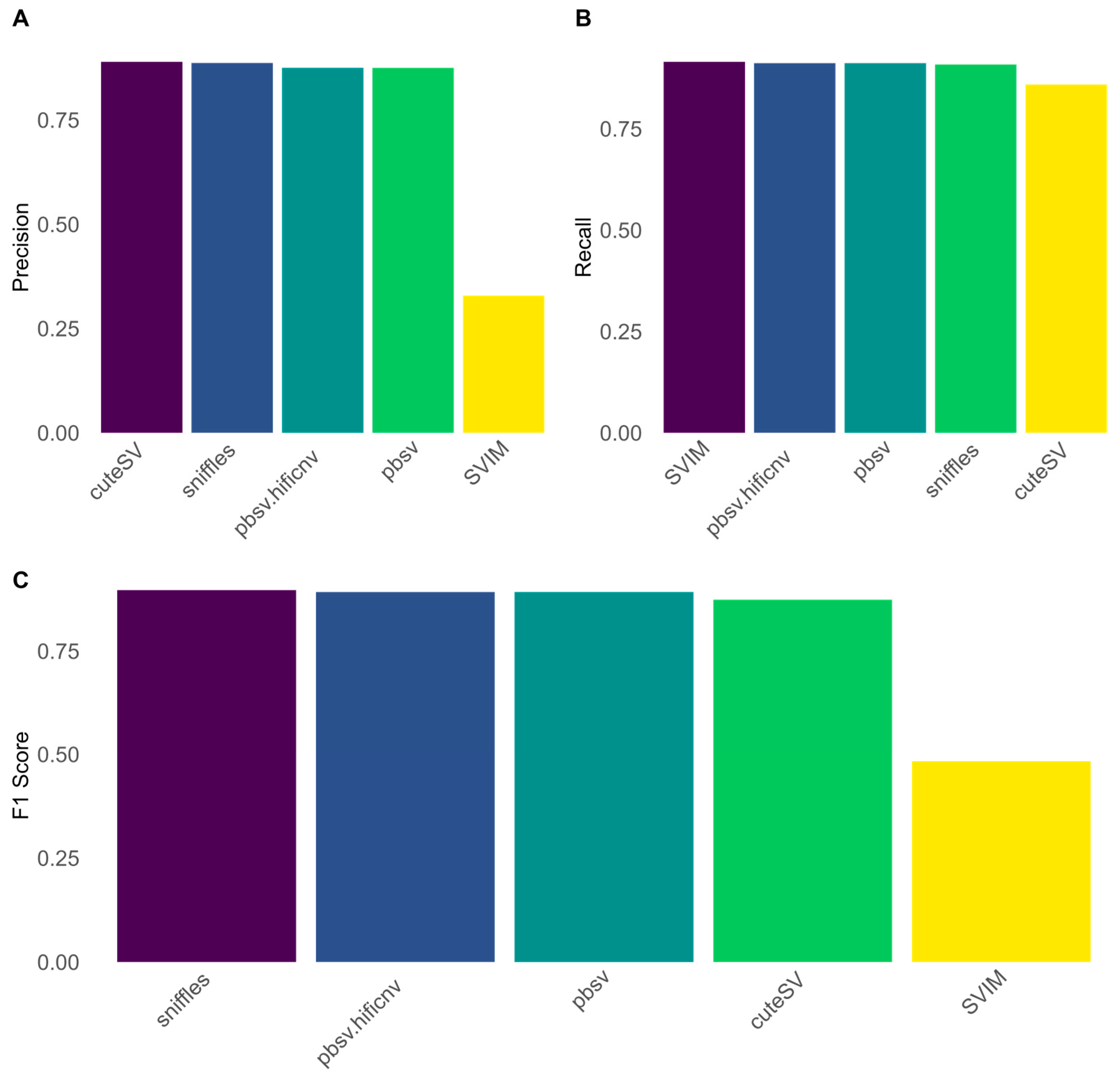
3.3. Evaluation of Structural Variant Calling in PacBio HiFi Reads
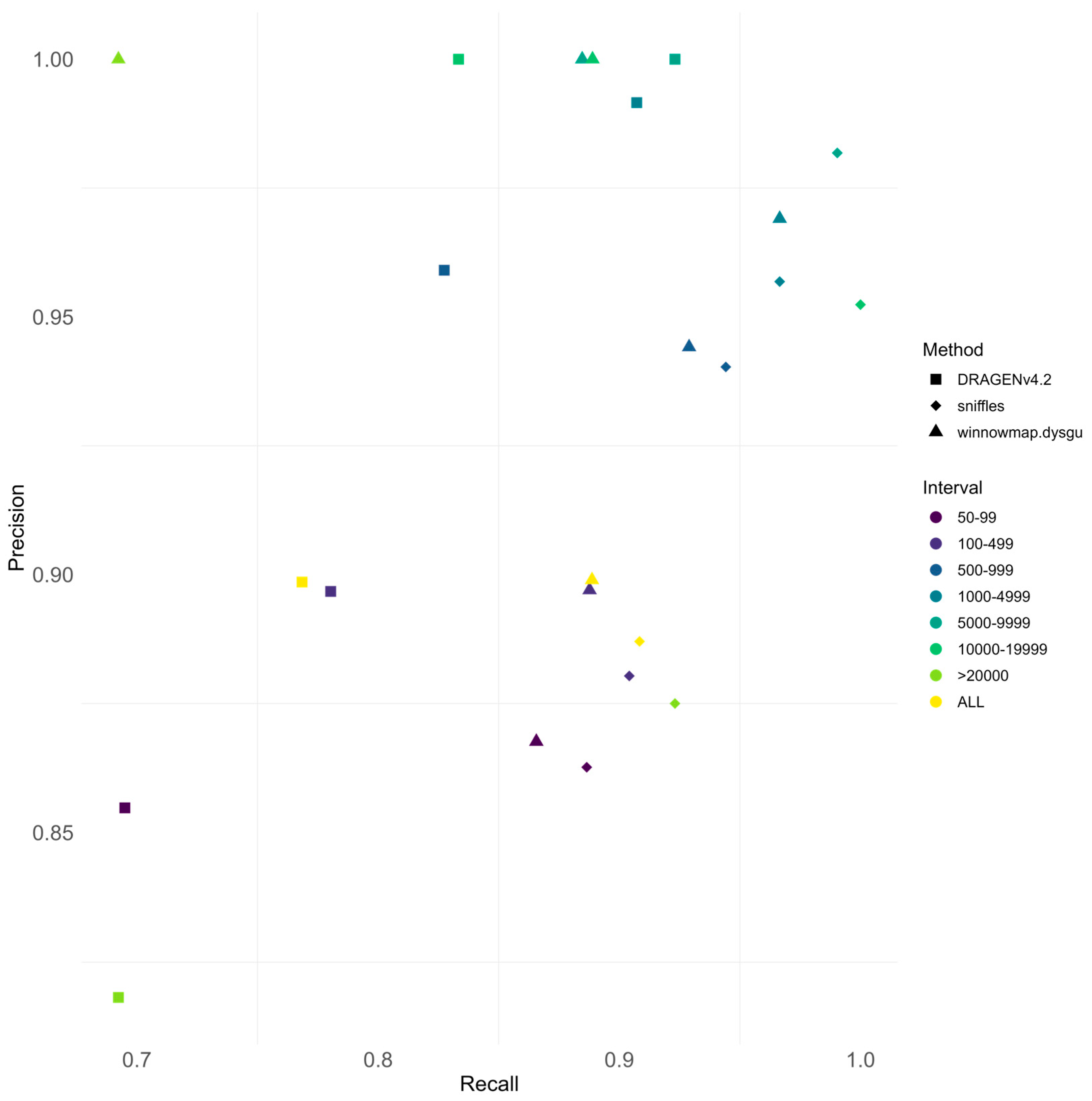
3.4. Performance Comparison Between Sequencing Technologies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mahmoud, M.; Gobet, N.; Cruz-Dávalos, D.I.; Mounier, N.; Dessimoz, C.; Sedlazeck, F.J. Structural Variant Calling: The Long and the Short of It. Genome Biol. 2019, 20, 246. [Google Scholar] [CrossRef]
- Pellestor, F.; Gaillard, J.B.; Schneider, A.; Puechberty, J.; Gatinois, V. Chromoanagenesis, the Mechanisms of a Genomic Chaos. Semin. Cell Dev. Biol. 2022, 123, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Dumas, L.J.; O’Bleness, M.S.; Davis, J.M.; Dickens, C.M.; Anderson, N.; Keeney, J.G.; Jackson, J.; Sikela, M.; Raznahan, A.; Giedd, J.; et al. DUF1220-Domain Copy Number Implicated in Human Brain-Size Pathology and Evolution. Am. J. Hum. Genet. 2012, 91, 444–454. [Google Scholar] [CrossRef]
- Perry, G.H.; Dominy, N.J.; Claw, K.G.; Lee, A.S.; Fiegler, H.; Redon, R.; Werner, J.; Villanea, F.A.; Mountain, J.L.; Misra, R.; et al. Diet and the Evolution of Human Amylase Gene Copy Number Variation. Nat. Genet. 2007, 39, 1256–1260. [Google Scholar] [CrossRef] [PubMed]
- Stankiewicz, P.; Lupski, J.R. Structural Variation in the Human Genome and Its Role in Disease. Annu. Rev. Med. 2010, 61, 437–455. [Google Scholar] [CrossRef]
- Campbell, P.J.; Stephens, P.J.; Pleasance, E.D.; O’Meara, S.; Li, H.; Santarius, T.; Stebbings, L.A.; Leroy, C.; Edkins, S.; Hardy, C.; et al. Identification of Somatically Acquired Rearrangements in Cancer Using Genome-Wide Massively Parallel Paired-End Sequencing. Nat. Genet. 2008, 40, 722–729. [Google Scholar] [CrossRef]
- Fahed, A.C.; Gelb, B.D.; Seidman, J.G.; Seidman, C.E. Genetics of Congenital Heart Disease. Circ. Res. 2013, 112, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Craddock, N.; Hurles, M.E.; Cardin, N.; Pearson, R.D.; Plagnol, V.; Robson, S.; Vukcevic, D.; Barnes, C.; Conrad, D.F.; Giannoulatou, E.; et al. Genome-Wide Association Study of CNVs in 16,000 Cases of Eight Common Diseases and 3,000 Shared Controls. Nature 2010, 464, 713–720. [Google Scholar] [CrossRef]
