Decoding Immunodeficiencies with Artificial Intelligence: A New Era of Precision Medicine
Abstract
1. Immunodeficiency’s Two Faces: Primary and Secondary
Artificial Intelligence: Revolutionizing the Medical Field
2. Early Immunodeficiency Diagnosis and AI
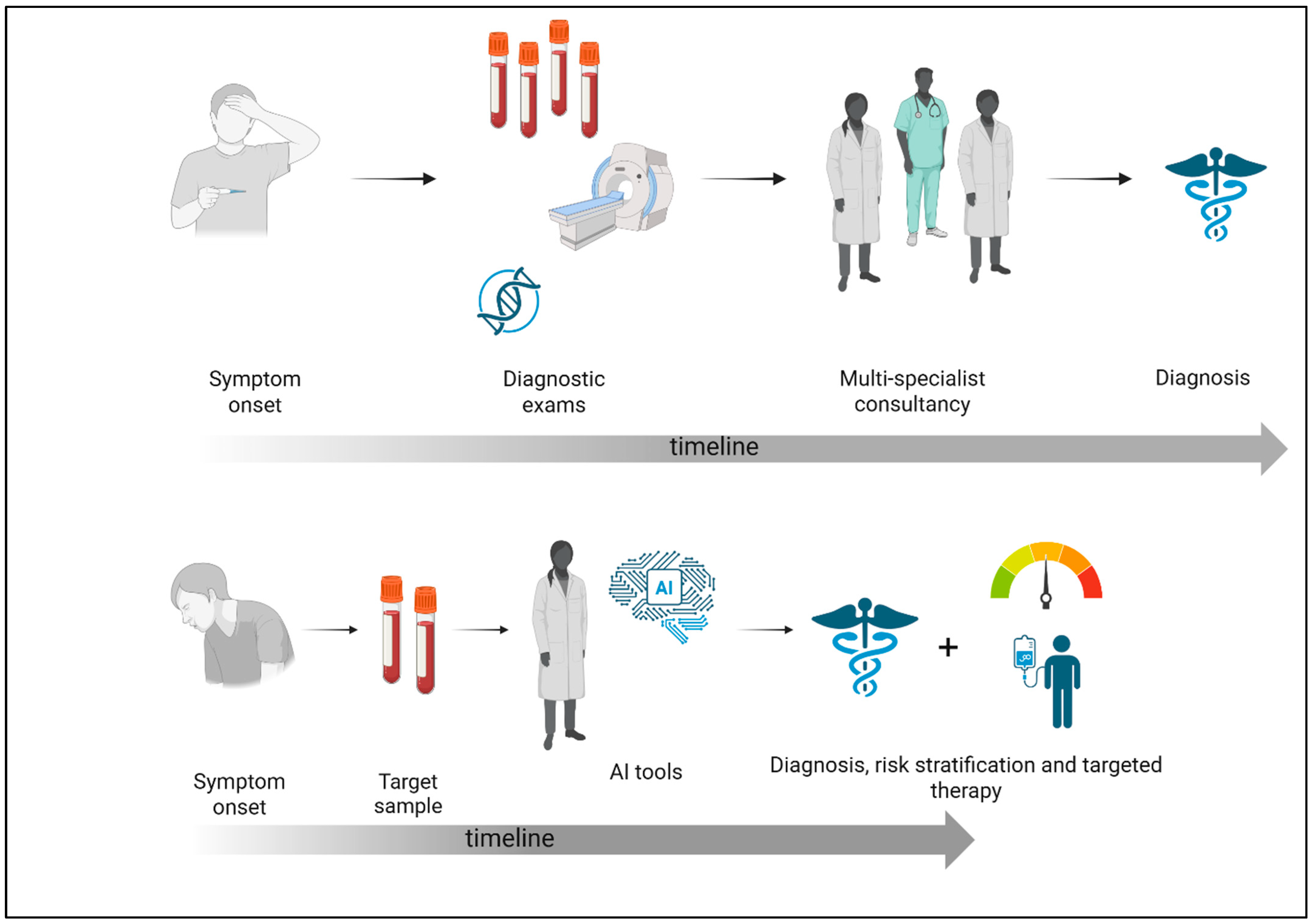
2.1. General Considerations
2.2. Warning Signs to AI-Powered Diagnosis: IEI Identification’s New Era
2.3. AI-Enhanced Flow Cytometry: Early PID Detection Precision Tools
2.4. AI Innovation in Immunopeptidomics and Phenotype Detection
3. Enhancing Immunodeficiency Management: The Role of AI in Risk Stratification
4. AI Improves Immunodeficiency Care: Pathogen Management
4.1. Anti-COVID-19 Vaccines, AI, and Immunodeficiency
4.2. Infections and Immunodeficiency: AI as a Diagnostic Tool
5. The AI Method for Diagnosing Late Immunodeficiencies
5.1. Immunodeficiencies and Cancer Susceptibility
5.2. Differential Diagnosis: PID vs. SID in Lymphoproliferative Disorders
6. AI in Secondary Immunodeficiency: Focus on MDS and CLL
6.1. Secondary Immunodeficiency in MDS
6.2. Secondary Immunodeficiency in CLL
7. Limitations and Biases of AI in Immunodeficiency Management
Challenges of AI in Immunodeficiencies
8. Future Perspectives and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sattler, S. The role of the immune system beyond the fight against infection. Adv. Exp. Med. Biol. 2017, 1003, 3–14. [Google Scholar]
- Justiz Vaillant, A.A.; Qurie, A. Immunodeficiency. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2023. [Google Scholar]
- Bonilla, F.A.; Khan, D.A.; Ballas, Z.K.; Chinen, J.; Frank, M.M.; Hsu, J.T.; Keller, M.; Kobrynski, L.J.; Komarow, H.D.; Mazer, B.; et al. Practice parameter for the diagnosis and management of primary immunodeficiency. J. Allergy Clin. Immunol. 2015, 136, 1186–1205.e78. [Google Scholar] [CrossRef]
- Picard, C.; Bobby Gaspar, H.; Al-Herz, W.; Bousfiha, A.; Casanova, J.L.; Chatila, T.; Crow, Y.J.; Cunningham-Rundles, C.; Etzioni, A.; Franco, J.L. International Union of Immunological Societies: 2017 Primary Immunodeficiency Diseases Committee Report on inborn errors of immunity. J. Clin. Immunol. 2018, 38, 96–128. [Google Scholar] [CrossRef] [PubMed]
- Buckley, R.H.; Orange, J.S. Primary immunodeficiency diseases. In Middleton’s Allergy Principles and Practice, 8th ed.; Adkinson, N.F., Bochner, B.S., Burks, A.W., Busse, W.W., Holgate, S.T., Lemanske, R.F., O’Hehir, R.E., Eds.; Saunders: Philadelphia, PA, USA, 2014; pp. 1144–1174. [Google Scholar]
- McDonald-McGinn, D.M.; Sullivan, K.E.; Marino, B.; Philip, N.; Swillen, A.; Vorstman, J.A.; Zackai, E.H.; Emanuel, B.S.; Vermeesch, J.R.; Morrow, B.E.; et al. 22q11.2 deletion syndrome. Nat. Rev. Dis. Primers 2015, 1, 1–19. [Google Scholar] [CrossRef]
- Kobrynski, L.; Powell, R.W.; Bowen, S. Prevalence and morbidity of primary immunodeficiency diseases, United States 2001–2007. J. Clin. Immunol. 2014, 34, 954–961. [Google Scholar] [CrossRef] [PubMed]
- Tangye, S.G.; Al-Herz, W.; Bousfiha, A.; Cunningham-Rundles, C.; Franco, J.L.; Holland, S.M.; Klein, C.; Morio, T.; Oksenhendler, E.; Picard, C.; et al. The ever-increasing array of novel inborn errors of immunity: An interim update by the IUIS committee. J. Clin. Immunol. 2021, 41, 666–679. [Google Scholar] [CrossRef] [PubMed]
- Roganović, J.; Bellesi, G. Inborn Errors of Immunity: New Insights. Acta Med. Acad. 2024, 53, 293–302. [Google Scholar]
- Anderson, J.T.; Cowan, J.; Condino-Neto, A.; Levy, D.; Prusty, S. Health-related quality of life in primary immunodeficiencies: Impact of delayed diagnosis and treatment burden. Clin. Immunol. 2022, 236, 108931. [Google Scholar] [CrossRef]
- Abolhassani, H.; Azizi, G.; Sharifi, L.; Yazdani, R.; Mohsenzadegan, M.; Delavari, S.; Sohani, M.; Shirmast, P.; Chavoshzadeh, Z.; Mahdaviani, S.A.; et al. Global systematic review of primary immunodeficiency registries. Expert Rev. Clin. Immunol. 2020, 16, 717–732. [Google Scholar] [CrossRef]
- Wood, P. UK Primary Immunodeficiency Network. Primary antibody deficiencies: Recognition, clinical diagnosis and referral of patients. Clin. Med. 2009, 9, 595–599. [Google Scholar] [CrossRef]
- Šedivá, A.; Milota, T.; Litzman, J.; Quinti, I.; Meyts, I.; Burns, S.; Jolles, S. Medical algorithm: Diagnosis and management of antibody immunodefciencies. Allergy 2021, 76, 3841–3844. [Google Scholar] [CrossRef]
- Wadhwa, P.D.; Morrison, V.A. Infectious complications of chronic lymphocytic leukemia. Semin. Oncol. 2006, 33, 240–249. [Google Scholar] [CrossRef]
