Non-Coding RNAs as Critical Modulators of Cholesterol Metabolism in Cancer
Abstract
1. Introduction
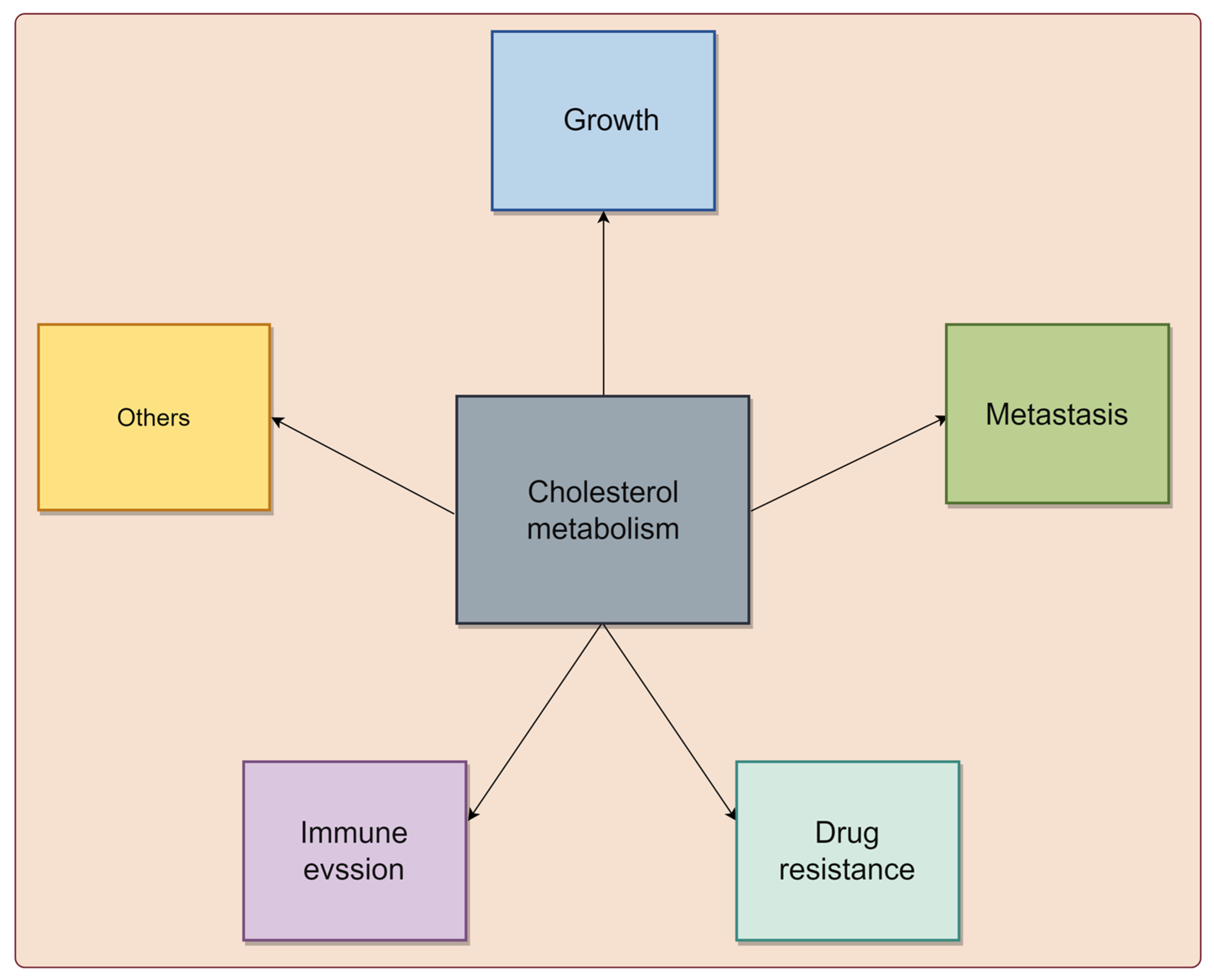
2. Cholesterol Metabolism Is Associated with Cancer Progression
2.1. Cholesterol Metabolism Is Involved in the Growth of Tumor Cells
2.2. Cholesterol Metabolism Regulates the Migration and Invasion of Tumor Cells
2.3. Cholesterol Metabolism Is Associated with the Drug Resistance of Tumor Cells
2.4. Cholesterol Metabolism Modulates the Immune Response in Cancer
3. ncRNAs Regulate Cholesterol Metabolism in Cancers
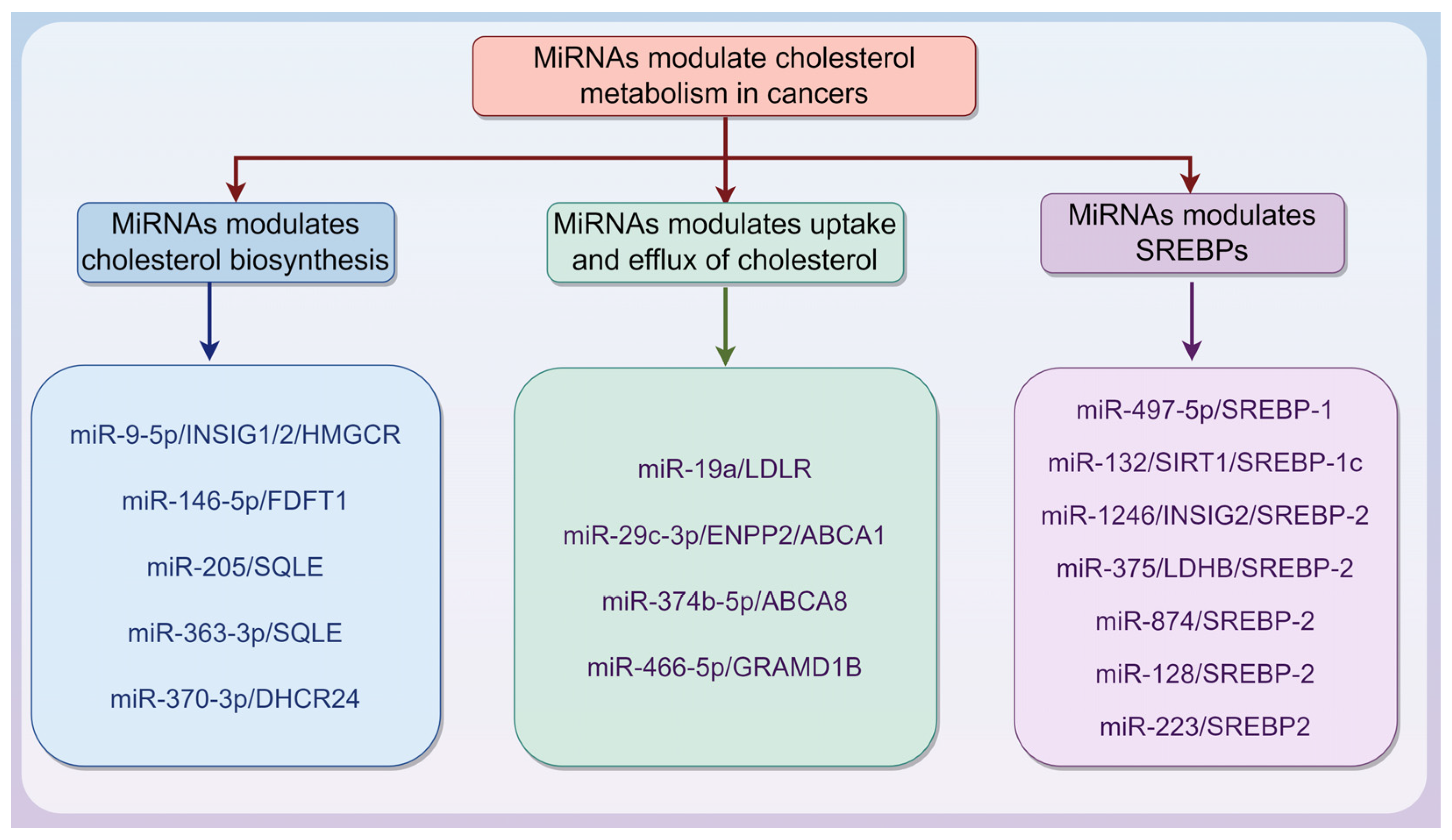
3.1. MiRNAs Are Involved in Modulating Cholesterol Metabolism in Cancers
3.1.1. MiRNAs Modulate Cholesterol Biosynthesis in Cancers
3.1.2. MiRNAs Modulate Uptake and Efflux of Cholesterol in Cancers
3.1.3. MiRNAs Modulate SREBPs in Cancers
| miRNAs | Cancer | Expression | Targets | Pathway | Phenotypes | Ref. |
|---|---|---|---|---|---|---|
| miR-874 | breast cancer | down | PMVK, SREBF2 | p53 | cell cycle arrest apoptosis | [76] |
| miR-29c-3p | melanoma | / | ENPP2 | ENPP2-PPARγ-LXRα/β/ ABCA1 | migration invasion ECM remodeling | [65] |
| miR-146b-5p | bladder cancer | up | FDFT1 | / | cisplatin sensitivity | [57] |
| miR-374b-5p | HCC | up | ABCA8 | ERK/ZEB1 | Proliferation metastasis | [66] |
| miR-375 | breast cancer | up | LDHB | LDHB/SREBP2 | proliferation | [75] |
| miR-4646-5p | TNBC | / | GRAMD1B | / | proliferation migration | [21] |
| miR-205 | PCa | down | SQLE | SQLE/AR | Proliferation AR inhibitor resistance | [58] |
| miR-9-5p | breast cancer | up | INSIG1, INSIG2 ATF3 | INSIG1/INSIG2/HMGCR ATF3/CH25H | metastasis | [55] |
| miR-370-3p | cervical cancer | / | DHCR24 | / | proliferation migration | [60] |
| miR-132 | glioma | / | SIRT1 | SIRT1/SREBP-1c/ HMGCR/FASN | proliferation invasion apoptosis | [73] |
| miR-1246 | CRC | up | INSIG1 | INSIG1/SREBP2 | proliferation metastasis | [74] |
| miR-128 | breast cancer | / | ABCC5 UGCG | / | drug resistance | [77] |
| miR-223 | breast cancer | / | ABCC5 UGCG | / | drug resistance | [77] |
| miR-137 | prostate cancer | / | SRC-1 SRC-2 SRC-3 | SRC1/SRC2/SRC3/AR | proliferation migration invasion | [78] |
| miR-497-5p | NSCLC | down | SREBP-1 SCAP | SREBP-1/hsa-miR-497/ SCAP/FASN | cell viability cisplatin resistance stemness | [72] |
| miR-19a | glioma | up | LDLR | PERK/miR-19a/LDLR | proliferation invasion | [63] |
| miR-363-3p | PAAD | down | SQLE | / | proliferation immune response | [59] |
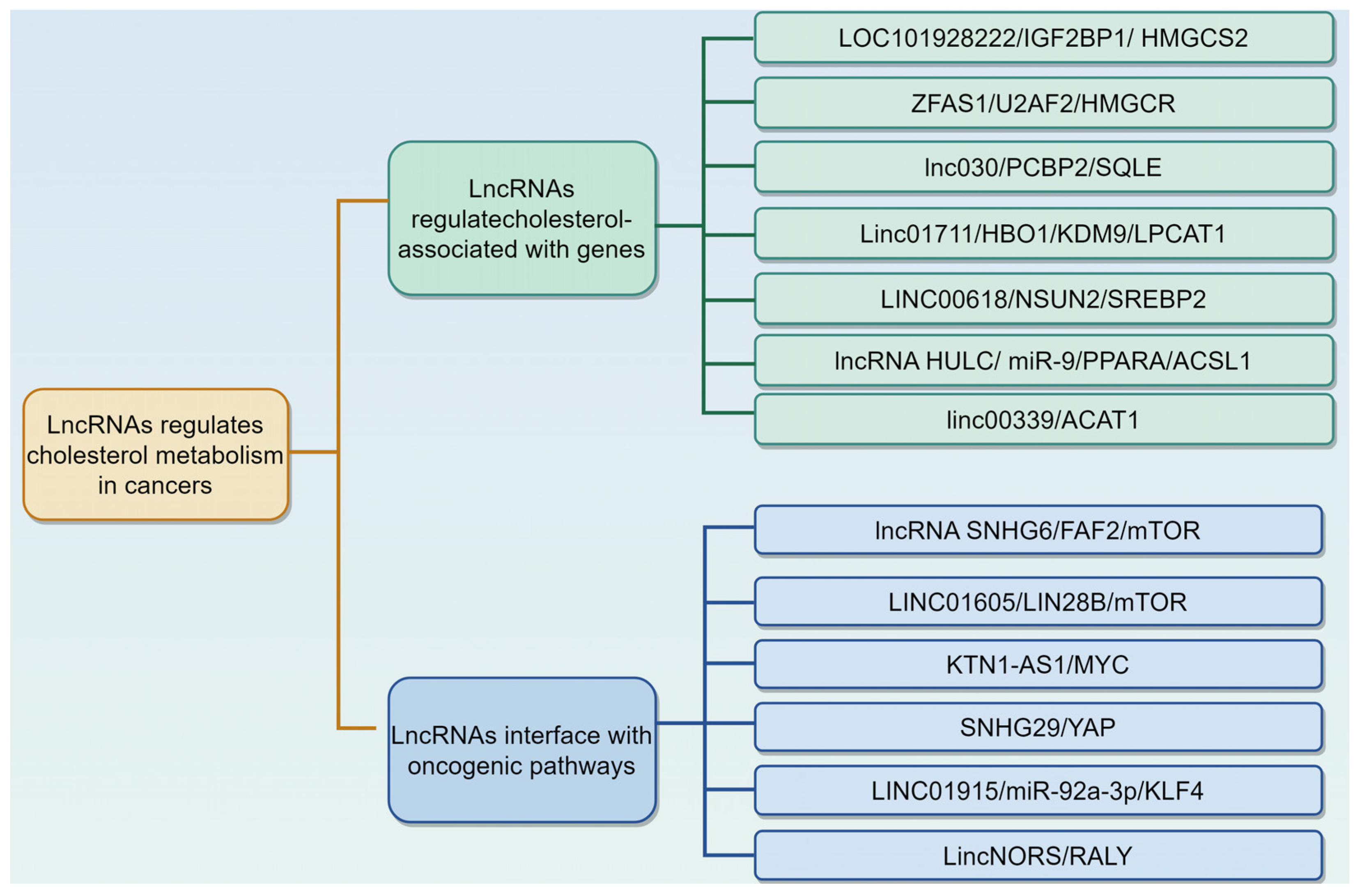
3.2. LncRNAs Regulate Cholesterol Metabolism in Cancers
3.2.1. LncRNAs Regulate Cholesterol Metabolism by Modulating Cholesterol Associated with Genes
3.2.2. LncRNAs Interface with Oncogenic Pathways to Reprogram Cholesterol Metabolism
| lncRNAs | Cancer | Expression | Regulatory Mechanism | Phenotypes | Ref. |
|---|---|---|---|---|---|
| LOC101928222 | CRC | up | collaborates with IGF2BP1 to stabilize HMGCS2 mRNA | migration, invasion, angiogenesis | [83] |
| HULC | HCC | up | ceRNA for miR-9 increases PPARA expression | proliferation | [88] |
| LINC00618 | HCC | up | Interacts with NSUN2 and regulates SREBP2 mRNA stability | proliferation, migration EMT | [87] |
| SNHG6 | HCC | up | Interacts with FAF2–mTOR complex | proliferation progression | [90] |
| linc00339 | glioma | / | / | proliferation, migration invasion | [89] |
| SNHG29 | CRC | up | Interacts with YAP and regulates PD-L1 expression | tumor immunity | [97] |
| lnc030 | breast cancer | up | cooperates with PCBP2 to stabilize SQLE mRNA | stemness | [85] |
| ZFAS1 | pancreatic carcinoma | up | interacts with U2AF2 to stabilize HMGCR mRNA | proliferation, invasion | [84] |
| KTN1-AS1 | Burkitt lymphoma | up | co-regulates Myc-target genes | proliferation | [95] |
| lincNORS | breast cancer | / | interacts with RALY | / | [100] |
| Linc01711 | gastric cancer | up | coordinates the localization of the HBO1/KDM9 complex | proliferation invasion metastasis | [86] |
| LINC01915 | CRC | down | ceRNA for miR-92a-3p to regulate the KLF4/CH25H axis | conversion of NF into CAF | [99] |
| LINC01605 | PDAC | up | interacts with LIN28B and activates the mTOR axis | proliferation migration | [93] |
3.3. CircRNAs Orchestrate Cholesterol Metabolic Reprogramming in Cancers
| circRNAs | Cancer | Expression | Regulatory Mechanism | Phenotypes | Ref. |
|---|---|---|---|---|---|
