Abstract
Background: Progressive Supranuclear Palsy (PSP) is a rare neurodegenerative disorder characterized by abnormal tau protein aggregation. The MAPT gene encodes for tau protein. The MAPT locus harbors two major haplotypes, H1 and H2, with H1 and its subhaplotypes being associated with an increased risk of PSP. Methods: In this study, we genotyped rs8070723 in a cohort of 73 PSP patients, including 47 PSP Richardson Syndrome (PSP-RS) and 27 PSP variants (vPSP), and 93 age-matched healthy controls (HC) from Southern Italy. Results: Haplotype analysis identified H1 and H2 haplotypes that conferred a risk (OR, 2.620; 95% CI, 1.399–5.140; p = 0.0035) and a protective effect (OR, 0.370; 95% CI, 0.196–0.695; p = 0.0015), respectively. In addition, we genotyped five MAPT variants (rs1467967, rs242557, rs3785883, rs2471738, and rs7521) that, together with rs8070723, defined H1 subhaplotypes. We identified 18 distinct MAPT H1 subhaplotypes, among which H1j displayed a nominally significant reduced risk of PSP (OR, 0.201; 95% CI, 0.044–0.915; p = 0.0265). Conclusions: These findings reinforce the role of MAPT genetic variation in PSP pathogenesis and highlight the potential impact of haplotype diversity on disease susceptibility.
1. Introduction
Progressive supranuclear palsy (PSP) is a rare neurodegenerative movement disorder with an estimated annual prevalence of 5.8–6.5 cases per 100,000 [1]. PSP is clinically heterogeneous and may manifest various phenotypes, in which the most common form is Richardson Syndrome (PSP-RS), with vertical supranuclear gaze palsy, postural instability, and motor and cognitive dysfunction [2,3]. Other recognized forms (termed PSP variant, vPSP) are PSP with predominant parkinsonism (PSP-P), PSP with progressive gait freezing (PSP-PGF), PSP with predominant corticobasal syndrome (PSP-CBS), PSP with a predominant speech/language disorder (PSP-SL), PSP with predominant frontal presentation (PSP-F), PSP with cerebellar ataxia (PSP-C), and oculomotor dysfunction (PSP-OM) [2,3].
Neuropathologically, PSP is characterized by abnormal accumulation of tau protein in the form of globose neurofibrillary tangles (NFTs), tufted astrocytes (TA), oligodendroglial coiled bodies (CB) and neuropil threads (NT) in the basal ganglia, brainstem, cortical regions, and cerebellum [4].
Tau, a microtubule-associated protein predominantly found in neuronal axons, plays a crucial role in regulating microtubule dynamics, modulating synaptic plasticity and supporting axonal transport [5]. It is organized into four main functional domains: a N-terminal region (NTR), a proline-rich domain (PRR), a microtubule-binding domain (MTBD), and the C-terminal assembly region [6].
Tau protein is encoded by MAPT, a long gene of 16 exons located on chromosome 17q2 [7]. In the adult human brain, six major tau protein isoforms are generated by alternative splicing of exons 2, 3, and 10, which determine the number of N-terminal inserts (0N, 1N, or 2N) and C-terminal repeat domains (3R or 4R) [8]. Alternative splicing of exons 2 and 3 generates isoforms 0N if both 2 and 3 exons are absent, 1N if only exon 2 is included, and 2N if both exons are included. The exclusion or inclusion of exon 10 determines whether an isoform has three (3R) or four (4R) repeat domains, respectively [6,9]. The ratio of 4R to 3R is approximately equal (1:1); however, an increased 4R:3R ratio has been linked to PSP-RS pathology. This imbalance enhances tau phosphorylation and aggregation, reduces its binding affinity to microtubules, and consequently impairs normal microtubule function, contributing to neurodegeneration [10].
MAPT is located in a region of chromosome 17 with extensive linkage disequilibrium (LD) that has undergone a 900 kb ancestral inversion, giving rise to two major haplotypes, H1 and H2 [11]. These haplotypes differ in orientation and do not recombine. While the H1 haplotype is widespread across all populations, the H2 haplotype is primarily found in Middle Eastern and European populations (~25%), but it is rare in Central Asia and nearly absent in African, East Asian, and Native American populations [12,13]. H1 haplotype, previously described as a risk factor for PSP, can be further divided into more than 20 subhaplotypes [14,15]. Among these, H1c, H1d, H1g, and H1o have consistently been associated with an increased risk of PSP [13,16,17,18]. In contrast, H2 haplotype appears to have a protective role [19,20,21].
