Abstract
Background: The relationship between oral contraceptive (OC) use and pancreatic cancer (PC) risk remains controversial, with inconsistent findings reported in observational studies. To clarify this relationship and better identify potential risk factors for PC prevention, more unbiased and robust approaches are needed. Methods: We investigated the potential causal relationship between OC use and PC risk using a two-sample Mendelian randomization (MR) analysis, with blood protein quantitative trait loci (pQTLs) as instrumental variables. To ensure the robustness of our findings, we performed a series of sensitivity analyses, colocalization analyses, and reverse MR. The causal effects of protein-coding genes on PC risk, as well as their expression patterns across different single-cell types, were subsequently investigated. To elucidate the potential pathogenic pathways, we conducted pathway enrichment analysis, protein–protein interaction (PPI) network analysis, and causal inference. Results: Our MR analysis identified five drug-targeted proteins significantly associated with PC risk. Higher levels of COMT, AGT, FN1, and UGT1A1, as well as lower levels of SERPINC1, were associated with an increased risk of PC. Among these, AGT, FN1, and COMT demonstrated consistent associations across sensitivity analyses and downstream analyses, providing robust evidence supporting their involvement in PC risk. Conclusions: This study provides genetic evidence suggesting, in European groups, a potential causal link between OC use and increased PC risk, possibly mediated through drug-targeted proteins such as AGT and FN1. These results highlight the importance for further research to elucidate the underlying mechanisms and assess the implications of OC use on PC risk.
1. Background
Pancreatic cancer (PC) is a highly aggressive and lethal malignancy, characterized by an alarmingly high mortality rate [1]. In the United States, the 5-year survival rate for PC remains approximately 10% [2], and its incidence is increasing annually by 0.5% to 1.0%. It is projected to become the second leading cause of cancer-related deaths by 2030 [3,4,5]. However, due to the absence of effective early diagnostic tools and preventive strategies, the public health burden of PC continues to grow. In recent years, the potential association between medication use and PC risk has garnered increasing attention. In particular, understanding how pharmacological agents may influence the development of PC through metabolic or hormonal regulation is critical for identifying risk factors and developing effective prevention strategies.
Oral contraceptives (OCs) are widely prescribed medications for both contraception and endocrine disorder management, particularly for patients with polycystic ovary syndrome (PCOS), where OCs are a common therapeutic option [6,7]. Studies have suggested that OCs influence cancer progression by altering the estradiol-to-progesterone ratio, modulating immune responses, and affecting one-carbon metabolism [8,9]. In recent years, numerous observational studies, including cohort and case–control designs, have investigated the potential association between OC use and PC risk. However, the results remain inconsistent: while some studies suggest that long-term OC use may increase PC risk [9,10], others indicate a potential protective effect [11,12], and some report no significant association at all [13]. These conflicting findings may result from various biases, including selection bias, information bias, and confounding by indication. The interpretation of the relationship between OC use and PC risk remains challenging due to the presence of multiple potential sources of bias. Given these discrepancies, there is a pressing need for more robust and rigorous approaches to assess the potential relationship between OCs and PC.
Mendelian randomization (MR) [14] offers a robust approach to address this issue. MR employs genetic variants as instrumental variables (IVs) to infer causal relationships between exposure factors and disease outcomes. Unlike conventional observational studies, MR minimizes the impact of confounding bias and reverse causation by leveraging the random assignment of genetic variants during conception. This method significantly enhances the validity of causal inference and has been extensively applied in pharmacological epidemiology to investigate drug–disease relationships [15,16,17,18,19,20,21]. Recent advancements in technology have led to significant breakthroughs in large-scale proteomics studies, particularly in the identification of protein quantitative trait loci (pQTLs), which have opened new possibilities for inferring the effect of plasma drug proteins on PC using a two-sample MR method [22,23].
This study aimed to investigate the potential causal relationship between OC use and PC risk. A two-sample MR analysis was employed to examine the relationship between OCs and PC risk at the protein level by identifying relevant drug targets as exposure. To strengthen the validation of the findings, colocalization and sensitivity analyses were conducted. Additionally, we conducted differential expression analysis to validate the gene expression differences of drug target genes in pancreatic cancer tissues compared with normal pancreatic tissues. Furthermore, we integrated single-cell RNA sequencing data with enrichment analyses to identify the specific cell types enriched for drug target genes in pancreatic tissues, as well as the biological pathways through which these genes may influence pancreatic cancer progression. These comprehensive analyses provide valuable insights into the potential impact of OC use on PC risk.
2. Methods
2.1. Overall Design
This study was based on publicly available summary-level datasets, and the overall study design is illustrated in Figure 1. Briefly, the study was divided into three major components: In the first part, we employed pQTLs of drug-targeted proteins as potential instrumental variables, with PC as the outcome for MR analysis. The second part involved a series of sensitivity analyses and Bayesian colocalization analyses to validate the robustness and specificity of the MR findings. In the third part, we conducted differential expression analysis at the bulk-tissue level, investigated the cell type-specific expression of target genes using single-cell transcriptomic data, and explored their functional interactions through a protein–protein interaction (PPI) network analysis. All reported p-values were adjusted for multiple testing by the Benjamini–Hochberg (BH) procedure.
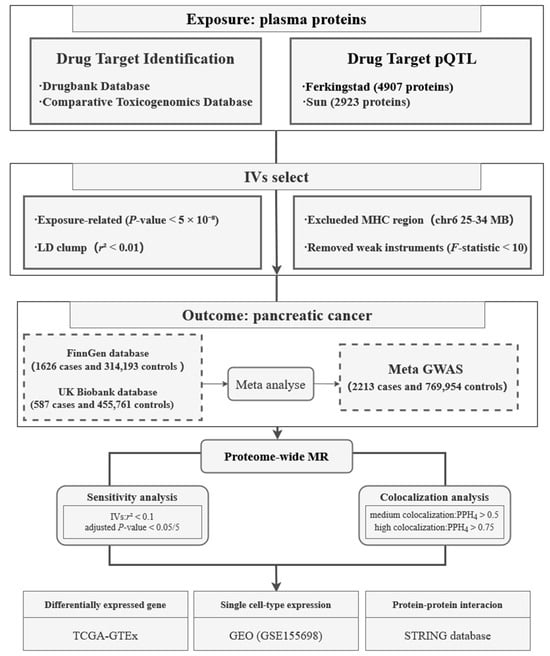
Figure 1.
Flowchart of the study design. pQTL, protein quantitative trait loci; IV, instrumental variable; LD, linkage disequilibrium; MHC, major histocompatibility complex; GWAS, genome-wide association study; MR, Mendelian randomization; GEO, Gene Expression Omnibus.
2.2. Study Population and Datasets
We defined PC as a malignant neoplasm of the pancreas, and identified patient cases based on the disease codes ICD-10 C25, ICD-9 157, and ICD-8 157. Summary-level genome-wide association study (GWAS) data for PC risk were obtained from the UK Biobank [23] and FinnGen [24] cohorts; after meta-analysis, the combined dataset included 772,167 individuals of European ancestry, comprising 2213 PC cases and 769,954 controls.
In this study, plasma proteins were considered as the primary endpoints, with drug-targeted proteins serving as exposures. pQTL data for plasma protein levels were obtained from two large-scale proteomic studies: deCODE (covering 4907 proteins from 35,559 individuals) and UKB-PPP (covering 2923 proteins from 54,219 individuals). For each protein, only variants with a reference SNP ID (rsID) were retained. We selected instrumental variables for drug targets based on pQTL data from these two databases.
