Expression Characteristics and Prognostic Value of KLRG2 in Endometrial Cancer: A Comprehensive Analysis Based on Multi-Omics Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Diagnostic Evaluation, Survival Analysis, and Nomogram Construction
2.3. Differential Expression and Functional Enrichment
2.4. Protein-Protein Interaction (PPI) Network
2.5. Genetic Mutations and Methylation Analysis
2.6. Immune Microenvironment Profiling
2.7. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.8. Statistical Methods
3. Results
3.1. High Expression of KLRG2 in EC
3.2. Association of KLRG2 Expression with Adverse Clinicopathological Features and Prognosis
3.3. Clinical Significance of KLRG2 in EC
3.4. DEGs and Functional Enrichment Analysis
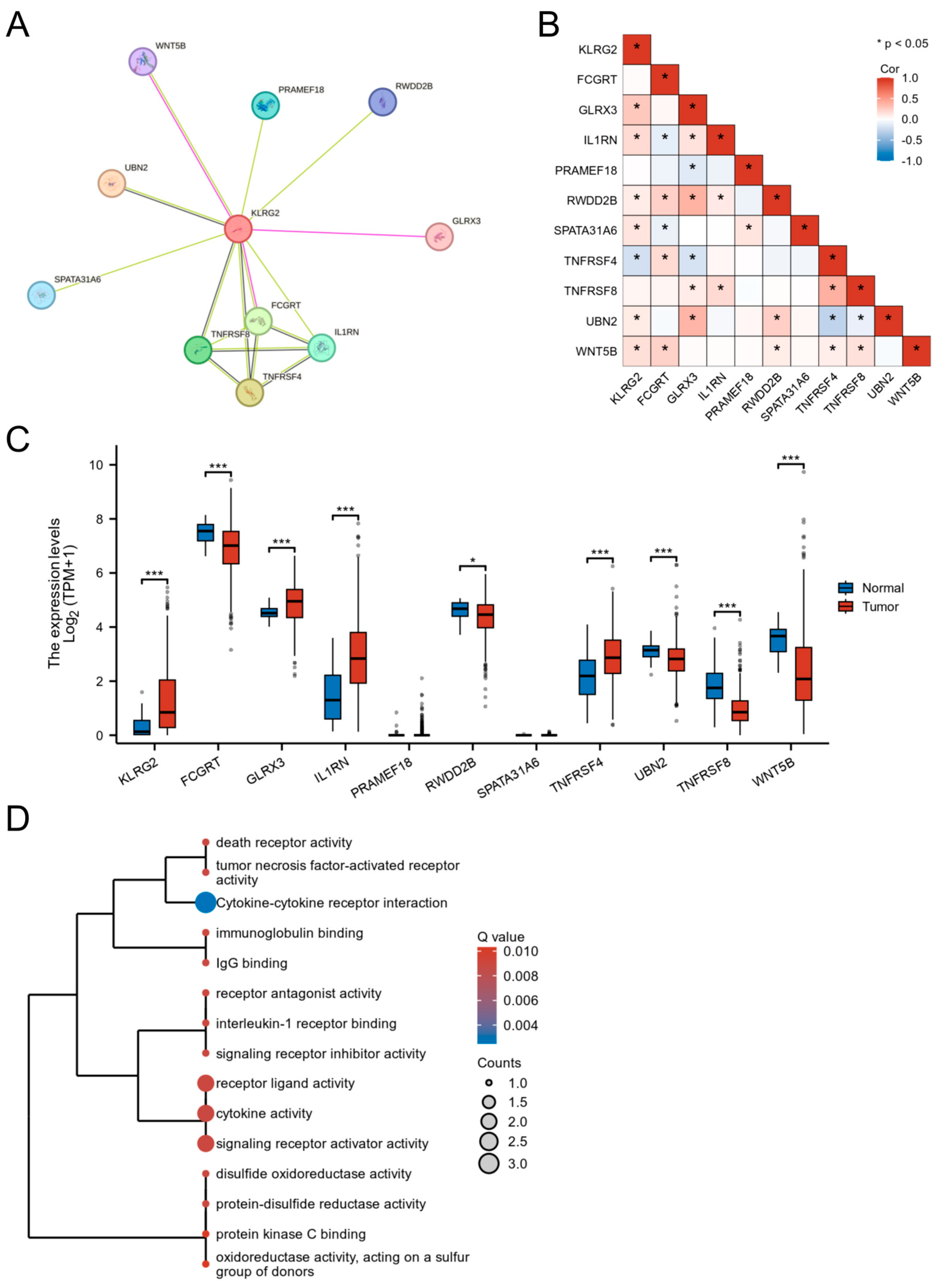
3.5. PPI Network and Functional Partners of KLRG2
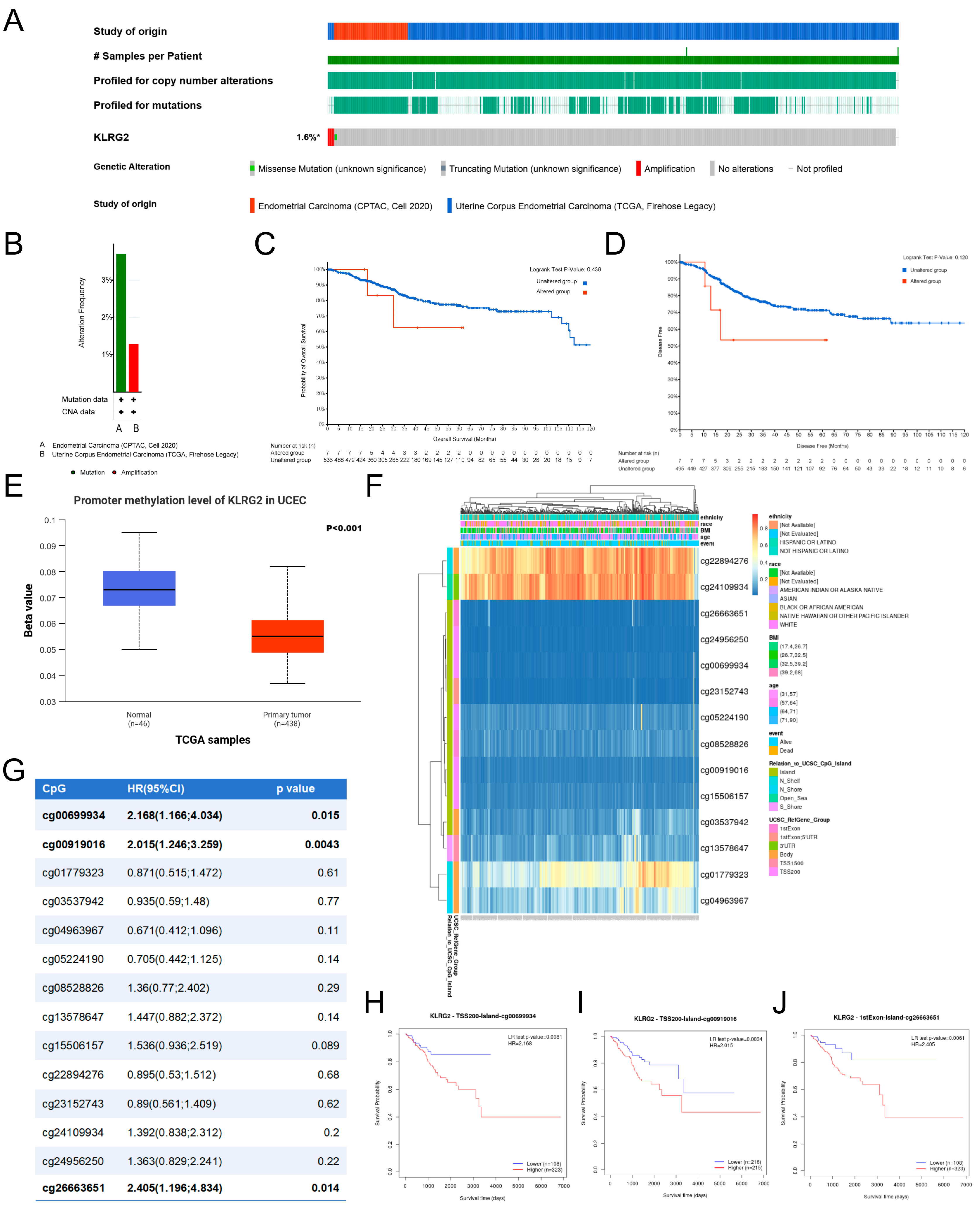
3.6. Genetic Mutation and Methylation of KLRG2
3.7. KLRG2 Correlation with Immune Infiltration and Checkpoints
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EC | Endometrial cancer |
| KLRG2 | Killer cell lectin-like receptor subfamily G member 2 |
| TME | Tumor microenvironment |
| UCEC | Uterine corpus endometrial carcinoma |
| TPM | Transcripts per million |
| GTEx | Genotype-Tissue Expression |
| GEO | Gene Expression Omnibus |
| IHC | Immunohistochemical |
| HPA | Human Protein Atlas |
| ROC | Receiver operating characteristic |
| AUC | Area under the curve |
| DEGs | Differentially expressed genes |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GSEA | Gene Set Enrichment Analysis |
| FDR | False discovery rate |
| PPI | Protein-Protein Interaction |
| CNVs | Copy number variations |
| ssGSEA | Single-sample gene set enrichment analysis |
