Improving Virological Monitoring of HDV Infection: A Proof-of-Concept Comparative Study of Bosphore and AltoStar® Assays in Patients Treated with Bulevirtide
Abstract
1. Introduction
2. Patients and Methods
2.1. Study Population
2.2. HDV RNA Quantification
2.3. Statistical Analysis
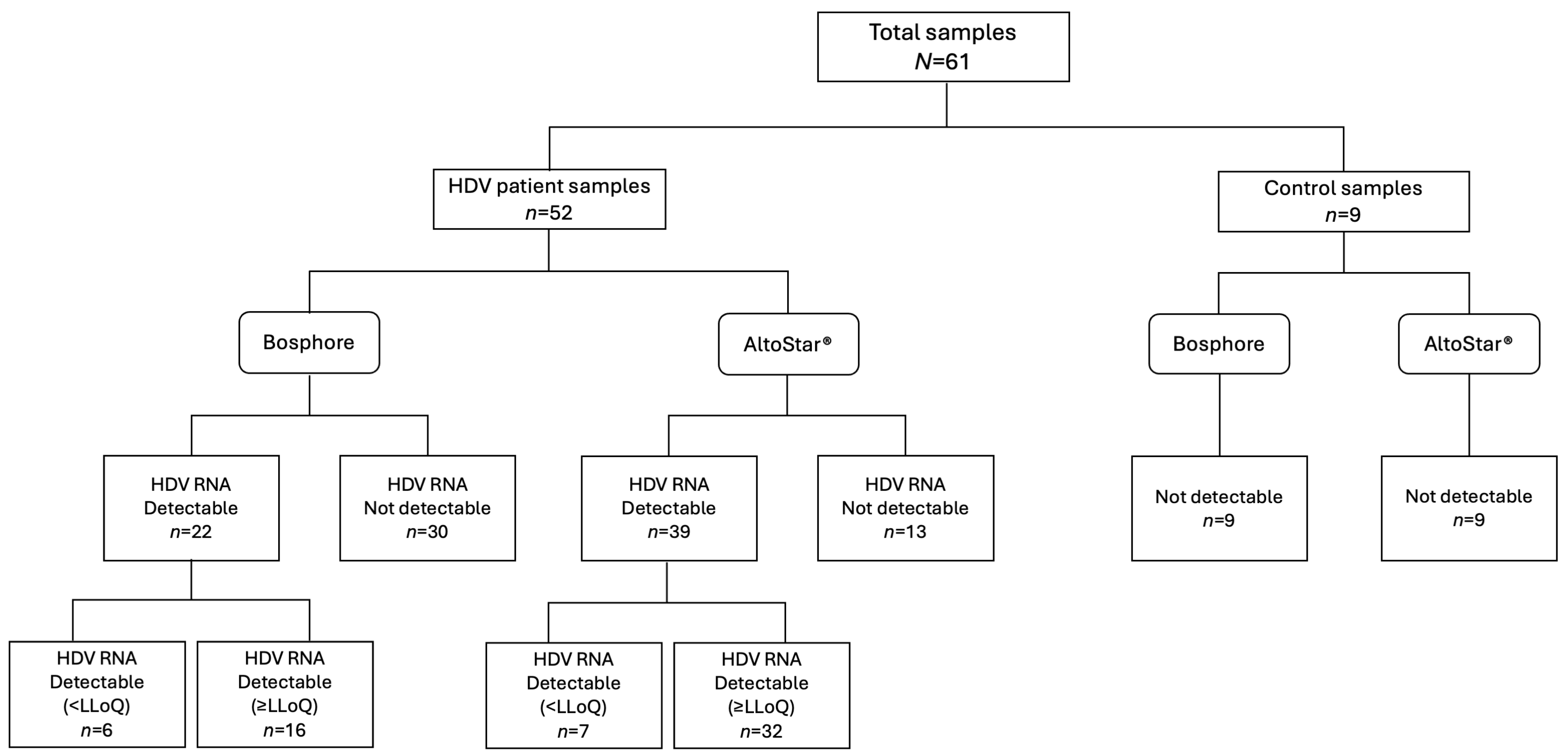
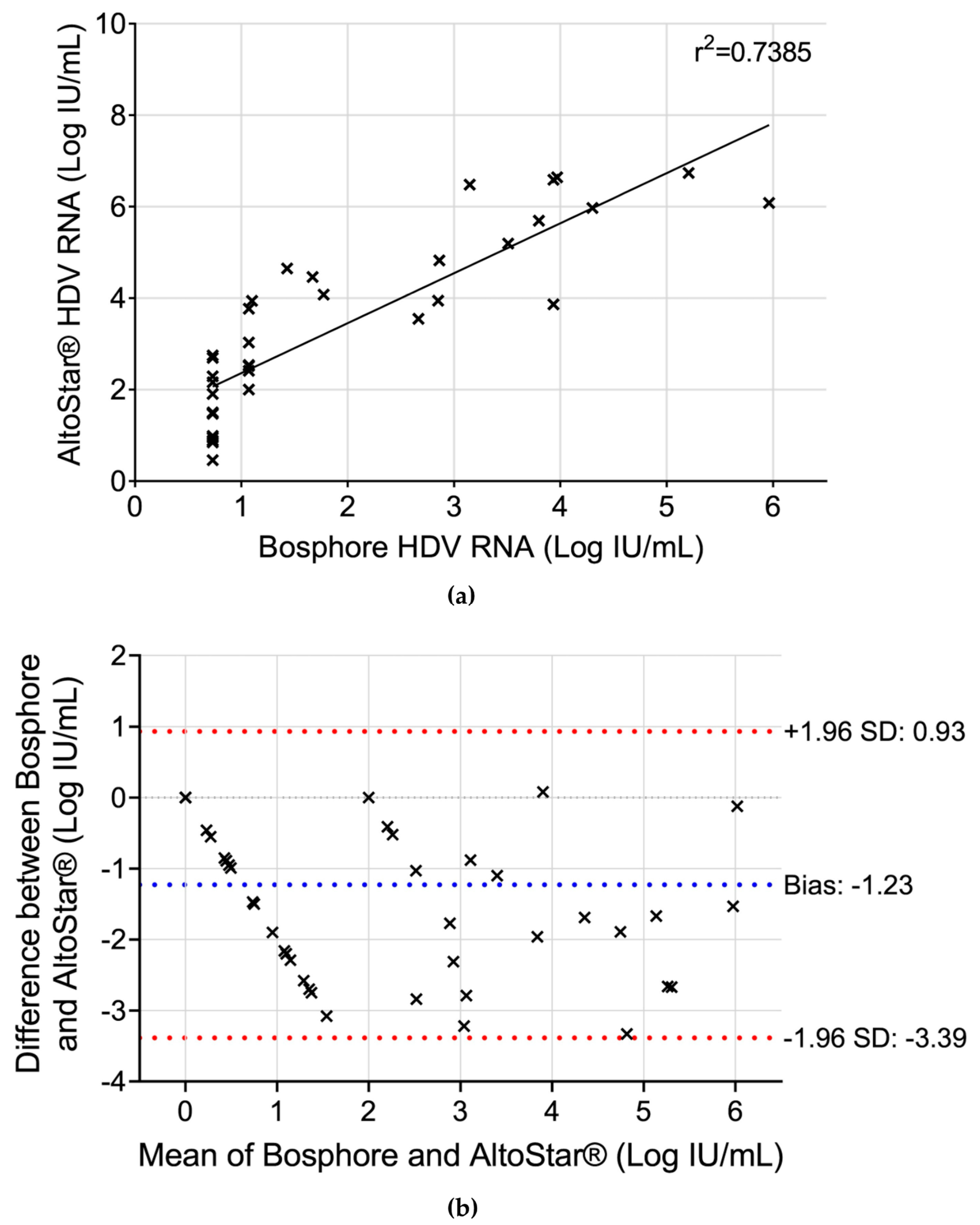
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Asselah, T.; Rizzetto, M. Hepatitis D Virus Infection. N. Engl. J. Med. 2023, 389, 58–70. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Hepatitis Delta Fact Sheet (WHO/CDS/CSR/NCS/2001.1); World Health Organization: Geneva, Switzerland, 2001; Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-d (accessed on 14 February 2025).