2.3. Identification of Drug Target Proteins
We included commonly prescribed OCs for analysis and categorized them into subtypes—including combined oral contraceptives, progestin-only pills, and emergency contraceptive pills—to minimize potential confounding factors and improve the precision of the association assessment. Drug targets were obtained using from the DrugBank database to ensure accurate identification of drug–protein interactions [25]. Given the widespread use and complex composition of combined oral contraceptives, we also considered additional proteins potentially affected by these formulations. To enhance coverage and ensure comprehensiveness, supplementary protein targets were retrieved from the Comparative Toxicogenomics Database (CTD) [26].
2.4. Selection of Instrumental Variables
To ensure the validity of MR, IVs must satisfy three core assumptions during selection: (1) the IVs are robustly associated with the exposure; (2) the effect of the IVs on the outcome is mediated only through the exposure variable; and (3) the effect of the IVs is not confounded by other variables. In this study, we defined the transcriptional start site of a protein-coding gene within 1 Mb as cis, and prioritized cis-pQTLs as IVs due to their direct relevance to protein expression. The following criteria were applied to screen for IVs: (1) Selection of single nucleotide polymorphisms (SNPs) commonly associated with the exposure (protein) as having genome-wide significance (p-value < 5 × 10−8) and with a minor allele frequency (MAF) > 0.01; (2) Independent SNPs were selected to reduce the risk of pleiotropy due to the linkage disequilibrium (LD) effect, with an LD threshold of r2 < 0.01, and the genetic distance was set at 10 Mb [27,28]; (3) SNPs within the major histocompatibility complex (MHC) region (chr 6: 25.5–34.0 Mb) were excluded due to their complex LD structure and high polymorphism; (4) For R2 and F-statistics, we used the following equations: R2 = 2 × MAF × (1 − MAF) × β2; F = R2 × (N − 2)/(1 − R2) [29], where the F-statistic measures the strength of the genetic instruments and R2 represents the proportion of variance in the exposure explained by the SNP. IVs with an F-statistic < 10 were removed to avoid weak instrument bias.
2.5. Mendelian Randomization
To ensure the scientific validity and robustness of the analysis, data preprocessing and harmonization were performed prior to the formal MR analysis. Specifically, we harmonized the protein IVs with PC meta-GWAS data, extracted overlapped IVs between the filtered exposure and the outcome datasets, and removed incompatible or palindromic SNPs. Subsequently, appropriate MR methods were selected based on the number of IVs available to estimate causal effects and assess potential pleiotropy. When only a single IV was available, the Wald ratio method was applied. For exposures with two or more IVs, the inverse-variance weighted (IVW) method was employed, as it is the most efficient approach under the assumption that all IVs are valid [30]. In cases with more than three IVs, the MR-Egger method was conducted to detect potential horizontal pleiotropy. If the MR-Egger intercept test results indicated no pleiotropy (p-value > 0.05), the IVW method was adopted. Otherwise, causal inference was based on MR-Egger results. Heterogeneity among IVs was assessed using Cochrane’s Q-test. When both the UKB and deCODE datasets provided significant results, the dataset with a greater number of IVs was selected for causal estimation. All MR analyses were conducted using the “TwoSampleMR” R package [31].
2.6. Sensitivity Analysis
MR results can be influenced by the selection of IVs. In the sensitivity analysis, we re-selected the IVs using a relaxed LD threshold of r2 < 0.1. Although using a larger set of IVs would increase statistical power, it can also introduce potential confounding effects [16,32]. Therefore, it is essential to ensure that the causal direction observed in the sensitivity analysis remained consistent with that of the primary MR analysis. To control for multiple testing, the Bonferroni correction was applied to identify significant associations. Furthermore, to minimize the risk of potential reverse causality bias, we conducted a reverse causality analysis by using PC as the exposure and cis-pQTLs as the outcome. For selecting IVs for PC, we set a p-value threshold of <5 × 10−8 and an r2 < 0.01 within a 10 MB window to ensure the validity of the IVs.
2.7. Colocalization Analysis
To further validate the results, we performed a Bayesian colocalization analysis, which estimates the posterior probability that a given genomic locus contains a single causal variant influencing both protein levels and disease risk. This approach helps assess whether the observed associations between drug target proteins and PC risk are confounded by LD. For each protein, we selected independent cis-pQTL within a 500 kb window as the candidate colocalized regions. The analysis evaluates five hypotheses: (1) H0: no causal variant for either drug target protein or PC; (2) H1: a causal variant only for protein; (3) H2: a causal variant only for diseases; (4) H3: two different causal variants for protein and diseases, respectively; and (5) H4: one shared causal variant influencing for both traits. The posterior probability (PP) for each hypothesis is denoted as PPH0, PPH1, PPH2, PPH3, and PPH4 [33].
For proteins with multiple independent loci, colocalization analysis was conducted separately for each locus using meta summary-level data. Regions with the highest posterior probability of H4 (PPH4) were considered to provide the strongest evidence for colocalization. We defined medium-support evidence of colocalization as a PPH4 between 0.50 and 0.75, and high-support colocalization as a PPH4 > 0.75 [34]. Bayesian colocalization analysis was conducted using the ‘coloc’ package.
2.8. Differentially Expressed Genes
To further validate the MR findings and explore whether the causal targets identified were differentially expressed in PC, we conducted a differential expression analysis between pancreatic tumor and normal pancreatic tissues. RNA transcriptomic data were obtained from 178 pancreatic cancer samples in The Cancer Genome Atlas (TCGA) “https://www.cancer.gov/tcga” (accessed on 19 September 2024) and 167 normal pancreatic tissue samples from the Genotype-Tissue Expression (GTEx) project. Normalized data were retrieved from the UCSC Xena platform [35]. Genes were defined as differentially expressed (DEGs) if they exhibited a Log2 fold change (Log2 FC) > 2 and an adjusted p-value < 0.05. Differential expression analysis was conducted using the Limma package to identify DEGs that met these criteria [36]. This analysis provided additional evidence supporting the causal relevance of the drug targets implicated by MR.
2.9. Single Cell-Type Expression Analysis
Single-cell RNA sequencing (scRNA-seq) provides valuable insights into the cell type-specific effects of drug targets and the underlying pathways involved in the development of PC. We analyzed scRNA-seq data derived from human pancreatic tumor tissues and adjacent normal tissues, available from the Gene Expression Omnibus (GEO) database [37] (GSE155698) [38,39]. This dataset included 16 pancreatic cancer samples and 3 adjacent normal samples. Prior to analysis, rigorous data preprocessing was performed to ensure high data quality for downstream applications. Specifically, using the “Seurat” package, we applied the following quality control criteria: retention of cells with gene counts between 600 and 4000, total RNA count per cell greater than 1000 but below the 97th percentile of the dataset, mitochondrial gene content below 10%, and hemoglobin gene percentage below 1%. After filtering, 23,432 cells and 24,725 genes were retained for analysis. Gene expression values were normalized and scaled using the NormalizeData and ScaleData functions in Seurat. Cell clustering was performed, and clusters were manually annotated based on canonical marker genes to facilitate cell type-specific downstream analyses.