| circFAM126A | PCa | up | ceRNA for miR-505-3p and increases CANX expression | proliferation, apoptosis invasion migration | [105] |
| Circ_0000182 | STAD | up | ceRNA for miR-579-3p and increases SQLE expression | proliferation | [106] |
| circ_0124346 | PAAD | up | ceRNA for miR-223-3p and increases ACSL3 expression | proliferation | [103] |
| circLDLR | CRC | up | ceRNA for miR-30a-3p and increases SOAT1 expression | proliferation invasion migration | [104] |
| circMyc | TNBC | up | interacts with HuR to stabilize SREBP1 mRNA | proliferation invasion | [107] |
| circRIC8B | CLL | up | ceRNA for miR-199b-5p and increases LPL expression | proliferation | [19] |
| circINSIG1 | CRC | up | promotes the degradation of INSIG1 | proliferation metastasis | [108] |
3.4. piRNAs and tRNAs Modulate Cholesterol Metabolism in Cancers
4. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fragner, T.; Belogianni, K.; Grabovac, I. Cancer care in people experiencing homelessness: Identifying key issues, challenges, and facilitators. Br. J. Gen. Pract. 2024, 74, 342–343. [Google Scholar] [CrossRef] [PubMed]
- Finley, L.W.S. What is cancer metabolism? Cell 2023, 186, 1670–1688. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Zhu, J.; Thompson, C.B. The hallmarks of cancer metabolism: Still emerging. Cell Metab. 2022, 34, 355–377. [Google Scholar] [CrossRef]
- Schiliro, C.; Firestein, B.L. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. Cells 2021, 10, 1056. [Google Scholar] [CrossRef] [PubMed]
- Ribas, V.; Garcia-Ruiz, C.; Fernandez-Checa, J.C. Mitochondria, cholesterol and cancer cell metabolism. Clin. Transl. Med. 2016, 5, 22. [Google Scholar] [CrossRef]
- Huang, B.; Song, B.-L.; Xu, C. Cholesterol metabolism in cancer: Mechanisms and therapeutic opportunities. Nat. Metab. 2020, 2, 132–141. [Google Scholar] [CrossRef]
- Wang, H.H.; Li, T.; Portincasa, P.; Ford, D.A.; Neuschwander-Tetri, B.A.; Tso, P.; Wang, D.Q.-H. New insights into the role of Lith genes in the formation of cholesterol-supersaturated bile. Liver Res. 2017, 1, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Ndhlala, A.R.; Kavaz Yüksel, A.; Çelebi, N.; Doğan, H.Ö. A General Review of Methodologies Used in the Determination of Cholesterol (C(27)H(46)O) Levels in Foods. Foods 2023, 12, 4424. [Google Scholar] [CrossRef]
- Luo, J.; Yang, H.; Song, B.-L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 225–245. [Google Scholar] [CrossRef]
- Ikonen, E.; Olkkonen, V.M. Intracellular Cholesterol Trafficking. Cold Spring Harb. Perspect. Biol. 2023, 15, a041404. [Google Scholar] [CrossRef]
- Guo, X.J.; Zhu, B.B.; Li, J.; Guo, P.; Niu, Y.B.; Shi, J.L.; Yokoyama, W.; Huang, Q.S.; Shao, D.Y. Cholesterol metabolism in tumor im-munity: Mechanisms and therapeutic opportunities for cancer. Biochem. Pharmacol. 2025, 234, 116802. [Google Scholar] [CrossRef] [PubMed]
- Ajoolabady, A.; Pratico, D.; Mazidi, M.; Davies, I.G.; Lip, G.Y.; Seidah, N.; Libby, P.; Kroemer, G.; Ren, J. PCSK9 in metabolism and diseases. Metabolism 2025, 163, 156064. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.H.; Tang, C.K. ABCA1, ABCG1, and Cholesterol Homeostasis. Adv. Exp. Med. Biol. 2022, 1377, 95–107. [Google Scholar]
- Garçon, D.; Berger, J.-M.; Cariou, B.; Le May, C. Transintestinal cholesterol excretion in health and disease. Curr. Atheroscler. Rep. 2022, 24, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, J.; Dimitrova, N. Transcription regulation by long non-coding RNAs: Mechanisms and disease relevance. Nat. Rev. Mol. Cell Biol. 2024, 25, 396–415. [Google Scholar] [CrossRef]
- Yan, H.; Bu, P. Non-coding RNA in cancer. Essays Biochem. 2021, 65, 625–639. [Google Scholar]
- Nemeth, K.; Bayraktar, R.; Ferracin, M.; Calin, G.A. Non-coding RNAs in disease: From mechanisms to therapeutics. Nat. Rev. Genet. 2023, 25, 211–232. [Google Scholar] [CrossRef]
- Hong, J.; Guo, F.; Lu, S.-Y.; Shen, C.; Ma, D.; Zhang, X.; Xie, Y.; Yan, T.; Yu, T.; Sun, T.; et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut 2020, 70, 2123–2137. [Google Scholar] [CrossRef]
- Wu, Z.; Gu, D.; Wang, R.; Zuo, X.; Zhu, H.; Wang, L.; Lu, X.; Xia, Y.; Qin, S.; Zhang, W.; et al. CircRIC8B regulates the lipid metabolism of chronic lymphocytic leukemia through miR199b-5p/LPL axis. Exp. Hematol. Oncol. 2022, 11, 51. [Google Scholar] [CrossRef]