- Pippucci, T.; Licchetta, L.; Baldassari, S.; Palombo, F.; Menghi, V.; D’Aurizio, R.; Leta, C.; Stipa, C.; Boero, G.; d’Orsi, G.; et al. Epilepsy with Auditory Features. Neurol. Genet. 2015, 1, e5. [Google Scholar] [CrossRef]
- Fanciulli, M.; Norsworthy, P.J.; Petretto, E.; Dong, R.; Harper, L.; Kamesh, L.; Heward, J.M.; Gough, S.C.L.; de Smith, A.; Blakemore, A.I.F.; et al. FCGR3B Copy Number Variation Is Associated with Susceptibility to Systemic, but Not Organ-Specific, Autoimmunity. Nat. Genet. 2007, 39, 721–723. [Google Scholar] [CrossRef]
- Metzker, M.L. Sequencing Technologies—the next Generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef]
- Podvalnyi, A.; Kopernik, A.; Sayganova, M.; Woroncow, M.; Zobkova, G.; Smirnova, A.; Esibov, A.; Deviatkin, A.; Volchkov, P.; Albert, E. Quantitative Analysis of Pseudogene-Associated Errors During Germline Variant Calling. Int. J. Mol. Sci. 2025, 26, 363. [Google Scholar] [CrossRef]
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B.; et al. Real-Time DNA Sequencing from Single Polymerase Molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.; Wu, H.-C.; Jayasinghe, L.; Patel, A.; Reid, S.; Bayley, H. Continuous Base Identification for Single-Molecule Nanopore DNA Sequencing. Nat. Nanotechnol. 2009, 4, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Logsdon, G.A.; Rozanski, A.N.; Ryabov, F.; Potapova, T.; Shepelev, V.A.; Catacchio, C.R.; Porubsky, D.; Mao, Y.; Yoo, D.; Rautiainen, M.; et al. The Variation and Evolution of Complete Human Centromeres. Nature 2024, 629, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Stephens, Z.; Kocher, J.-P. Characterization of Telomere Variant Repeats Using Long Reads Enables Allele-Specific Telomere Length Estimation. BMC Bioinform. 2024, 25, 194. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Momozawa, Y.; Liu, X.; Terao, C.; Kubo, M.; Kamatani, Y. Comprehensive Evaluation of Structural Variation Detection Algorithms for Whole Genome Sequencing. Genome Biol. 2019, 20, 117. [Google Scholar] [CrossRef]
- Ho, S.S.; Urban, A.E.; Mills, R.E. Structural Variation in the Sequencing Era. Nat. Rev. Genet. 2020, 21, 171–189. [Google Scholar] [CrossRef]