| qRT-PCR | Quantitative Real-Time PCR |
| OS | Overall survival |
| DSS | Disease-specific survival |
| PFI | Progression-free interval |
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA A Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Cronin, K.A.; Scott, S.; Firth, A.U.; Sung, H.; Henley, S.J.; Sherman, R.L.; Siegel, R.L.; Anderson, R.N.; Kohler, B.A.; Benard, V.B.; et al. Annual report to the nation on the status of cancer, part 1: National cancer statistics. Cancer 2022, 128, 4251–4284. [Google Scholar] [CrossRef]
- Doll, K.M.; Winn, A.N. Assessing endometrial cancer risk among US women: Long-term trends using hysterectomy-adjusted analysis. Am. J. Obstet. Gynecol. 2019, 221, e311–e318. [Google Scholar] [CrossRef]
- Corr, B.R.; Erickson, B.K.; Barber, E.L.; Fisher, C.M.; Slomovitz, B. Advances in the management of endometrial cancer. BMJ 2025, 388, e080978. [Google Scholar] [CrossRef] [PubMed]
- Clarke, M.A.; Devesa, S.S.; Harvey, S.V.; Wentzensen, N. Hysterectomy-Corrected Uterine Corpus Cancer Incidence Trends and Differences in Relative Survival Reveal Racial Disparities and Rising Rates of Nonendometrioid Cancers. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 37, 1895–1908. [Google Scholar] [CrossRef]
- Ito, Y.; Kanda, M.; Sasahara, M.; Tanaka, C.; Shimizu, D.; Umeda, S.; Inokawa, Y.; Hattori, N.; Hayashi, M.; Nakayama, G.; et al. Killer cell lectin-like receptor G2 facilitates aggressive phenotypes of gastric cancer cells via dual activation of the ERK1/2 and JAK/STAT pathways. Gastric Cancer Off. J. Int. Gastric Cancer Assoc. Jpn. Gastric Cancer Assoc. 2024, 27, 506–518. [Google Scholar] [CrossRef]