- Allegra, A.; Tonacci, A.; Musolino, C.; Pioggia, G.; Gangemi, S. Secondary Immunodeficiency in Hematological Malignancies: Focus on Multiple Myeloma and Chronic Lymphocytic Leukemia. Front. Immunol. 2021, 12, 738915. [Google Scholar] [CrossRef] [PubMed]
- Tuano, K.S.; Seth, N.; Chinen, J. Secondary immunodefciencies: An overview. Ann. Allergy Asthma Immunol. 2021, 127, 617–626. [Google Scholar] [CrossRef]
- Otani, I.M.; Lehman, H.K.; Jongco, A.M.; Tsao, L.R.; Azar, A.E.; Tarrant, T.K.; Engel, E.; Walter, J.E.; Truong, T.Q.; Khan, D.A.; et al. Practical guidance for the diagnosis and management of secondary hypogammaglobulinemia: A Work Group Report of the AAAAI Primary Immunodeficiency and Altered Immune Response Committees. J. Allergy Clin. Immunol. 2022, 149, 1525–1560. [Google Scholar] [CrossRef] [PubMed]
- Fitzmaurice, C.; Allen, C.; Barber, R.M.; Barregard, L.; Bhutta, Z.A.; Brenner, H.; Dicker, D.J.; Chimed-Orchir, O.; Dandona, R.; Dandona, L.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2017, 3, 524–548. [Google Scholar] [PubMed]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Whicher, D.; Ahmed, M.; Israni, S.T.; Matheny, M. (Eds.) Artificial Intelligence in Health Care: The Hope, the Hype, the Promise, the Peril; National Academy of Medicine: Washington, DC, USA, 2019. [Google Scholar]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef]
- Witten, I.H.; Frank, E.; Hall, M.A.; Pal, C.J. Data Mining Practical Machine Learning Tools and Techniques, 4th ed.; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Rider, N.L.; Srinivasan, R.; Khoury, P. Artificial intelligence and the hunt for immunological disorders. Curr. Opin. Allergy Clin. Immunol. 2020, 20, 565–573. [Google Scholar] [CrossRef]
- Rudrapatna, V.A.; Butte, A.J. Opportunities and challenges in using real-world data for health care. J. Clin. Investig. 2020, 130, 565–574. [Google Scholar] [CrossRef]
- Banda, J.M.; Seneviratne, M.; Hernandez-Boussard, T.; Shah, N.H. Advances in Electronic Phenotyping: From Rule-Based Definitions to Machine Learning Models. Annu. Rev. Biomed. Data Sci. 2018, 1, 53–68. [Google Scholar] [CrossRef]
- Bastarache, L.; Hughey, J.J.; Goldstein, J.A.; Bastraache, J.A.; Das, S.; Zaki, N.C.; Zeng, C.; Tang, L.A.; Roden, D.M.; Denny, J.C. Improving the phenotype risk score as a scalable approach to identifying patients with Mendelian disease. J. Am. Med. Inform. Assoc. 2019, 26, 1437–1447. [Google Scholar] [CrossRef]
- Pacheco, J.A.; Rasmussen, L.V.; Kiefer, R.C.; Campion, T.R.; Speltz, P.; Carroll, R.J.; Stallings, S.C.; Mo, H.; Ahuja, M.; Jiang, G.; et al. A case study evaluating the portability of an executable computable phenotype algorithm across multiple institutions and electronic health record environments. J. Am. Med. Inform. Assoc. 2018, 25, 1540–1546. [Google Scholar] [CrossRef] [PubMed]
- Darcy, A.M.; Louie, A.K.; Roberts, L.W. Machine Learning and the Profession of Medicine. JAMA 2016, 315, 551–552. [Google Scholar] [CrossRef] [PubMed]
- Kantor, P. Foundations of Statistical Natural Language Processing; MIT Press: Cambridge, MA, USA, 1999; pp. 91–92. [Google Scholar]
- Murff, H.J.; FitzHenry, F.; Matheny, M.E.; Gentry, N.; Kotter, K.L.; Crimin, K.; Dittus, R.S.; Rosen, A.K.; Elkin, P.L.; Brown, S.H.; et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA 2011, 306, 848–855. [Google Scholar] [PubMed]
- Afzal, N.; Sohn, S.; Abram, S.; Scott, C.G.; Chaudhry, R.; Liu, H.; Kullo, I.J.; Arruda-Olson, A.M. Mining peripheral arterial disease cases from narrative clinical notes using natural language processing. J. Vasc. Surg. 2017, 65, 1753–1761. [Google Scholar] [CrossRef]
- Odnoletkova, I.; Kindle, G.; Quinti, I.; Grimbacher, B.; Knerr, V.; Gathmann, B.; Ehl, S.; Mahlaoui, N.; Van Wilder, P.; Bogaerts, K.; et al. Plasma Protein Therapeutics Association (PPTA) Taskforce. The burden of common variable immunodeficiency disorders: A retrospective analysis of the European Society for Immunodeficiency (ESID) registry data. Orphanet J. Rare Dis. 2018, 13, 201. [Google Scholar]
- Seth, N.; Tuano, K.S.; Chinen, J. Inborn errors of immunity: Recent progress. J. Allergy Clin. Immunol. 2021, 148, 1442–1450. [Google Scholar] [CrossRef]
- Mayampurath, A.; Ajith, A.; Anderson-Smits, C.; Chang, S.C.; Brouwer, E.; Johnson, J.; Baltasi, M.; Volchenboum, S.; Devercelli, G.; Ciaccio, C.E. Early Diagnosis of Primary Immunodeficiency Disease Using Clinical Data and Machine Learning. J. Allergy Clin. Immunol. Pract. 2022, 10, 3002–3007.e5. [Google Scholar] [CrossRef]
- Dunphy, C.H. Applications of flow cytometry and immunohistochemistry to diagnostic hematopathology. Arch. Pathol. Lab. Med. 2004, 128, 1004–1022. [Google Scholar] [CrossRef]