To investigate the role of genes encoding target proteins in specific cell types, we performed a differential expression analysis between pancreatic cancer and adjacent normal tissue cells within each cell cluster. This analysis was conducted using the Wilcoxon rank-sum test, implemented in the FindMarkers function in Seurat. Genes were considered differentially expressed if they exhibited an average |Log2 FC| > 1 and an adjusted p-value < 0.05. Subsequently, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses on the identified differentially expressed genes. Particular attention was given to the biological process (BP) category in the GO enrichment to elucidate the potential biological functions and mechanisms underlying the cell type-specific DEGs [40].
To further explore the cell type-specific expression of genes encoding pathogenic proteins within pancreatic cancer tissues, we performed differential expression analysis across single-cell clusters. Specifically, we compared gene expression levels between different cell types using the Wilcoxon rank-sum test. Genes with an average |Log2 FC| > 1 and an adjusted p-value < 0.05 were considered significantly enriched in specific cell types.
2.10. Protein Interaction Networks and Protein Function Queries
To explore the known functions and biological roles of the target proteins, we utilized the STRING database https://string-db.org/ (accessed on 26 December 2024) and conducted a comprehensive literature review. This approach enabled the exploration of protein–protein interactions (PPIs), the identification of potential functional associations, and the recognition of key molecules involved in relevant biological processes. By integrating evidence from both experimental studies and computational predictions, this provided valuable insights into the involvement of these proteins in cellular mechanisms and disease pathophysiology.
3. Results
3.1. MR Results Identified PC-Related OC Target Proteins
In this study, the oral contraceptives investigated include ethinyl estradiol, levonorgestrel, desogestrel, cyproterone acetate, medroxyprogesterone acetate, dienogest, estradiol valerate, gestodene, spironolactone, finasteride, letrozole, and drospirenone. Drug-related targets were identified using the DrugBank database, resulting in 118 drug-related targets. Additionally, combined oral contraceptives, which are defined as compound prescriptions containing estrogen and progesterone, were also considered, and 48 interaction targets were retrieved from the CTD. Detailed drug target information is provided in Supplementary Tables S1 and S2. After applying thresholds for p-values and r2, we calculated F-statistics, with all genetic instruments having F-statistics > 10. Following the selection of IVs, a total of 319 SNPs were available to proxy 32 proteins in deCODE, and 256 SNPs were available to proxy 35 proteins in UKB-PPP.
The primary analysis identified five proteins that were causally associated with PC at the blood level, as determined using IVW as the main MR method. These significant findings, along with the analytical approach, are illustrated in Figure 2. Specifically, the genetically predicted levels of four proteins (AGT, COMT, FN1, and UGT1A1) were positively associated with increased PC risk, while SERPINC1 was negatively associated with PC risk. Detailed information on the corresponding drug target interactions and their effects on PC risk are provided in Table 1. COMT and UGT1A1 are involved as substrates or inducers in the metabolism of hormonal drugs such as ethinyl estradiol, desogestrel, and estradiol valerate. In contrast, AGT, FN1, and SERPINC1 are known targets modulated by ethinyl estradiol [41] and combined oral contraceptives, including formulations co-administered with levonorgestrel, gestodene, norgestimate, and dienogest [42,43,44,45,46,47]. Notably, while ethinyl estradiol combined with dienogest has been reported to influence SERPINC1 activity [48], other COC formulations have been associated with either the upregulation or downregulation of these protein expression levels, depending on the specific drug combination and context.
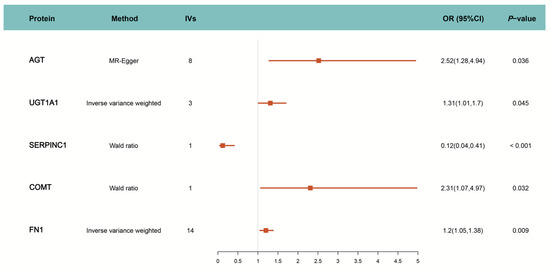
Figure 2.
MR estimation of causal relationship between drug target and PC. IVs: Number of instrumental variables; OR: Odds Ratio; CI: Confidence Interval.

Table 1.
Drug Target interactions.
3.2. Sensitivity Analysis and Colocalization Analysis
We conducted a series of sensitivity analyses to assess the robustness and reliability of our MR findings. First, the MR was repeated using additional sets of threshold-screened IVs (r2 < 0.1 within 10 Mb). The directions of causal effects for all initially significant proteins remained consistent with the primary MR result. To account for multiple testing, all p-values were adjusted using the BH procedure, and four targets (AGT, COMT, FN1, and SERPINC1) passed the corrected significance threshold of 0.01 (calculated as 0.05/5). It is worth noting that although UGT1A1 was significant in the primary MR analysis, its association did not remain significant after multiple testing correction (adjusted p-value > 0.05), likely due to the large number of comparisons. Accordingly, we prioritized the exclusion of targets—such as UGT1A1—that failed to pass multiple testing correction or exhibited signs of heterogeneity or directional pleiotropy. We further performed reverse MR analysis using 1401 IVs for PC, and found no significant evidence of reverse causality influencing drug target protein levels, suggesting that the observed associations were not biased by reverse causal effects. Moreover, no heterogeneity was detected using Cochran’s Q test, and no horizontal pleiotropy was observed through the MR-Egger intercept test. These analyses provide strong evidence that the MR results are both robust and reliable.
Despite the consistency across sensitivity analyses, a causal relationship between genetically determined protein levels and PC could still be influenced by LD, or it could reflect true shared causal variation between the two. To address these possibilities, we performed a colocalization analysis, with the results summarized in Table 2. Among the five candidate proteins, the PPH4 values for AGT (PPH4 = 0.93) and SERPINC1 (PPH4 = 0.99) showed high-support evidence for an association linked to a shared causal variant. Additionally, medium-support evidence of colocalization (PPH4 > 0.5) was observed for FN1 (PPH4 = 0.51) and UGT1A1 (PPH4 = 0.59) with PC. These results suggest a high likelihood of shared causal variants between AGT, SERPINC1, FN1, and UGT1A1 protein levels and PC risk, indicating that the MR findings for these proteins not attributable to LD. However, the absence of significant colocalization results does not undermine the validity of the MR analysis [49].

Table 2.
Sensitivity analysis and colocalization analysis.
3.3. Identified Protein-Coding Genes Differentially Expressed Between Tumor and Normal Tissue
To investigate whether the identified protein-coding genes are differentially expressed in cancer and normal tissues, we conducted a comprehensive differentially expressed analysis. We applied a stringent correction for the FDR to control for multiple testing, with an adjusted p-value threshold of <0.05. The analysis revealed marked differences in expression levels for several target genes. Specifically, FN1 (Log2 FC = 6.2), UGT1A1 (Log2 FC = 6.1), AGT (Log2 FC = 3.31), and COMT (Log2 FC = 2.4) were significantly upregulated in tumor tissues compared to normal tissues. In contrast, SERPINC1 showed no significant differential expression, indicating that its expression levels remained relatively stable between tumor and normal samples. As shown in Figure 3A, nearly all of the identified protein-coding genes exhibited significant differential expression between tumor and normal tissues, with the notable exception of SERPINC1.