- Yuan, S.M.; Chen, X.; Qu, Y.Q.; Zhang, M.Y. C6 and KLRG2 are pyroptosis subtype-related prognostic biomarkers and correlated with tumor-infiltrating lymphocytes in lung adenocarcinoma. Sci. Rep. 2024, 14, 24861. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, I.; Plummer, S.J.; Suarez, B.K.; Casey, G.; Catalona, W.J.; Witte, J.S. Fine-mapping of prostate cancer aggressiveness loci on chromosome 7q22-35. Prostate 2011, 71, 682–689. [Google Scholar] [CrossRef]
- Łasut-Szyszka, B.; Gdowicz-Kłosok, A.; Małachowska, B.; Krześniak, M.; Będzińska, A.; Gawin, M.; Pietrowska, M.; Rusin, M. Transcriptomic and proteomic study of cancer cell lines exposed to actinomycin D and nutlin-3a reveals numerous, novel candidates for p53-regulated genes. Chem.-Biol. Interact. 2024, 392, 110946. [Google Scholar] [CrossRef]
- Li, S.; Wang, W.; Yu, H.; Zhang, S.; Bi, W.; Sun, S.; Hong, B.; Fang, Z.; Chen, X. Characterization of genomic instability-related genes predicts survival and therapeutic response in lung adenocarcinoma. BMC Cancer 2023, 23, 1115. [Google Scholar] [CrossRef]
- Lysenko, O.; Schulte, D.; Mittelbronn, M.; Steinle, A. BACL is a novel brain-associated, non-NKC-encoded mammalian C-type lectin-like receptor of the CLEC2 family. PLoS ONE 2013, 8, e65345. [Google Scholar] [CrossRef]
- Wang, S.; Hu, D.; Wang, C.; Tang, X.; Du, M.; Gu, X.; Suo, J.; Hu, M.; Fang, R.; Zhu, X.; et al. Transcriptional profiling of innate immune responses in sheep PBMCs induced by Haemonchus contortus soluble extracts. Parasites Vectors 2019, 12, 182. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Sugiyama, Y.; Gotoh, O.; Fukui, N.; Tanaka, N.; Hasumi, K.; Takazawa, Y.; Noda, T.; Mori, S. Two Distinct Tumorigenic Processes in Endometrial Endometrioid Adenocarcinoma. Am. J. Pathol. 