- Liu, S.; Jiao, B.; Zhao, H.; Liang, X.; Jin, F.; Liu, X.; Hu, J. LncRNAs-circRNAs as Rising Epigenetic Binary Superstars in Regulating Lipid Metabolic Reprogramming of Cancers. Adv. Sci. 2023, 11, e2303570. [Google Scholar] [CrossRef]
- Jonas, K.; Prinz, F.; Ferracin, M.; Krajina, K.; Deutsch, A.; Madl, T.; Rinner, B.; Slaby, O.; Klec, C.; Pichler, M. MiR-4646-5p Acts as a Tumor-Suppressive Factor in Triple Negative Breast Cancer and Targets the Cholesterol Transport Protein GRAMD1B. Non-Coding RNA 2023, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Hassen, C.B.; Goupille, C.; Vigor, C.; Durand, T.; Guéraud, F.; Silvente-Poirot, S.; Poirot, M.; Frank, P.G. Is cholesterol a risk factor for breast cancer incidence and outcome? J. Steroid Biochem. Mol. Biol. 2023, 232, 106346. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Li, P.; Cheng, C.; Zhao, Y.; Li, D.; Du, C. Cholesterol Levels in Blood and the Risk of Prostate Cancer: A Meta-analysis of 14 Prospective Studies. Cancer Epidemiol. Biomark. Prev. 2015, 24, 1086–1093. [Google Scholar]
- Zhang, Y.; Wu, K.; Chan, A.T.; Meyerhardt, J.A.; Giovannucci, E.L. Giovannucci. Long-Term Statin Use, Total Cholesterol Level, and Risk of Colorectal Cancer: A Prospective Cohort Study. Am. J. Gastroenterol. 2022, 117, 158–166. [Google Scholar] [PubMed]
- Bu, L.; Zhang, Z.; Chen, J.; Fan, Y.; Guo, J.; Su, Y.; Wang, H.; Zhang, X.; Wu, X.; Jiang, Q.; et al. High-fat diet promotes liver tumorigenesis via palmitoylation and activation of AKT. Gut 2024, 73, 1156–1168. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Liu, L.; Fu, Y.; Gao, J.; He, Y.; Wu, Y.; Lian, X. Dietary Cholesterol Intake and Risk of Lung Cancer: A Meta-Analysis. Nutrients 2018, 10, 185. [Google Scholar] [CrossRef]
- Ren, Y.-M.; Zhuang, Z.-Y.; Xie, Y.-H.; Yang, P.-J.; Xia, T.-X.; Xie, Y.-L.; Liu, Z.-H.; Kang, Z.-R.; Leng, X.-X.; Lu, S.-Y.; et al. BCAA-producing Clostridium symbiosum promotes colorectal tumorigenesis through the modulation of host cholesterol metabolism. Cell Host Microbe 2024, 32, 1519–1535.e7. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lv, M.; Zhang, W.; Zhan, Q. Dysregulation of cholesterol metabolism in cancer progression. Oncogene 2023, 42, 3289–3302. [Google Scholar] [CrossRef]
- A Riobo, N. Cholesterol and its derivatives in Sonic Hedgehog signaling and cancer. Curr. Opin. Pharmacol. 2012, 12, 736–741. [Google Scholar] [CrossRef]
- Zheng, S.; Lin, J.; Pang, Z.; Zhang, H.; Wang, Y.; Ma, L.; Zhang, H.; Zhang, X.; Chen, M.; Zhang, X.; et al. Aberrant Cholesterol Metabo-lism and Wnt/beta-Catenin Signaling Coalesce via Frizzled5 in Supporting Cancer Growth. Adv. Sci. (Weinh.) 2022, 9, e2200750. [Google Scholar]
- Jung, Y.Y.; Ko, J.; Um, J.; Chinnathambi, A.; Alharbi, S.A.; Sethi, G.; Ahn, K.S. LDL cholesterol promotes the proliferation of prostate and pancreatic cancer cells by activating the STAT3 pathway. J. Cell. Physiol. 2021, 236, 5253–5264. [Google Scholar] [CrossRef]
- Su, H.; Chen, L.; Wu, J.; Cheng, Z.; Li, J.; Ren, Y.; Xu, J.; Dang, Y.; Zheng, M.; Cao, Y.; et al. Proteogenomic characterization reveals tumorigenesis and progression of lung cancer manifested as subsolid nodules. Nat. Commun. 2025, 16, 2414. [Google Scholar] [CrossRef]
- Liang, Z.; Chen, Y.; Wang, L.; Li, D.; Yang, X.; Ma, G.; Wang, Y.; Li, Y.; Zhao, H.; Liang, Y.; et al. CYP27A1 inhibits bladder cancer cells proliferation by regulating cholesterol homeostasis. Cell Cycle 2019, 18, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Qiu, Z.; Yang, Z.; Xu, L.; Pearce, T.M.; Wu, Q.; Yang, K.; Li, F.; Saulnier, O.; Fei, F.; et al. Lymphatic endothelial-like cells promote glioblastoma stem cell growth through cytokine-driven cholesterol metabolism. Nat. Cancer 2024, 5, 147–166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.-L.; Zhu, W.-W.; Wang, S.-H.; Gao, C.; Pan, J.-J.; Du, Z.-G.; Lu, L.; Jia, H.-L.; Dong, Q.-Z.; Chen, J.-H.; et al. Organ-specific cholesterol metabolic aberration fuels liver metastasis of colorectal cancer. Theranostics 2021, 11, 6560–6572. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Chakraborty, B.; Safi, R.; Kazmin, D.; Chang, C.-Y.; McDonnell, D.P. Dysregulated cholesterol homeostasis results in resistance to ferroptosis increasing tumorigenicity and metastasis in cancer. Nat. Commun. 2021, 12, 5103. [Google Scholar] [CrossRef]