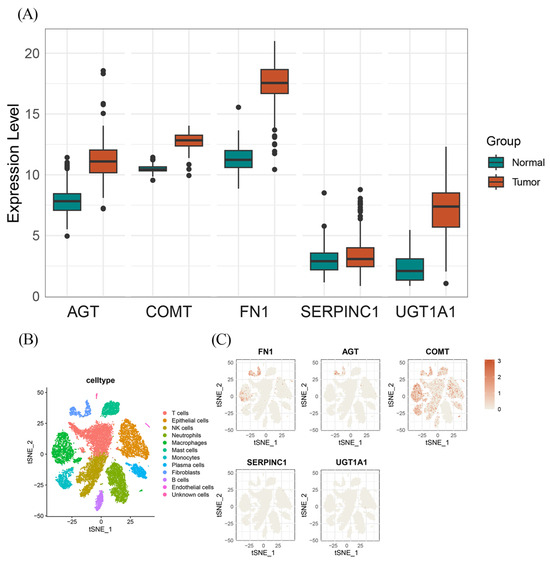
Figure 3.
The results of DEGs and specific expression protein coding genes. (A) The DEGs in PC tissue for the protein-coding genes of the identified OCs; (B) Single cell-type expression in PC tissue represented the 11 cell clusters labeled and annotated; (C) The expression of identified protein coding gene in each cell type.
3.4. Single-Cell Type Expression in PC Tissues and Pathway Enrichment
To investigate the cell type-specific expression patterns of the identified protein-coding genes and their potential roles in PC development within distinct cellular contexts, we analyzed scRNA-seq data from 16 samples obtained from the GSE155698 dataset [38,39]. As shown in Figure 3B, all cells were manually annotated and classified into 11 clusters: T cells, epithelial cells, NK cells, neutrophils, monocytes, mast cells, plasma cells, fibroblasts, B cells, endothelial cells, and unknown cells. Cell type-specific gene expression profiles are shown in Figure 3C. After adjusting for p-value and performing the Wilcoxon rank-sum test, COMT was found to be ubiquitously expressed across all cell types, with the exception of monocytes. In contrast, AGT and FN1 were observed to exhibit highly cell type-specific expression patterns, with both genes predominantly expressed in fibroblasts compared to other cell clusters (Log2 FC values of 1.28 and 3.94, respectively). Additionally, FN1 exhibited notable expression in endothelial cells. No significant or specific expression of SERPINC1 or UGT1A1 was detected in any particular cell type in the single-cell dataset.
To further explore how the identified cell type-specific protein-coding gene influenced the progression of PC, we conducted a differential expression analysis between tumor and adjacent normal samples for each cell type, followed by KEGG and GO enrichment analyses. As displayed in Figure 4A, the KEGG analysis revealed that DEGs were broadly involved in pancreatic secretion pathways across all cell types. Additionally, protein digestion and uptake pathways were enriched in nearly all cell types, except for acinar cells. Focusing on the BP enriched within each cell type, we observed distinct functional signatures. In B cells, mast cells, macrophages, neutrophils, NK cells, plasma cells, T cells, and endothelial cells, DEGs were significantly enriched in processes related to digestion, leukotriene metabolism, regulation of lipase activity, lipid catabolism, and the positive regulation of lipase activity. Notably, fibroblasts exhibited a distinct enrichment pattern, with DEGs predominantly associated with structural and extracellular organization, including collagen fibril organization and extracellular matrix (ECM) organization. These pathways have been well-documented in pancreatic cancer progression, particularly in facilitating tumor growth, metastasis, and chemoresistance [38,50,51].
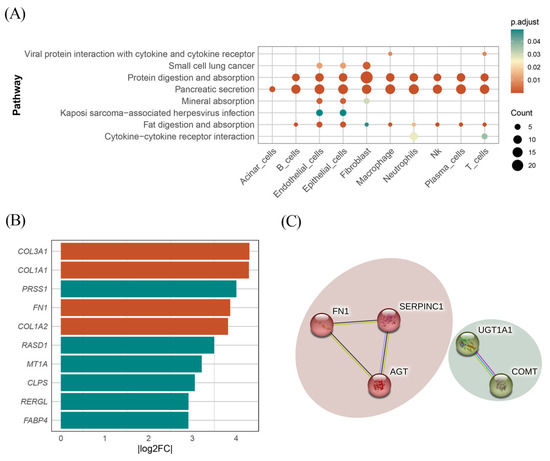
Figure 4.
Results of KEGG and PPI network. (A) Cluster-specific DEGs KEGG pathway. (B) The top 10 DEGs identified in fibroblasts ranked by |Log2 FC|. Green indicates that Log2 FC is negative, and red indicates Log2 FC is positive. (C) Identified protein interaction network.
To further evaluate the involvement of the identified protein-coding genes in specific biological pathways, we calculated the Log2 FC of DEGs in fibroblasts. Among these DEGs, two identified genes (AGT and FN1) demonstrated Log2 FC values of 1.16 and 3.86, respectively. Notably, as shown in Figure 4B, FN1 exhibited an exceptionally high expression level, ranking fourth in terms of |Log2 FC| among all fibroblast DEGs. These findings suggest that FN1 and AGT may play critical roles in the progression of pancreatic cancer, particularly within the tumor microenvironment, and may represent promising therapeutic targets.
3.5. Protein PPI Network and Gene Function
To explore the interactions between the identified plasma proteins, PPI network analysis was performed using the STRING database. As shown in Figure 4C, with a confidence score of 0.4, interactions were observed between AGT, FN1, and SERPINC1, as well as between UGT1A1 and COMT.
To further examine the roles of the identified proteins in specific biological pathways, we investigated their known functions. COMT is involved in neurotransmitter metabolism, specifically degrading catecholamine transmitters through methylation [52]. AGT plays a critical role in maintaining fluid and electrolyte homeostasis and has been implicated in the pathogenesis of essential hypertension [53]. UGT1A1 participates in the glucuronidation pathway, which is essential for detoxification and the regulation of various substances in metabolism [54,55]. SERPINC1 is associated with coagulation, functioning primarily by inhibiting the activation of serine proteases and thrombin. Importantly, FN1 is crucial for cell adhesion and migration, playing a vital role in the ECM [56,57,58]; it contributes to the formation of structural fibers that facilitate tissue repair and supports the continuous remodeling of the ECM, thereby maintaining tissue strength and structural integrity [59].
4. Discussion
PC remains a major global health challenge due to its high mortality rate and limited improvements in survival, despite recent medical advances. This highlights the urgent need for effective preventive strategies [60]. In this context, identifying and mitigating risk factors, particularly the potential impact of OCs on PC, has become increasingly important. In recent years, numerous studies have investigated the relationship between OCs on PC risk. However, findings from observational studies have been inconsistent, largely due to inherent limitations such as selection bias, information bias, and confounding by indication [13,61,62]. These methodological weaknesses impede the ability to draw reliable causal inferences. Given these issues, using advanced analytical methods is essential to gain a more accurate understanding of the causal relationship between OCs and PC. MR offers a robust method of addressing confounding by leveraging genetic variants as instrumental variables, allowing for a more accurate assessment of the causal relationship between OCs and PC [63,64].