- Negro, F.; Lok, A.S. Hepatitis D: A Review. JAMA 2023, 330, 2376–2387. [Google Scholar] [CrossRef]
- Sureau, C.; Negro, F. The Hepatitis Delta Virus: Replication and Pathogenesis. J. Hepatol. 2016, 64, S102–S116. [Google Scholar] [CrossRef]
- Amini, N.; Alavian, S.M.; Kabir, A.; Aalaei-Andabili, S.H.; Saiedi Hosseini, S.Y.; Rizzetto, M. Prevalence of Hepatitis D in the Eastern Mediterranean Region: Systematic Review and Meta Analysis. Hepat. Mon. 2013, 13, e8210. [Google Scholar] [CrossRef]
- Sandmann, L.; Wedemeyer, H. Interferon-based Treatment of Chronic Hepatitis D. Liver Int. 2023, 43, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Degasperi, E.; Anolli, M.P.; Lampertico, P. Bulevirtide-Based Treatment Strategies for Chronic Hepatitis Delta: A Review. J. Viral Hepat. 2023, 30 (Suppl. 1), 26–32. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.; Schieck, A.; Ni, Y.; Mier, W.; Urban, S. Fine Mapping of Pre-S Sequence Requirements for Hepatitis B Virus Large Envelope Protein-Mediated Receptor Interaction. J. Virol. 2010, 84, 1989–2000. [Google Scholar] [CrossRef]
- Yan, H.; Peng, B.; He, W.; Zhong, G.; Qi, Y.; Ren, B.; Gao, Z.; Jing, Z.; Song, M.; Xu, G.; et al. Molecular Determinants of Hepatitis B and D Virus Entry Restriction in Mouse Sodium Taurocholate Cotransporting Polypeptide. J. Virol. 2013, 87, 7977–7991. [Google Scholar] [CrossRef]
- Ni, Y.; Lempp, F.A.; Mehrle, S.; Nkongolo, S.; Kaufman, C.; Fälth, M.; Stindt, J.; Königer, C.; Nassal, M.; Kubitz, R.; et al. Hepatitis B and D Viruses Exploit Sodium Taurocholate Co-Transporting Polypeptide for Species-Specific Entry into Hepatocytes. Gastroenterology 2014, 146, 1070–1083. [Google Scholar] [CrossRef]
- Durník, R.; Šindlerová, L.; Babica, P.; Jurček, O. Bile Acids Transporters of Enterohepatic Circulation for Targeted Drug Delivery. Molecules 2022, 27, 2961. [Google Scholar] [CrossRef]
- Jachs, M.; Panzer, M.; Hartl, L.; Schwarz, M.; Balcar, L.; Camp, J.V.; Munda, P.; Mandorfer, M.; Trauner, M.; Aberle, S.W.; et al. Long-Term Follow-up of Patients Discontinuing Bulevirtide Treatment upon Long-Term HDV-RNA Suppression. JHEP Rep. 2023, 5, 100751. [Google Scholar] [CrossRef] [PubMed]
- Anolli, M.P.; Renteria, S.U.; Degasperi, E.; Borghi, M.; Facchetti, F.; Sambarino, D.; Perbellini, R.; Monico, S.; Ceriotti, F.; Lampertico, P. Quantification of Serum HDV RNA by Robogene 2.0 in HDV Patients Is Significantly Influenced by the Extraction Methods. Liver Int. 2024, 44, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Wedemeyer, H.; Leus, M.; Battersby, T.R.; Glenn, J.; Gordien, E.; Kamili, S.; Kapoor, H.; Kessler, H.H.; Lenz, O.; Lütgehetmann, M.; et al. HDV RNA Assays: Performance Characteristics, Clinical Utility, and Challenges. Hepatology 2025, 81, 637–650. [Google Scholar] [CrossRef]
- Piermatteo, L.; Salpini, R.; Caviglia, G.; Bertoli, A.; Brunetto, M.R.; Bruzzone, B.; Callegaro, A.; Caudai, C.; Cavallone, D.; Chessa, L.; et al. Comparison of Diagnostic Performances of HDV RNA Quantification Assays Used in Clinical Practice in Italy: Data from the First National Quality Control Multicenter Study. Dig. Liver Dis. 2025, 57, S53–S54. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. Measuring Agreement in Method Comparison Studies. Stat. Methods Med. Res. 1999, 8, 135–160. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; The R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: https://www.r-project.org (accessed on 14 February 2025).
- Zulian, V.; Salichos, L.; Taibi, C.; Pauciullo, S.; Dong, L.; D’Offizi, G.; Biliotti, E.; Rianda, A.; Federici, L.; Bibbò, A.; et al. Exploring Predictive Factors for Bulevirtide Treatment Response in Hepatitis Delta-Positive Patients. Biomedicines 2025, 13, 280. [Google Scholar] [CrossRef]