Beyond the haplotypic architecture, several single nucleotide polymorphisms (SNPs) within MAPT have been implicated in modulating disease susceptibility and tau-related pathophysiology. For instance, the A allele of rs242557 has been linked to increased plasma tau levels and enhanced transcriptional activity of the MAPT promoter, indicating a regulatory role in gene expression [22] Similarly, rs3785883 has been associated with higher cerebrospinal fluid (CSF) tau concentrations and accelerated functional decline in patients with Alzheimer’s disease, suggesting a broader role in tau-related neurodegeneration [23]. The T allele of rs2471738 has been correlated with elevated MAPT expression in the human hippocampus, supporting its involvement in tissue-specific gene regulation [24]. Notably, rs8070723 is in strong LD with rs242561, which resides within an antioxidant response element (ARE) of MAPT that binds NRF2/sMAF transcription factors. This genomic context implies that rs8070723 may influence the transcriptional regulation of MAPT in response to oxidative stress, thereby modulating tau expression [25]. Additionally, rs1467967 has been associated with increased levels of total tau (t-tau) and phosphorylated tau (p-tau181) in the CSF, particularly among individuals carrying the AA or AG genotypes [26]. Lastly, rs7521 has been shown to affect the expression of the 4R tau isoform, particularly in carriers of the H1c haplotype—an isoform that predominates in PSP and other tauopathies [27].
In this study, we performed for the first time a genetic association analysis to investigate the link between MAPT haplotypes and PSP susceptibility evaluated in the whole PSP group and separately in the different phenotypes (PSP-RS and vPSP). We analyzed six MAPT SNPs in a cohort of 73 PSP patients and 93 age-matched healthy controls from Southern Italy.
2. Materials and Methods
2.1. Patients
We enrolled unrelated 73 PSP cases (46 RS and 27 Variant) and HC subjects (n = 93) at the Neuroscience Research Center, University Magna Graecia of Catanzaro, Italy. All of them were Caucasian, originated in Calabria, Southern Italy. The two groups were age matched, with PSP cases having an average age at blood collection of 69.90 (65.75% male) while healthy controls of 72.85 (43.01% male). Demographic information of the patient and control groups is summarized in Table 1. The study was conducted in accordance with the Declaration of Helsinki and approved by the Calabria Region Ethics Committee under protocol code 143 on 13 May 2024. All participants provided informed consent after receiving a detailed explanation of the study’s purpose.

Table 1.
Age and sex characteristics of the PSP and HC populations. The sample mean ± standard deviation is given for age at blood collection of PSP and HC. Number and percentage of females and males are given for both populations.
2.2. Genetic Analysis
Genomic DNA was extracted from peripheral blood samples of both PSP patients and healthy controls using Maxwell® RSC Blood DNA kit. Genotyping was conducted in triplicate using TaqMan SNP genotyping assays on QuantStudio 5 instrument (Applied Biosystems, Waltham, MA, USA), adhering to the manufacturer’s instructions. Genotype calling was performed using QuantStudioTM Design & Analysis software v.1.5.3 (Applied Biosystems) to ensure accurate variant identification. Genotyping of six MAPT variants (rs1467967, rs242557, rs3785883, rs2471738, rs8070723, and rs7521) was performed to evaluate the most common MAPT haplotypes [13,14,28,29,30]. Among them, the MAPT H2 haplotype was tagged by the variant rs8070723, which was genotyped in all patients and controls. The minor allele G of rs8070723 corresponds to the MAPT H2 haplotype, while the major allele A is associated with the MAPT H1 haplotype [31]. The rate of genotype calls was 100% in each population. A total of 19 distinct MAPT haplotypes, each observed in at least 1% of individuals across the association analyses conducted in this study, are presented in Table 2.

Table 2.
MAPT H2 haplotype and H1 subhaplotypes that were observed in 1% or more of PSP and HC populations in any of the 19 association analyses.