- Fu, R.; Xue, W.; Liang, J.; Li, X.; Zheng, J.; Wang, L.; Zhang, M.; Meng, J. SOAT1 regulates cholesterol metabolism to induce EMT in hepatocellular carcinoma. Cell Death Dis. 2024, 15, 325. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, X.; Liu, S.; Cao, Q. Cholesterol promotes the migration and invasion of renal carcinoma cells by regulating the KLF5/miR-27a/FBXW7 pathway. Biochem. Biophys. Res. Commun. 2018, 502, 69–75. [Google Scholar] [CrossRef]
- Shen, Z.; Zhu, D.; Liu, J.; Chen, J.; Liu, Y.; Hu, C.; Li, Z.; Li, Y. 27-Hydroxycholesterol induces invasion and migration of breast cancer cells by increasing MMP9 and generating EMT through activation of STAT-3. Environ. Toxicol. Pharmacol. 2017, 51, 1–8. [Google Scholar] [CrossRef]
- Kopecka, J.; Trouillas, P.; Gašparović, A.Č.; Gazzano, E.; Assaraf, Y.G.; Riganti, C. Phospholipids and cholesterol: Inducers of cancer multidrug resistance and therapeutic targets. Drug Resist. Updat. 2020, 49, 100670. [Google Scholar] [CrossRef]
- Yan, A.; Jia, Z.; Qiao, C.; Wang, M.; Ding, X. Cholesterol metabolism in drug-resistant cancer (Review). Int. J. Oncol. 2020, 57, 1103–1115. [Google Scholar] [CrossRef] [PubMed]
- Palma, G.B.H.; Kaur, M. Cholesterol Depletion Modulates Drug Resistance Pathways to Sensitize Resistant Breast Cancer Cells to Tamoxifen. Anticancer Res. 2022, 42, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Wang, K.; Wang, X.; Jia, Z.; Yang, Y.; Duan, Y.; Huang, L.; Wu, Z.-X.; Zhang, J.-Y.; Ding, X. Cholesterol promotes EGFR-TKIs resistance in NSCLC by inducing EGFR/Src/Erk/SP1 signaling-mediated ERRα re-expression. Mol. Cancer 2022, 21, 77. [Google Scholar] [CrossRef] [PubMed]
- Mok, E.H.K.; Leung, C.O.N.; Zhou, L.; Lei, M.M.L.; Leung, H.W.; Tong, M.; Wong, T.L.; Lau, E.Y.T.; Ng, I.O.L.; Ding, J.; et al. Caspase-3–Induced Activation of SREBP2 Drives Drug Resistance via Promotion of Cholesterol Biosynthesis in Hepatocellular Carcinoma. Cancer Res. 2022, 82, 3102–3115. [Google Scholar] [CrossRef]
- King, R.J.; Singh, P.K.; Mehla, K. The cholesterol pathway: Impact on immunity and cancer. Trends Immunol. 2022, 43, 78–92. [Google Scholar] [CrossRef]
- Xiao, M.; Xu, J.; Wang, W.; Zhang, B.; Liu, J.; Li, J.; Xu, H.; Zhao, Y.; Yu, X.; Shi, S. Functional significance of cholesterol metabolism in cancer: From threat to treatment. Exp. Mol. Med. 2023, 55, 1982–1995. [Google Scholar] [CrossRef]
- Ma, X.; Bi, E.; Lu, Y.; Su, P.; Huang, C.; Liu, L.; Wang, Q.; Yang, M.; Kalady, M.F.; Qian, J.; et al. Cholesterol Induces CD8(+) T Cell Ex-haustion in the Tumor Microenvironment. Cell Metab. 2019, 30, 143–156.e5. [Google Scholar] [CrossRef]
- Yan, C.; Zheng, L.; Jiang, S.; Yang, H.; Guo, J.; Jiang, L.Y.; Li, T.; Zhang, H.; Bai, Y.; Lou, Y.; et al. Exhaustion-associated cholesterol de-ficiency dampens the cytotoxic arm of antitumor immunity. Cancer Cell 2023, 41, 1276–1293.e11. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, Y.; Zhao, H.; Zhou, Y.; Zhang, J.; Lei, J.; Wu, L.; Zhou, M.; Wang, J.; Yang, S.; et al. Myeloid-derived suppressor cells deficient in cholesterol biosynthesis promote tumor immune evasion. Cancer Lett. 2023, 564, 216208. [Google Scholar] [CrossRef]
- Sun, H.; Yang, W.; Tian, Y.; Zeng, X.; Zhou, J.; Mok, M.T.S.; Tang, W.; Feng, Y.; Xu, L.; Chan, A.W.H.; et al. An inflammatory-CCRK circuitry drives mTORC1-dependent metabolic and immunosuppressive reprogramming in obesity-associated hepatocellular carcinoma. Nat. Commun. 2018, 9, 5214. [Google Scholar] [CrossRef]
- Kim, H.; Lee, Y.-Y.; Kim, V.N. The biogenesis and regulation of animal microRNAs. Nat. Rev. Mol. Cell Biol. 2025, 26, 276–296. [Google Scholar] [CrossRef]
- Hussen, B.M.; Hidayat, H.J.; Salihi, A.; Sabir, D.K.; Taheri, M.; Ghafouri-Fard, S. MicroRNA: A signature for cancer progression. Biomed. Pharmacother. 2021, 138, 111528. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Q.; Wang, S.; Wang, B.-J.; Jin, Y.; Hu, H.; Fu, Q.-S.; Wang, J.-W.; Wu, Q.; Qian, L.; et al. The role of noncoding RNAs in cancer lipid metabolism. Front. Oncol. 2022, 12, 1026257. [Google Scholar] [CrossRef] [PubMed]
- Monchusi, B.; Kaur, M. miRNAs as Modulators of Cholesterol in Breast Cancer Stem Cells: An Approach to Overcome Drug Resistance in Cancer. Curr. Drug Targets 2022, 23, 656–677. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-X.; Hu, S.; Lei, H.-H.; Yuan, M.; Li, X.; Hou, W.-K.; Huang, X.-J.; Xiao, B.-W.; Yu, T.-X.; Zhang, X.-H.; et al. Tumor-derived miR-9-5p-loaded EVs regulate cholesterol homeostasis to promote breast cancer liver metastasis in mice. Nat. Commun. 2024, 15, 10539. [Google Scholar] [CrossRef]
