Deciphering Medulloblastoma: Epigenetic and Metabolic Changes Driving Tumorigenesis and Treatment Outcomes
Abstract
1. Introduction
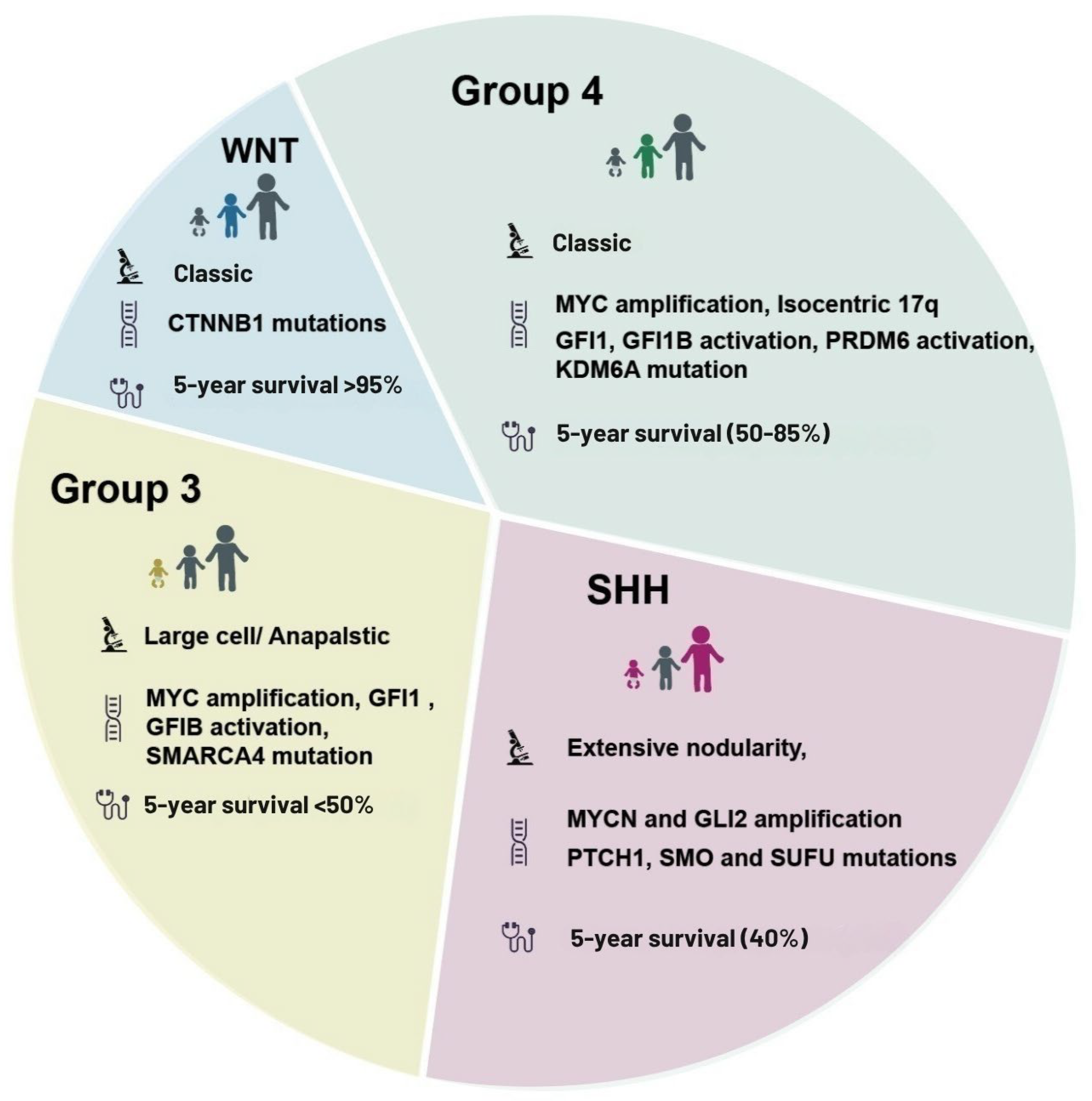
2. Medulloblastoma Subtypes and Molecular Classification
2.1. Current Classification
2.2. Subtype-Specific Pathways: Metabolic and Epigenetic Mechanisms
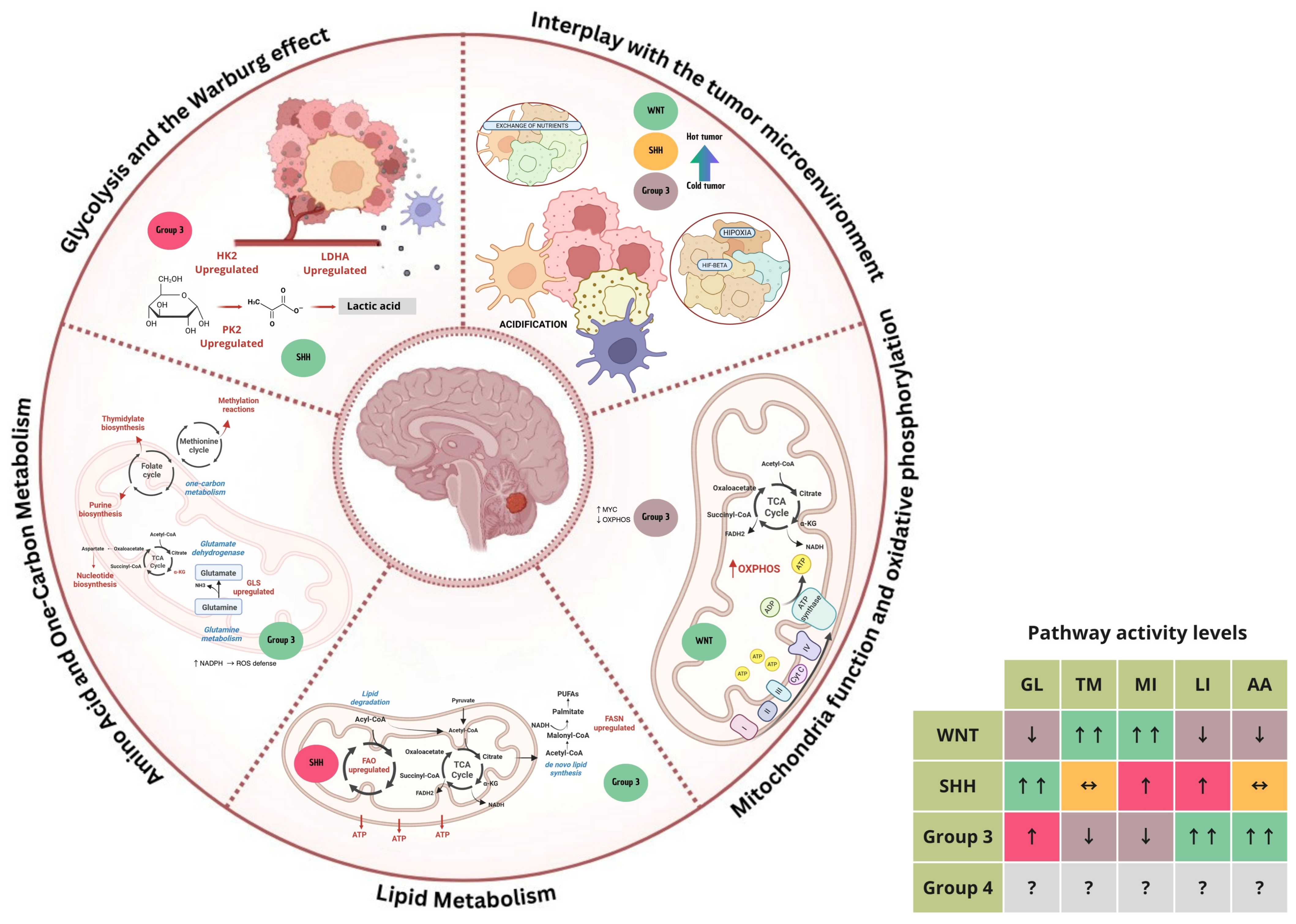
3. Metabolic Mechanisms in Medulloblastoma
3.1. Glycolysis and the Warburg Effect
3.2. Mitochondrial Function and Oxidative Phosphorylation
3.3. Lipid Metabolism
3.4. Amino Acid and One-Carbon Metabolism
3.5. Interplay with the Tumor Microenvironment
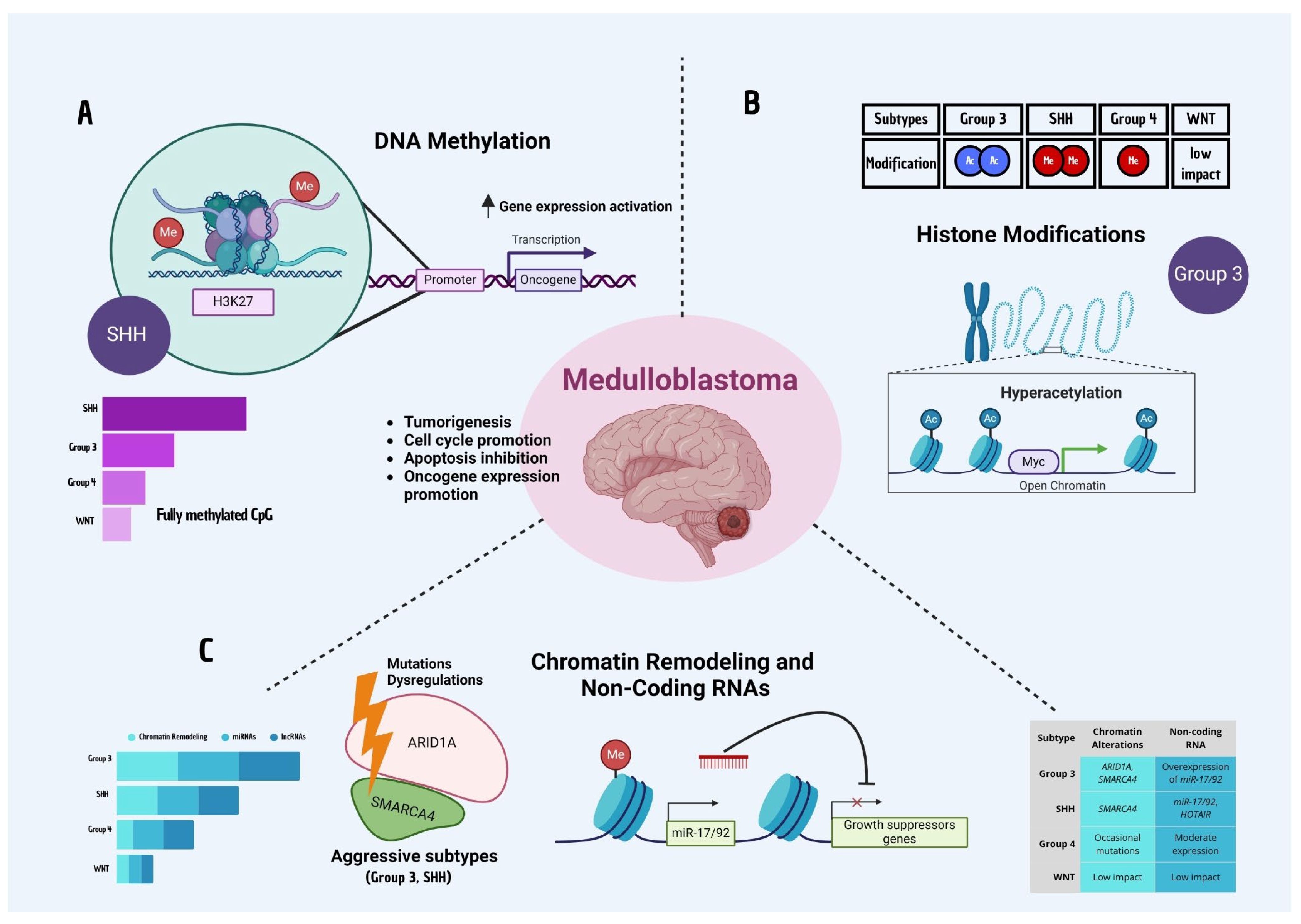
4. Epigenetic Mechanisms in Medulloblastoma
4.1. DNA Methylation
4.2. Histone Modifications
4.3. Chromatin Remodeling and Non-Coding RNAs
4.4. Epigenetic Heterogeneity and Tumor Adaptation
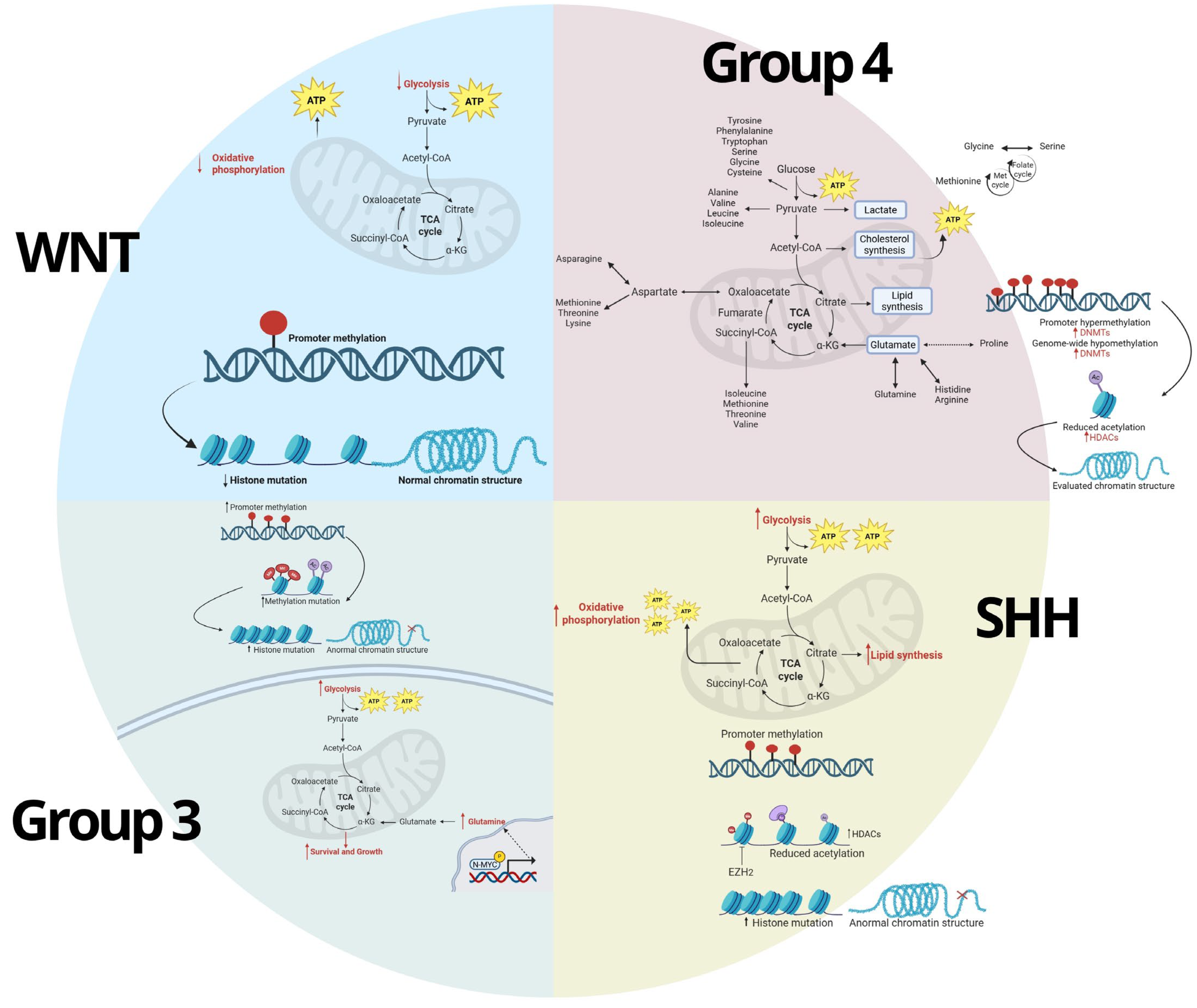
5. Interconnection Between Metabolic and Epigenetic Pathways
5.1. Metabolic Regulation of Epigenetics
5.2. Epigenetic Regulation of Metabolism
5.3. Metabolic and Epigenetic Crosstalk in MB
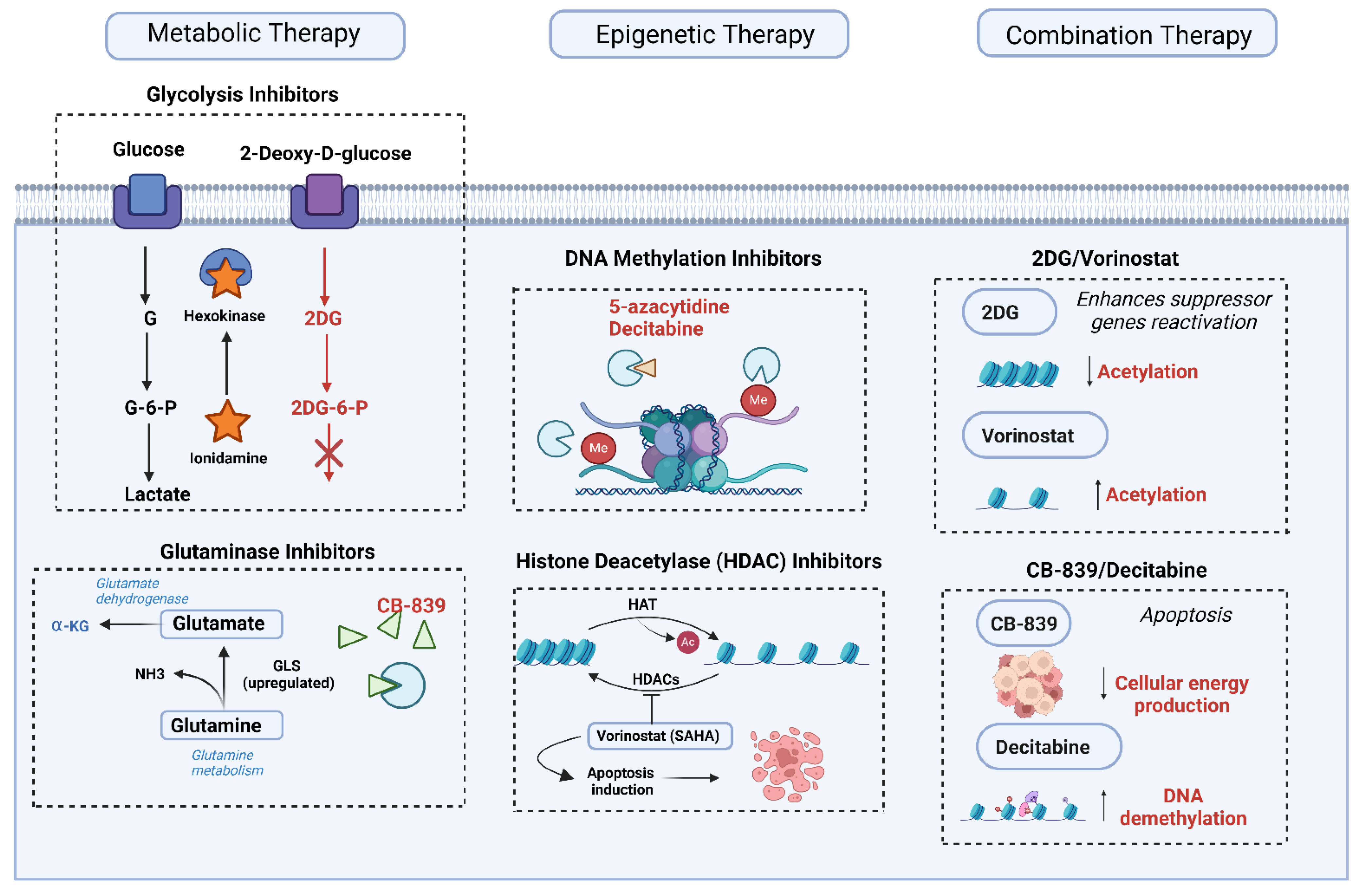
6. Current Therapeutic Approaches Targeting Metabolic and Epigenetic Mechanisms
6.1. Metabolic Therapies
6.2. Epigenetic Therapies
6.3. Combination Therapies
7. Challenges and Future Directions
7.1. Heterogeneity in Tumor Response
7.2. Biomarker Development
7.3. Potential for Immunometabolism and Immunoepigenetics
7.4. Preclinical Models and Translational Studies
8. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Rossi, A.; Caracciolo, V.; Russo, G.; Reiss, K.; Giordano, A. Medulloblastoma: From molecular pathology to therapy. Clin. Cancer Res. 2008, 14, 971–976. [Google Scholar] [CrossRef]
- Choi, J.Y. Medulloblastoma: Current Perspectives and Recent Advances. Brain Tumor Res. Treat. 2023, 11, 28–38. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Northcott, P.A.; Dubuc, A.M.; Pfister, S.; Taylor, M.D. Molecular subgroups of medulloblastoma. Expert Rev. Neurother. 2012, 12, 871–884. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Roussel, M.F.; Robinson, G.W. Role of MYC in Medulloblastoma. Cold Spring Harb. Perspect. Med. 2013, 3, a014308. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Marabitti, V.; Giansanti, M.; De Mitri, F.; Gatto, F.; Mastronuzzi, A.; Nazio, F. Pathological implications of metabolic reprogramming and its therapeutic potential in medulloblastoma. Front. Cell Dev. Biol. 2022, 10, 1007641. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Navarro, C.; Ortega, Á.; Santeliz, R.; Garrido, B.; Chacín, M.; Galban, N.; Vera, I.; De Sanctis, J.B.; Bermúdez, V. Metabolic Reprogramming in Cancer Cells: Emerging Molecular Mechanisms and Novel Therapeutic Approaches. Pharmaceutics 2022, 14, 1303. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Slika, H.; Alimonti, P.; Raj, D.; Caraway, C.; Alomari, S.; Jackson, E.M.; Tyler, B. The Neurodevelopmental and Molecular Landscape of Medulloblastoma Subgroups: Current Targets and the Potential for Combined Therapies. Cancers 2023, 15, 3889. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yi, J.; Wu, J. Epigenetic regulation in medulloblastoma. Mol. Cell Neurosci. 2018, 87, 65–76. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ray, S.; Chaturvedi, N.K.; Bhakat, K.K.; Rizzino, A.; Mahapatra, S. Subgroup-Specific Diagnostic, Prognostic, and Predictive Markers Influencing Pediatric Medulloblastoma Treatment. Diagnostics 2021, 12, 61. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jin, M.L.; Jeong, K.W. Histone modifications in drug-resistant cancers: From a cancer stem cell and immune evasion perspective. Exp. Mol. Med. 2023, 55, 1333–1347. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Joshi, P.; Katsushima, K.; Zhou, R.; Meoded, A.; Stapleton, S.; Jallo, G.; Raabe, E.; Eberhart, C.G.; Perera, R.J. The therapeutic and diagnostic potential of regulatory noncoding RNAs in medulloblastoma. Neurooncol. Adv. 2019, 1, vdz023. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Williamson, D.; Schwalbe, E.C.; Hicks, D.; Aldinger, K.A.; Lindsey, J.C.; Crosier, S.; Richardson, S.; Goddard, J.; Hill, R.M.; Castle, J.; et al. Medulloblastoma group 3 and 4 tumors comprise a clinically and biologically significant expression continuum reflecting human cerebellar development. Cell Rep. 2022, 40, 111162. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Patmore, D.M.; Jassim, A.; Nathan, E.; Gilbertson, R.J.; Tahan, D.; Hoffmann, N.; Tong, Y.; Smith, K.S.; Kanneganti, T.D.; Suzuki, H.; et al. DDX3X Suppresses the Susceptibility of Hindbrain Lineages to Medulloblastoma. Dev. Cell. 2020, 54, 455–470.e5. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ganesh, K.; Massagué, J. Targeting metastatic cancer. Nat. Med. 2021, 27, 34–44. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Smit, M.J.; Martini, T.E.I.; Armandari, I.; Bočkaj, I.; Zomerman, W.W.; de Camargo Magalhães, E.S.; Siragna, Z.; Meeuwsen, T.G.J.; Scherpen, F.J.G.; Schoots, M.H.; et al. The developmental stage of the medulloblastoma cell-of-origin restricts Sonic hedgehog pathway usage and drug sensitivity. J. Cell Sci. 2022, 135, jeb258608. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Menyhárt, O.; Győrffy, B. Principles of tumorigenesis and emerging molecular drivers of SHH-activated medulloblastomas. Ann. Clin. Transl. Neurol. 2019, 6, 990–1005. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lazow, M.A.; Palmer, J.D.; Fouladi, M.; Salloum, R. Medulloblastoma in the Modern Era: Review of Contemporary Trials, Molecular Advances, and Updates in Management. Neurotherapeutics 2022, 19, 1733–1751. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nguyen, N.M.; Cho, J. Hedgehog Pathway Inhibitors as Targeted Cancer Therapy and Strategies to Overcome Drug Resistance. Int. J. Mol. Sci. 2022, 23, 1733. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- ClinicalTrials.gov. ID NCT01878617. A Clinical and Molecular Risk-Directed Therapy for Newly Diagnosed Medulloblastoma; St. Jude Children’s Research Hospital., 2025. Available online: https://clinicaltrials.gov/study/NCT01878617#study-overview (accessed on 20 February 2025).