2020, 190, 234–251. [Google Scholar] [CrossRef]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas--a tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Blanche, P.; Dartigues, J.F.; Jacqmin-Gadda, H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat. Med. 2013, 32, 5381–5397. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Modhukur, V.; Iljasenko, T.; Metsalu, T.; Lokk, K.; Laisk-Podar, T.; Vilo, J. MethSurv: A web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018, 10, 277–288. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Carrassa, L.; Damia, G. DNA damage response inhibitors: Mechanisms and potential applications in cancer therapy. Cancer Treat. Rev. 2017, 60, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Bucher, N.; Britten, C.D. G2 checkpoint abrogation and checkpoint kinase-1 targeting in the treatment of cancer. Br. J. Cancer 2008, 98, 523–528. [Google Scholar] [CrossRef]
- Ciardo, D.; Goldar, A.; Marheineke, K. On the Interplay of the DNA Replication Program and the Intra-S Phase Checkpoint Pathway. Genes 2019, 10, 94. [Google Scholar] [CrossRef]
- Luczak, M.W.; Jagodziński, P.P. The role of DNA methylation in cancer development. Folia Histochem. Cytobiol. 2006, 44, 143–154. [Google Scholar]
- Yakoob, J.; Fan, X.G.; Hu, G.L.; Zhang, Z. DNA methylation and carcinogenesis in digestive neoplasms. World J. Gastroenterol. 1998, 4, 174–177. [Google Scholar] [CrossRef] [PubMed]
- Su, J.Q.; Lai, P.Y.; Hu, P.H.; Hu, J.M.; Chang, P.K.; Chen, C.Y.; Wu, J.J.; Lin, Y.J.; Sun, C.A.; Yang, T.; et al. Differential DNA methylation analysis of SUMF2, ADAMTS5, and PXDN provides novel insights into colorectal cancer prognosis prediction in Taiwan. World J. Gastroenterol. 