- Oliveira, J.B.; Fleisher, T.A. Molecular- and flow cytometry-based diagnosis of primary immunodeficiency disorders. Curr. Allergy Asthma Rep. 2010, 10, 460–467. [Google Scholar] [CrossRef]
- Robinson, J.P.; Ostafe, R.; Iyengar, S.N.; Rajwa, B.; Fischer, R. Flow Cytometry: The Next Revolution. Cells 2023, 12, 1875. [Google Scholar] [CrossRef]
- Vandenberghe, M.E.; Scott, M.L.; Scorer, P.W.; Söderberg, M.; Balcerzak, D.; Barker, C. Relevance of deep learning to facilitate the diagnosis of HER2 status in breast cancer. Sci. Rep. 2017, 7, 45938. [Google Scholar] [CrossRef] [PubMed]
- Pavillon, N.; Hobro, A.J.; Akira, S.; Smith, N.I. Noninvasive detection of macrophage activation with single-cell resolution through machine learning. Proc. Nat. Acad. Sci. USA 2018, 115, E2676–E2685. [Google Scholar] [CrossRef] [PubMed]
- Modell, F.; Puente, D.; Modell, V. From genotype to phenotype. Further studies measuring the impact of a physician education and public awareness campaign on early diagnosis and management of primary immunodeficiencies. Immunol. Res. 2009, 44, 132–149. [Google Scholar] [CrossRef]
- Thalhammer, J.; Kindle, G.; Nieters, A.; Rusch, S.; Seppänen, M.R.J.; Fischer, A.; Grimbacher, B.; Edgar, D.; Buckland, M.; Mahlaoui, N.; et al. Initial presenting manifestations in 16,486 patients with inborn errors of immunity include infections and noninfectious manifestations. J. Allergy Clin. Immunol. 2021, 148, 1332–1341.e5. [Google Scholar] [CrossRef]
- Cunningham-Rundles, C.; Sidi, P.; Estrella, L.; Doucette, J. Identifying undiagnosed primary immunodeficiency diseases in minority subjects by using computer sorting of diagnosis codes. J. Allergy Clin. Immunol. 2004, 113, 747–755. [Google Scholar] [CrossRef]
- Modell, V.; Gee, B.; Lewis, D.B.; Orange, J.S.; Roifman, C.M.; Routes, J.M.; Sorensen, R.U.; Notarangelo, L.D.; Modell, F. Global study of primary immunodeficiency diseases (PI)-diagnosis, treatment, and economic impact: An updated report from the Jeffrey Modell Foundation. Immunol. Res. 2011, 51, 61–70. [Google Scholar] [CrossRef]
- Bastarache, L. Using phecodes for research with the electronic health record: From PheWAS to PheRS. Annu. Rev. Biomed. Data Sci. 2021, 4, 1–19. [Google Scholar] [CrossRef]
- Rivière, J.G.; Soler Palacín, P.; Butte, M.J. Proceedings from the inaugural Artificial Intelligence in Primary Immune Deficiencies (AIPID) conference. J. Allergy Clin. Immunol. 2024, 153, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.; Stephens, A.V.; Mester, R.; Knyazev, S.; Kohn, L.A.; Freund, M.K.; Bondhus, L.; Hill, B.L.; Schwarz, T.; Zaitlen, N.; et al. Electronic health record signatures identify undiagnosed patients with common variable immunodeficiency disease. Sci. Transl. Med. 2024, 16, eade4510. [Google Scholar] [CrossRef]
- Schiavo, E.; Martini, B.; Attardi, E.; Consonni, F.; Ciullini Mannurita, S.; Coniglio, M.L.; Tellini, M.; Chiocca, E.; Fotzi, I. Autoimmune cytopenias and dysregulated immunophenotype act as warning signs of inborn errors of immunity: Results from a prospective study. Front. Immunol. 2021, 12, 790455. [Google Scholar] [CrossRef] [PubMed]
- Rider, N.L.; Li, Y.; Chin, A.T.; DiGiacomo, D.V.; Dutmer, C.; Farmer, J.R.; Roberts, K.; Savova, G.; Ong, M.S. Evaluating large language model performance to support the diagnosis and management of patients with primary immune disorders. J. Allergy Clin. Immunol. 2025, 156, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Sarikonda, A.; Isch, E.; Self, M.; Sambangi, A.; Carreras, A.; Sivaganesan, A.; Harrop, J.; Jallo, J. Evaluating the Adherence of Large Language Models to Surgical Guidelines: A Comparative Analysis of Chatbot Recommendations and North American Spine Society (NASS) Coverage Criteria. Cureus 2024, 16, e68521. [Google Scholar] [CrossRef]
- Tangye, S.G.; Al-Herz, W.; Bousfiha, A.; Cunningham-Rundles, C.; Franco, J.L.; Holland, S.M.; Klein, C.; Morio, T.; Oksenhendler, E.; Picard, C.; et al. Human inborn errors of immunity: 2022 update on the classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 2022, 42, 1473–1475. [Google Scholar] [CrossRef] [PubMed]
- Futatani, T.; Miyawaki, T.; Tsukada, S.; Hashimoto, S.; Kunikata, T.; Arai, S.; Kurimoto, M.; Niida, Y.; Matsuoka, H.; Sakiyama, Y.; et al. Deficient expression of Bruton’s tyrosine kinase in monocytes from X-linked agammaglobulinemia as evaluated by a flow cytometric analysis and its clinical application to carrier detection. Blood 1998, 91, 595–602. [Google Scholar]