- Adiamah, M.; Poole, B.; Lindsey, J.C.; Kohe, S.; Morcavallo, A.; Burté, F.; Hill, R.M.; Blair, H.; Thompson, D.; Singh, M.; et al. MYC-dependent upregulation of the de novo serine and glycine synthesis pathway is a targetable metabolic vulnerability in group 3 medulloblastoma. Neuro-Oncol. 2025, 27, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Koczkodaj, D.; Muzyka-Kasietczuk, J.; Chocholska, S.; Podhorecka, M. Prognostic significance of isochromosome 17q in hematologic malignancies. Oncotarget 2021, 12, 708–718. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mlakar, V.; Dupanloup, I.; Gonzales, F.; Papangelopoulou, D.; Ansari, M.; Gumy-Pause, F. 17q Gain in Neuroblastoma: A Review of Clinical and Biological Implications. Cancers 2024, 16, 338. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, T.; Liu, L.; Chen, X.; Shen, Y.; Lian, G.; Shah, N.; Davidoff, A.M.; Yang, J.; Wang, R. MYCN drives glutaminolysis in neuroblastoma and confers sensitivity to an ROS augmenting agent. Cell Death Dis. 2018, 9, 220. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, L.; He, X.; Liu, X.; Zhang, F.; Huang, L.F.; Potter, A.S.; Xu, L.; Zhou, W.; Zheng, T.; Luo, Z.; et al. Single-Cell Transcriptomics in Medulloblastoma Reveals Tumor-Initiating Progenitors and Oncogenic Cascades during Tumorigenesis and Relapse. Cancer Cell 2019, 36, 302–318.e7. [Google Scholar] [CrossRef]
- Gorini, F.; Miceli, M.; de Antonellis, P.; Amente, S.; Zollo, M.; Ferrucci, V. Epigenetics and immune cells in medulloblastoma. Front. Genet. 2023, 14, 1135404. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huhtala, L.; Karabiyik, G.; Rautajoki, K.J. Development and epigenetic regulation of Atypical teratoid/rhabdoid tumors in the context of cell-of-origin and halted cell differentiation. Neurooncol. Adv. 2024, 6, vdae162. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rugo, H.S.; Jacobs, I.; Sharma, S.; Scappaticci, F.; Paul, T.A.; Jensen-Pergakes, K.; Malouf, G.G. The Promise for Histone Methyltransferase Inhibitors for Epigenetic Therapy in Clinical Oncology: A Narrative Review. Adv. Ther. 2020, 37, 3059–3082. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gu, M.; Ren, B.; Fang, Y.; Ren, J.; Liu, X.; Wang, X.; Zhou, F.; Xiao, R.; Luo, X.; You, L.; et al. Epigenetic regulation in cancer. MedComm 2024, 5, e495. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pérez-Salvia, M.; Esteller, M. Bromodomain inhibitors and cancer therapy: From structures to applications. Epigenetics 2017, 12, 323–339. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xu, L.; Yan, X.; Wang, J.; Zhao, Y.; Liu, Q.; Fu, J.; Shi, X.; Su, J. The Roles of Histone Deacetylases in the Regulation of Ovarian Cancer Metastasis. Int. J. Mol. Sci. 2023, 24, 15066. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, Y.; Li, Q.; Huang, Z.; Li, B.; Nice, E.C.; Huang, C.; Wei, L.; Zou, B. Targeting Glucose Metabolism Enzymes in Cancer Treatment: Current and Emerging Strategies. Cancers 2022, 14, 4568. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liao, M.; Yao, D.; Wu, L.; Luo, C.; Wang, Z.; Zhang, J.; Liu, B. Targeting the Warburg effect: A revisited perspective from molecular mechanisms to traditional and innovative therapeutic strategies in cancer. Acta Pharm. Sin. B 2024, 14, 953–1008. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nong, S.; Han, X.; Xiang, Y.; Qian, Y.; Wei, Y.; Zhang, T.; Tian, K.; Shen, K.; Yang, J.; Ma, X. Metabolic reprogramming in cancer: Mechanisms and therapeutics. MedComm 2023, 4, e218. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schiliro, C.; Firestein, B.L. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. Cells 2021, 10, 1056. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jin, H.R.; Wang, J.; Wang, Z.J.; Xi, M.J.; Xia, B.H.; Deng, K.; Yang, J.L. Lipid metabolic reprogramming in tumor microenvironment: From mechanisms to therapeutics. J. Hematol. Oncol. 2023, 16, 103. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Talapatra, J.; Reddy, M.M. Lipid Metabolic Reprogramming in Embryonal Neoplasms with MYCN Amplification. Cancers 2023, 15, 2144. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ducker, G.S.; Rabinowitz, J.D. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017, 25, 27–42. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- van Bree, N.F.H.N.; Wilhelm, M. The Tumor Microenvironment of Medulloblastoma: An Intricate Multicellular Network with Therapeutic Potential. Cancers 2022, 14, 5009. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Strejczek, A.; Woszczyk, D.; Urbaniak, H.; Różańska, M.; Robak, M.; Matuszewska, Z.; Barciszewska, A.M. Epigenetic-Based Therapy-A Prospective Chance for Medulloblastoma Patients’ Recovery. Int. J. Mol. Sci. 2021, 22, 4925. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Thomson, J.P.; Skene, P.J.; Selfridge, J.; Clouaire, T.; Guy, J.; Webb, S.; Kerr, A.R.; Deaton, A.; Andrews, R.; James, K.D.; et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 2010, 464, 1082–1086. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA methylation landscape of cancer. Trends Genet. 2021, 37, 1012–1027. [Google Scholar] [CrossRef] [PubMed]
- Hovestadt, V.; Ayrault, O.; Swartling, F.J.; Robinson, G.W.; Pfister, S.M.; Northcott, P.A. Medulloblastomics revisited: Biological and clinical insights from thousands of patients. Nat. Rev. Cancer 2020, 20, 42–56. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Parsons, M.J.; Tammela, T.; Dow, L.E. WNT as a Driver and Dependency in Cancer. Cancer Discov. 2021, 11, 2413–2429. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Swartling, F.J.; Čančer, M.; Frantz, A.; Weishaupt, H.; Persson, A.I. Deregulated proliferation and differentiation in brain tumors. Cell Tissue Res. 2015, 359, 225–254. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hutter, S.; Bolin, S.; Weishaupt, H.; Swartling, F.J. Modeling and Targeting MYC Genes in Childhood Brain Tumors. Genes 2017, 8, 107. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schwalbe, E.C.; Williamson, D.; Lindsey, J.C.; Hamilton, D.; Ryan, S.L.; Megahed, H.; Garami, M.; Hauser, P.; Dembowska-Baginska, B.; Perek, D.; et al. DNA methylation profiling of medulloblastoma allows robust subclassification and improved outcome prediction using formalin-fixed biopsies. Acta Neuropathol. 2013, 125, 359–371. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Silva-Hurtado, T.J.; Inocencio, J.F.; Yong, R.L. Emerging applications of hypomethylating agents in the treatment of glioblastoma (Review). Mol. Clin. Oncol. 2024, 21, 59. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Luo, Z.; Xin, D.; Liao, Y.; Berry, K.; Ogurek, S.; Zhang, F.; Zhang, L.; Zhao, C.; Rao, R.; Dong, X.; et al. Loss of phosphatase CTDNEP1 potentiates aggressive medulloblastoma by triggering MYC amplification and genomic instability. Nat. Commun. 2023, 14, 762. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rechberger, J.S.; Toll, S.A.; Vanbilloen, W.J.F.; Daniels, D.J.; Khatua, S. Exploring the Molecular Complexity of Medulloblastoma: Implications for Diagnosis and Treatment. Diagnostics 2023, 13, 2398. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Navickas, S.M.; Giles, K.A.; Brettingham-Moore, K.H.; Taberlay, P.C. The role of chromatin remodeler SMARCA4/BRG1 in brain cancers: A potential therapeutic target. Oncogene 2023, 42, 2363–2373. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Toden, S.; Goel, A. Non-coding RNAs as liquid biopsy biomarkers in cancer. Br. J. Cancer 2022, 126, 351–360. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Slika, H.; Shahani, A.; Wahi, R.; Miller, J.; Groves, M.; Tyler, B. Overcoming Treatment Resistance in Medulloblastoma: Underlying Mechanisms and Potential Strategies. Cancers 2024, 16, 2249. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cristalli, C.; Scotlandi, K. Targeting DNA Methylation Machinery in Pediatric Solid Tumors. Cells 2024, 13, 1209. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lei, Z.N.; Tian, Q.; Teng, Q.X.; Wurpel, J.N.D.; Zeng, L.; Pan, Y.; Chen, Z.S. Understanding and targeting resistance mechanisms in cancer. MedComm 2023, 4, e265. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Paredes, F.; Williams, H.C.; San Martin, A. Metabolic adaptation in hypoxia and cancer. Cancer Lett. 2021, 502, 133–142. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, N.; Ma, T.; Yu, B. Targeting epigenetic regulators to overcome drug resistance in cancers. Signal Transduct. Target. Ther. 2023, 8, 69. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yun, J.; Johnson, J.L.; Hanigan, C.L.; Locasale, J.W. Interactions between epigenetics and metabolism in cancers. Front. Oncol. 2012, 2, 163. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, G.; Bao, B.; Cheng, Y.; Tian, M.; Song, J.; Zheng, L.; Tong, Q. Acetyl-CoA metabolism as a therapeutic target for cancer. Biomed. Pharmacother. 2023, 168, 115741. [Google Scholar] [CrossRef] [PubMed]
- Martell, E.; Kuzmychova, H.; Kaul, E.; Senthil, H.; Chowdhury, S.R.; Morrison, L.C.; Fresnoza, A.; Zagozewski, J.; Venugopal, C.; Anderson, C.M.; et al. Metabolism-based targeting of MYC via MPC-SOD2 axis-mediated oxidation promotes cellular differentiation in group 3 medulloblastoma. Nat. Commun. 2023, 14, 2502. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tran, K.A.; Dillingham, C.M.; Sridharan, R. The role of α-ketoglutarate-dependent proteins in pluripotency acquisition and maintenance. J. Biol. Chem. 2019, 294, 5408–5419. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, R.; Zhao, E.; Yu, H.; Yuan, C.; Abbas, M.N.; Cui, H. Methylation across the central dogma in health and diseases: New therapeutic strategies. Signal Transduct. Target. Ther. 2023, 8, 310. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Testa, U.; Castelli, G.; Pelosi, E. Genetic Abnormalities, Clonal Evolution, and Cancer Stem Cells of Brain Tumors. Med. Sci. 2018, 6, 85. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kratz, E.M.; Sołkiewicz, K.; Kubis-Kubiak, A.; Piwowar, A. Sirtuins as Important Factors in Pathological States and the Role of Their Molecular Activity Modulators. Int. J. Mol. Sci. 2021, 22, 630. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Weyandt, J.D.; Thompson, C.B.; Giaccia, A.J.; Rathmell, W.K. Metabolic Alterations in Cancer and Their Potential as Therapeutic Targets. Am. Soc. Clin. Oncol. Educ. Book 2017, 37, 825–832. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lu, J.; Tan, M.; Cai, Q. The Warburg effect in tumor progression: Mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. 2015, 356 Pt A, 156–164. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Alshahrani, S.H.; Ibrahim, Y.S.; Jalil, A.T.; Altoum, A.A.; Achmad, H.; Zabibah, R.S.; Gabr, G.A.; Ramírez-Coronel, A.A.; Alameri, A.A.; Qasim, Q.A.; et al. Metabolic reprogramming by miRNAs in the tumor microenvironment: Focused on immunometabolism. Front. Oncol. 2022, 12, 1042196. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, X.Y.; Lin, J.D. Long Noncoding RNAs: A New Regulatory Code in Metabolic Control. Trends Biochem. Sci. 2015, 40, 586–596. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ge, T.; Gu, X.; Jia, R.; Ge, S.; Chai, P.; Zhuang, A.; Fan, X. Crosstalk between metabolic reprogramming and epigenetics in cancer: Updates on mechanisms and therapeutic opportunities. Cancer Commun. 2022, 42, 1049–1082. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, F.; Zhang, Z.W.; Zhang, J.; Zhang, S.; Zhang, H.; Zhao, C.; Chen, Y.; Luo, L.; Tong, W.M.; Li, C.; et al. Loss of 5-Hydroxymethylcytosine as an Epigenetic Signature That Correlates with Poor Outcomes in Patients with Medulloblastoma. Front. Oncol. 2021, 11, 603686. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Thienpont, B.; Steinbacher, J.; Zhao, H.; D’Anna, F.; Kuchnio, A.; Ploumakis, A.; Ghesquière, B.; Van Dyck, L.; Boeckx, B.; Schoonjans, L.; et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature 2016, 537, 63–68. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Taylor, C.T.; Scholz, C.C. The effect of HIF on metabolism and immunity. Nat. Rev. Nephrol. 2022, 18, 573–587. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huo, M.; Zhang, J.; Huang, W.; Wang, Y. Interplay Among Metabolism, Epigenetic Modifications, and Gene Expression in Cancer. Front. Cell Dev. Biol. 2021, 9, 793428. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cooney, T.M.; Cohen, K.J.; Guimaraes, C.V.; Dhall, G.; Leach, J.; Massimino, M.; Erbetta, A.; Chiapparini, L.; Malbari, F.; Kramer, K.; et al. Response assessment in diffuse intrinsic pontine glioma: Recommendations from the Response Assessment in Pediatric Neuro-Oncology (RAPNO) working group. Lancet Oncol. 2020, 21, e330–e336. [Google Scholar] [CrossRef] [PubMed]
- Stine, Z.E.; Schug, Z.T.; Salvino, J.M.; Dang, C.V. Targeting cancer metabolism in the era of precision oncology. Nat. Rev. Drug Discov. 2022, 21, 141–162. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, Y.; Sun, G.; Sun, X.; Li, F.; Zhao, L.; Zhong, R.; Peng, Y. The Potential of Lonidamine in Combination with Chemotherapy and Physical Therapy in Cancer Treatment. Cancers 2020, 12, 3332. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kelly, W.; Diaz Duque, A.E.; Michalek, J.; Konkel, B.; Caflisch, L.; Chen, Y.; Pathuri, S.C.; Madhusudanannair-Kunnuparampil, V.; Floyd, J.; Brenner, A. Phase II Investigation of TVB-2640 (Denifanstat) with Bevacizumab in Patients with First Relapse High-Grade Astrocytoma. Clin. Cancer Res. 2023, 29, 2419–2425. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Claiborne, M.D.; Leone, R. Differential glutamine metabolism in the tumor microenvironment—Studies in diversity and heterogeneity: A mini-review. Front. Oncol. 2022, 12, 1011191. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, Z.; Wang, G.; Li, Y.; Lei, D.; Xiang, J.; Ouyang, L.; Wang, Y.; Yang, J. Recent progress in DNA methyltransferase inhibitors as anticancer agents. Front. Pharmacol. 2022, 13, 1072651. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- El Omari, N.; Khalid, A.; Makeen, H.A.; Alhazmi, H.A.; Albratty, M.; Mohan, S.; Tan, C.S.; Ming, L.C.; Chook, J.B.; Bouyahya, A. Stochasticity of anticancer mechanisms underlying clinical effectiveness of vorinostat. Heliyon 2024, 10, e33052. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fu, L.L.; Tian, M.; Li, X.; Li, J.J.; Huang, J.; Ouyang, L.; Zhang, Y.; Liu, B. Inhibition of BET bromodomains as a therapeutic strategy for cancer drug discovery. Oncotarget 2015, 6, 5501–5516. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Laneve, P.; Caffarelli, E. The Non-coding Side of Medulloblastoma. Front. Cell Dev. Biol. 2020, 8, 275. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tufail, M.; Jiang, C.H.; Li, N. Altered metabolism in cancer: Insights into energy pathways and therapeutic targets. Mol. Cancer. 2024, 23, 203. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Okabe, S.; Tanaka, Y.; Moriyama, M.; Gotoh, A. Inhibition of glutaminolysis alone and in combination with HDAC inhibitor has anti-myeloma therapeutic effects. Cancer Drug Resist. 2024, 7, 25. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hay, J.F.; Lappin, K.; Liberante, F.; Kettyle, L.M.; Matchett, K.B.; Thompson, A.; Mills, K.I. Integrated analysis of the molecular action of Vorinostat identifies epi-sensitised targets for combination therapy. Oncotarget 2017, 8, 67891–67903. [Google Scholar] [CrossRef][Green Version]
- Robbins, C.J.; Bou-Dargham, M.J.; Sanchez, K.; Rosen, M.C.; Sang, Q.A. Decoding Somatic Driver Gene Mutations and Affected Signaling Pathways in Human Medulloblastoma Subgroups. J. Cancer 2018, 9, 4596–4610. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Connal, S.; Cameron, J.M.; Sala, A.; Brennan, P.M.; Palmer, D.S.; Palmer, J.D.; Perlow, H.; Baker, M.J. Liquid biopsies: The future of cancer early detection. J. Transl. Med. 2023, 21, 118. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhu, Y.; Li, X.; Wang, L.; Hong, X.; Yang, J. Metabolic reprogramming and crosstalk of cancer-related fibroblasts and immune cells in the tumor microenvironment. Front. Endocrinol. 2022, 13, 988295. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, J.; Xu, J.; Wang, W.; Zhang, B.; Yu, X.; Shi, S. Epigenetic regulation in the tumor microenvironment: Molecular mechanisms and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 210. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Eisemann, T.; Wechsler-Reya, R.J. Coming in from the cold: Overcoming the hostile immune microenvironment of medulloblastoma. Genes Dev. 2022, 36, 514–532. [Google Scholar] [CrossRef]
- Boutin, L.; Liu, M.; Déchanet Merville, J.; Bedoya-Reina, O.; Wilhelm, M.T. EphA2 and phosphoantigen-mediated selective killing of medulloblastoma by γδT cells preserves neuronal and stem cell integrity. Oncoimmunology 2025, 14, 2485535. [Google Scholar] [CrossRef]
