The Role of Pharmacogenetic Biomarkers in Pain
Abstract
1. Introduction
2. Pharmacogenetic Biomarkers in Analgesia
2.1. Non-Steroidal Anti-Inflammatory Drugs (NSAIDs)
- Pharmacogenetic Biomarkers and Clinical Implications
- CYP2C9 gene
- CYP2C8 gene
- Other Pharmacogenes: UGTs, ABCB1, and SLCO1B1
- Summary of Evidence
2.2. Opioids
- Pharmacogenetic Biomarkers and Clinical Implications
- CYP2D6 gene
- OPRM1 gene
- COMT gene
- Other Pharmacogenes: CYP3A, CYP2B6, ABCB1, UGT2B7
- Summary of Evidence
2.3. Antidepressants, Anticonvulsants, and Gabapentinoids
- Pharmacogenetic Biomarkers and Clinical Implications
2.3.1. Tricyclic Antidepressants (TCAs)
- CYP2D6 gene
- CYP2C19 gene
- ABCB1 gene
2.3.2. Anticonvulsants: Carbamazepine and Oxcarbazepine
- HLA-A*31:01 and HLA-B*15:02 genes
2.3.3. Selective Serotonin and Norepinephrine Reuptake Inhibitors (SNRIs) and Selective Serotonin Reuptake Inhibitors (SSRIs)
- CYP2D6 and CYP2C19 genes
- Other Pharmacogenes: ABCB1, SLC6A4, and HTR2A
2.3.4. Gabapentinoids: Gabapentin and Pregabalin
- Summary of Evidence
3. Clinical Implications and Future Perspectives
3.1. From Single-Gene Testing to Multi-Omic Pain Signatures
3.2. Implementation Science and Real-World Pragmatism
3.3. Clinical Decision Support (CDS) That Clinicians Will Use
3.4. Equity, Ancestry, and Allele Frequency Gaps
3.5. Digital Phenotyping and AI-Driven Dosing
3.6. Real-World Evidence (RWE) and Health–Economic Data
- -
- Pre-emptive panel testing becoming part of routine pre-operative assessments, starting with high-risk surgeries.
- -
- Joint PGx + opioid-sparing protocols (regional anesthesia, NSAID rotation) assessed in factorial designs.
- -
- Cost effectiveness and payer coverage analyses embedded as secondary outcomes to accelerate reimbursement decisions.
- -
- Regulatory alignment as CPIC/DPWG tables converge and updated NSAID and opioid labels incorporate genotype-based dosing ranges.
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gan, T.J.; Habib, A.S.; Miller, T.E.; White, W.; Apfelbaum, J.L. Incidence, patient satisfaction, and perceptions of post-surgical pain: Results from a US national survey. Curr. Med. Res. Opin. 2014, 30, 149–160. [Google Scholar] [CrossRef]
- Zhao, M.; Ma, J.; Li, M.; Zhang, Y.; Jiang, B.; Zhao, X.; Huai, C.; Shen, L.; Zhang, N.; He, L.; et al. Cytochrome P450 Enzymes and Drug Metabolism in Humans. Int. J. Mol. Sci. 2021, 22, 12808. [Google Scholar] [CrossRef] [PubMed]
- Rollason, V.; Samer, C.; Piguet, V.; Dayer, P.; Desmeules, J. Pharmacogenetics of analgesics: Toward the individualization of prescription. Pharmacogenomics 2008, 9, 905–933. [Google Scholar] [CrossRef] [PubMed]