- Chaisson, M.J.P.; Sanders, A.D.; Zhao, X.; Malhotra, A.; Porubsky, D.; Rausch, T.; Gardner, E.J.; Rodriguez, O.L.; Guo, L.; Collins, R.L.; et al. Multi-Platform Discovery of Haplotype-Resolved Structural Variation in Human Genomes. Nat. Commun. 2019, 10, 1784. [Google Scholar] [CrossRef]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Altemose, N.; Uralsky, L.; Gershman, A.; et al. The Complete Sequence of a Human Genome. Science 2022, 376, 44–53. [Google Scholar] [CrossRef]
- Liao, W.-W.; Asri, M.; Ebler, J.; Doerr, D.; Haukness, M.; Hickey, G.; Lu, S.; Lucas, J.K.; Monlong, J.; Abel, H.J.; et al. A Draft Human Pangenome Reference. Nature 2023, 617, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Poplin, R.; Ruano-Rubio, V.; DePristo, M.A.; Fennell, T.J.; Carneiro, M.O.; der Auwera, G.A.V.; Kling, D.E.; Gauthier, L.D.; Levy-Moonshine, A.; Roazen, D.; et al. Scaling Accurate Genetic Variant Discovery to Tens of Thousands of Samples. bioRxiv 2018. [Google Scholar] [CrossRef]
- Zook, J.M.; Hansen, N.F.; Olson, N.D.; Chapman, L.; Mullikin, J.C.; Xiao, C.; Sherry, S.; Koren, S.; Phillippy, A.M.; Boutros, P.C.; et al. A Robust Benchmark for Detection of Germline Large Deletions and Insertions. Nat. Biotechnol. 2020, 38, 1347–1355. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Sun, Z.; Wang, J.; Huang, H.; Kocher, J.-P.; Wang, L. CrossMap: A Versatile Tool for Coordinate Conversion between Genome Assemblies. Bioinformatics 2014, 30, 1006–1007. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Källberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid Detection of Structural Variants and Indels for Germline and Cancer Sequencing Applications. Bioinformatics 2016, 32, 1220–1222. [Google Scholar] [CrossRef]
- Wu, Y.; Tian, L.; Pirastu, M.; Stambolian, D.; Li, H. MATCHCLIP: Locate Precise Breakpoints for Copy Number Variation Using CIGAR String by Matching Soft Clipped Reads. Front. Genet. 2013, 4, 157. [Google Scholar] [CrossRef]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stütz, A.M.; Benes, V.; Korbel, J.O. DELLY: Structural Variant Discovery by Integrated Paired-End and Split-Read Analysis. Bioinformatics 2012, 28, i333–i339. [Google Scholar] [CrossRef]