In this study, we systematically investigated the causal relationship between OC drug target proteins and PC risk using MR. Our analysis was structured into three kay sections, each addressing a specific objective: First, we aimed to identify drug target proteins that are causally linked to PC and evaluate the potential effects of OCs on these targets using MR. Second, we performed sensitivity analyses and colocalization analyses to assess the robustness of our MR findings. Third, we conducted additional validation through differential expression analysis, single-cell transcriptomic analysis, and enrichment analysis to explore the cell-specific expression of the identified protein targets and their potential biological functions, particularly in pathways relevant to PC progression. Our results identified key drug targets, including FN1, AGT, and COMT, that may mediate the effect of OCs on PC risk. Specifically, our findings suggest that OCs increase the expression of FN1 and AGT and utilize COMT as a substrate. FN1, a crucial component of extracellular matrix (ECM) organization, plays a pivotal role in cancer progression and metastasis [65,66,67,68,69,70,71,72,73]. Its upregulation in fibroblasts contributes to the fibrotic stroma of PC [74,75], thereby promoting tumor progression. Additionally, FN1 has been associated with drug resistance in PC and is considered a potential therapeutic target [76,77]. AGT has similarly been implicated in tumor growth in various cancers [78,79,80], and our findings further support its role in PC. Our findings suggest that co-treatment with different contraceptive drugs upregulates the expression of both FN1 and AGT, which may contribute to an increased risk of PC. Although Bayesian colocalization analysis for COMT showed negative results, both tissue and single-cell analyses indicated a strong correlation between COMT expression and PC. COMT overexpression has been linked to early-stage carcinogenesis in PC and is markedly elevated in PC tissues, correlating with features such as early T stage [81,82,83]. While the role of OCs in modulating COMT remains uncertain, evidence from animal models suggests that ethinyl estradiol can upregulate COMT expression, highlighting the need for further validation in human studies [84,85].
In conclusion, our study provides compelling evidence that OCs, particularly combined oral contraceptives, may increase PC risk by upregulating key drug target proteins, including FN1 and AGT. By leveraging MR to minimize confounding, we establish a robust causal relationship between OC use and elevated PC risk, supporting and extending findings from previous research. These results offer important insights into the underlying biological mechanisms of OC-associated PC risk. Further studies are warranted to validate these findings and to elucidate the precise molecular pathways involved.
However, several limitations should be considered: First, our analyses were limited to individuals of European ancestry, which may restrict the generalizability of the findings to other ethnic populations. Second, we focused on the pQTLs of drug-targeted proteins by leveraging the most comprehensive and up-to-date datasets currently available, aiming to ensure maximal coverage. However, we acknowledge that some relevant pQTLs may have been excluded, which may provide further insights into the pathogenesis of PC. Third, although the use of stringent criteria for instrument selection enhances validity, it may also reduce statistical power and potentially overlook relevant associations. For example, while UGT1A1 and SERPINC1 showed significant associations in the initial MR analysis, these findings were not supported by sensitivity analyses or DEG analyses. In addition, we did not include trans-pQTLs, which may have limited a more comprehensive understanding of the roles of these proteins in PC.
To address the limitations of our study, future research should prioritize the inclusion of ethnically diverse populations, particularly non-European groups, to improve the generalizability of the findings. Expanding the analysis to incorporate a wider range of pQTLs, including trans-pQTLs, may uncover additional drug targets involved in PC progression. Broader IV selection criteria and larger datasets will enhance the statistical power and may identify previously overlooked targets. Moreover, experimental studies are necessary to validate the regulation effects of OCs on COMT expression, especially in humans. Lastly, large-scale longitudinal cohort or case–control studies are also essential to confirm the causal relationship between OC use and PC risk, and to differentiate the effects of various OC formulations. Ultimately, these efforts will strengthen the evidence base for refining prevention strategies and therapeutic interventions.
In summary, our study utilized MR to investigate the causal effects of plasma drug targets associated with OCs on PC risk. Among the identified targets, FN1 and AGT demonstrated the strongest association, suggesting that combined oral contraceptives may contribute to increased PC risk through the modulation of these proteins. These findings suggest that oral contraceptives, particularly combined oral contraceptives, may influence the risk of PC development. However, further experimental and clinical validation is necessary to confirm these associations and clarify the underlying mechanisms.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines13061351/s1, Table S1: drug target with Drugbank; Table S2: drug target with CTD.
Author Contributions
Conceptualization, Y.T.; Validation, Y.T. and S.W.; Formal analysis, Y.T., Y.Z. and X.S.; Investigation, Y.T., S.W., X.S. and Y.C.; Writing—original draft, Y.T.; Writing—review & editing, Y.Z., S.W., X.S., X.R. and Y.C.; Visualization, Y.T. and X.R.; Supervision, F.Y. and T.L.; Project administration, F.Y. and T.L.; Funding acquisition, F.Y. and T.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Fundamental Research Funds for the Central Universities of China Pharmaceutical University (2632023FY04) and of Shanghai Jiao Tong University (YG2023QNA28), the National Natural Science Foundation of China (82404380; 82273735; 82473737), Key Laboratory of Scientific and Engineering Computing (Ministry of Education), and the Shanghai Frontiers Science Center of Modern Analysis.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All analysis was carried out in Rstudio 4.1.1. UKB-PPP pQTL https://www.synapse.org/Synapse:syn51364943/wiki/622119 (accessed on 12 December 2024). UKB PC GWAS (GWAS catalog: GCST90041814). FinnGen PC GWAS https://risteys.finngen.fi (accessed on 5 June 2024). deCODE pQTL https://www.decode.com/summarydata/ (accessed on 16 December 2024). GEO database: https://www.ncbi.nlm.nih.gov/geo/ (accessed on 26 December 2024). Single-cell data: GSE155698. GTEx-TCGA data: https://xenabrowser.net/datapages/ (accessed on 26 December 2024).