- Grosser, T.; Theken, K.N.; FitzGerald, G.A. Cyclooxygenase inhibition: Pain, inflammation, and the cardiovascular system. Clin. Pharmacol. Ther. 2017, 102, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Zobdeh, F.; Eremenko, I.I.; Akan, M.A.; Tarasov, V.V.; Chubarev, V.N.; Schiöth, H.B.; Mwinyi, J. Pharmacogenetics and pain treatment with a focus on non-steroidal anti-inflammatory drugs (NSAIDs) and antidepressants: A systematic review. Pharmaceutics 2022, 14, 1190. [Google Scholar] [CrossRef]
- Theken, K.N.; Lee, C.R.; Gong, L.; Caudle, K.E.; Formea, C.M.; Gaedigk, A.; Klein, T.E.; Agúndez, J.A.G.; Grosser, T. Clinical Pharmacogenetics Implementation Consortium guideline (CPIC) for CYP2C9 and nonsteroidal anti-inflammatory drugs. Clin. Pharmacol. Ther. 2020, 108, 191–200. [Google Scholar] [CrossRef]
- Vogl, S.; Lutz, R.W.; Schönfelder, G.; Lutz, W.K. CYP2C9 genotype vs. metabolic phenotype for individual drug dosing—A correlation analysis using flurbiprofen as probe drug. PLoS ONE 2015, 10, e0120403. [Google Scholar]
- Kusama, M.; Maeda, K.; Chiba, K.; Aoyama, A.; Sugiyama, Y. Prediction of the effects of genetic polymorphism on the pharmacokinetics of CYP2C9 substrates from in vitro data. Pharm. Res. 2009, 26, 822–835. [Google Scholar] [CrossRef]
- Ochoa, D.; Prieto-Pérez, R.; Román, M.; Talegón, M.; Rivas, A.; Galicia, I.; Abad-Santos, F. Effect of gender and CYP2C9 and CYP2C8 polymorphisms on the pharmacokinetics of ibuprofen enantiomers. Pharmacogenomics 2015, 16, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Martínez, C.; Blanco, G.; Ladero, J.M.; García-Martín, E.; Taxonera, C.; Gamito, F.G.; Agúndez, J.A.G. Genetic predisposition to acute gastrointestinal bleeding after NSAIDs use. Br. J. Pharmacol. 2004, 141, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.I.; Bae, J.W.; Choi, C.I.; Lee, Y.J.; Byeon, J.Y.; Jang, C.G.; Shin, J.G. Strongly increased exposure of meloxicam in CYP2C9*3/*3 individuals. Pharmacogenomics 2014, 24, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Mejía-Abril, G.; Zubiaur, P.; Navares-Gómez, M.; Villapalos-García, G.; Román, M.; Ochoa, D.; Abad-Santos, F. Dexketoprofen pharmacokinetics is not significantly altered by genetic polymorphism. Front. Pharmacol. 2021, 12, 660639. [Google Scholar] [CrossRef]
- Calvo, A.M.; Zupelari-Gonçalves, P.; Dionísio, T.J.; Brozoski, D.T.; Faria, F.A.; Santos, C.F. Efficacy of piroxicam for postoperative pain after lower third molar surgery associated with CYP2C8*3 and CYP2C9. J. Pain. Res. 2017, 10, 1581–1589. [Google Scholar] [CrossRef] [PubMed]
- Světlík, S.; Hronová, K.; Bakhouche, H.; Matoušková, O.; Slanař, O. Pharmacogenetics of chronic pain and its treatment. Mediat. Inflamm. 2013, 2013, 864319. [Google Scholar] [CrossRef] [PubMed]
- Daly, A.K.; Aithal, G.P.; Leathart, J.B.S.; Swainsbury, R.A.; Dang, T.S.; Day, C.P. Genetic susceptibility to diclofenac-induced hepatotoxicity: Contribution of UGT2B7, CYP2C8, and ABCC2 genotypes. Gastroenterology 2007, 132, 272–281. [Google Scholar] [CrossRef]