2.3. Statistical Analysis
Statistical assessment for the allele and genotype frequencies and Hardy–Weinberg equilibrium (HWE) was made using TaqManTM Genotyper Software v1.7 (Applied Biosystems). For each SNP, the allele and genotype distributions in PSP cases were compared with those of the HC group. Pairwise LD across MAPT for each SNP was then evaluated using both D’ and the squared correlation coefficient (r2) with Haploview v.4.2 (Broad Institute, Cambridge, MA, USA). The Expectation-Maximization (EM) algorithm, adjusted for sex, was employed to infer haplotypes and estimate their frequencies from genotype data. Odds ratios (ORs) and 95% confidence intervals (CIs) of inferred haplotypes were calculated using the epi.2by2 function from the epiR package, while odds ratios and CIs of A and G alleles on the rs8070723 SNP were evaluated using logistic regression, adjusting for age and sex as covariates. ORs and CIs were computed in the R statistical computing environment, version 4.2.2 (The R Foundation for Statistical Computing, Vienna, Austria) to assess the association between six variant MAPT haplotypes and risk of PSP. Specifically, association tests were conducted for 18 or 19 haplotypes, depending on the patient group under analysis. Comparisons were performed between the following groups: PSP versus HC, PSP-RS versus HC, vPSP versus HC. A Bonferroni correction for multiple testing established statistical significance at p ≤ 0.0026 (0.05/19) or p ≤ 0.0027 (0.05/18).
3. Results
3.1. Linkage Disequilibrium Analysis
No significant differences in allele and genotype frequencies were found between PSP and controls (Table 3). None of the six SNPs departed from HWE in both populations (Table 3).

Table 3.
Allele and genotype distributions. The calculated HWE p-value is the probability of the differences in observed and expected genotype calls accounted for by chance alone. No deviations of the genotype frequencies from HWE equilibrium were detected in either cases or controls.
Our analysis of LD patterns at the MAPT locus supports the presence of two distinct haplotype lineages, H1 and H2, which have remained evolutionarily separate for approximately 3 million years [11]. By examining D’ and r2 in HC and PSP cases, we identified SNP pairs (rs1467967-rs8070723, rs242557-rs8070723, rs3785883-rs8070723, rs2471738-rs8070723, rs8070723-rs7521) that exhibit high LD in both groups, suggesting that they define stable haplotype structures (H1 and H2) rather than random associations (Table 4). Furthermore, the absence of recombination between H1 and H2 supports the use of H2-specific SNP alleles (rs8070723) as reliable surrogate markers for inversion status. This finding has important implications for genetic association studies, as it ensures that the differentiation between H1 and H2 haplotypes can be accurately captured without requiring direct structural analysis.

Table 4.
Pairwise LD analysis. LD was calculated as D’ and r2. A D’ of 0.0 indicates that the markers are assorting independently, while a D’ of 1.0 implies that the less common allele is always found paired with a specific allele at the other marker. r2 = 1.0 only when the marker loci also have identical allele frequencies.
3.2. Association of MAPT rs8070723 Alleles with PSP
In our association analysis, the MAPT rs8070723 H2 allele was significantly associated with a reduced risk of PSP (OR, 0.382; 95% CI, 0.195–0.715; p = 0.0035) and PSP-RS subtype (OR, 0.250; 95% CI, 0.098–0.556; p = 0.0015), with minor allele frequencies of 11.0% in the 73 PSP, and 8.7% in the 46 PSP-RS patient subgroup, compared to 24.7% in the 93 HC. Conversely, the rs8070723 H1 allele was significantly associated with an increased risk of PSP (OR, 2.620; 95% CI, 1.399–5.140; p = 0.0035) and PSP-RS (OR, 4.004; 95% CI, 1.798–10.237; p = 0.0015), with major allele frequencies of 89.0% and 91.3% in PSP cases and PSP-RS patient subgroup, respectively, compared to 75.3% in HC (Table 5 and Table S1).

Table 5.
Association of A and G alleles in rs8070723 SNP with risk of PSP and PSP subtypes. ORs (95% CIs) were evaluated using logistic regression models, adjusted for sex and age, in the R statistical computing environment. ORs represent the effect per additional copy of the respective allele. Power values are reported for each OR, with significant effects consistently showing power >0.8. A Bonferroni correction for multiple testing (two association tests) established statistical significance at p ≤ 0.025.