- Oliveira, J.B.; Bleesing, J.J.; Dianzani, U.; Fleisher, T.A.; Jaffe, E.S.; Lenardo, M.J.; Rieux-Laucat, F.; Siegel, R.M.; Su, H.C.; Teachey, D.T.; et al. Revised diagnostic criteria and classification for the autoimmune lymphoproliferative syndrome (ALPS): Report from the 2009 NIH International Workshop. Blood 2010, 116, e35–e40. [Google Scholar] [CrossRef]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef]
- Lima, K.; Abrahamsen, T.G.; Foelling, I.; Natvig, S.; Ryder, L.P.; Olaussen, R.W. Low thymic output in the 22q11.2 deletion syndrome measured by CCR9+CD45RA+ T cell counts and T cell receptor rearrangement excision circles. Clin. Exp. Immunol. 2010, 161, 98–107. [Google Scholar] [CrossRef]
- Mallesh, N.; Zhao, M.; Meintker, L.; Höllein, A.; Elsner, F.; Lüling, H.; Haferlach, T.; Kern, W.; Westermann, J.; Brossart, P.; et al. Knowledge transfer to enhance the performance of deep learning models for automated classification of B cell neoplasms. Patterns 2021, 2, 100351. [Google Scholar] [CrossRef]
- Monaghan, S.A.; Li, J.L.; Liu, Y.C.; Ko, M.Y.; Boyiadzis, M.; Chang, T.Y.; Wang, Y.F.; Lee, C.C.; Swerdlow, S.H.; Ko, B.S. A Machine Learning Approach to the Classification of Acute Leukemias and Distinction From Nonneoplastic Cytopenias Using Flow Cytometry Data. Am. J. Clin. Pathol. 2022, 157, 546–553. [Google Scholar] [CrossRef]
- Stagno, F.; Mirabile, G.; Rizzotti, P.; Bottaro, A.; Pagana, A.; Gangemi, S.; Allegra, A. Using Artificial Intelligence to Enhance Myelodysplastic Syndrome Diagnosis, Prognosis, and Treatment. Biomedicines 2025, 13, 835. [Google Scholar] [CrossRef]
- Lu, Z.; Morita, M.; Yeager, T.S.; Lyu, Y.; Wang, S.Y.; Wang, Z.; Fan, G. Validation of Artificial Intelligence (AI)-Assisted Flow Cytometry Analysis for Immunological Disorders. Diagnostics 2024, 14, 420. [Google Scholar] [CrossRef]
- Sun, R.; Limkin, E.J.; Vakalopoulou, M.; Dercle, L.; Champiat, S.; Han, S.R.; Verlingue, L.; Brandao, D.; Lancia, A.; Ammari, S.; et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: An imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018, 19, 1180–1191. [Google Scholar] [CrossRef] [PubMed]
- Moghram, B.A.; Nabil, E.; Badr, A. Ab-initio conformational epitope structure prediction using genetic algorithm and SVM for vaccine design. Comput. Methods Programs Biomed. 2018, 153, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Degoot, A.M.; Chirove, F.; Ndifon, W. Trans-Allelic Model for Prediction of Peptide: MHC-II Interactions. Front. Immunol. 2018, 9, 1410. [Google Scholar] [CrossRef]
- Nagpal, G.; Gupta, S.; Chaudhary, K.; Dhanda, S.K.; Prakash, S.; Raghava, G.P. VaccineDA: Prediction, design and genome-wide screening of oligodeoxynucleotide-based vaccine adjuvants. Sci. Rep. 2015, 5, 12478. [Google Scholar] [CrossRef] [PubMed]
- Rider, N.L.; Cahill, G.; Motazedi, T.; Wei, L.; Kurian, A.; Noroski, L.M.; Seeborg, F.O.; Chinn, I.K.; Roberts, K. PI Prob: A risk prediction and clinical guidance system for evaluating patients with recurrent infections. PLoS ONE 2021, 16, e0237285. [Google Scholar] [CrossRef]
- Arora, P.; Boyne, D.; Slater, J.J.; Gupta, A.; Brenner, D.R.; Druzdzel, M.J. Bayesian Networks for Risk Prediction Using Real-World Data: A Tool for Precision Medicine. Value Health 2019, 22, 439–445. [Google Scholar] [CrossRef]
- Sesen, M.B.; Nicholson, A.E.; Banares-Alcantara, R.; Kadir, T.; Brady, M. Bayesian networks for clinical decision support in lung cancer care. PLoS ONE 2013, 8, e82349. [Google Scholar] [CrossRef]
- Loghmanpour, N.A.; Kormos, R.L.; Kanwar, M.K.; Teuteberg, J.J.; Murali, S.; Antaki, J.F. A Bayesian Model to Predict Right Ventricular Failure Following Left Ventricular Assist Device Therapy. JACC Heart Fail. 2016, 4, 711–721. [Google Scholar] [CrossRef]
- Constantinou, A.C.; Fenton, N.; Marsh, W.; Radlinski, L. From complex questionnaire and interviewing data to intelligent Bayesian network models for medical decision support. Artif. Intell. Med. 2016, 67, 75–93. [Google Scholar] [CrossRef]
- Wasyluk, H.; Onisko, A.; Druzdzel, M.J. Support of diagnosis of liver disorders based on a causal Bayesian network model. Med. Sci. Monit. 2001, 7 (Suppl. S1), 327–332. [Google Scholar] [PubMed]
- Heckerman, D.E.; Horvitz, E.J.; Nathwani, B.N. Toward normative expert systems: Part, I. The Pathfinder project. Methods Inf. Med. 1992, 31, 90–105. [Google Scholar] [CrossRef]
- Heckerman, D.E.; Nathwani, B.N. Toward normative expert systems: Part II. Probability-based representations for efficient knowledge acquisition and inference. Methods Inf. Med. 1992, 31, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Vega, F.; Younes, L. Learning multivariate distributions by competitive assembly of marginals. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 398–410. [Google Scholar] [CrossRef]