2022, 28, 825–839. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Utsunomiya, T.; Ikemoto, T.; Yamada, S.; Morine, Y.; Imura, S.; Arakawa, Y.; Takasu, C.; Ishikawa, D.; Imoto, I.; et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) is associated with poor prognosis via activation of c-MET in hepatocellular carcinoma. Ann. Surg. Oncol. 2014, 21 (Suppl. S4), S729–S735. [Google Scholar] [CrossRef] [PubMed]
- Schulz, W.A. DNA methylation in urological malignancies (review). Int. J. Oncol. 1998, 13, 151–167. [Google Scholar] [CrossRef]
- Wang, H.; Yung, M.M.; Xuan, Y.; Chen, F.; Chan, W.; Siu, M.K.; Long, R.; Jia, S.; Liang, Y.; Xu, D.; et al. Polyunsaturated fatty acids promote M2-like TAM deposition via dampening RhoA-YAP1 signaling in the ovarian cancer microenvironment. Exp. Hematol. Oncol. 2024, 13, 90. [Google Scholar] [CrossRef]
- Meng, J.; Jiang, Y.Z.; Zhao, S.; Tao, Y.; Zhang, T.; Wang, X.; Zhang, Y.; Sun, K.; Yuan, M.; Chen, J.; et al. Tumor-derived Jagged1 promotes cancer progression through immune evasion. Cell reports 2022, 38, 110492. [Google Scholar] [CrossRef]
- Myers, J.A.; Miller, J.S. Exploring the NK cell platform for cancer immunotherapy. Nat. Rev. Clin. Oncol. 2021, 18, 85–100. [Google Scholar] [CrossRef]
- Frafjord, A.; Buer, L.; Hammarström, C.; Aamodt, H.; Woldbæk, P.R.; Brustugun, O.T.; Helland, Å.; Øynebråten, I.; Corthay, A. The Immune Landscape of Human Primary Lung Tumors Is Th2 Skewed. Front. Immunol. 2021, 12, 764596. [Google Scholar] [CrossRef]
- Murray, P.J. Macrophage Polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef]
- Mamrot, J.; Balachandran, S.; Steele, E.J.; Lindley, R.A. Molecular model linking Th2 polarized M2 tumour-associated macrophages with deaminase-mediated cancer progression mutation signatures. Scand. J. Immunol. 2019, 89, e12760. [Google Scholar] [CrossRef]