3.3. Genetic Association of MAPT Haplotypes with PSP
A subhaplotype (H1j) was associated with a nominally significant (p < 0.05) decreased risk of PSP in our cohort (H1j: OR, 0.201; 95% CI, 0.044–0.915; p = 0.0265) (Table 6 and Table S2), whereas another study reported a significant association with increased risk in a different PSP cohort [14].

Table 6.
Association of MAPT haplotypes with risk of PSP. ORs (95% CIs) were derived from epiR package in the R statistical computing environment. ORs represent the effect per additional copy of the respective haplotype. Power values are reported for each OR, indicating the probability of correctly detecting a true effect; significant associations correspond to power >0.8. A Bonferroni correction for multiple testing (19 association tests) established statistical significance at p ≤ 0.0026.
Consistently with previously findings [14,20], H2 haplotype showed a significant protective effect on PSP risk (OR, 0.370; 95% CI, 0.196–0.695; p = 0.0015) since it was overrepresented in healthy controls (Table 6). Despite the small sample size, H2 frequency was high in both PSP and HC groups as expected in the European population [12].
3.4. Haplotype-Based Genetic Stratification of PSP-RS and vPSP
A similar analysis was conducted in two different PSP phenotypes: PSP-RS and vPSP, compared to the HC group. A total of 18 haplotypes (H2 haplotype and 17 H1 subhaplotypes) were found in the PSP-RS/HC group (Table S3), but only H2 had a significant protective effect on the risk of PSP-RS (OR, 0.274; 95% CI, 0.118–0.636; p = 0.0014) (Table 7 and Table S4). In the vPSP/HC group, 18 haplotypes (H2 haplotype, one H2 subhaplotype and 16 H1 subhaplotypes) were found (Table S5), but no significant effects were observed (Table 8 and Table S6).

Table 7.
Association of MAPT haplotypes with risk of PSP-RS. Statistical significance was defined as p ≤ 0.0027 after Bonferroni adjustment for 18 tests. ORs with power > 0.8 are considered reliably detectable and statistically robust.

Table 8.
Association of MAPT haplotypes with risk of vPSP. Bonferroni correction (18 tests) set significance at p ≤ 0.0027. ORs with power > 0.8 are considered statistically reliable.
When stratified by phenotype (PSP-RS and vPSP), the protective trend of H1j haplotype persisted but did not achieve statistical significance, likely due to limited sample sizes. Distribution of haplotype frequencies across the three groups is illustrated in the bar plot (Figure 1), highlighting differences in the prevalence of specific subhaplotypes.
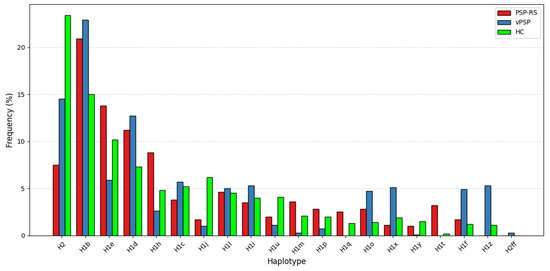
Figure 1.
Distribution of haplotype frequencies. The bar plot shows the distribution of MAPT haplotypes across PSP-RS, vPSP, and HC groups. Several H1 subhaplotypes, such as H1e, H1q, H1t, H1z, show different distribution in PSP-RS and vPSP, while the H2 haplotype shows a markedly higher frequency in the HC group, supporting its potential protective role.
4. Discussion
In this study, the association between MAPT haplotypes and the risk of PSP was investigated for the first time in a cohort of Italian PSP patients with two different phenotypes (RS and vPSP). Our findings confirm the strong LD at the MAPT locus, driven by the 900-kb inversion at 17q21.31, which defines the H1 and H2 haplotypes [11,14]. The absence of recombination between these haplotypes preserves their distinct genetic architecture. Notably, rs8070723 tags the H2 haplotype, enabling efficient haplotype differentiation without structural analysis. This reinforces the evolutionary stability of the MAPT inversion and its relevance for genetic association studies.
The analysis of major allele distribution in the rs8070723 SNP among different groups confirmed the association between H1 and the risk of PSP and, in particular, PSP-RS, while the overrepresentation of minor allele in HC confirmed the protective effect of H2 [13,18,28,29].