- Kanmalar, M.; Sani, S.F.A.; Kamri, N.I.N.B.; Said, N.A.B.M.; Jamil, A.H.B.A.; Kuppusamy, S.; Mun, K.S.; Bradley, D.A. Raman spectroscopy biochemical characterisation of bladder cancer cisplatin resistance regulated by FDFT1: A review. Cell. Mol. Biol. Lett. 2022, 27, 9. [Google Scholar] [CrossRef]
- Azhar, N.A.; Paramanantham, Y.; Nor, W.F.; Said, N.A. MicroRNA-146b-5p/FDFT1 mediates cisplatin sensitivity in bladder cancer by redirecting cholesterol biosynthesis to the non-sterol branch. Int. J. Biochem. Cell Biol. 2024, 176, 106652. [Google Scholar] [CrossRef]
- Kalogirou, C.; Linxweiler, J.; Schmucker, P.; Snaebjornsson, M.T.; Schmitz, W.; Wach, S.; Krebs, M.; Hartmann, E.; Puhr, M.; Müller, A.; et al. MiR-205-driven downregulation of cholesterol biosynthesis through SQLE-inhibition identifies therapeutic vulnerability in aggressive prostate cancer. Nat. Commun. 2021, 12, 5066. [Google Scholar] [CrossRef]
- You, W.; Ke, J.; Chen, Y.; Cai, Z.; Huang, Z.-P.; Hu, P.; Wu, X. SQLE, A Key Enzyme in Cholesterol Metabolism, Correlates With Tumor Immune Infiltration and Immunotherapy Outcome of Pancreatic Adenocarcinoma. Front. Immunol. 2022, 13, 864244. [Google Scholar] [CrossRef]
- Li, W.; Zhang, C.; Gao, T.; Sun, Y.; Yang, H.; Liu, L.; Shi, M.; Ding, L.; Zhang, C.; Deng, D.Y.B.; et al. Human umbilical cord mesenchymal stem cells small extracellular vesicles-derived miR-370-3p inhibits cervical precancerous lesions by targeting DHCR24. Stem CELLS Transl. Med. 2025, 14, szae087. [Google Scholar] [CrossRef]
- Davidson, M.H. Therapies Targeting Exogenous Cholesterol Uptake: New Insights and Controversies. Curr. Atheroscler. Rep. 2010, 13, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Reue, K.; Fong, L.G.; Young, S.G.; Tontonoz, P. Feedback regulation of cholesterol uptake by the LXR-IDOL-LDLR axis. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 2541–2546. [Google Scholar] [CrossRef]
- Wang, T.; Zhou, Y.; Fan, Y.; Duan, H.; Guo, X.; Chang, J.; Jiang, Y.; Li, C.; Fu, Z.; Gao, Y.; et al. PERK-Mediated Cholesterol Excretion from IDH Mutant Glioma Determines Anti-Tumoral Polarization of Microglia. Adv. Sci. 2023, 10, e2205949. [Google Scholar] [CrossRef]
- Segrest, J.P.; Davidson, W.S.; Heinecke, J.W. Phospholipid transport by ABCA1: The extracellular translocase or alternating access model? Curr. Opin. Lipidol. 2023, 34, 208–213. [Google Scholar] [CrossRef]
- An, B.; Shin, C.-H.; Kwon, J.W.; Tran, N.L.; Kim, A.H.; Jeong, H.; Kim, S.-H.; Park, K.; Oh, S.J. M1 macrophage-derived exosomal microRNA-29c-3p suppresses aggressiveness of melanoma cells via ENPP. Cancer Cell Int. 2024, 24, 325. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Liang, S.; Zhang, S.; Zhang, C.; Zhao, Y.; Wu, D.; Wang, J.; Song, R.; Wang, J.; Yin, D.; et al. ABCA8 is regulated by miR-374b-5p and inhibits proliferation and metastasis of hepatocellular carcinoma through the ERK/ZEB1 pathway. J. Exp. Clin. Cancer Res. 2020, 39, 90. [Google Scholar] [CrossRef]
- Li, Y.; Wu, S.; Zhao, X.; Hao, S.; Li, F.; Wang, Y.; Liu, B.; Zhang, D.; Wang, Y.; Zhou, H. Key events in cancer: Dysregulation of SREBPs. Front. Pharmacol. 2023, 14, 1130747. [Google Scholar] [CrossRef]
- Shen, S.; Shen, M.; Kuang, L.; Yang, K.; Wu, S.; Liu, X.; Wang, Y.; Wang, Y. SIRT1/SREBPs-mediated regulation of lipid metabolism. Pharmacol. Res. 2024, 199, 107037. [Google Scholar] [CrossRef] [PubMed]
- Witkin, J. Aging changes in synaptology of luteinizing hormone-releasing hormone neurons in male rat preoptic area. Neuroscience 1987, 22, 1003–1013. [Google Scholar] [CrossRef]
- Kawamura, S.; Matsushita, Y.; Kurosaki, S.; Tange, M.; Fujiwara, N.; Hayata, Y.; Hayakawa, Y.; Suzuki, N.; Hata, M.; Tsuboi, M.; et al. Inhibiting SCAP/SREBP exacerbates liver injury and carcinogenesis in murine nonalcoholic steatohepatitis. J. Clin. Investig. 2022, 132, e151895. [Google Scholar] [CrossRef]
- Zheng, Z.-G.; Zhou, Y.-P.; Zhang, X.; Thu, P.M.; Xie, Z.-S.; Lu, C.; Pang, T.; Xue, B.; Xu, D.-Q.; Chen, Y.; et al. Anhydroicaritin improves diet-induced obesity and hyperlipidemia and alleviates insulin resistance by suppressing SREBPs activation. Biochem. Pharmacol. 2016, 122, 42–61. [Google Scholar] [CrossRef] [PubMed]