- Layer, R.M.; Chiang, C.; Quinlan, A.R.; Hall, I.M. LUMPY: A Probabilistic Framework for Structural Variant Discovery. Genome Biol. 2014, 15, R84. [Google Scholar] [CrossRef]
- Bartenhagen, C.; Dugas, M. Robust and Exact Structural Variation Detection with Paired-End and Soft-Clipped Alignments: SoftSV Compared with Eight Algorithms. Brief. Bioinform. 2016, 17, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Zhao, F.; Buboltz, A.; Schuster, S.C. inGAP: An Integrated next-Generation Genome Analysis Pipeline. Bioinformatics 2010, 26, 127–129. [Google Scholar] [CrossRef]
- Kronenberg, Z.N.; Osborne, E.J.; Cone, K.R.; Kennedy, B.J.; Domyan, E.T.; Shapiro, M.D.; Elde, N.C.; Yandell, M. Wham: Identifying Structural Variants of Biological Consequence. PLoS Comput. Biol. 2015, 11, e1004572. [Google Scholar] [CrossRef]
- Abyzov, A.; Urban, A.E.; Snyder, M.; Gerstein, M. CNVnator: An Approach to Discover, Genotype, and Characterize Typical and Atypical CNVs from Family and Population Genome Sequencing. Genome Res. 2011, 21, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Sedlazeck, F.J.; Rescheneder, P.; Smolka, M.; Fang, H.; Nattestad, M.; von Haeseler, A.; Schatz, M.C. Accurate Detection of Complex Structural Variations Using Single Molecule Sequencing. Nat. Methods 2018, 15, 461–468. [Google Scholar] [CrossRef]
- Jiang, T.; Liu, Y.; Jiang, Y.; Li, J.; Gao, Y.; Cui, Z.; Liu, Y.; Liu, B.; Wang, Y. Long-Read-Based Human Genomic Structural Variation Detection with cuteSV. Genome Biol. 2020, 21, 189. [Google Scholar] [CrossRef]
- Heller, D.; Vingron, M. SVIM: Structural Variant Identification Using Mapped Long Reads. Bioinformatics 2019, 35, 2907–2915. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Jain, C.; Rhie, A.; Hansen, N.F.; Koren, S.; Phillippy, A.M. Long-Read Mapping to Repetitive Reference Sequences Using Winnowmap2. Nat. Methods 2022, 19, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Chaisson, M.J.P. Lra: A Long Read Aligner for Sequences and Contigs. PLoS Comput. Biol. 2021, 17, e1009078. [Google Scholar] [CrossRef] [PubMed]
- Cleal, K.; Baird, D.M. Dysgu: Efficient Structural Variant Calling Using Short or Long Reads. Nucleic Acids Res. 2022, 50, e53. [Google Scholar] [CrossRef] [PubMed]