In our haplotype association analysis, the H1j haplotype, previously identified [14] only in a UK population as being more prevalent in controls than in PSP cases, showed a nominally significant association with a reduced risk of PSP (OR, 0.201; 95% CI, 0.044–0.915; p = 0.0265). Although the p-value does not reach significance after Bonferroni correction due to sample size limitations, the confidence intervals fall below 1, suggesting a potential protective effect. This trend is further supported by a similarly low frequency in cases compared to controls, even when analyzing PSP subgroups. The observed discrepancy with the UK findings may reflect differences in population-specific genetic backgrounds (Southern Italian vs. British), LD structures, or methodological aspects of cohort design. Nevertheless, additional studies are warranted to better clarify and validate this potential association.
The H1 haplotype has been linked to increased expression of 4-repeat (4R) tau isoforms, which are predominant in PSP pathology, while the H2 haplotype is associated with reduced MAPT expression and a lower 4R/3R tau ratio [10]. The protective effects of H2 and H1j may stem from their association with decreased 4R-tau expression, potentially mitigating tau aggregation and neurodegeneration. These functional differences may help explain the observed associations with PSP risk and support further studies to clarify the molecular mechanisms involved.
Our study also examined the distribution of MAPT haplotypes in different PSP subtypes, including PSP-RS and vPSP patients. We identified 18 haplotypes in this stratified analysis, but only H2 haplotype reached statistical significance in the PSP-RS/HC after multiple testing corrections, and it appeared to confer greater protective effects in PSP-RS compared to vPSP. Although these associations did not reach statistical significance probably because of small sample size, our analysis revealed distinct H1 subhaplotype distributions between PSP-RS and vPSP subtypes and HC subjects. The exclusive presence of H1t and H1q in PSP-RS and H1z in vPSP suggests that these haplotypes may contribute to different genetic background in PSP subtypes. The observed overrepresentation of subhaplotype H1e in PSP-RS patients, compared to those with vPSP (Figure 1), may reflect underlying genetic factors that contribute to the differentiation of these clinical subtypes. However, future studies on larger international cohorts, including on the whole PSP spectrum and specifically looking at distinct PSP phenotypes, are needed to validate our findings and shed further light on this point.
Despite our study providing valuable insights into MAPT haplotypes in PSP, several limitations should be acknowledged. First, our sample size, though comparable to other genetic studies of PSP, remains relatively small, potentially limiting statistical power. Second, our cohort consists exclusively of individuals of Southern Italian ancestry, which may restrict the generalizability of our findings to other populations. Future studies in larger patient cohorts should include diverse ethnic groups to explore potential population-specific effects.
In conclusion, our results reinforce the role of MAPT haplotypes in PSP susceptibility and highlight the need for further research to validate and expand upon these findings. Understanding the role of haplotypes in influencing the risk of PSP is a crucial step toward unrevealing the complex genetic underpinnings of this neurodegenerative disorder.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines13061405/s1, Table S1: Allele frequency and association of rs8070723 with PSP and its subtypes; Table S2: Association of MAPT Haplotypes with risk of PSP; Table S3: MAPT H2 haplotypes and H1 sub-haplotypes that were observed in 1% or more of 46 PSP-RS cases and 93 HC in any of the 18 association analyses; Table S4: Association of MAPT Haplotypes with risk of PSP-RS; Table S5: MAPT H2 haplotypes and H1 sub-haplotypes that were observed in 1% or more of 27 vPSP cases and 93 HC in any of the 18 association analyses; Table S6: Association of MAPT Haplotypes with risk of vPSP.
Author Contributions
Conceptualization, A.Q. (Aldo Quattrone) and A.Q. (Andrea Quattrone); formal analysis, investigation, and data curation, M.G., R.P., A.F., G.A., M.T., and B.V.; writing–original draft preparation, M.G, R.P. and A.F.; writing–review and editing, G.A, A.Q. (Aldo Quattrone) and A.Q. (Andrea Quattrone). All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the ethics committee of University Magna Graecia of Catanzaro. (No.143; 13 May 2024).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author (R.P.) upon reasonable request.