Acknowledgments
We would like to thank all colleagues and collaborators who provided valuable input and support during the course of this work. We are particularly grateful to Jingya Fang for their insightful discussions and helpful suggestions.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Abbreviations
| PC | Pancreatic cancer |
| pQTL | Protein quantitative trait loci |
| Cis-pQTL | Cis-acting protein quantitative trait loci |
| MHC | Major histocompatibility complex |
| MAF | Minor allele frequency |
| GWAS | Genome-wide association study |
| UKB-PPP | UK Biobank Pharma Proteomics Project |
| MR | Mendelian randomization |
| LD | Linkage disequilibrium |
| IVs | Instrumental variables |
| SNP | Single nucleotide polymorphism |
| PPI | Protein–protein interaction |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| STRING | Search Tool for the Retrieval of Interacting Genes |
| OR | Odds ratio |
| CI | Confidence interval |
| GEO | Gene Expression Omnibus |
| scRNA-seq | Single-cell RNA sequencing |
References
- Luo, W.; Tao, J.; Zheng, L.; Zhang, T. Current epidemiology of pancreatic cancer: Challenges and opportunities. Chin. J. Cancer Res. Chung-Kuo Yen Cheng Yen Chiu 2020, 32, 705–719. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi, J.D.; Surana, R.; Valle, J.W.; Shroff, R.T. Pancreatic cancer. Lancet Lond. Engl. 2020, 395, 2008–2020. [Google Scholar] [CrossRef] [PubMed]
- Park, W.; Chawla, A.; O’Reilly, E.M. Pancreatic Cancer: A Review. JAMA 2021, 326, 851–862. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef]
- Collaborative Group on Hormonal Factors in Breast Cancer Breast cancer and hormonal contraceptives: Collaborative reanalysis of individual data on 53 297 women with breast cancer and 100 239 women without breast cancer from 54 epidemiological studies. Lancet Lond. Engl. 1996, 347, 1713–1727. [CrossRef] [PubMed]
- Havrilesky, L.J.; Gierisch, J.M.; Moorman, P.G.; Coeytaux, R.R.; Urrutia, R.P.; Lowery, W.J.; Dinan, M.; McBroom, A.J.; Wing, L.; Musty, M.D.; et al. Oral contraceptive use for the primary prevention of ovarian cancer. Evid. Rep. Assess. 2013, 212, 1–514. [Google Scholar]
- Bouman, A.; Heineman, M.J.; Faas, M.M. Sex hormones and the immune response in humans. Hum. Reprod. Update 2005, 11, 411–423. [Google Scholar] [CrossRef]
- Michels, K.A.; Brinton, L.A.; Pfeiffer, R.M.; Trabert, B. Oral Contraceptive Use and Risks of Cancer in the NIH-AARP Diet and Health Study. Am. J. Epidemiol. 2018, 187, 1630–1641. [Google Scholar] [CrossRef]
- Zhang, Y.; Coogan, P.F.; Palmer, J.R.; Strom, B.L.; Rosenberg, L. A case-control study of reproductive factors, female hormone use, and risk of pancreatic cancer. Cancer Causes Control CCC 2010, 21, 473–478. [Google Scholar] [CrossRef]
- Lee, E.; Horn-Ross, P.L.; Rull, R.P.; Neuhausen, S.L.; Anton-Culver, H.; Ursin, G.; Henderson, K.D.; Bernstein, L. Reproductive factors, exogenous hormones, and pancreatic cancer risk in the CTS. Am. J. Epidemiol. 2013, 178, 1403–1413. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.-M. Use of oral contraceptives and risk of pancreatic cancer in women: A recalculated meta-analysis of prospective cohort studies. World J. Gastroenterol. 2021, 27, 8374–8377. [Google Scholar] [CrossRef]
- Butt, S.A.; Lidegaardi, Ø.; Skovlund, C.; Hannaford, P.C.; Iversen, L.; Fielding, S.; Mørch, L.S. Hormonal contraceptive use and risk of pancreatic cancer-A cohort study among premenopausal women. PLoS ONE 2018, 13, e0206358. [Google Scholar] [CrossRef]
- Smith, G.D.; Ebrahim, S. Mendelian randomization: Prospects, potentials, and limitations. Int. J. Epidemiol. 2004, 33, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Larsson, S.C.; Butterworth, A.S.; Burgess, S. Mendelian randomization for cardiovascular diseases: Principles and applications. Eur. Heart J. 2023, 44, 4913–4924. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.F.; Finan, C.; Gordillo-Marañón, M.; Asselbergs, F.W.; Freitag, D.F.; Patel, R.S.; Tyl, B.; Chopade, S.; Faraway, R.; Zwierzyna, M.; et al. Genetic drug target validation using Mendelian randomisation. Nat. Commun. 2020, 11, 3255. [Google Scholar] [CrossRef]
- Burgess, S.; Butterworth, A.; Malarstig, A.; Thompson, S.G. Use of Mendelian randomisation to assess potential benefit of clinical intervention. BMJ 2012, 345, e7325. [Google Scholar] [CrossRef]
- Gill, D.; Georgakis, M.K.; Walker, V.M.; Schmidt, A.F.; Gkatzionis, A.; Freitag, D.F.; Finan, C.; Hingorani, A.D.; Howson, J.M.M.; Burgess, S.; et al. Mendelian randomization for studying the effects of perturbing drug targets. Wellcome Open Res. 2021, 6, 16. [Google Scholar] [CrossRef]
- Walker, V.M.; Davey Smith, G.; Davies, N.M.; Martin, R.M. Mendelian randomization: A novel approach for the prediction of adverse drug events and drug repurposing opportunities. Int. J. Epidemiol. 2017, 46, 2078–2089. [Google Scholar] [CrossRef]
- Patel, A.; Gill, D.; Shungin, D.; Mantzoros, C.S.; Knudsen, L.B.; Bowden, J.; Burgess, S. Robust use of phenotypic heterogeneity at drug target genes for mechanistic insights: Application of cis-multivariable Mendelian randomization to GLP1R gene region. Genet. Epidemiol. 2024, 48, 151–163. [Google Scholar] [CrossRef]
- Zheng, G.; Chattopadhyay, S.; Sundquist, J.; Sundquist, K.; Ji, J. Antihypertensive drug targets and breast cancer risk: A two-sample Mendelian randomization study. Eur. J. Epidemiol. 2024, 39, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Ferkingstad, E.; Sulem, P.; Atlason, B.A.; Sveinbjornsson, G.; Magnusson, M.I.; Styrmisdottir, E.L.; Gunnarsdottir, K.; Helgason, A.; Oddsson, A.; Halldorsson, B.V.; et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 2021, 53, 1712–1721. [Google Scholar] [CrossRef]
- Sun, B.B.; Chiou, J.; Traylor, M.; Benner, C.; Hsu, Y.-H.; Richardson, T.G.; Surendran, P.; Mahajan, A.; Robins, C.; Vasquez-Grinnell, S.G.; et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature 2023, 622, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Kurki, M.I.; Karjalainen, J.; Palta, P.; Sipilä, T.P.; Kristiansson, K.; Donner, K.M.; Reeve, M.P.; Laivuori, H.; Aavikko, M.; Kaunisto, M.A.; et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 2023, 613, 508–518. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Davis, A.P.; Wiegers, T.C.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2023. Nucleic Acids Res. 2023, 51, D1257–D1262. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Gu, X.; Dou, M.; Duan, Q.; Jiang, Z.; Yin, K.; Cai, W.; Cao, B.; Wang, Y.; Chen, Y. Systematic druggable genome-wide Mendelian randomisation identifies therapeutic targets for Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry 2023, 94, 954–961. [Google Scholar] [CrossRef]