- Tiong, T.-Y.; Weng, P.-W.; Wang, C.-H.; Setiawan, S.A.; Yadav, V.K.; Pikatan, N.W.; Fong, I.-H.; Yeh, C.-T.; Hsu, C.-H.; Kuo, K.-T. Targeting the SREBP-1/Hsa-Mir-497/SCAP/FASN Oncometabolic Axis Inhibits the Cancer Stem-like and Chemoresistant Phenotype of Non-Small Cell Lung Carcinoma Cells. Int. J. Mol. Sci. 2022, 23, 7283. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; He, J.; Zhou, W.; Xiang, G.; Xu, R. MicroRNA-132 cause apoptosis of glioma cells through blockade of the SREBP-1c metabolic pathway related to SIRT1. Biomed. Pharmacother. 2016, 78, 177–184. [Google Scholar] [CrossRef]
- Liu, X.; Liu, J.; Wang, X.; Zou, Y.; Tao, X.; Li, J.; Ye, M.; Xu, W.; Deng, Y.; Liu, L.; et al. Cancer-secreted exosomal miR-1246 promotes colorectal cancer liver metastasis by activating hepatic stellate cells. Mol. Med. 2025, 31, 68. [Google Scholar] [CrossRef]
- Frank, A.-C.; Raue, R.; Fuhrmann, D.C.; Sirait-Fischer, E.; Reuse, C.; Weigert, A.; Lütjohann, D.; Hiller, K.; Syed, S.N.; Brüne, B. Lactate dehydrogenase B regulates macrophage metabolism in the tumor microenvironment. Theranostics 2021, 11, 7570–7588. [Google Scholar] [CrossRef] [PubMed]
- Aersilan, A.; Hashimoto, N.; Yamagata, K.; Yokoyama, M.; Nakayama, A.; Shi, X.; Nagano, H.; Sakuma, I.; Nohata, N.; Kinoshita, T.; et al. MicroRNA-874 targets phosphomevalonate kinase and inhibits cancer cell growth via the mevalonate pathway. Sci. Rep. 2022, 12, 18443. [Google Scholar] [CrossRef]
- Palma, G.B.H.; Kaur, M. miRNA-128 and miRNA-223 regulate cholesterol-mediated drug resistance in breast cancer. IUBMB Life 2023, 75, 743–764. [Google Scholar] [CrossRef]
- Pimenta, R.; Mioshi, C.M.; Gonçalves, G.L.; Candido, P.; Camargo, J.A.; Guimarães, V.R.; Chiovatto, C.; Ghazarian, V.; Romão, P.; da Silva, K.S.; et al. Intratumoral Restoration of miR-137 Plus Cholesterol Favors Homeostasis of the miR-137/Coactivator p160/AR Axis and Negatively Modulates Tumor Progression in Advanced Prostate Cancer. Int. J. Mol. Sci. 2023, 24, 9633. [Google Scholar] [CrossRef]
- Herman, A.B.; Tsitsipatis, D.; Gorospe, M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol. Cell 2022, 82, 2252–2266. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Lei, H.; Xiang, T.; Zhu, H.; Hu, X. A Novel Cholesterol Metabolism-Related lncRNA Signature Predicts the Prognosis of Patients with Hepatocellular Carcinoma and Their Response to Immunotherapy. Front. Biosci. 2024, 29, 129. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Park, S.; Kim, J.H.; Bang, S.-B.; Kim, H.-J.; Ka, N.-L.; Ko, Y.; Kim, S.-S.; Lim, G.Y.; Lee, S.; et al. Targeting HMG-CoA synthase 2 suppresses tamoxifen-resistant breast cancer growth by augmenting mitochondrial oxidative stress-mediated cell death. Life Sci. 2023, 328, 121827. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Ding, J.; Pu, J.; Zhu, J.; Zhou, X.; Luo, Q.; Li, J.; Qian, M.; Lin, S.; Li, J.; et al. A novel lncRNA LOC101928222 promotes colorectal cancer angiogenesis by stabilizing HMGCS2 mRNA and increasing cholesterol synthesis. J. Exp. Clin. Cancer Res. 2024, 43, 185. [Google Scholar] [CrossRef]
- Wang, L.; Ruan, Y.; Wu, X.; Zhou, X.; Wang, F. lncRNA ZFAS1 Promotes HMGCR mRNA Stabilization via Binding U2AF2 to Modulate Pancreatic Carcinoma Lipometabolism. J. Immunol. Res. 2022, 2022, 4163198. [Google Scholar] [CrossRef]
- Qin, Y.; Hou, Y.; Liu, S.; Zhu, P.; Wan, X.; Zhao, M.; Peng, M.; Zeng, H.; Li, Q.; Jin, T.; et al. A Novel Long Non-Coding RNA lnc030 Maintains Breast Cancer Stem Cell Stemness by Stabilizing SQLE mRNA and Increasing Cholesterol Synthesis. Adv. Sci. 2022, 9, e2204046. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.; Chen, J.; Bao, T.; Zhang, Y.; Yang, L.; Zhang, Z.; Wang, Z.; Zhu, C. Chromosomal copy number amplification-driven Linc01711 contributes to gastric cancer progression through histone modification-mediated reprogramming of cholesterol metabolism. Gastric Cancer 2024, 27, 308–323. [Google Scholar] [CrossRef]
- Li, R.; Li, S.; Shen, L.; Li, J.; Zhang, D.; Yu, J.; Huang, L.; Liu, N.; Lu, H.; Xu, M. LINC00618 facilitates growth and metastasis of hepatocellular carcinoma via elevating cholesterol synthesis by promoting NSUN2-mediated SREBP2 m5C modification. Ecotoxicol. Environ. Saf. 2024, 285, 117064. [Google Scholar] [CrossRef]
- Cui, M.; Xiao, Z.; Wang, Y.; Zheng, M.; Song, T.; Cai, X.; Sun, B.; Ye, L.; Zhang, X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res 2015, 75, 846–857. [Google Scholar] [CrossRef]