- Heckerman, D.E. A Tutorial on Learning With Bayesian Networks. In Innovations in Bayesian Networks; Microsoft Corporation, Division AT; 1995. Contract No.: MSR-TR-95-06; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Rider, N.L.; Miao, D.; Dodds, M.; Modell, V.; Modell, F.; Quinn, J.; Schwarzwald, H.; Orange, J.S. Calculation of a Primary Immunodeficiency “Risk Vital Sign” via Population-Wide Analysis of Claims Data to Aid in Clinical Decision Support. Front. Pediatr. 2019, 7, 70. [Google Scholar] [CrossRef]
- Rider, N.L.; Coffey, M.; Kurian, A.; Quinn, J.; Orange, J.S.; Modell, V.; Modell, F. A validated artificial intelligence-based pipeline for population-wide primary immunodeficiency screening. J. Allergy Clin. Immunol. 2023, 151, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Modell, V.; Quinn, J.; Ginsberg, G.; Gladue, R.; Orange, J.; Modell, F. Modeling strategy to identify patients with primary immunodeficiency utilizing risk management and outcome measurement. Immunol. Res. 2017, 65, 713–720. [Google Scholar] [CrossRef]
- American Academy of Allergy, Asthma, and Immunology. Immunodeficiencies Disease Codes. Available online: https://www.aaaai.org/Aaaai/media/MediaLibrary/PDF%20Documents/Practice%20Management/finances-coding/ICD-10-Codes-Immunodeficiencies.pdf (accessed on 15 May 2025).
- Bastarache, L.; Hughey, J.J.; Hebbring, S.; Marlo, J.; Zhao, W.; Ho, W.T.; Van Driest, S.L.; McGregor, T.L.; Mosley, J.D.; Wells, Q.S. Phenotype risk scores identify patients with unrecognized mendelian disease patterns. Science 2018, 359, 1233–1239. [Google Scholar] [CrossRef]
- Cassini, T.; Bastarache, L.; Zeng, C.; Han, S.T.; Wang, J.; He, J.; Denny, J.C. A test of automated use of electronic health records to aid in diagnosis of genetic disease. Genet. Med. 2023, 25, 100966. [Google Scholar] [CrossRef]
- Langerbeins, P.; Hallek, M. COVID-19 in patients with hematologic malignancy. Blood 2022, 140, 236–252. [Google Scholar] [CrossRef]
- Piñana, J.L.; López-Corral, L.; Martino, R.; Vazquez, L.; Pérez, A.; Martin-Martin, G.; Gago, B.; Sanz-Linares, G.; Sanchez-Salinas, A.; Villalon, L.; et al. SARS-CoV-2 vaccine response and rate of breakthrough infection in patients with hematological disorders. J. Hematol. Oncol. 2022, 15, 54. [Google Scholar] [CrossRef]
- Piñana, J.L.; Vazquez, L.; Calabuig, M.; López-Corral, L.; Martin-Martin, G.; Villalon, L.; Sanz-Linares, G.; Conesa-Garcia, V.; Sanchez-Salinas, A.; Gago, B.; et al. One-year breakthrough SARS-CoV-2 infection and correlates of protection in fully vaccinated hematological patients. Blood Cancer J. 2023, 13, 8. [Google Scholar] [CrossRef]
- La, J.; Wu, J.T.; Branch-Elliman, W.; Huhmann, L.; Han, S.S.; Brophy, M.; Do, N.V.; Lin, A.Y.; Fillmore, N.R.; Munshi, N.C. Increased COVID-19 breakthrough infection risk in patients with plasma cell disorders. Blood 2022, 140, 782–785. [Google Scholar] [CrossRef]
- Potter, A.L.; Vaddaraju, V.; Venkateswaran, S.; Mansur, A.; Bajaj, S.S.; Kiang, M.V.; Jena, A.B.; Yang, C.J. Deaths Due to COVID-19 in Patients With Cancer During Different Waves of the Pandemic in the US. JAMA Oncol. 2023, 9, 1417–1422. [Google Scholar] [CrossRef]
- Piñana, J.L.; Heras, I.; Aiello, T.F.; García-Cadenas, I.; Vazquez, L.; Lopez-Jimenez, J.; Chorão, P.; Aroca, C.; García-Vidal, C.; Arroyo, I.; et al. Remdesivir or Nirmatrelvir/Ritonavir Therapy for Omicron SARS-CoV-2 Infection in Hematological Patients and Cell Therapy Recipients. Viruses 2023, 15, 2066. [Google Scholar] [CrossRef] [PubMed]
- Piñana, J.L.; Guerreiro, M.; Solano, C. SARS-CoV-2 Immunity in Hematopoietic Stem Cell Transplant and Cell Therapy Recipients: What Do We Know, and What Remains to Be Determined? Hemato 2023, 4, 170–183. [Google Scholar] [CrossRef]
- Cesaro, S.; Ljungman, P.; Mikulska, M.; Hirsch, H.H.; von Lilienfeld-Toal, M.; Cordonnier, C.; Meylan, S.; Mehra, V.; Styczynski, J.; Marchesi, F.; et al. Recommendations for the management of COVID-19 in patients with haematological malignancies or haematopoietic cell transplantation, from the 2021 European Conference on Infections in Leukaemia (ECIL 9). Leukemia 2022, 36, 1467–1480. [Google Scholar] [CrossRef] [PubMed]