- Maina, J.G.; Balkhiyarova, Z.; Nouwen, A.; Pupko, I.; Ulrich, A.; Boissel, M.; Bonnefond, A.; Froguel, P.; Khamis, A.; Prokopenko, I.; et al. Bidirectional Mendelian Randomization and Multiphenotype GWAS Show Causality and Shared Pathophysiology Between Depression and Type 2 Diabetes. Diabetes Care 2023, 46, 1707–1714. [Google Scholar] [CrossRef]
- Papadimitriou, N.; Dimou, N.; Tsilidis, K.K.; Banbury, B.; Martin, R.M.; Lewis, S.J.; Kazmi, N.; Robinson, T.M.; Albanes, D.; Aleksandrova, K.; et al. Physical activity and risks of breast and colorectal cancer: A Mendelian randomisation analysis. Nat. Commun. 2020, 11, 597. [Google Scholar] [CrossRef]
- Burgess, S.; Dudbridge, F.; Thompson, S.G. Combining information on multiple instrumental variables in Mendelian randomization: Comparison of allele score and summarized data methods. Stat. Med. 2016, 35, 1880–1906. [Google Scholar] [CrossRef]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Zuber, V.; Valdes-Marquez, E.; Sun, B.B.; Hopewell, J.C. Mendelian randomization with fine-mapped genetic data: Choosing from large numbers of correlated instrumental variables. Genet. Epidemiol. 2017, 41, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Giambartolomei, C.; Vukcevic, D.; Schadt, E.E.; Franke, L.; Hingorani, A.D.; Wallace, C.; Plagnol, V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014, 10, e1004383. [Google Scholar] [CrossRef]
- Kia, D.A.; Zhang, D.; Guelfi, S.; Manzoni, C.; Hubbard, L.; Reynolds, R.H.; Botía, J.; Ryten, M.; Ferrari, R.; Lewis, P.A.; et al. Identification of Candidate Parkinson Disease Genes by Integrating Genome-Wide Association Study, Expression, and Epigenetic Data Sets. JAMA Neurol. 2021, 78, 464–472. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets--update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Steele, N.G.; Carpenter, E.S.; Kemp, S.B.; Sirihorachai, V.R.; The, S.; Delrosario, L.; Lazarus, J.; Amir, E.-A.D.; Gunchick, V.; Espinoza, C.; et al. Multimodal Mapping of the Tumor and Peripheral Blood Immune Landscape in Human Pancreatic Cancer. Nat. Cancer 2020, 1, 1097–1112. [Google Scholar] [CrossRef] [PubMed]
- Halbrook, C.J.; Thurston, G.; Boyer, S.; Anaraki, C.; Jiménez, J.A.; McCarthy, A.; Steele, N.G.; Kerk, S.A.; Hong, H.S.; Lin, L.; et al. Differential integrated stress response and asparagine production drive symbiosis and therapy resistance of pancreatic adenocarcinoma cells. Nat. Cancer 2022, 3, 1386–1403. [Google Scholar] [CrossRef]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef]
- Sitruk-Ware, R.; Plu-Bureau, G.; Menard, J.; Conard, J.; Kumar, S.; Thalabard, J.-C.; Tokay, B.; Bouchard, P. Effects of oral and transvaginal ethinyl estradiol on hemostatic factors and hepatic proteins in a randomized, crossover study. J. Clin. Endocrinol. Metab. 2007, 92, 2074–2079. [Google Scholar] [CrossRef] [PubMed]
- van Rooijen, M.; Silveira, A.; Hamsten, A.; Bremme, K. Sex hormone--binding globulin--a surrogate marker for the prothrombotic effects of combined oral contraceptives. Am. J. Obstet. Gynecol. 2004, 190, 332–337. [Google Scholar] [CrossRef] [PubMed]
- United Nations Development Programme; United Nations Population Fund; World Health Organization; World Bank Special Programme of Research, Development and Research Training in Human Reproduction. Task Force on Long-acting Systemic Agents for Fertility Regulaion Comparative study of the effects of two once-a-month injectable contraceptives (Cyclofem and Mesigyna) and one oral contraceptive (Ortho-Novum 1/35) on coagulation and fibrinolysis. Contraception 2003, 68, 159–176. [Google Scholar] [CrossRef]
- Lippi, G.; Manzato, F.; Brocco, G.; Franchini, M.; Guidi, G. Prothrombotic effects and clinical implications of third-generation oral contraceptives use. Blood Coagul. Fibrinolysis Int. J. Haemost. Thromb. 2002, 13, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Oral Contraceptive and Hemostasis Study Group The effects of seven monophasic oral contraceptive regimens on hemostatic variables: Conclusions from a large randomized multicenter study. Contraception 2003, 67, 173–185. [CrossRef]
- Sitruk-Ware, R.L.; Menard, J.; Rad, M.; Burggraaf, J.; de Kam, M.L.; Tokay, B.A.; Sivin, I.; Kluft, C. Comparison of the impact of vaginal and oral administration of combined hormonal contraceptives on hepatic proteins sensitive to estrogen. Contraception 2007, 75, 430–437. [Google Scholar] [CrossRef]
- Machado, R.B.; Fabrini, P.; Cruz, A.M.; Maia, E.; da Cunha Bastos, A. Clinical and metabolic aspects of the continuous use of a contraceptive association of ethinyl estradiol (30 microg) and gestodene (75 microg). Contraception 2004, 70, 365–370. [Google Scholar] [CrossRef]
- Wiegratz, I.; Lee, J.H.; Kutschera, E.; Winkler, U.H.; Kuhl, H. Effect of four oral contraceptives on hemostatic parameters. Contraception 2004, 70, 97–106. [Google Scholar] [CrossRef]
- Zuber, V.; Grinberg, N.F.; Gill, D.; Manipur, I.; Slob, E.A.W.; Patel, A.; Wallace, C.; Burgess, S. Combining evidence from Mendelian randomization and colocalization: Review and comparison of approaches. Am. J. Hum. Genet. 2022, 109, 767–782. [Google Scholar] [CrossRef]
- Wang, D.; Li, Y.; Ge, H.; Ghadban, T.; Reeh, M.; Güngör, C. The Extracellular Matrix: A Key Accomplice of Cancer Stem Cell Migration, Metastasis Formation, and Drug Resistance in PDAC. Cancers 2022, 14, 3998. [Google Scholar] [CrossRef]
- Mancini, A.; Gentile, M.T.; Pentimalli, F.; Cortellino, S.; Grieco, M.; Giordano, A. Multiple aspects of matrix stiffness in cancer progression. Front. Oncol. 2024, 14, 1406644. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.T. Catechol-O-Methyltransferase (COMT)-mediated methylation metabolism of endogenous bioactive catechols and modulation by endobiotics and xenobiotics: Importance in pathophysiology and pathogenesis. Curr. Drug Metab. 2002, 3, 321–349. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Cassis, L.A.; Kooi, C.W.V.; Daugherty, A. Structure and functions of angiotensinogen. Hypertens. Res. Off. J. Jpn. Soc. Hypertens. 2016, 39, 492–500. [Google Scholar] [CrossRef]
- Rowland, A.; Miners, J.O.; Mackenzie, P.I. The UDP-glucuronosyltransferases: Their role in drug metabolism and detoxification. Int. J. Biochem. Cell Biol. 2013, 45, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Mennillo, E.; Yang, X.; Weber, A.A.; Maruo, Y.; Verreault, M.; Barbier, O.; Chen, S.; Tukey, R.H. Intestinal UDP-Glucuronosyltransferase 1A1 and Protection against Irinotecan-Induced Toxicity in a Novel UDP-Glucuronosyltransferase 1A1 Tissue-Specific Humanized Mouse Model. Drug Metab. Dispos. Biol. Fate Chem. 2022, 50, 33–42. [Google Scholar] [CrossRef]
- Magnusson, M.K.; Mosher, D.F. Fibronectin: Structure, assembly, and cardiovascular implications. Arterioscler. Thromb. Vasc. Biol. 1998, 18, 1363–1370. [Google Scholar] [CrossRef]
- Soikkeli, J.; Podlasz, P.; Yin, M.; Nummela, P.; Jahkola, T.; Virolainen, S.; Krogerus, L.; Heikkilä, P.; von Smitten, K.; Saksela, O.; et al. Metastatic outgrowth encompasses COL-I, FN1, and POSTN up-regulation and assembly to fibrillar networks regulating cell adhesion, migration, and growth. Am. J. Pathol. 2010, 177, 387–403. [Google Scholar] [CrossRef] [PubMed]