Conflicts of Interest
Author Basilio Vescio was partially employed by the company Biotecnomed SCaRL, which is a non-profit private research organization. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| PSP | Progressive Supranuclear Palsy |
| PSP-RS | Progressive Supranuclear Palsy Richardson syndrome |
| vPSP | Progressive Supranuclear Palsy variants |
| PSP-P | Progressive Supranuclear Palsy with predominant parkinsonism |
| PSP-PGF | Progressive Supranuclear Palsy with progressive gait freezing |
| PSP-CBS | Progressive Supranuclear Palsy with predominant corticobasal syndrome |
| PSP-SL | Progressive Supranuclear Palsy with a predominant speech/language disorder |
| PSP-F | Progressive Supranuclear Palsy with predominant frontal presentation |
| PSP-C | Progressive Supranuclear Palsy with cerebellar ataxia |
| PSP-OM | Progressive Supranuclear Palsy with oculomotor dysfunction |
| NFTs | Neurofibrillary tangles |
| TA | Tufted astrocytes |
| CB | Oligodendroglial coiled bodies |
| NT | Neuropil threads |
| NTR | N-terminal region |
| PRR | Proline-rich domain |
| MTBD | Microtubule-binding domain |
| LD | Linkage disequilibrium |
| HC | Healthy control |
| r2 | Squared correlation coefficient |
| EM | Expectation-Maximization algorithm |
| ORs | Odds ratios |
| CI | Confidence intervals |
| HWE | Hardy–Weinberg equilibrium |
References
- Schrag, A.; Ben-Shlomo, Y.; Quinn, N.P. Prevalence of progressive supranuclear palsy and multiple system atrophy: A cross-sectional study. Lancet 1999, 354, 1771–1775. [Google Scholar] [CrossRef] [PubMed]
- Höglinger, G.U.; Respondek, G.; Stamelou, M.; Kurz, C.; Josephs, K.A.; Lang, A.E.; Mollenhauer, B.; Müller, U.; Nilsson, C.; Whitwell, J.L.; et al. Clinical diagnosis of progressive supranuclear palsy: The movement disorder society criteria. Mov. Disord. 2017, 32, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Boxer, A.L.; Yu, J.-T.; Golbe, L.I.; Litvan, I.; Lang, A.E.; Höglinger, G.U. Advances in progressive supranuclear palsy: New diagnostic criteria, biomarkers, and therapeutic approaches. Lancet Neurol. 2017, 16, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Stamelou, M. Sensitivity and specificity of diagnostic criteria for progressive supranuclear palsy. Mov. Disord. 2019, 34, 1087–1088. [Google Scholar] [CrossRef]
- Barbier, P.; Zejneli, O.; Martinho, M.; Lasorsa, A.; Belle, V.; Smet-Nocca, C.; Tsvetkov, P.O.; Devred, F.; Landrieu, I. Role of Tau as a Microtubule-Associated Protein: Structural and Functional Aspects. Front. Aging Neurosci. 2019, 11, 204. [Google Scholar] [CrossRef]
- Corsi, A.; Bombieri, C.; Valenti, M.T.; Romanelli, M.G. Tau Isoforms: Gaining Insight into MAPT Alternative Splicing. Int. J. Mol. Sci. 2022, 23, 15383. [Google Scholar] [CrossRef]
- Wen, Y.; Zhou, Y.; Jiao, B.; Shen, L. Genetics of Progressive Supranuclear Palsy: A Review. J. Park. Dis. 2021, 11, 93–105. [Google Scholar] [CrossRef]
- Vaquer-Alicea, J.; Diamond, M.I.; Joachimiak, L.A. Tau strains shape disease. Acta Neuropathol. 2021, 142, 57–71. [Google Scholar] [CrossRef]
- Zhong, Q.; Congdon, E.E.; Nagaraja, H.N.; Kuret, J. Tau isoform composition influences rate and extent of filament formation. J. Biol. Chem. 2012, 287, 20711–20719. [Google Scholar] [CrossRef]