- Mezheyeuski, A.; Backman, M.; Mattsson, J.; Martín-Bernabé, A.; Larsson, C.; Hrynchyk, I.; Hammarström, K.; Ström, S.; Ekström, J.; Mauchanski, S.; et al. An immune score reflecting pro- and anti-tumoural balance of tumour microenvironment has major prognostic impact and predicts immunotherapy response in solid cancers. EBioMedicine 2023, 88, 104452. [Google Scholar] [CrossRef]
- Hamoud, B.H.; Sima, R.M.; Vacaroiu, I.A.; Georgescu, M.T.; Bobirca, A.; Gaube, A.; Bobirca, F.; Georgescu, D.E. The Evolving Landscape of Immunotherapy in Uterine Cancer: A Comprehensive Review. Life 2023, 13, 1502. [Google Scholar] [CrossRef] [PubMed]
- Di Tucci, C.; Capone, C.; Galati, G.; Iacobelli, V.; Schiavi, M.C.; Di Donato, V.; Muzii, L.; Panici, P.B. Immunotherapy in endometrial cancer: New scenarios on the horizon. J. Gynecol. Oncol. 2019, 30, e46. [Google Scholar] [CrossRef] [PubMed]
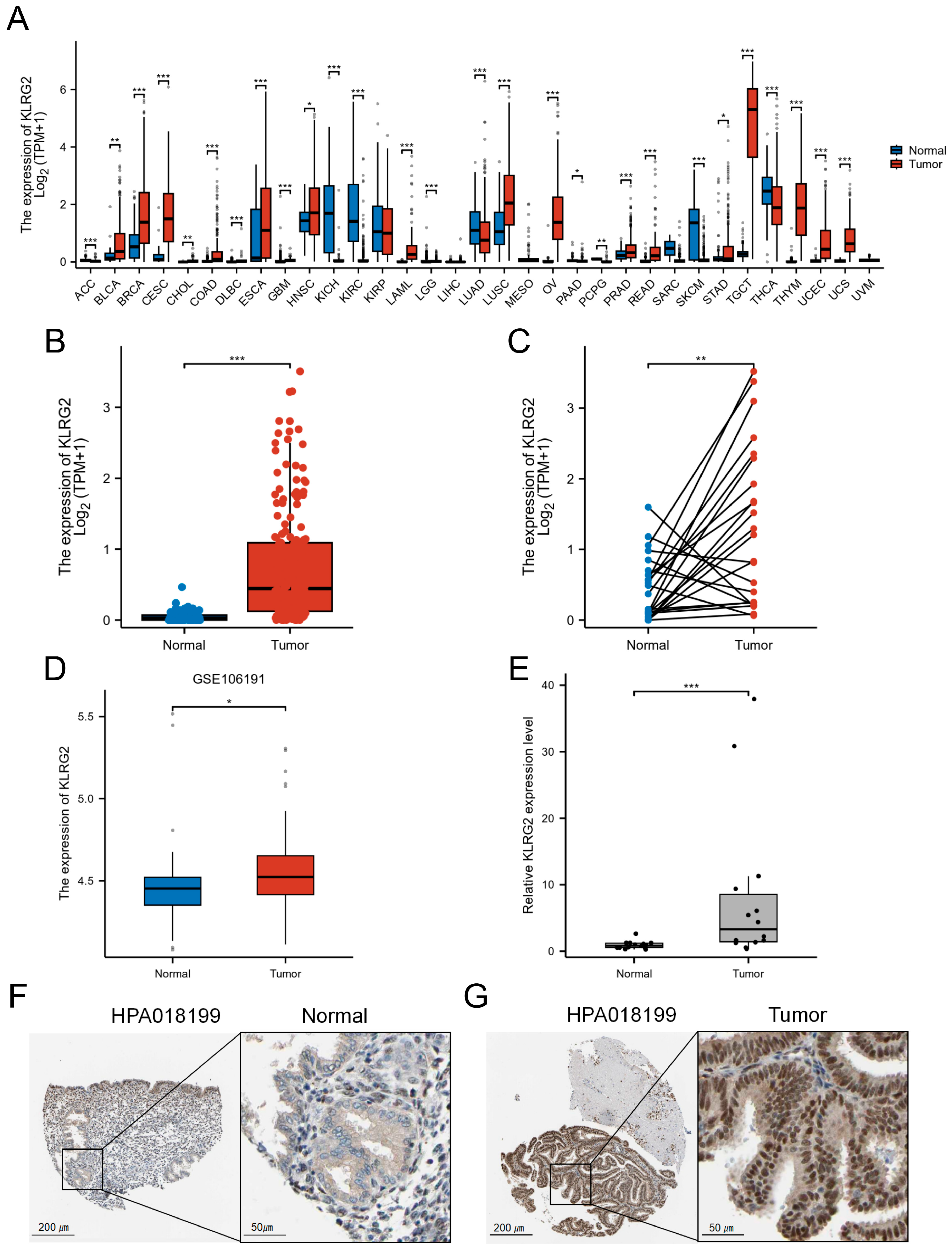
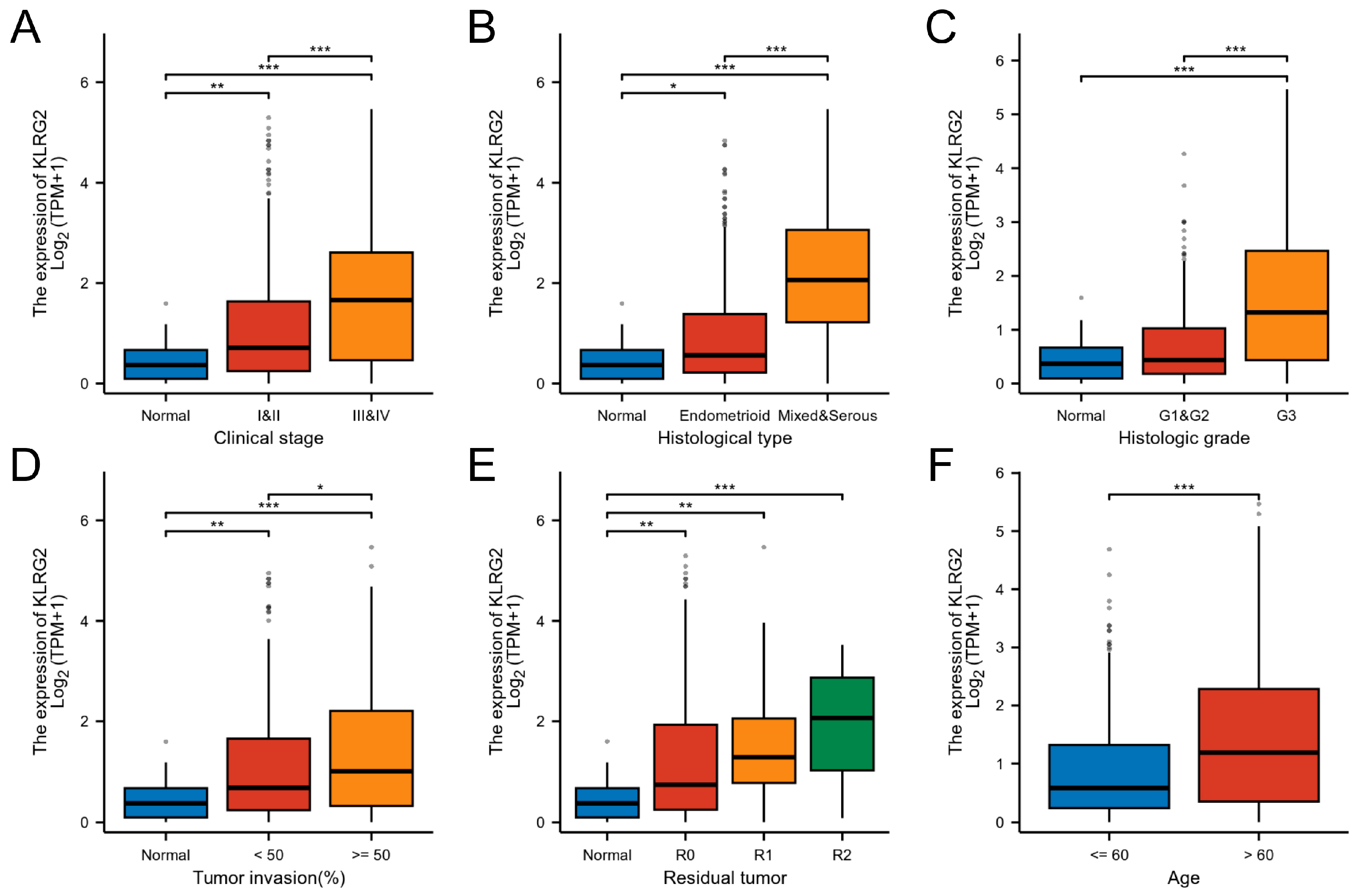

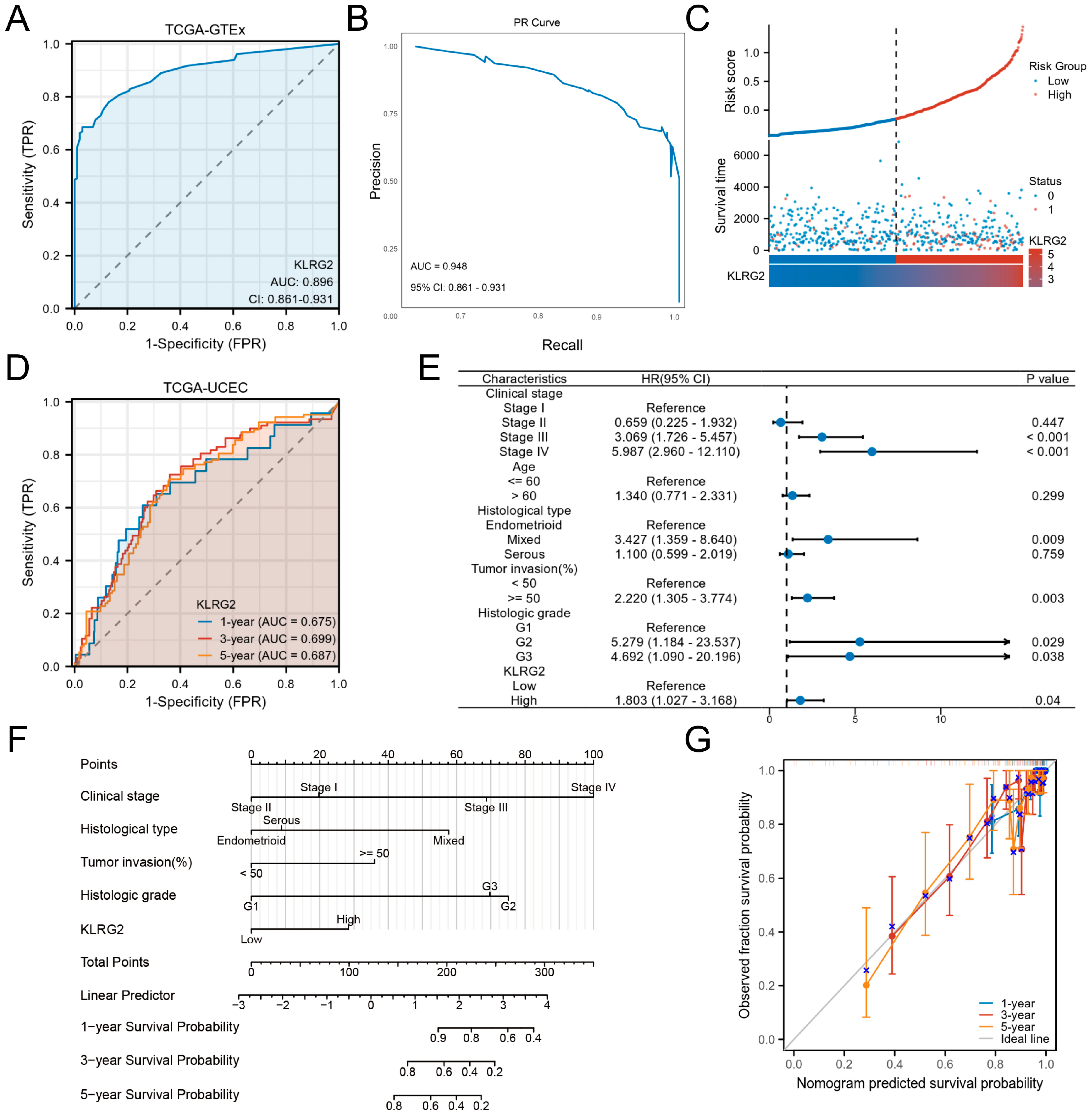
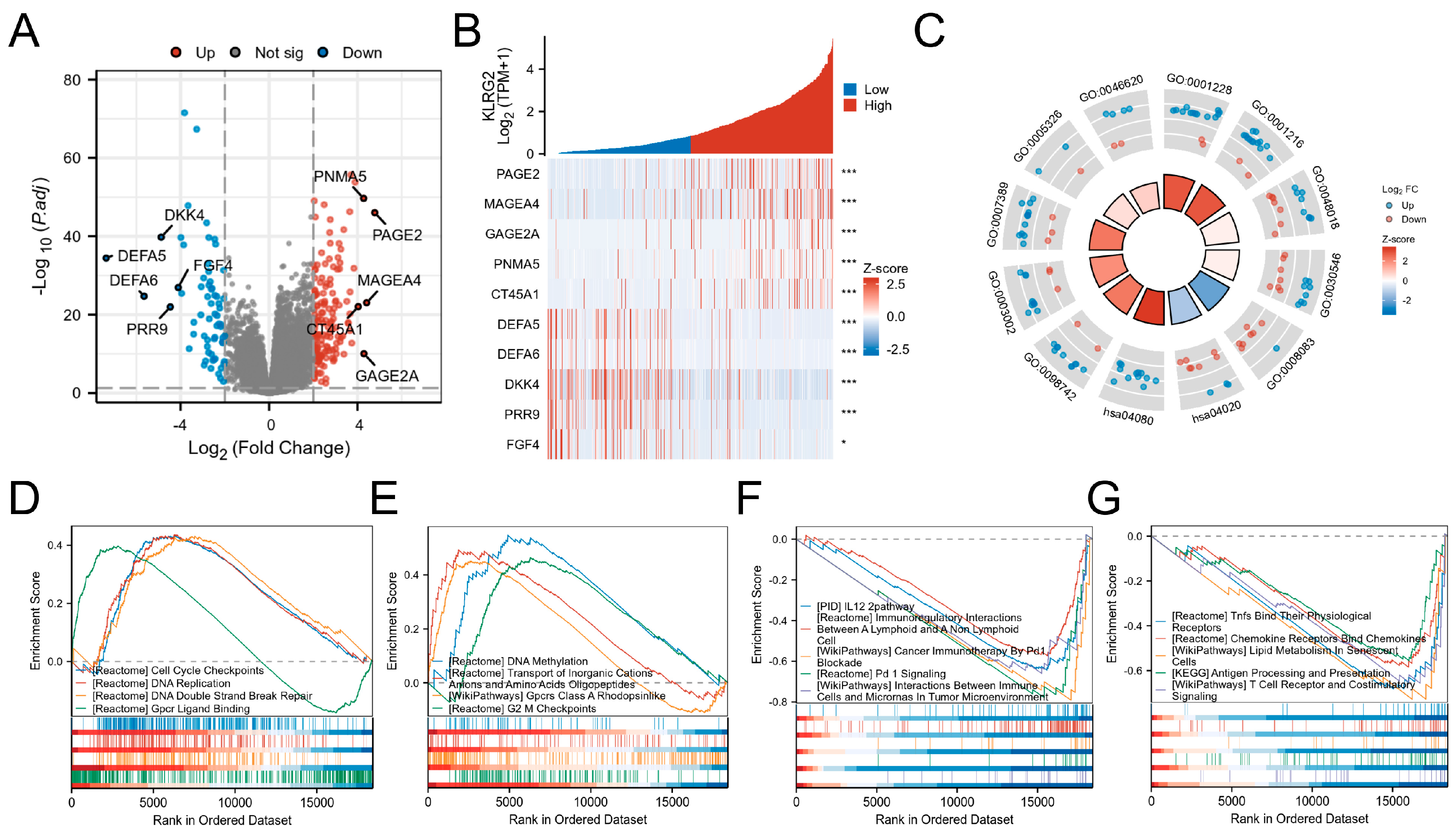

| Characteristics | Low Expression of KLRG2 | High Expression of KLRG2 | p-Value 1 |
|---|---|---|---|
| n | 277 | 277 | |
| Age, n (%) | <0.001 | ||
| <=60 | 128 (61.8%) | 79 (38.2%) | |
| >60 | 146 (42.4%) | 198 (57.6%) | |
| Race, n (%) | 0.070 | ||
| Asian | 10 (50%) | 10 (50%) | |
| Black or African American | 44 (40.4%) | 65 (59.6%) | |
| White | 201 (52.9%) | 179 (47.1%) | |
| BMI, n (%) | 0.054 | ||
| <=30 | 95 (44.8%) | 117 (55.2%) | |
| >30 | 165 (53.4%) | 144 (46.6%) | |
| Menopause status, n (%) | 0.034 | ||
| Pre | 24 (68.6%) | 11 (31.4%) | |