- Treede, R.D. The International Association for the Study of Pain definition of pain: As valid in 2018 as in 1979, but in need of regularly updated footnotes. Pain Rep. 2018, 3, e643. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, P.A.; Conchon Costa, A.C.; Lauretti, G.R.; de Moraes, N.V. Pharmacogenomics in chronic pain therapy: From disease to treatment and challenges for clinical practice. Pharmacogenomics 2019, 20, 971–982. [Google Scholar] [CrossRef] [PubMed]
- Nofziger, C.; Turner, A.J.; Sangkuhl, K.; Whirl-Carrillo, M.; Klein, T.E.; Caudle, K.E. PharmVar GeneFocus: CYP2D6. Clin. Pharmacol. Ther. 2020, 107, 154–170. [Google Scholar] [CrossRef]
- Caudle, K.E.; Sangkuhl, K.; Whirl-Carrillo, M.; Swen, J.J.; Klein, T.E.; Gammal, R.S.; Relling, M.V. Standardizing CYP2D6 genotype to phenotype translation: Consensus recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin. Transl. Sci. 2020, 13, 116–124. [Google Scholar] [CrossRef]
- Crews, K.R.; Monte, A.A.; Huddart, R.; Caudle, K.E.; Kharasch, E.D.; Gaedigk, A.; Dunnenberger, H.M. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6, OPRM1, and COMT genotypes and select opioid therapy. Clin. Pharmacol. Ther. 2021, 110, 888–896. [Google Scholar] [CrossRef]
- Ruano, G.; Kost, J.A. Fundamental considerations for genetically-guided pain management with opioids based on CYP2D6 and OPRM1 polymorphisms. Pain Physician 2018, 21, E611–E621. [Google Scholar] [CrossRef] [PubMed]
- Matic, M.; Nijenhuis, M.; Soree, B.; de Boer-Veger, N.J.; Buunk, A.M.; Houwink, E.J.F.; van der Wouden, C.H. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene–drug interaction between CYP2D6 and opioids (codeine, tramadol and oxycodone). Eur. J. Hum. Genet. 2022, 30, 1105–1113. [Google Scholar] [CrossRef]
- Ferreira do Couto, M.L.; Fonseca, S.; Pozza, D.H. Pharmacogenetic approaches in personalized medicine for postoperative pain management. Biomedicines 2024, 12, 729. [Google Scholar] [CrossRef]
- Boswell, M.V.; Stauble, M.E.; Loyd, G.E.; Langman, L.; Ramey-Hartung, B.; Baumgartner, R.N.; Davis, M.P. The role of hydromorphone and OPRM1 in postoperative pain relief with hydrocodone. Pain Physician 2013, 16, E227–E235. [Google Scholar]
- Tan, E.C.; Lim, E.C.; Teo, Y.Y.; Lim, Y.; Law, H.Y.; Sia, A.T. Ethnicity and OPRM variant independently predict pain perception and patient-controlled analgesia usage for post-operative pain. Mol. Pain. 2009, 5, 32. [Google Scholar] [CrossRef]
- Vetterlein, A.; Monzel, M.; Reuter, M. Are catechol-O-methyltransferase gene polymorphisms genetic markers for pain sensitivity after all? A review and meta-analysis. Neurosci. Biobehav. Rev. 2023, 148, 105112. [Google Scholar] [CrossRef] [PubMed]
- Korczeniewska, O.A.; Kuo, F.; Huang, C.Y.; Nasri-Heir, C.; Khan, J.; Benoliel, R.; Greenspan, J.D. Genetic variation in catechol-O-methyltransferase is associated with individual differences in conditioned pain modulation in healthy subjects. J. Gene Med. 2021, 23, e3374. [Google Scholar] [CrossRef]