- Schoch, K.M.; DeVos, S.L.; Miller, R.L.; Chun, S.J.; ∙Norrbom, M.; Wozniak, D.F.; ∙Dawson, H.N.; Bennett, C.F.; Rigo, F.; Miller, T.M. Increased 4R-Tau Induces Pathological Changes in a Human-Tau Mouse Model. Neuron 2016, 90, 941–947. [Google Scholar] [CrossRef]
- Stefansson, H.; Helgason, A.; Thorleifsson, G.; Steinthorsdottir, V.; Masson, G.; Barnard, J.; Baker, A.; Jonasdottir, A.; Ingason, A.; Gudnadottir, V.G.; et al. A common inversion under selection in Europeans. Nat. Genet. 2005, 37, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Evans, W.; Fung, H.C.; Steele, J.; Eerola, J.; Tienari, P.; Pittman, A.; de Silva, R.; Myers, A.; Vrieze, F.W.-D.; Singleton, A.; et al. The tau H2 haplotype is almost exclusively Caucasian in origin. Neurosci. Lett. 2004, 369, 183–185. [Google Scholar] [CrossRef]
- Baker, M.; Litvan, I.; Houlden, H.; Adamson, J.; Dickson, D.; Perez-Tur, J.; Hardy, J.; Lynch, T.; Bigio, E.; Hutton, M. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum. Mol. Genet. 1999, 8, 711–715. [Google Scholar] [CrossRef] [PubMed]
- Pittman, A.M.; Myers, A.J.; Abou-Sleiman, P.; Fung, H.C.; Kaleem, M.; Marlowe, L.; Duckworth, J.; Leung, D.; Williams, D.; Kilford, L.; et al. Linkage disequilibrium fine mapping and haplotype association analysis of the tau gene in progressive supranuclear palsy and corticobasal degeneration. J. Med. Genet. 2005, 42, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Pittman, A.M.; Myers, A.J.; Duckworth, J.; Bryden, L.; Hanson, M.; Abou-Sleiman, P.; Wood, N.W.; Hardy, J.; Lees, A.; de Silva, R.; et al. The structure of the tau haplotype in controls and in progressive supranuclear palsy. Hum. Mol. Genet. 2004, 13, 1267–1274. [Google Scholar] [CrossRef][Green Version]
- Myers, A.J.; Pittman, A.M.; Zhao, A.S.; Rohrer, K.; Kaleem, M.; Marlowe, L.; Lees, A.; Leung, D.; McKeith, I.G.; Perry, R.H.; et al. The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol. Dis. 2007, 25, 561–570. [Google Scholar] [CrossRef]
- Boettger, L.M.; Handsaker, R.E.; Zody, M.C.; McCarroll, S.A. Structural haplotypes and recent evolution of the human 17q21.31 region. Nat. Genet. 2012, 44, 881–885. [Google Scholar] [CrossRef]
- Heckman, M.G.; Brennan, R.R.; Labbé, C.; Soto, A.I.; Koga, S.; DeTure, M.A.; Murray, M.E.; Petersen, R.C.; Boeve, B.F.; van Gerpen, J.A.; et al. Association of MAPT Subhaplotypes with Risk of Progressive Supranuclear Palsy and Severity of Tau Pathology. JAMA Neurol. 2019, 76, 710–717. [Google Scholar] [CrossRef]
- Pastor, P.; Ezquerra, M.; Perez, J.C.; Chakraverty, S.; Norton, J.; Racette, B.A.; McKeel, D.; Perlmutter, J.S.; Tolosa, E.; Goate, A.M. Novel haplotypes in 17q21 are associated with progressive supranuclear palsy. Ann. Neurol. 2004, 56, 249–258. [Google Scholar] [CrossRef]
- Zhang, C.-C.; Zhu, J.-X.; Wan, Y.; Tan, L.; Wang, H.-F.; Yu, J.-T.; Tan, L. Meta-analysis of the association between variants in MAPT and neurodegenerative diseases. Oncotarget 2017, 8, 44994–45007. [Google Scholar] [CrossRef]