| Peri | 11 (64.7%) | 6 (35.3%) | |
| Post | 220 (48.4%) | 235 (51.6%) | |
| Diabetes, n (%) | 0.585 | ||
| No | 166 (50.5%) | 163 (49.5%) | |
| Yes | 59 (47.6%) | 65 (52.4%) | |
| Clinical stage, n (%) | <0.001 | ||
| Stage I | 199 (58%) | 144 (42%) | |
| Stage II | 23 (44.2%) | 29 (55.8%) | |
| Stage III | 50 (38.5%) | 80 (61.5%) | |
| Stage IV | 5 (17.2%) | 24 (82.8%) | |
| Histological type, n (%) | <0.001 | ||
| Endometrioid | 253 (61.4%) | 159 (38.6%) | |
| Serous | 13 (11%) | 105 (89%) | |
| Mixed | 11 (45.8%) | 13 (54.2%) | |
| Histologic grade, n (%) | <0.001 | ||
| G1 | 75 (75.8%) | 24 (24.2%) | |
| G2 | 78 (64.5%) | 43 (35.5%) | |
| G3 | 123 (38.1%) | 200 (61.9%) | |
| Tumor invasion (%), n (%) | 0.021 | ||
| <50 | 149 (57.1%) | 112 (42.9%) | |
| >=50 | 100 (46.5%) | 115 (53.5%) | |
| Residual tumor, n (%) | 0.002 | ||
| R0 | 201 (53.3%) | 176 (46.7%) | |
| R1 | 6 (27.3%) | 16 (72.7%) | |
| R2 | 3 (18.8%) | 13 (81.2%) | |
| Primary therapy outcome, n (%) | 0.041 2 | ||
| PD | 6 (30%) | 14 (70%) | |
| SD | 3 (50%) | 3 (50%) | |
| PR | 3 (25%) | 9 (75%) | |
| CR | 241 (54.3%) | 203 (45.7%) | |
| Hormones therapy, n (%) | 0.521 | ||
| No | 155 (51.8%) | 144 (48.2%) | |
| Yes | 22 (46.8%) | 25 (53.2%) | |
| Radiation therapy, n (%) | 0.367 | ||
| No | 147 (52.3%) | 134 (47.7%) | |
| Yes | 120 (48.4%) | 128 (51.6%) | |
| OS event, n (%) | <0.001 | ||
| Alive | 253 (55%) | 207 (45%) | |
| Dead | 24 (25.5%) | 70 (74.5%) | |
| DSS event, n (%) | <0.001 | ||
| No | 264 (54%) | 225 (46%) | |
| Yes | 12 (19%) | 51 (81%) | |
| PFI event, n (%) | <0.001 | ||
| No | 234 (55.1%) | 191 (44.9%) | |
| Yes | 43 (33.3%) | 86 (66.7%) |
| Characteristics | Total (N) | OR (95% CI) | p-Value |