- Henker, R.A.; Lewis, A.; Dai, F.; Lariviere, W.R.; Meng, L.; Gruen, G.S.; Pape, H.C. The associations between OPRM1 and COMT genotypes and postoperative pain, opioid use, and opioid-induced sedation. Biol. Res. Nurs. 2013, 15, 309–317. [Google Scholar] [CrossRef]
- Robinson, K.M.; Eum, S.; Desta, Z.; Tyndale, R.F.; Gaedigk, A.; Crist, R.C.; Haidar, C.E.; Myers, A.L.; Samer, C.F.; Somogyi, A.A.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2B6 Genotype and Methadone Therapy. Clin. Pharmacol. Ther. 2024, 116, 932–938. [Google Scholar] [CrossRef] [PubMed]
- Sadhasivam, S.; Chidambaran, V.; Zhang, X.; Meller, J.; Esslinger, H.; Zhang, K.; Martin, L.J. Opioid-induced respiratory depression: ABCB1 transporter pharmacogenetics. Pharmacogenom. J. 2015, 15, 119–126. [Google Scholar] [CrossRef]
- Benavides, R.; Vsevolozhskaya, O.; Cattaneo, S.; Zaykin, D.; Brenton, A.; Parisien, M.; Diatchenko, L. A functional polymorphism in the ATP-binding cassette B1 transporter predicts pharmacologic response to combination of nortriptyline and morphine in neuropathic pain patients. Pain 2020, 161, 619–629. [Google Scholar] [CrossRef]
- De Gregori, M.; Garbin, G.; De Gregori, S.; Minella, C.E.; Bugada, D.; Lisa, A.; Govoni, S. Genetic variability at COMT but not at OPRM1 and UGT2B7 loci modulates morphine analgesic response in acute postoperative pain. Eur. J. Clin. Pharmacol. 2013, 69, 1651–1658. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.K.; Sangkuhl, K.; Swen, J.J.; Ellingrod, V.L.; Müller, D.J.; Shimoda, K.; Bishop, J.R. Clinical Pharmacogenetics Implementation Consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin. Pharmacol. Ther. 2017, 102, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Dean, L.; Kane, M. Imipramine therapy and CYP2D6 and CYP2C19 genotype. In Medical Genetics Summaries; Pratt, V.M., Scott, S.A., Pirmohamed, M., Esquivel, B., Kattman, B.L., Malheiro, A.J., Eds.; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2017. [Google Scholar]
- Kim, S.H.; Lee, K.W.; Song, W.J.; Kwon, H.C.; Lee, S.M.; Kang, H.R. Carbamazepine-induced severe cutaneous adverse reactions and HLA genotypes in Koreans. Epilepsy Res. 2011, 97, 190–197. [Google Scholar] [CrossRef]
- Caudle, K.E.; Thorn, C.F.; Klein, T.E.; Swen, J.J.; McLeod, H.L.; Dias da Silva, M.R.; Gammal, R.S. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for HLA-B genotype and carbamazepine dosing: 2014 update. Clin. Pharmacol. Ther. 2014, 96, 376–382. [Google Scholar]