- Chaturvedi, N.K.; Kling, M.J.; Griggs, C.N.; Kesherwani, V.; Shukla, M.; McIntyre, E.M.; Ray, S.; Liu, Y.; McGuire, T.R.; Sharp, J.G.; et al. A Novel Combination Approach Targeting an Enhanced Protein Synthesis Pathway in MYC-driven (Group 3) Medulloblastoma. Mol. Cancer Ther. 2020, 19, 1351–1362. [Google Scholar] [CrossRef]
- Oberholtzer, N.; Quinn, K.M.; Chakraborty, P.; Mehrotra, S. New Developments in T Cell Immunometabolism and Implications for Cancer Immunotherapy. Cells 2022, 11, 708. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lovett, M.L.; Nieland, T.J.F.; Dingle, Y.L.; Kaplan, D.L. Innovations in 3-Dimensional Tissue Models of Human Brain Physiology and Diseases. Adv. Funct. Mater. 2020, 30, 1909146. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Joseph, P.D.; Craig, J.C.; Caldwell, P.H. Clinical trials in children. Br. J. Clin. Pharmacol. 2015, 79, 357–369. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| Subtype | Key Metabolic Features | Key Epigenetic Features | Therapeutic Targets (Examples) |
|---|---|---|---|
| WNT | Minimal metabolic reprogramming; low glycolytic and OXPHOS activity [7] | Stable DNA methylation and histone profile [10,26] | Limited metabolic targeting; possible use of HDAC inhibitors [26] |
| SHH | Elevated glycolysis, fatty acid synthesis, and mitochondrial OXPHOS [6,19] | EZH2 and HDAC activity; SHH-specific DNA methylation [27,28] | SMO inhibitors (e.g., vismodegib) [19], EZH2 inhibitors [28], HDACi [27] |
| Group 3 | High glycolytic flux, glutamine dependency; MYC-driven metabolism [6,24] | Widespread histone acetylation/methylation; BRD4 dependence [29,30] | Glycolysis inhibitors [32], GLS inhibitors [8], BET inhibitors [30] |
| Group 4 | Amino acid metabolism (e.g., BCAT1), mitochondrial respiration; SLC transporter upregulation [6,25] | Distinct DNA methylation profiles; HDAC sensitivity [26,31] | Targeting amino acid metabolism or mitochondrial function [25]; HDACi [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonifacio-Mundaca, J.; Casavilca-Zambrano, S.; Desterke, C.; Casafont, Í.; Mata-Garrido, J. Deciphering Medulloblastoma: Epigenetic and Metabolic Changes Driving Tumorigenesis and Treatment Outcomes. Biomedicines 2025, 13, 1898. https://doi.org/10.3390/biomedicines13081898
Bonifacio-Mundaca J, Casavilca-Zambrano S, Desterke C, Casafont Í, Mata-Garrido J. Deciphering Medulloblastoma: Epigenetic and Metabolic Changes Driving Tumorigenesis and Treatment Outcomes. Biomedicines. 2025; 13(8):1898. https://doi.org/10.3390/biomedicines13081898
Chicago/Turabian StyleBonifacio-Mundaca, Jenny, Sandro Casavilca-Zambrano, Christophe Desterke, Íñigo Casafont, and Jorge Mata-Garrido. 2025. "Deciphering Medulloblastoma: Epigenetic and Metabolic Changes Driving Tumorigenesis and Treatment Outcomes" Biomedicines 13, no. 8: 1898. https://doi.org/10.3390/biomedicines13081898
APA StyleBonifacio-Mundaca, J., Casavilca-Zambrano, S., Desterke, C., Casafont, Í., & Mata-Garrido, J. (2025). Deciphering Medulloblastoma: Epigenetic and Metabolic Changes Driving Tumorigenesis and Treatment Outcomes. Biomedicines, 13(8), 1898. https://doi.org/10.3390/biomedicines13081898