- Luo, Y.; Liu, L.; Li, X.; Shi, Y. Avasimibe inhibits the proliferation, migration and invasion of glioma cells by suppressing linc00339. Biomed. Pharmacother. 2020, 130, 110508. [Google Scholar] [CrossRef]
- Liu, F.; Tian, T.; Zhang, Z.; Xie, S.; Yang, J.; Zhu, L.; Wang, W.; Shi, C.; Sang, L.; Guo, K.; et al. Long non-coding RNA SNHG6 couples cholesterol sensing with mTORC1 activation in hepatocellular carcinoma. Nat. Metab. 2022, 4, 1022–1040. [Google Scholar] [CrossRef]
- Mei, X.; Xiong, J.; Liu, J.; Huang, A.; Zhu, D.; Huang, Y.; Wang, H. DHCR7 promotes lymph node metastasis in cervical cancer through cholesterol reprogramming-mediated activation of the KANK4/PI3K/AKT axis and VEGF-C secretion. Cancer Lett. 2024, 584, 216609. [Google Scholar] [CrossRef]
- Benatzy, Y.; Palmer, M.A.; Lütjohann, D.; Ohno, R.-I.; Kampschulte, N.; Schebb, N.H.; Fuhrmann, D.C.; Snodgrass, R.G.; Brüne, B. ALOX15B controls macrophage cholesterol homeostasis via lipid peroxidation, ERK1/2 and SREBP2. Redox Biol. 2024, 72, 103149. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-H.; Jia, Q.-Y.; Yao, H.-F.; Duan, Z.-H.; Ma, X.-S.; Zheng, J.-H.; Yin, Y.-F.; Liu, W.; Zhang, J.-F.; Hua, R.; et al. The lncRNA LINC01605 promotes the progression of pancreatic ductal adenocarcinoma by activating the mTOR signaling pathway. Cancer Cell Int. 2024, 24, 262. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Pyun, W.Y.; Park, H.W. Cancer Metabolism: Phenotype, Signaling and Therapeutic Targets. Cells 2020, 9, 2308. [Google Scholar] [CrossRef]
- Winkle, M.; Tayari, M.M.; Kok, K.; Duns, G.; Grot, N.; Kazimierska, M.; Seitz, A.; de Jong, D.; Koerts, J.; Diepstra, A.; et al. The lncRNA KTN1-AS1 co-regulates a variety of Myc-target genes and enhances proliferation of Burkitt lymphoma cells. Hum. Mol. Genet. 2022, 31, 4193–4206. [Google Scholar] [CrossRef] [PubMed]
- Shu, Z.; Gao, Y.; Zhang, G.; Zhou, Y.; Cao, J.; Wan, D.; Zhu, X.; Xiong, W. A functional interaction between Hippo-YAP signalling and SREBPs mediates hepatic steatosis in diabetic mice. J. Cell. Mol. Med. 2019, 23, 3616–3628. [Google Scholar] [CrossRef]
- Ni, W.; Mo, H.; Liu, Y.; Xu, Y.; Qin, C.; Zhou, Y.; Li, Y.; Li, Y.; Zhou, A.; Yao, S.; et al. Targeting cholesterol biosynthesis promotes anti-tumor immunity by inhibiting long noncoding RNA SNHG29-mediated YAP activation. Mol. Ther. 2021, 29, 2995–3010. [Google Scholar] [CrossRef]
- He, Z.; He, J.; Xie, K. KLF4 transcription factor in tumorigenesis. Cell Death Discov. 2023, 9, 118. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, S.; Liu, D.; Zhou, J. LINC01915 Facilitates the Conversion of Normal Fibroblasts into Cancer-Associated Fi-broblasts Induced by Colorectal Cancer-Derived Extracellular Vesicles through the miR-92a-3p/KLF4/CH25H Axis. ACS Biomater. Sci. Eng. 2021, 7, 5255–5268. [Google Scholar] [CrossRef]
- Wu, X.; Niculite, C.M.; Preda, M.B.; Rossi, A.; Tebaldi, T.; Butoi, E.; White, M.K.; Tudoran, O.M.; Petrusca, D.N.; Jannasch, A.S.; et al. Regulation of cellular sterol homeostasis by the oxygen responsive noncoding RNA lincNORS. Nat. Commun. 2020, 11, 172. [Google Scholar] [CrossRef]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol Cancer 2020, 19, 172. [Google Scholar] [CrossRef] [PubMed]
- Conn, V.M.; Chinnaiyan, A.M.; Conn, S.J. Circular RNA in cancer. Nat. Rev. Cancer 2024, 24, 597–613. [Google Scholar] [CrossRef] [PubMed]
- Shu, M.-L.; Yang, W.-T.; Li, H.-M.; Qian, C.-J.; Teng, X.-S.; Yao, J. Circ_0124346 facilitates cell proliferation of pancreatic adenocarcinoma cells by regulating lipid metabolism via miR-223-3p/ACSL3 axis. Discov. Oncol. 2024, 15, 670. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, J.; Chen, Y.; Chen, Y.; Xi, Q.; Sun, L.; Zhang, X.; Zhang, G.; Ding, X.; Shi, T.; et al. Circular RNA circLDLR facilitates cancer progression by altering the miR-30a-3p/SOAT1 axis in colorectal cancer. Cell Death Discov. 2022, 8, 314. [Google Scholar] [CrossRef]
- Luo, L.; Li, P.; Xie, Q.; Wu, Y.; Qin, F.; Liao, D.; Zeng, K.; Wang, K. n6-methyladenosine-modified circular RNA family with sequence similarity 126, member A affects cholesterol synthesis and malignant progression of prostate cancer cells by targeting mi-croRNA-505-3p to mediate calnexin. J. Cancer 2024, 15, 966–980. [Google Scholar] [CrossRef]