- Manson, L.E.N.; Nijenhuis, M.; Soree, B.; de Boer-Veger, N.J.; Buunk, A.M.; Houwink, E.J.F.; van der Wouden, C.H. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene–drug interaction of CYP2C9, HLA-A and HLA-B with anti-epileptic drugs. Eur. J. Hum. Genet. 2024, 32, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tobe, J.; Au, E.; Tran, C.; Jomy, J.; Oparin, Y.; Ladha, K.S. Selective serotonin reuptake inhibitors and serotonin-norepinephrine reuptake inhibitors as adjuncts for postoperative pain management: Systematic review and meta-analysis of randomised controlled trials. Br. J. Anaesth. 2022, 128, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Aldrich, S.L.; Poweleit, E.A.; Prows, C.A.; Martin, L.J.; Strawn, J.R.; Ramsey, L.B. Influence of CYP2C19 metabolizer status on escitalopram/citalopram tolerability and response in youth with anxiety and depressive disorders. Front. Pharmacol. 2019, 10, 99. [Google Scholar] [CrossRef]
- Swen, J.J.; Nijenhuis, M.; de Boer, A.; Grandia, L.; Maitland-van der Zee, A.H.; Mulder, H.; Guchelaar, H.J. Pharmacogenetic recommendations for selective serotonin reuptake inhibitors and serotonin-norepinephrine reuptake inhibitors: Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline. Clin. Pharmacol. Ther. 2023, 113, 1249–1257. [Google Scholar]
- Manson, L.E.N.; Nijenhuis, M.; Soree, B.; de Boer-Veger, N.J.; Buunk, A.M.; Houwink, E.J.F.; van der Wouden, C.H. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene–drug interactions of CYP2D6 and CYP2C19 with other antidepressants. Eur. J. Hum. Genet. 2024, 32, 585–595. [Google Scholar] [CrossRef]
- Hung, C.C.; Chen, P.L.; Huang, W.M.; Tai, J.J.; Hsieh, T.J.; Ding, S.T.; Hsu, T.C. Gene-wide tagging study of the effects of common genetic polymorphisms in the α subunits of the GABA(A) receptor on epilepsy treatment response. Pharmacogenomics 2013, 14, 1849–1856. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, A.; Alam, S.M.; Azam, F.; Khan, M.; Saleem, S.A.; Liaquat, A.; Riaz, H. Influence of single nucleotide polymorphism of LAT1 on therapeutic response to gabapentinoids in Pakistani patients with neuropathic pain. Basic Clin. Pharmacol. Toxicol. 2021, 128, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.M.; Beyene, R.; Kolm, P.; Young, T.A.; Zifa, S.; Natividad, V.; Licata, A.; Podolsky, R.H.; Moore, T.; Walsh, R.; et al. A randomized hybrid-effectiveness trial comparing pharmacogenomics (PGx) to standard care: The PGx Applied to Chronic Pain Treatment in Primary Care (PGx-ACT) trial. Clin. Transl. Sci. 2025, 18, e70154. [Google Scholar] [CrossRef]
- Wake, D.T.; Smith, D.M.; Kazi, S.; Dunnenberger, H.M. Pharmacogenomic clinical decision support: A review, how-to guide, and future vision. Clin. Pharmacol. Ther. 2022, 112, 44–57. [Google Scholar] [CrossRef]
- Maulana, Y.; Toro Jimenez, R.; Twesigomwe, D.; Sani, L.; Irwanto, A.; Bertin, N.; Gonzalez-Porta, M. The variation landscape of CYP2D6 in a multi-ethnic Asian population. Sci. Rep. 2024, 14, 16725. [Google Scholar] [CrossRef] [PubMed]
- Soley, N.; Speed, T.J.; Xie, A.; Taylor, C.O. Predicting postoperative pain and opioid use with machine learning applied to longitudinal electronic health record and wearable data. Appl. Clin. Inform. 2024, 15, 569–582. [Google Scholar] [CrossRef]