- Cesaro, S.; Mikulska, M.; Hirsch, H.H.; Styczynski, J.; Meylan, S.; Cordonnier, C.; Navarro, D.; von Lilienfeld-Toal, M.; Mehra, V.; Marchesi, F.; et al. Update of recommendations for the management of COVID-19 in patients with haematological malignancies, haematopoietic cell transplantation and CAR T therapy, from the 2022 European Conference on Infections in Leukaemia (ECIL 9). Leukemia 2023, 37, 1933–1938. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Belenguer, P.; Piñana, J.L.; Sánchez-Montañés, M.; Soria-Olivas, E.; Martínez-Sober, M.; Serrano-López, A.J. A machine learning approach to identify groups of patients with hematological malignant disorders. Comput. Methods Programs Biomed. 2024, 246, 10801. [Google Scholar] [CrossRef] [PubMed]
- Furman, C.D.; Rayner, A.V.; Tobin, E.P. Pneumonia in older residents of long- term care facilities. Am. Fam. Physician 2004, 70, 1495–1500. [Google Scholar]
- Di Pasquale, M.F.; Sotgiu, G.; Gramegna, A.; Radovanovic, D.; Terraneo, S.; Reyes, L.F.; Rupp, J.; González Del Castillo, J.; Blasi, F.; Aliberti, S.; et al. Prevalence and etiology of community-acquired pneumonia in Immunocompromised patients. Clin. Infect. Dis. 2019, 68, 1482–1493. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Chen, Y.; Xie, P.; Luo, Y.; Wang, B.; Meng, Y.; Zhong, J.; Mei, J.; Zou, W. Development of a predictive machine learning model for pathogen profiles in patients with secondary immunodeficiency. BMC Med. Inform. Decis. Mak. 2024, 24, 48. [Google Scholar] [CrossRef]
- Jain, S.; Nehra, M.; Kumar, R.; Dilbaghi, N.; Hu, T.; Kumar, S.; Kaushik, A.; Li, C.Z. Internet of medical things (IoMT)-integrated biosensors for point-of-care testing of infectious diseases. Biosens. Bioelectron. 2021, 179, 113074. [Google Scholar] [CrossRef] [PubMed]
- Tiri, A.; Masetti, R.; Conti, F.; Tignanelli, A.; Turrini, E.; Bertolini, P.; Esposito, S.; Pession, A. Inborn errors of immunity and cancer. Biology 2021, 10, 313. [Google Scholar] [CrossRef]
- Gangemi, S.; Allegra, A.; Musolino, C. Lymphoproliferative disease and cancer among patients with common variable immunodeficiency. Leuk. Res. 2015, 39, 389–396. [Google Scholar] [CrossRef]
- Riaz, I.B.; Faridi, W.; Patnaik, M.M.; Abraham, R.S. A Systematic Review on Predisposition to Lymphoid (B and T cell) Neoplasias in Patients With Primary Immunodefciencies and Immune Dysregulatory Disorders (Inborn Errors of Immunity). Front. Immunol. 2019, 16, 777. [Google Scholar]
- Kolijn, P.M.; Langerak, A.W. Immune dysregulation as a leading principle for lymphoma development in diverse immunological backgrounds. Immunol. Lett. 2023, 263, 46–59. [Google Scholar] [CrossRef]
- Voskoboinik, I.; Trapani, J.A. Perforinopathy: A spectrum of human immune disease caused by defective perforin delivery or function. Front. Immunol. 2013, 4, 441. [Google Scholar] [CrossRef]
- Compagno, N.; Cinetto, F.; Boscaro, E.; Semenzato, G.; Agostini, C. Serum free light chains in the differential diagnosis and prognosis of primary and secondary hypogammaglobulinemia. J. Allergy Clin. Immunol. 2015, 135, 1075–1077.e6. [Google Scholar] [CrossRef] [PubMed]
- Knight, A.K.; Cunningham-Rundles, C. Inflammatory and autoimmune complications of common variable immune deficiency. Autoimmun. Rev. 2006, 5, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Kano, G.; Nakatani, T.; Yagi, K.; Sakamoto, I.; Imamura, T. Complicated pathophysiology behind rituximab-induced persistent hypogammaglobulinemia. Immunol. Lett. 2014, 159, 76–78. [Google Scholar] [CrossRef]
- Caeser, R.; Di Re, M.; Krupka, J.A.; Gao, J.; Lara-Chica, M.; Dias, J.M.L.; Cooke, S.L.; Fenner, R.; Usheva, Z.; Runge, H.F.P.; et al. Genetic modification of primary human B cells to model high-grade lymphoma. Nat. Commun. 2019, 10, 4543. [Google Scholar] [CrossRef]
- Xu, F.; Wu, L.Y.; He, Q.; Wu, D.; Zhang, Z.; Song, L.X.; Zhao, Y.S.; Su, J.Y.; Zhou, L.Y.; Guo, J.; et al. Exploration of the role of gene mutations in myelodysplastic syndromes through a sequencing design involving a small number of target genes. Sci. Rep. 2017, 7, 43113. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, L. Identification of latent core genes and pathways associated with myelodysplastic syndromes based on integrated bioinformatics analysis. Hematology 2020, 25, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Elhadary, M.; Elshoeibi, A.M.; Badr, A.; Elsayed, B.; Metwally, O.; Elshoeibi, A.M.; Mattar, M.; Alfarsi, K.; AlShammari, S.; Alshurafa, A.; et al. Revolutionizing chronic lymphocytic leukemia diagnosis: A deep dive into the diverse applications of machine learning. Blood Rev. 2023, 62, 101134. [Google Scholar] [CrossRef]
- Zhu, Y.; Gan, X.; Qin, R.; Lin, Z. Identification of Six Diagnostic Biomarkers for Chronic Lymphocytic Leukemia Based on Machine Learning Algorithms. J. Oncol. 2022, 2022, 3652107. [Google Scholar] [CrossRef]