- Tham, C.Y.; Tirado-Magallanes, R.; Goh, Y.; Fullwood, M.J.; Koh, B.T.H.; Wang, W.; Ng, C.H.; Chng, W.J.; Thiery, A.; Tenen, D.G.; et al. NanoVar: Accurate Characterization of Patients’ Genomic Structural Variants Using Low-Depth Nanopore Sequencing. Genome Biol. 2020, 21, 56. [Google Scholar] [CrossRef]
- Zhou, Y.; Leung, A.W.-S.; Ahmed, S.S.; Lam, T.-W.; Luo, R. Duet: SNP-Assisted Structural Variant Calling and Phasing Using Oxford Nanopore Sequencing. BMC Bioinform. 2022, 23, 465. [Google Scholar] [CrossRef]
- Joe, S.; Park, J.-L.; Kim, J.; Kim, S.; Park, J.-H.; Yeo, M.-K.; Lee, D.; Yang, J.O.; Kim, S.-Y. Comparison of Structural Variant Callers for Massive Whole-Genome Sequence Data. BMC Genom. 2024, 25, 318. [Google Scholar] [CrossRef]
- Zheng, Z.; Li, S.; Su, J.; Leung, A.W.-S.; Lam, T.-W.; Luo, R. Symphonizing Pileup and Full-Alignment for Deep Learning-Based Long-Read Variant Calling. Nat. Comput. Sci. 2022, 2, 797–803. [Google Scholar] [CrossRef]
- Martin, M.; Ebert, P.; Marschall, T. Read-Based Phasing and Analysis of Phased Variants with WhatsHap. In Methods in Molecular Biology; Humana Press: Clifton, NJ, USA, 2023; Volume 2590, pp. 127–138. [Google Scholar] [CrossRef]
- Bolognini, D.; Magi, A. Evaluation of Germline Structural Variant Calling Methods for Nanopore Sequencing Data. Front. Genet. 2021, 12, 761791. [Google Scholar] [CrossRef]
- Helal, A.A.; Saad, B.T.; Saad, M.T.; Mosaad, G.S.; Aboshanab, K.M. Benchmarking Long-Read Aligners and SV Callers for Structural Variation Detection in Oxford Nanopore Sequencing Data. Sci. Rep. 2024, 14, 6160. [Google Scholar] [CrossRef] [PubMed]
- Glotov, O.S.; Chernov, A.N.; Glotov, A.S. Human Exome Sequencing and Prospects for Predictive Medicine: Analysis of International Data and Own Experience. J. Pers. Med. 2023, 13, 1236. [Google Scholar] [CrossRef] [PubMed]

| Software | Software Type | Sequencing Type | Reference |
|---|---|---|---|
| DRAGENv4.2 | SV calling | srWGS | [44] |
| DRAGENv4.0 | SV calling | srWGS | [44] |
| Manta | SV calling | srWGS | [26] |
| MATCHCLIP | SV calling | srWGS | [27] |
| DELLY | SV calling | srWGS | [28] |
| Lumpy | SV calling | srWGS | [29] |
| SoftSV | SV calling | srWGS | [30] |