- Qian, C.-J.; Zhou, Y.-X.; Wu, L.-K.; Wang, Y.-C.; Teng, X.-S.; Yao, J. Circ_0000182 promotes cholesterol synthesis and proliferation of stomach adenocarcinoma cells by targeting miR-579-3p/SQLE axis. Discov. Oncol. 2023, 14, 22. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y.; Wang, Y.; Li, Q.; Zeng, K.; Li, X.; Feng, X. Myc derived circRNA promotes triple-negative breast cancer progression via reprogramming fatty acid metabolism. Discov. Oncol. 2023, 14, 67. [Google Scholar] [CrossRef]
- Xiong, L.; Liu, H.-S.; Zhou, C.; Yang, X.; Huang, L.; Jie, H.-Q.; Zeng, Z.-W.; Zheng, X.-B.; Li, W.-X.; Liu, Z.-Z.; et al. A novel protein encoded by circINSIG1 reprograms cholesterol metabolism by promoting the ubiquitin-dependent degradation of INSIG1 in colorectal cancer. Mol. Cancer 2023, 22, 72. [Google Scholar] [CrossRef]
- Wang, X.; Ramat, A.; Simonelig, M.; Liu, M.-F. Emerging roles and functional mechanisms of PIWI-interacting RNAs. Nat. Rev. Mol. Cell Biol. 2023, 24, 123–141. [Google Scholar] [CrossRef]
- Wu, P.-H.; Zamore, P.D. Defining the functions of PIWI-interacting RNAs. Nat. Rev. Mol. Cell Biol. 2021, 22, 239–240. [Google Scholar] [CrossRef]
- Lin, Y.; Zheng, J.; Lin, D. PIWI-interacting RNAs in human cancer. Semin. Cancer Biol. 2021, 75, 15–28. [Google Scholar] [CrossRef]
- Liu, Y.; Dou, M.; Song, X.; Dong, Y.; Liu, S.; Liu, H.; Tao, J.; Li, W.; Yin, X.; Xu, W. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 2019, 18, 123. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, T.; Gupta, P.; Nayak, R.; Mallick, B. Genome-wide profiling of dysregulated piRNAs and their target genes implicated in oncogenicity of tongue squamous cell carcinoma. Gene 2023, 849, 146919. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Tian, F.; Fan, W.; Li, X.; Wang, X.; Zhang, H.; Hong, X.; Wang, X.; Cai, L.; Song, Y.; et al. Targeting piRNA-137463 Inhibits Tumor Progression and Boosts Sensitivity to Immune Checkpoint Blockade via De Novo Cholesterol Biosynthesis in Lung Adenocarcinoma. Adv. Sci. 2025, 12, e2414100. [Google Scholar] [CrossRef] [PubMed]
- Ohira, T.; Suzuki, T. Transfer RNA modifications and cellular thermotolerance. Mol. Cell 2024, 84, 94–106. [Google Scholar] [CrossRef]
- Wang, Y.; Tao, E.-W.; Tan, J.; Gao, Q.-Y.; Chen, Y.-X.; Fang, J.-Y. tRNA modifications: Insights into their role in human cancers. Trends Cell Biol. 2023, 33, 1035–1048. [Google Scholar] [CrossRef]
- Pinzaru, A.M.; Tavazoie, S.F. Transfer RNAs as dynamic and critical regulators of cancer progression. Nat. Rev. Cancer 2023, 23, 746–761. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Li, X.; Xiong, X.; Wang, J.; Zhou, Z.; Zhu, X.; Gu, Y.; Dominissini, D.; He, L.; et al. N1-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat. Commun. 2021, 12, 6314. [Google Scholar] [CrossRef]
- Miao, S.; Li, H.; Song, X.; Liu, Y.; Wang, G.; Kan, C.; Ye, Y.; Liu, R.-J.; Li, H.-B. tRNA m1A modification regulates cholesterol biosynthesis to promote antitumor immunity of CD8+ T cells. J. Exp. Med. 2025, 222, e20240559. [Google Scholar] [CrossRef]
- Holý, P.; Brynychová, V.; Šeborová, K.; Haničinec, V.; Koževnikovová, R.; Trnková, M.; Vrána, D.; Gatěk, J.; Kopečková, K.; Mrhalová, M.; et al. Integrative analysis of mRNA and miRNA expression profiles and somatic variants in oxysterol signaling in early-stage luminal breast cancer. Mol. Oncol. 2023, 17, 2074–2089. [Google Scholar] [CrossRef]
- Liu, H.; Wu, X.; Wang, D.; Li, Q.; Zhang, X.; Xu, L. Unveiling the role of miR-137-3p/miR-296-5p/SERPINA3 signaling in colorectal cancer progression: Integrative analysis of gene expression profiles and in vitro studies. BMC Med. Genom. 2023, 16, 327. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Miao, Z.; Xu, Y.; Shi, T. Non-Coding RNAs as Critical Modulators of Cholesterol Metabolism in Cancer. Biomedicines 2025, 13, 1631. https://doi.org/10.3390/biomedicines13071631
Zhang C, Miao Z, Xu Y, Shi T. Non-Coding RNAs as Critical Modulators of Cholesterol Metabolism in Cancer. Biomedicines. 2025; 13(7):1631. https://doi.org/10.3390/biomedicines13071631
Chicago/Turabian StyleZhang, Chunyu, Zhiwei Miao, Yan Xu, and Tongguo Shi. 2025. "Non-Coding RNAs as Critical Modulators of Cholesterol Metabolism in Cancer" Biomedicines 13, no. 7: 1631. https://doi.org/10.3390/biomedicines13071631
APA StyleZhang, C., Miao, Z., Xu, Y., & Shi, T. (2025). Non-Coding RNAs as Critical Modulators of Cholesterol Metabolism in Cancer. Biomedicines, 13(7), 1631. https://doi.org/10.3390/biomedicines13071631