- Efthymiou, G.; Saint, A.; Ruff, M.; Rekad, Z.; Ciais, D.; Van Obberghen-Schilling, E. Shaping Up the Tumor Microenvironment With Cellular Fibronectin. Front. Oncol. 2020, 10, 641. [Google Scholar] [CrossRef]
- Dalton, C.J.; Lemmon, C.A. Fibronectin: Molecular Structure, Fibrillar Structure and Mechanochemical Signaling. Cells 2021, 10, 2443. [Google Scholar] [CrossRef]
- Khalaf, N.; El-Serag, H.B.; Abrams, H.R.; Thrift, A.P. Burden of Pancreatic Cancer: From Epidemiology to Practice. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2021, 19, 876–884. [Google Scholar] [CrossRef]
- Alvarez, A.; Benjaminsen Borch, K.; Rylander, C. Reproductive Factors, Use of Exogenous Hormones, and Pancreatic Cancer Incidence: The Norwegian Women and Cancer Study. Clin. Epidemiol. 2021, 13, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Kabat, G.C.; Kamensky, V.; Rohan, T.E. Reproductive factors, exogenous hormone use, and risk of pancreatic cancer in postmenopausal women. Cancer Epidemiol. 2017, 49, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Richmond, R.C.; Davey Smith, G. Mendelian Randomization: Concepts and Scope. Cold Spring Harb. Perspect. Med. 2022, 12, a040501. [Google Scholar] [CrossRef]
- Sekula, P.; Del Greco, M.F.; Pattaro, C.; Köttgen, A. Mendelian Randomization as an Approach to Assess Causality Using Observational Data. J. Am. Soc. Nephrol. JASN 2016, 27, 3253–3265. [Google Scholar] [CrossRef]
- Zhang, X.; Luo, Y.; Cen, Y.; Qiu, X.; Li, J.; Jie, M.; Yang, S.; Qin, S. MACC1 promotes pancreatic cancer metastasis by interacting with the EMT regulator SNAI1. Cell Death Dis. 2022, 13, 923. [Google Scholar] [CrossRef]
- Jiang, P.; Li, Z.; Tian, F.; Li, X.; Yang, J. Fyn/heterogeneous nuclear ribonucleoprotein E1 signaling regulates pancreatic cancer metastasis by affecting the alternative splicing of integrin β1. Int. J. Oncol. 2017, 51, 169–183. [Google Scholar] [CrossRef]
- Lei, X.; Chen, G.; Li, J.; Wen, W.; Gong, J.; Fu, J. Comprehensive analysis of abnormal expression, prognostic value and oncogenic role of the hub gene FN1 in pancreatic ductal adenocarcinoma via bioinformatic analysis and in vitro experiments. PeerJ 2021, 9, e12141. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Messina-Pacheco, J.; Corredor, A.; Gregorieff, A.; Liu, J.; Nehme, A.; Najafabadi, H.; Riazalhosseini, Y.; Gao, B.; Gao, Z. An integrated model of acinar to ductal metaplasia-related N7-methyladenosine regulators predicts prognosis and immunotherapy in pancreatic carcinoma based on digital spatial profiling. Front. Immunol. 2022, 13, 961457. [Google Scholar] [CrossRef]
- Hiroshima, Y.; Kasajima, R.; Kimura, Y.; Komura, D.; Ishikawa, S.; Ichikawa, Y.; Bouvet, M.; Yamamoto, N.; Oshima, T.; Morinaga, S.; et al. Novel targets identified by integrated cancer-stromal interactome analysis of pancreatic adenocarcinoma. Cancer Lett. 2020, 469, 217–227. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, J.; Li, H.; Yu, H.; Chen, S.; Liu, S.; Zhang, C.; He, Y. FN1 is a prognostic biomarker and correlated with immune infiltrates in gastric cancers. Front. Oncol. 2022, 12, 918719. [Google Scholar] [CrossRef]
- Zhang, X.-X.; Luo, J.-H.; Wu, L.-Q. FN1 overexpression is correlated with unfavorable prognosis and immune infiltrates in breast cancer. Front. Genet. 2022, 13, 913659. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, C.; Chen, B.; Guo, T. High FN1 expression correlates with gastric cancer progression. Pathol. Res. Pract. 2022, 239, 154179. [Google Scholar] [CrossRef]
- Pan, H.; Luo, Z.; Lin, F.; Zhang, J.; Xiong, T.; Hong, Y.; Sun, B.; Yang, Y. FN1, a reliable prognostic biomarker for thyroid cancer, is associated with tumor immunity and an unfavorable prognosis. Oncol. Lett. 2024, 28, 510. [Google Scholar] [CrossRef]
- Eble, J.A.; Niland, S. The extracellular matrix in tumor progression and metastasis. Clin. Exp. Metastasis 2019, 36, 171–198. [Google Scholar] [CrossRef]
- Chakkera, M.; Foote, J.B.; Farran, B.; Nagaraju, G.P. Breaking the stromal barrier in pancreatic cancer: Advances and challenges. Biochim. Biophys. Acta Rev. Cancer 2024, 1879, 189065. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, T.; Sun, C.; Du, Y.; Chen, L.; Du, C.; Shi, J.; Wang, W. Identification and prognostic analysis of biomarkers to predict the progression of pancreatic cancer patients. Mol. Med. 2022, 28, 43. [Google Scholar] [CrossRef] [PubMed]
- Xavier, C.P.R.; Castro, I.; Caires, H.R.; Ferreira, D.; Cavadas, B.; Pereira, L.; Santos, L.L.; Oliveira, M.J.; Vasconcelos, M.H. Chitinase 3-like-1 and fibronectin in the cargo of extracellular vesicles shed by human macrophages influence pancreatic cancer cellular response to gemcitabine. Cancer Lett. 2021, 501, 210–223. [Google Scholar] [CrossRef]
- Zhu, L.; Ma, M.; Zhang, L.; Wang, S.; Guo, Y.; Ling, X.; Lin, H.; Lai, N.; Lin, S.; Du, L.; et al. System Analysis Based on Lipid-Metabolism-Related Genes Identifies AGT as a Novel Therapy Target for Gastric Cancer with Neoadjuvant Chemotherapy. Pharmaceutics 2023, 15, 810. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, L.; Wang, L.; Zhang, D. AGT May Serve as a Prognostic Biomarker and Correlated with Immune Infiltration in Gastric Cancer. Int. J. Gen. Med. 2022, 15, 1865–1878. [Google Scholar] [CrossRef]
- Chen, W.; Chen, Y.; Zhang, K.; Yang, W.; Li, X.; Zhao, J.; Liu, K.; Dong, Z.; Lu, J. AGT serves as a potential biomarker and drives tumor progression in colorectal carcinoma. Int. Immunopharmacol. 2021, 101, 108225. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, J.; Zhou, L.; You, L.; Zhao, Y.; Li, J. Increased COMT expression in pancreatic cancer and correlation with clinicopathologic parameters. Sci. China Life Sci. 2012, 55, 747–752. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dai, H.; Wang, X.; Hong, X.; Wu, H.; Di, W.; Wang, W.; Xu, P.; Jia, C.; Wang, J.; Chen, H.; et al. Overexpression of catechol-O-methyltransferase occurs early in the progression of pancreatic cancer. J. Pancreatol. 2018, 1, 39. [Google Scholar] [CrossRef]
- Creveling, C.R. The role of catechol-O-methyltransferase in the inactivation of catecholestrogen. Cell. Mol. Neurobiol. 2003, 23, 289–291. [Google Scholar] [CrossRef] [PubMed]
- Boverhof, D.R.; Burgoon, L.D.; Williams, K.J.; Zacharewski, T.R. Inhibition of estrogen-mediated uterine gene expression responses by dioxin. Mol. Pharmacol. 2008, 73, 82–93. [Google Scholar] [CrossRef]
- Yadetie, F.; Brun, N.R.; Vieweg, I.; Nahrgang, J.; Karlsen, O.A.; Goksøyr, A. Transcriptome responses in polar cod (Boreogadus saida) liver slice culture exposed to benzo[a]pyrene and ethynylestradiol: Insights into anti-estrogenic effects. Toxicol. Vitr. Int. J. Publ. Assoc. BIBRA 2021, 75, 105193. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).