| inGAP | SV calling | srWGS | [31] |
| Wham | SV calling | srWGS | [32] |
| CNVnator | SV calling | srWGS | [33] |
| CuteSV | SV calling | lrWGS | [36] |
| Nanovar | SV calling | lrWGS | [42] |
| dysgu | SV calling | lrWGS | [41] |
| duet | SV calling | lrWGS | [43] |
| sniffles | SV calling | lrWGS | [35] |
| pbsv | SV calling | lrWGS | https://github.com/PacificBiosciences/pbsv |
| SVIM | SV calling | lrWGS | [37] |
| minimap2 | Alignment | srWGS, lrWGS | [34] |
| winnowmap | Alignment | lrWGS | [39] |
| ngmlr | Alignment | lrWGS | [35] |
| lra | Alignment | lrWGS | [40] |
| bwa-mem2 | Alignment | srWGS | [25] |
| dragmap | Alignment | srWGS | https://github.com/Illumina/DRAGMAP |
| bowtie2 | Alignment | srWGS | [45] |
| DRAGEN pipeline alignment | Alignment | srWGS | [44] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nardone, G.G.; Andrioletti, V.; Santin, A.; Morgan, A.; Spedicati, B.; Concas, M.P.; Gasparini, P.; Girotto, G.; Limongelli, I. A Hitchhiker Guide to Structural Variant Calling: A Comprehensive Benchmark Through Different Sequencing Technologies. Biomedicines 2025, 13, 1949. https://doi.org/10.3390/biomedicines13081949
Nardone GG, Andrioletti V, Santin A, Morgan A, Spedicati B, Concas MP, Gasparini P, Girotto G, Limongelli I. A Hitchhiker Guide to Structural Variant Calling: A Comprehensive Benchmark Through Different Sequencing Technologies. Biomedicines. 2025; 13(8):1949. https://doi.org/10.3390/biomedicines13081949
Chicago/Turabian StyleNardone, Giuseppe Giovanni, Valentina Andrioletti, Aurora Santin, Anna Morgan, Beatrice Spedicati, Maria Pina Concas, Paolo Gasparini, Giorgia Girotto, and Ivan Limongelli. 2025. "A Hitchhiker Guide to Structural Variant Calling: A Comprehensive Benchmark Through Different Sequencing Technologies" Biomedicines 13, no. 8: 1949. https://doi.org/10.3390/biomedicines13081949
APA StyleNardone, G. G., Andrioletti, V., Santin, A., Morgan, A., Spedicati, B., Concas, M. P., Gasparini, P., Girotto, G., & Limongelli, I. (2025). A Hitchhiker Guide to Structural Variant Calling: A Comprehensive Benchmark Through Different Sequencing Technologies. Biomedicines, 13(8), 1949. https://doi.org/10.3390/biomedicines13081949