- Patterson, D.G.; Wilson, D.; Fishman, M.A.; Neufeld, E.; Butera, G.; Cornwell, H.L.; Engle, A.; Minshall, M.E. Objective wearable measures correlate with self-reported chronic pain levels in people with spinal cord stimulation systems. NPJ Digit. Med. 2023, 6, 146. [Google Scholar] [CrossRef] [PubMed]
- Agulló, L.; Aguado, I.; Muriel, J.; Margarit, C.; Gómez, A.; Escorial, M.; Sánchez, A.; Fernández, A.; Peiró, A.M. Pharmacogenetic guided opioid therapy improves chronic pain outcomes and comorbid mental health: A randomized, double-blind, controlled study. Int. J. Mol. Sci. 2023, 24, 10754. [Google Scholar] [CrossRef]
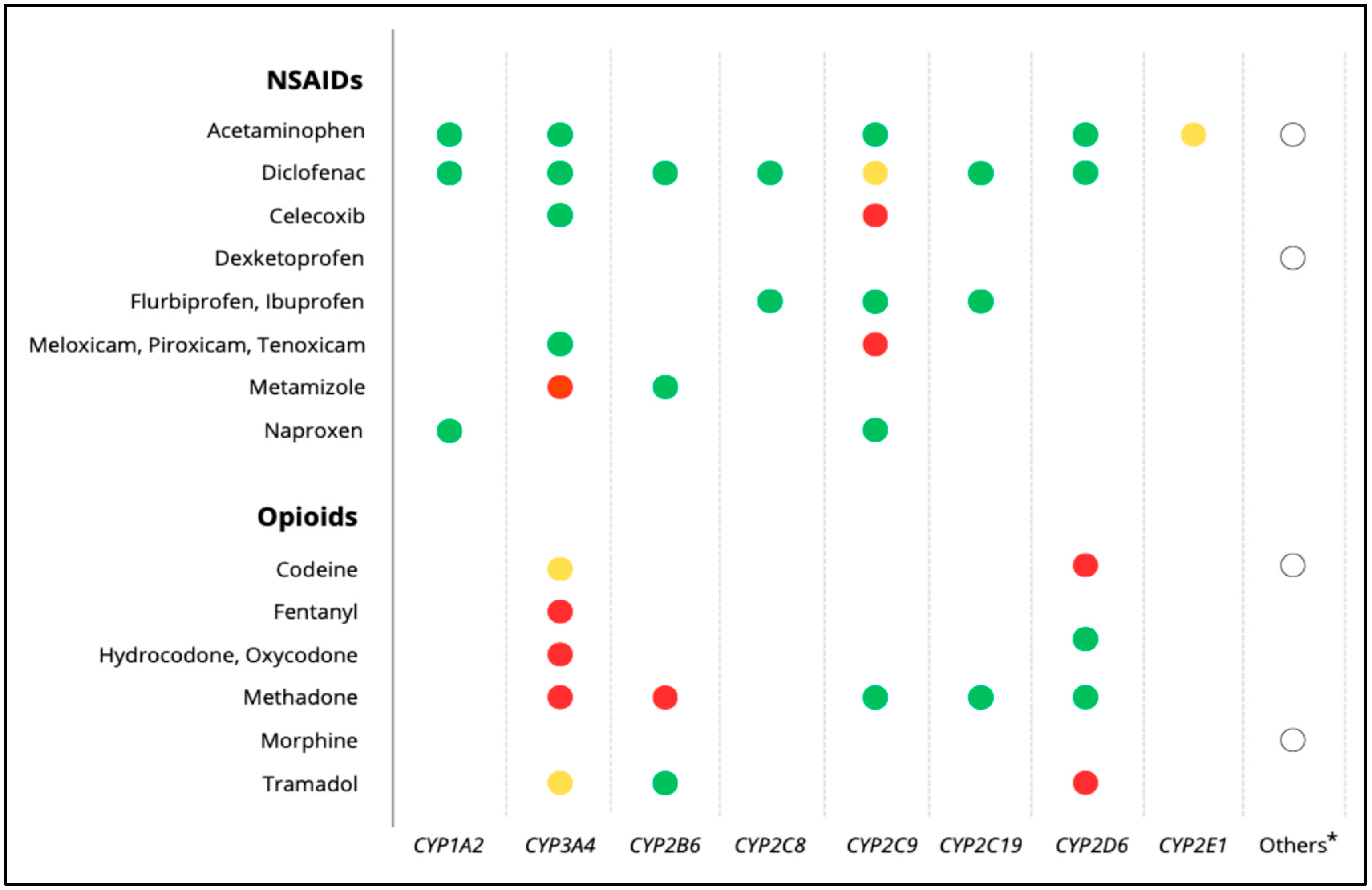
| Genes | Level of Evidence † | Drug(s) | CPIC Recommendation | DPWG Recommendation | |
|---|---|---|---|---|---|
| NSAIDs | CYP2C9 | Level 1B | Celecoxib, Flurbiprofen, Ibuprofen, Lornoxicam | PMs (AS 0) should begin therapy at just 25–50% of the lowest recommended dose or use alternative analgesics | No recommendation |
| Level 1B | Meloxicam | IMs (AS 1) should either start at half the lowest dose or choose an alternative; PMs should avoid it | No recommendation | ||
| Level 1B | Piroxicam, Tenoxicam | IMs (AS 1) and PMs (AS 0) are best managed with alternative NSAIDs | No recommendation | ||
| CYP2C8 | Level 2 | Ibuprofen, Piroxicam, Diclofenac | Not actionable | No recommendation | |
| UGTs, ABCB1 and SLCO1B1 | Level 3 | NSAIDs | No recommendation | No recommendation | |
| Opioids | CYP2D6 | Level 1A | Codeine, Tramadol, Oxycodone, Hydrocodone | Avoid codeine/tramadol in PMs/UMs, and monitor in IM patients | Avoid codeine/tramadol in PMs/UMs |
| OPRM1 | Level 2 | Morphine, Hydrocodone, Fentanyl | Not actionable | No recommendation | |
| COMT | Level 2 | All opioids | Not actionable | No recommendation | |
| CYP3A4/5 | Level 3 | Fentanyl, Alfentanil, Oxycodone | No recommendation | No recommendation | |
| CYP2B6 | Level 2 | Methadone | Not actionable | No recommendation | |
| ABCB1 and UGT2B7 | Level 3 | Morphine, Methadone | No recommendation | No recommendation | |
| Anticonvulsants | HLA-B*15:02 | Level 1A | Carbamazepine, Oxcarbazepine | Avoid if possible | Carbamazepine is contraindicated |
| HLA-A*31:01 | Level 1A | Carbamazepine | Considering alternative therapies. | Avoid if possible. | |
| Antidepressants | CYP2C19 | Level 1A | Amitriptyline, Nortriptyline, | Dose reduction or alternative therapy in UMs/PMs | 70% of the standard dose in UMs/PMs patients |
| Level 1A | Citalopram, Escitalopram, Sertraline | Dose reduction or alternative therapy in PMs and avoidance of these agents in UMs | In PMs escitalopram/citalopram dose should not exceed 50% of the maximum dose, and it should be avoided in UMs (not citalopram) | ||
| CYP2D6 | Level 1A | Amitriptyline, Nortriptyline | 50% dose reduction in PMs and the use of alternative agents in UMs | 30% dose reduction in IMs/PMs, and 1.7 times dose increase in UMs | |