- Pedicone, C.; Weitzman, S.A.; Renton, A.E.; Goate, A.M. Unraveling the complex role of MAPT-containing H1 and H2 haplotypes in neurodegenerative diseases. Mol. Neurodegener. 2024, 19, 43. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yu, J.-T.; Wojta, K.; Wang, H.-F.; Zetterberg, H.; Blennow, K.; Yokoyama, J.S.; Weiner, M.W.; Kramer, J.H.; Rosen, H.; et al. Genome-wide association study identifies MAPT locus influencing human plasma tau levels. Neurology 2017, 88, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Peterson, D.; Munger, C.; Crowley, J.; Corcoran, C.; Cruchaga, C.; Goate, A.M.; Norton, M.C.; Green, R.C.; Munger, R.G.; Breitner, J.C.S.; et al. Variants in PPP3R1 and MAPT are associated with more rapid functional decline in Alzheimer’s disease: The Cache County Dementia Progression Study. Alzheimers Dement. 2014, 10, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Luo, G.; Ding, X.; Sun, G.; Zhang, M.; Dong, J.; Xu, H.; Lu, J.; Li, Z.; Ning, B.; et al. A comprehensive analysis of MAPT-related genetic risk in Alzheimer’s disease. IBRO Neurosci. Rep. 2025, 18, 300–305. [Google Scholar] [CrossRef]
- Wang, X.; Campbell, M.R.; Lacher, S.E.; Cho, H.-Y.; Wan, M.; Crowl, C.L.; Chorley, B.N.; Bond, G.L.; Kleeberger, S.R.; Slattery, M.; et al. A polymorphic antioxidant response element links NRF2/sMAF binding to enhanced MAPT expression and reduced risk of Parkinsonian disorders. Cell Rep. 2016, 15, 830–842. [Google Scholar] [CrossRef]
- Leko, M.B.; Popovački, E.Š.; Willumsen, N.; Perković, M.N.; Pleić, N.; Zubčić, K.; Horvat, L.L.; Vogrinc, Ž.; Boban, M.; Borovečki, F.; et al. Further validation of the association between MAPT haplotype-tagging polymorphisms and Alzheimer’s disease: Neuropsychological tests, cerebrospinal fluid biomarkers, and APOE genotype. Front. Mol. Neurosci. 2024, 17, 1456670. [Google Scholar] [CrossRef]
- Majounie, E.; Cross, W.; Newsway, V.; Dillman, A.; Vandrovcova, J.; Morris, C.M.; Nalls, M.A.; Ferrucci, L.; Owen, M.J.; O’Donovan, M.C.; et al. Variation in tau isoform expression in different brain regions and disease states. Neurobiol. Aging 2013, 34, 1922.e7–1922.e12. [Google Scholar] [CrossRef]
- Höglinger, G.U.; Melhem, N.M.; Dickson, D.W.; Sleiman, P.M.A.; Wang, L.-S.; Klei, L.; Rademakers, R.; de Silva, R.; Litvan, I.; Riley, D.E.; et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat. Genet. 2011, 43, 699–705. [Google Scholar] [CrossRef]
- de Silva, R.; Weiler, M.; Morris, H.R.; Martin, E.R.; Wood, N.W.; Lees, A.J. Strong association of a novel Tau promoter haplotype in progressive supranuclear palsy. Neurosci. Lett. 2001, 311, 145–148. [Google Scholar] [CrossRef]
- Rademakers, R.; Melquist, S.; Cruts, M.; Theuns, J.; Del-Favero, J.; Poorkaj, P.; Baker, M.; Sleegers, K.; Crook, R.; De Pooter, T.; et al. High-density SNP haplotyping suggests altered regulation of tau gene expression in progressive supranuclear palsy. Hum. Mol. Genet. 2005, 14, 3281–3292. [Google Scholar] [CrossRef]
- Valentino, R.R.; Scotton, W.J.; Roemer, S.F.; Lashley, T.; Heckman, M.G.; Shoai, M.; Martinez-Carrasco, A.; Tamvaka, N.; Walton, R.L.; Baker, M.C.; et al. MAPT H2 haplotype and risk of Pick’s disease in the Pick’s disease International Consortium: A genetic association study. Lancet Neurol. 2024, 23, 487–499. [Google Scholar] [CrossRef]
- Leko, M.B.; Willumsen, N.; Perković, M.N.; Klepac, N.; Borovečki, F.; Hof, P.R.; Sonicki, Z.; Pivac, N.; de Silva, R.; Šimić, G. Association of MAPT haplotype-tagging polymorphisms with cerebrospinal fluid biomarkers of Alzheimer’s disease: A preliminary study in a Croatian cohort. Brain Behav. 2018, 8, e01128. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).