- Xia, D.; Leon, A.J.; Yan, J.; Silva, A.; Bakhtiari, M.; Tremblay-LeMay, R.; Selvarajah, S.; Sabatini, P.; Diamandis, P.; Pugh, T.; et al. DNA Methylation-Based Classification of Small B-Cell Lymphomas: A Proof-of-Principle Study. J. Mol. Diagn. 2021, 23, 1774–1786. [Google Scholar] [CrossRef]
- Ng, D.P.; Zuromski, L.M. Augmented human intelligence and automated diagnosis in flow cytometry for hematologic malignancies. Am. J. Clin. Pathol. 2021, 155, 597–605. [Google Scholar] [CrossRef]
- Féré, M.; Gobinet, C.; Liu, L.H.; Beljebbar, A.; Untereiner, V.; Gheldof, D.; Chollat, M.; Klossa, J.; Chatelain, B.; Piot, O. Implementation of a classification strategy of Raman data collected in different clinical conditions: Application to the diagnosis of chronic lymphocytic leukemia. Anal. Bioanal. Chem. 2020, 412, 949–962. [Google Scholar] [CrossRef]
- Prince, B.T.; Thielen, B.K.; Williams, K.W.; Kellner, E.S.; Arnold, D.E.; Cosme-Blanco, W.; Redmond, M.T.; Hartog, N.L.; Chong, H.J.; Holland, S.M. Geographic Variability and Pathogen-Specific Considerations in the Diagnosis and Management of Chronic Granulomatous Disease. Pediatr. Health Med. Ther. 2020, 11, 257–268. [Google Scholar] [CrossRef]
- Maurya, R.; Rajput, N.; Diviit, M.G.; Mahapatra, S.; Ojha, M.K. Exploring the potential of lightweight large language models for AI-based mental health counselling task: A novel comparative study. Sci. Rep. 2025, 15, 22463. [Google Scholar] [CrossRef] [PubMed]
- Orange, J.S.; Seeborg, F.O.; Boyle, M.; Scalchunes, C.; Hernandez-Trujillo, V. Family Physician Perspectives on Primary Immunodeficiency Diseases. Front. Med. 2016, 3, 12. [Google Scholar] [CrossRef]
- Gao, S.; Fang, A.; Huang, Y.; Giunchiglia, V.; Noori, A.; Schwarz, J.R.; Ektefaie, Y.; Kondic, J.; Zitnik, M. Empowering biomedical discovery with AI agents. Cell 2024, 187, 6125–6151. [Google Scholar] [CrossRef]
- Khoury, P.; Srinivasan, R.; Kakumanu, S.; Ochoa, S.; Keswani, A.; Sparks, R.; Rider, N.L. A Framework for Augmented Intelligence in Allergy and Immunology Practice and Research-A Work Group Report of the AAAAI Health Informatics, Technology, and Education Committee. J. Allergy Clin. Immunol. Pract. 2022, 10, 1178–1188. [Google Scholar] [CrossRef]
- You, Y.; Lai, X.; Pan, Y.; Zheng, H.; Vera, J.; Liu, S.; Deng, S.; Zhang, L. Artificial intelligence in cancer target identification and drug discovery. Signal Transduct. Target. Ther. 2022, 7, 156. [Google Scholar] [CrossRef]
- Subramanian, M.; Wojtusciszyn, A.; Favre, L.; Boughorbel, S.; Shan, J.; Letaief, K.B.; Pitteloud, N.; Chouchane, L. Precision medicine in the era of artificial intelligence: Implications in chronic disease management. J. Transl. Med. 2020, 18, 472. [Google Scholar] [CrossRef]
- Liu, G.Y.; Yu, D.; Fan, M.M.; Zhang, X.; Jin, Z.Y.; Tang, C.; Liu, X.F. Antimicrobial resistance crisis: Could artificial intelligence be the solution? Mil. Med. Res. 2024, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Goktas, P.; Karakaya, G.; Kalyoncu, A.F.; Damadoglu, E. Artificial Intelligence Chatbots in Allergy and Immunology Practice: Where Have We Been and Where Are We Going? J. Allergy Clin. Immunol. Pract. 2023, 11, 2697–2700. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, Y.I.; Schuman, J.S.; Shabsigh, R.; Caplan, A.; Al-Aswad, L.A. Ethics of Artificial Intelligence in Medicine and Ophthalmology. Asia Pac. J. Ophthalmol. 2021, 10, 289–298. [Google Scholar] [CrossRef] [PubMed]
| Immunodeficiency Category | Key Characteristics | Representative Disorders |
|---|---|---|
| Antibody Deficiencies | Decreased or absent antibody synthesis, resulting in recurrent bacterial infections | Common Variable Immunodeficiency (CVID), X-linked Agammaglobulinemia (XLA) |
| Combined Immunodeficiencies | Deficiencies impacting both T and B lymphocytes, resulting in severe and early-onset infections | Severe Combined Immunodeficiency (SCID), DiGeorge Syndrome |
| Phagocyte Disorders | Reduced efficacy of phagocytes in eradicating infections | Chronic Granulomatous Disease |
| Complement Deficiencies | Deficiencies in the complement cascade hinder opsonization and lysis of microorganisms | C1q, C2, C3 Deficiencies |
| Diseases of Immune Dysregulation | Unregulated immunological activation resulting in autoimmunity and lymphoproliferation | Autoimmune Lymphoproliferative Syndrome (ALPS), IPEX Syndrome |
| Defects of Innate Immunity | Failures in innate immune signaling pathways | MyD88 Deficiency, IRAK-4 Deficiency |
| Autoinflammatory Disorders | Spontaneous inflammation resulting from dysfunction of the innate immune system and absence of autoantibodies | Familial Mediterranean Fever, TNF Receptor-Associated Periodic Syndrome (TRAPS) |