| Level 1A | Paroxetine, Fluoxetine | Considering alternative agents or genotype-informed dose adjustment | Paroxetine should be avoided in PMs patients | ||
| Level 2 | Duloxetine | Not actionable | Not actionable | ||
| Level 1B | Venlafaxine | Not actionable | IMs/PMs are at risk of toxicity and suboptimal response; avoidance is advised | ||
| SLC6A4 | Level 2 | SSRIs/SNRIs | Not actionable | No recommendation | |
| HTR2A | Level 2 | SSRIs/SNRIs | Not actionable | No recommendation | |
| Gabapentinoids | GABRA1 | Level 3 | Gabapentin, Pregabalin | No recommendation | No recommendation |
| SLC7A5 | Level 3 | Gabapentin, Pregabalin | No recommendation | No recommendation |
| Trial/Year | Population and Setting | Genetic Focus and Intervention | Primary Outcome(s)/Status |
|---|---|---|---|
| NCT05452694/2022 | 235 adults after lumbar fusion or decompression surgery (UPMC, USA) | 16-gene opioid/NSAID panel (CYP2D6/3A4/2B6, OPRM1, ABCB1 ± risk score) returned pre-discharge | Composite opioid-related adverse events (sedation, respiratory depression, PONV) to 72 h; recruiting |
| NCT05525923/2023 | 200 adults undergoing thoracotomy/VATS lung resection | Same 16-gene panel guiding oxycodone dosing and rescue choices | 90-day chronic post-surgical pain; opioid-AE rate; recruiting |
| NCT04685304/2023 † | 315 primary care adults on tramadol/codeine/hydrocodone (PGx-ACT, USA) | Immediate vs. 6 mo-delayed PGx (CYP2D6 ± CYP2C19) + pharmacist CDS | Δ Pain-intensity at 3 mo; active, not recruiting |
| NCT06669650/2024 | 208 opioid-naïve adults, mixed surgeries (UTenn, USA) | Rapid saliva panel—CYP2D6 phenotype branches to hydromorphone vs. oxycodone regimen | Persistent opioid use at 90 days; active, not recruiting |
| NCT01140724/2022 | 1200 pediatric tonsillectomy patients, multi-center USA | Multi-gene morphine panel (COMT, CYP2D6, OPRM1, ABCC3) with opioid-avoidance algorithm | Genotype-morphine dose–response and opioid-AE composite; active, not recruiting |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin da Silva, I.; Plaza-Díaz, A.; Ruiz-Ramos, J.; Juanes-Borrego, A.; Riera, P. The Role of Pharmacogenetic Biomarkers in Pain. Biomedicines 2025, 13, 1935. https://doi.org/10.3390/biomedicines13081935
Martin da Silva I, Plaza-Díaz A, Ruiz-Ramos J, Juanes-Borrego A, Riera P. The Role of Pharmacogenetic Biomarkers in Pain. Biomedicines. 2025; 13(8):1935. https://doi.org/10.3390/biomedicines13081935
Chicago/Turabian StyleMartin da Silva, Ivan, Adrián Plaza-Díaz, Jesus Ruiz-Ramos, Ana Juanes-Borrego, and Pau Riera. 2025. "The Role of Pharmacogenetic Biomarkers in Pain" Biomedicines 13, no. 8: 1935. https://doi.org/10.3390/biomedicines13081935
APA StyleMartin da Silva, I., Plaza-Díaz, A., Ruiz-Ramos, J., Juanes-Borrego, A., & Riera, P. (2025). The Role of Pharmacogenetic Biomarkers in Pain. Biomedicines, 13(8), 1935. https://doi.org/10.3390/biomedicines13081935