- Illescas-López, M.; Chaves-Blanco, L.; de Salazar, A.; Hernández-Febles, M.; Carracedo, R.; Lagarejos, E.; Fuentes, A.; Pereira, S.; Cea, M.; De La Iglesia, A.; et al. Assessment of Performance and Comparison of Three Commercial HDV RNA Detection Assays: Implications for Diagnosis and Treatment Monitoring. Front. Cell Infect. Microbiol. 2024, 14, 1422299. [Google Scholar] [CrossRef]
- Sandmann, L.; Bremer, B.; Deterding, K.; Port, K.; Gey, B.; Früchtel, C.; Reinhardt, A.; Lachmann, I.; Cornberg, M.; Kefalakes, H.; et al. Letter to the Editor: The WHO HDV RNA International Standard Does Not Reflect Variability of Real-World Samples. Hepatology 2025, 81, E32–E33. [Google Scholar] [CrossRef]
- Salichos, L.; Minosse, C.; Visco-Comandini, U.; Taibi, C.; Zulian, V.; D’Offizi, G.; Pallothu, N.; McPhee, F.; Garbuglia, A.R. Phylogenetic and Phylodynamic Analysis of Delta Strains Circulating in Italy. Viruses 2023, 15, 1791. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines on Hepatitis Delta Virus. J. Hepatol. 2023, 79, 433–460. [Google Scholar] [CrossRef]
- Caviglia, G.P.; Loglio, A.; Viganò, M.; Fagiuoli, S.; Colombatto, P.; Coco, B.; Dileo, E.; Manetta, T.; Olivero, A.; Troshina, G.; et al. Burden of Hepatitis D Virus Infection in Italy: Final Results from the HDV Describe Study. Dig. Liver Dis. 2025, 57, S6–S7. [Google Scholar] [CrossRef]
| Parameter | N = 15 |
|---|---|
| Age at recruitment, years | 50 (47–62) |
| Male sex | 4 (26.7) |
| Body-mass index | 24 (23.0–26.5) |
| Cirrhosis | 11 (73.3) |
| Previous interferon therapy | 9 (60.0) |
| Concomitant NUC therapy | 15 (100.0) |
| ALT, U/L | 78.0 (46.5–100.0) |
| AST, U/L | 70.0 (59.8–76.5) |
| Albumin, g/dL | 4.2 (4.0–4.4) |
| Bile acids, μmol/L | 9.1 (6.4–16.7) |
| Total bilirubin, mg/dL | 0.9 (0.7–1.6) |
| Platelet count, ×103/μL | 90.0 (68.0–152.0) |
| HDV RNA, Log cp/mL | 5.0 (3.4–5.7) |
| HBV DNA detectable * | 0 (0.0) |
| HBsAg, Log IU/mL | 4.0 (3.6–4.2) |
| HBeAg positive | 0 (0.0) |
| AltoStar® HDV RNA (IU/mL) | Bosphore HDV RNA (IU/mL) | ||||
|---|---|---|---|---|---|
| Patient ID | HDV Genotype | T1 * | T2 ** | T1 | T2 |
| PT1 | 1e | ND a | ND | ND | ND |
| PT2 | 1 unsubtyped | ND | ND | ND | ND |
| PT3 | 1e | 30 | 9 | ND | ND |
| PT4 | 1e | 10 | ND | ND | ND |
| PT5 | 1e | 3 | ND | ND | ND |
| PT6 | 1b | 497 | 146 | ND | ND |
| PT7 | 1e | 559 | 32 | ND | ND |
| PT8 | 1e | 1190 | 1069 | ND | <LLoQ b |
| PT9 | 1e | 158 | 345 | ND | <LLoQ |
| PT10 | 1e | 383 | 259 | ND | <LLoQ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zulian, V.; Taibi, C.; Coppola, A.; Bibbò, A.; Federici, L.; De Sanctis, M.; Pauciullo, S.; D’Offizi, G.; Biliotti, E.; McPhee, F.; et al. Improving Virological Monitoring of HDV Infection: A Proof-of-Concept Comparative Study of Bosphore and AltoStar® Assays in Patients Treated with Bulevirtide. Biomedicines 2025, 13, 1564. https://doi.org/10.3390/biomedicines13071564
Zulian V, Taibi C, Coppola A, Bibbò A, Federici L, De Sanctis M, Pauciullo S, D’Offizi G, Biliotti E, McPhee F, et al. Improving Virological Monitoring of HDV Infection: A Proof-of-Concept Comparative Study of Bosphore and AltoStar® Assays in Patients Treated with Bulevirtide. Biomedicines. 2025; 13(7):1564. https://doi.org/10.3390/biomedicines13071564
Chicago/Turabian StyleZulian, Verdiana, Chiara Taibi, Antonio Coppola, Angela Bibbò, Luigi Federici, Martina De Sanctis, Silvia Pauciullo, Gianpiero D’Offizi, Elisa Biliotti, Fiona McPhee, and et al. 2025. "Improving Virological Monitoring of HDV Infection: A Proof-of-Concept Comparative Study of Bosphore and AltoStar® Assays in Patients Treated with Bulevirtide" Biomedicines 13, no. 7: 1564. https://doi.org/10.3390/biomedicines13071564
APA StyleZulian, V., Taibi, C., Coppola, A., Bibbò, A., Federici, L., De Sanctis, M., Pauciullo, S., D’Offizi, G., Biliotti, E., McPhee, F., & Garbuglia, A. R. (2025). Improving Virological Monitoring of HDV Infection: A Proof-of-Concept Comparative Study of Bosphore and AltoStar® Assays in Patients Treated with Bulevirtide. Biomedicines, 13(7), 1564. https://doi.org/10.3390/biomedicines13071564