| Syndromic Immunodeficiencies | Immunodeficiency linked to developmental anomalies or multisystem disorders | Wiskott–Aldrich Syndrome, Ataxia–Telangiectasia |
| Bone Marrow Failure | Hematopoietic defects lead to cytopenias and heightened vulnerability to infections | Fanconi Anemia, Dyskeratosis Congenita, Hematologic Neoplasia |
| AI Tool/Technique | Application in Immunodeficiency |
|---|---|
| Predictive Algorithms | Early detection, risk stratification |
| Electronic Phenotyping (EP) | Automated illness diagnosis |
| Automated Flow Cytometry Analysis (the DeepFlowTM software) | Detection of immune cell abnormalities |
| Machine Learning-Based Tool | Identification of high-risk individuals, reducing diagnostic delay |
| Deep Learning (DL)/Artificial Neural Networks (ANNs) | Analysis of high-dimensional data |
| Natural Language Processing (NLP) | Extracting information from unstructured text |
| SPIRIT Analyzer | Primary immunodeficiency tracking |
| Phenotype Capture Tool | CVID diagnosis |
| Regression Model | Identification of CVID patients |
| Logistic Regression (LR)/Elastic Nets (ENs)/Random Forests (RFs) | Estimating PIDD probability |
| Large Language Models (LLMs) | Assisting in diagnosis and management |
| AI Tool/Technique | Key Features | Limitations/Biases |
|---|---|---|
| Predictive Algorithms | Analyze large EHR or claims data to generate risk scores for PIDs | May depend on coding practices; lacks sensitivity to atypical phenotypes; needs validation across cohorts |
| Electronic Phenotyping (EP) | Uses computable phenotypes to identify diseases in large datasets | Portability across institutions varies; limited by data quality and completeness |
| Automated Flow Cytometry (DeepFlow™) | High-dimensional clustering, objective gating, standardized reports | Requires robust instrument data; cannot fully replace expert morphological assessment |
| Machine Learning-Based Tools | Learns hidden associations; processes complex clinical variables | Sensitive to data heterogeneity; generalizability may be limited |
| Deep Learning (DL)/Artificial Neural Networks (ANNs) | Models complex nonlinear relationships (e.g., flow data, images) | Often “black box”; interpretation challenges for clinicians |
| Natural Language Processing (NLP) | Transforms free text into structured data for ML | Depending on quality/standardization of documentation; may miss nuance |
| SPIRIT Analyzer | Uses ICD and pharmacy codes to classify patients into risk categories | Rely heavily on billing data, potentially missing under-coded presentations |
| Phenotype Capture Tool | Collects HPO-coded phenotypes; builds weighted risk scores | Expert weighting can introduce subjective bias; needs continuous update with new phenotypes |
| Regression Models | Integrate multiple phenotypes + lab data to predict risk | May overfit to local patient profiles; performance varies by healthcare system |
| Logistic Regression (LR)/Elastic Nets (ENs)/Random Forests (RFs) | Transparent, interpretable; can rank variable importance | Needs well-curated, balanced data; may miss nonlinear interactions |
| Large Language Models (LLMs) | Generate differential diagnoses, summarize complex histories | Prone to suggesting common over rare diseases; non-reproducible outputs; requires expert oversight |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sciaccotta, R.; Barone, P.; Murdaca, G.; Fazio, M.; Stagno, F.; Gangemi, S.; Genovese, S.; Allegra, A. Decoding Immunodeficiencies with Artificial Intelligence: A New Era of Precision Medicine. Biomedicines 2025, 13, 1836. https://doi.org/10.3390/biomedicines13081836
Sciaccotta R, Barone P, Murdaca G, Fazio M, Stagno F, Gangemi S, Genovese S, Allegra A. Decoding Immunodeficiencies with Artificial Intelligence: A New Era of Precision Medicine. Biomedicines. 2025; 13(8):1836. https://doi.org/10.3390/biomedicines13081836
Chicago/Turabian StyleSciaccotta, Raffaele, Paola Barone, Giuseppe Murdaca, Manlio Fazio, Fabio Stagno, Sebastiano Gangemi, Sara Genovese, and Alessandro Allegra. 2025. "Decoding Immunodeficiencies with Artificial Intelligence: A New Era of Precision Medicine" Biomedicines 13, no. 8: 1836. https://doi.org/10.3390/biomedicines13081836
APA StyleSciaccotta, R., Barone, P., Murdaca, G., Fazio, M., Stagno, F., Gangemi, S., Genovese, S., & Allegra, A. (2025). Decoding Immunodeficiencies with Artificial Intelligence: A New Era of Precision Medicine. Biomedicines, 13(8), 1836. https://doi.org/10.3390/biomedicines13081836