|---|---|---|---|
| Clinical stage (Stage III and Stage IV vs. Stage I and Stage II) | 554 | 2.426 (1.655–3.557) | <0.001 |
| Age (>60 vs. <=60) | 551 | 2.197 (1.544–3.126) | <0.001 |
| BMI (>30 vs. <=30) | 521 | 0.709 (0.499–1.007) | 0.054 |
| Histological type (Mixed and Serous vs. Endometrioid) | 554 | 7.823 (4.833–12.664) | <0.001 |
| Tumor invasion(%) (>=50 vs. <50) | 476 | 1.530 (1.064–2.200) | 0.022 |
| Diabetes (Yes vs. No) | 453 | 1.122 (0.742–1.696) | 0.585 |
| Residual tumor (R1 and R2 vs. R0) | 415 | 3.680 (1.696–7.985) | <0.001 |
| Menopause status (Post vs. Pre and Peri) | 507 | 2.199 (1.197–4.039) | 0.011 |
| Histologic grade (G3 vs. G1 and G2) | 543 | 3.713 (2.579–5.346) | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Li, A.; Xu, D. Expression Characteristics and Prognostic Value of KLRG2 in Endometrial Cancer: A Comprehensive Analysis Based on Multi-Omics Data. Biomedicines 2025, 13, 1592. https://doi.org/10.3390/biomedicines13071592
Huang X, Li A, Xu D. Expression Characteristics and Prognostic Value of KLRG2 in Endometrial Cancer: A Comprehensive Analysis Based on Multi-Omics Data. Biomedicines. 2025; 13(7):1592. https://doi.org/10.3390/biomedicines13071592
Chicago/Turabian StyleHuang, Xiaoyan, Ailian Li, and Dianbo Xu. 2025. "Expression Characteristics and Prognostic Value of KLRG2 in Endometrial Cancer: A Comprehensive Analysis Based on Multi-Omics Data" Biomedicines 13, no. 7: 1592. https://doi.org/10.3390/biomedicines13071592
APA StyleHuang, X., Li, A., & Xu, D. (2025). Expression Characteristics and Prognostic Value of KLRG2 in Endometrial Cancer: A Comprehensive Analysis Based on Multi-Omics Data. Biomedicines, 13(7), 1592. https://doi.org/10.3390/biomedicines13071592






