Developments and Applications of Liver-on-a-Chip Technology—Current Status and Future Prospects
Abstract
1. Introduction
2. Early Developments and Initial Applications
Evolution of Fabrication Techniques
3. Hepatic Organoids
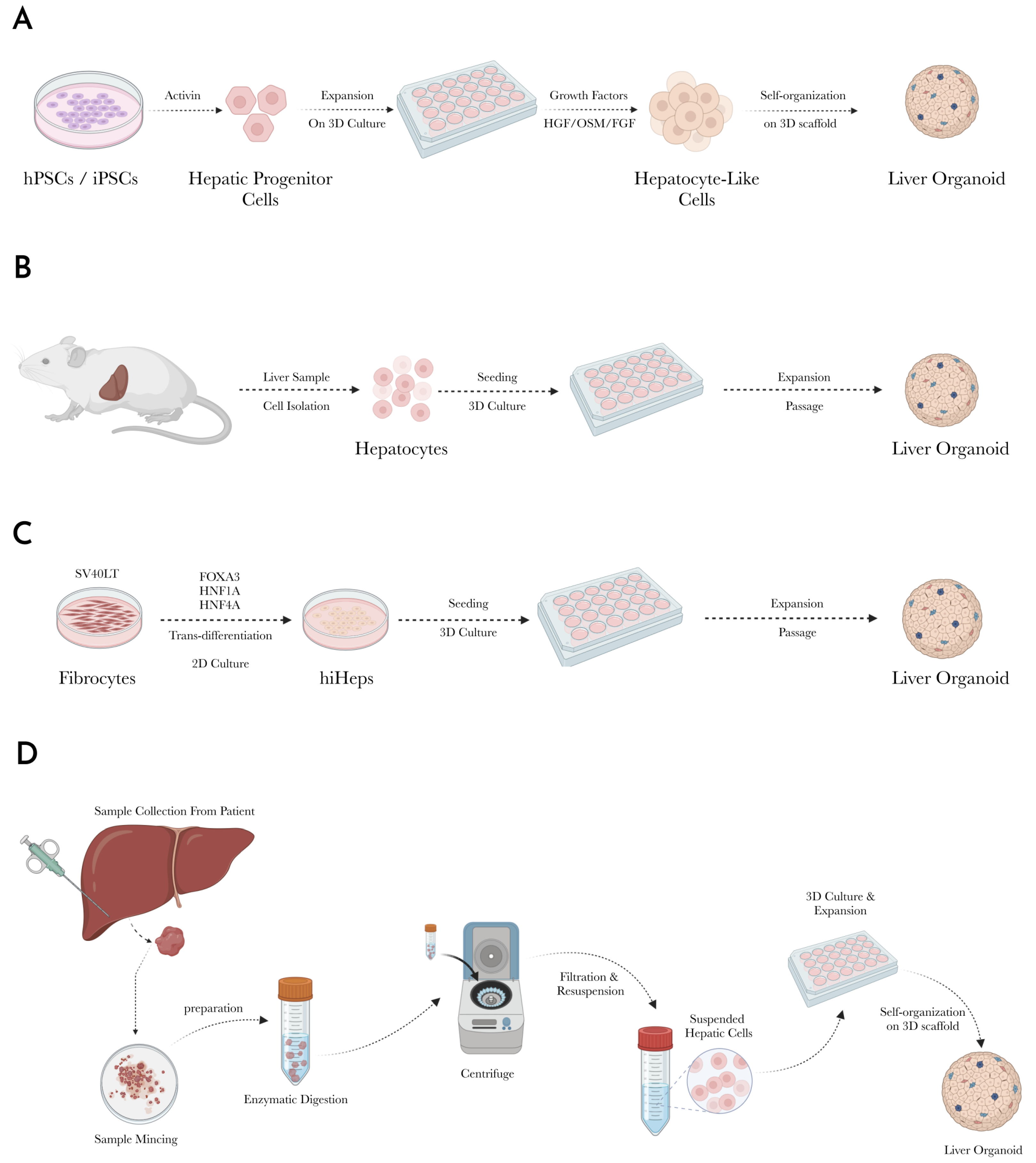
3.1. Stem Cell-Derived Liver Organoids
3.1.1. Isolation and Differentiation of Stem Cells into Hepatic Progenitors
3.1.2. Maturation into Hepatocyte-like Cells
3.1.3. Incorporation of Non-Parenchymal Cells
3.1.4. 3D Culture and Organoid Formation
3.2. Liver Tissue-Derived Liver Organoids
3.2.1. Sample Collection
3.2.2. Tissue Preparation and Cell Isolation
3.2.3. Cell Expansion
3.2.4. Sub-Culture and Maintenance
3.2.5. Organoid Formation
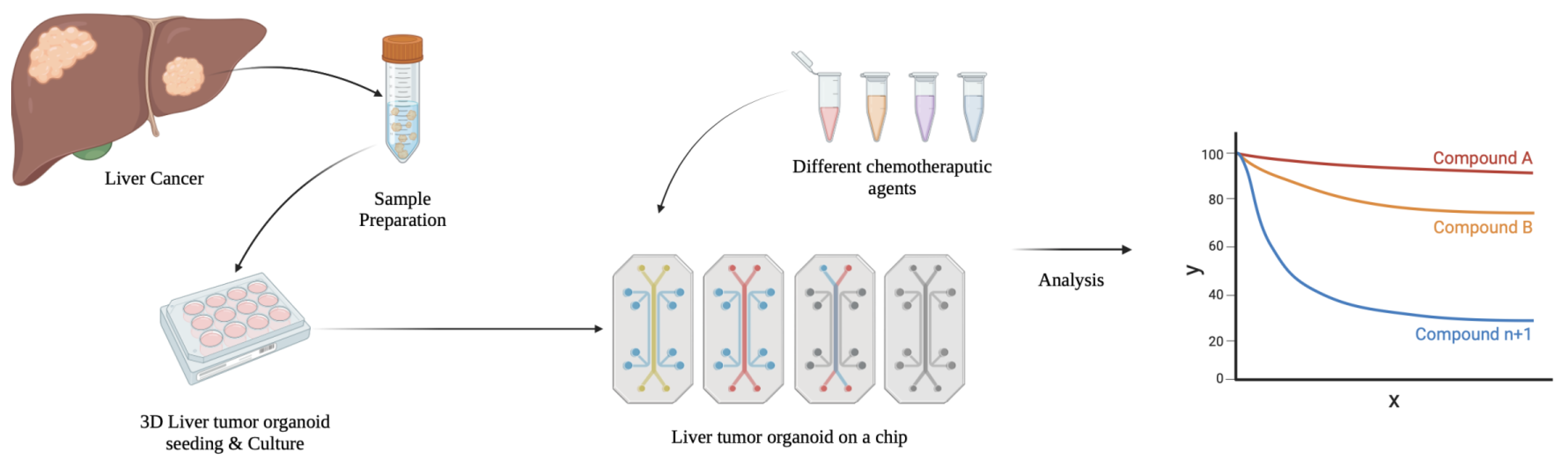
3.3. Tumor-Derived Liver Organoids
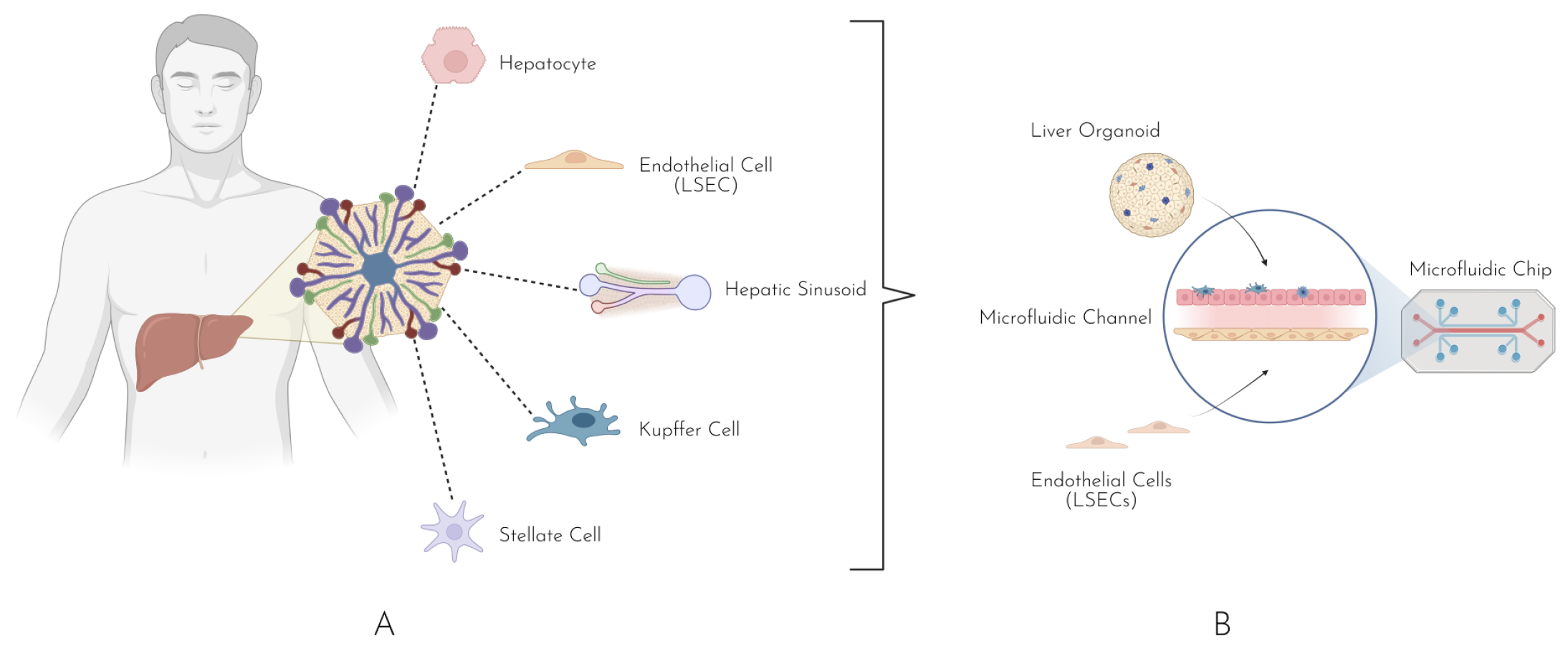
4. Liver-on-a-Chip
Diversity of Liver-on-a-Chip Models
5. Applications of Liver Organoids and Liver-on-a-Chip
5.1. Cancer Research
5.2. Drug Development and Toxicity Testing
5.3. Biological Mechanism Research
5.4. Disease Modeling
5.5. Regenerative Medicine
5.6. Commercialization and Regulatory Relevance
6. Future Prospects
- (1)
- Scalability and High-Throughput Screening: Scalability is a critical priority for the future of LiOC to meet the needs of the pharmaceutical industry and research institutions. Current LiOC systems are often limited by the number of replicates that can be run simultaneously. This significantly restricts their utility in large-scale drug screening applications. Efforts are underway to automate LiOC production and operation, allowing for high-throughput screening [134,135].
- (2)
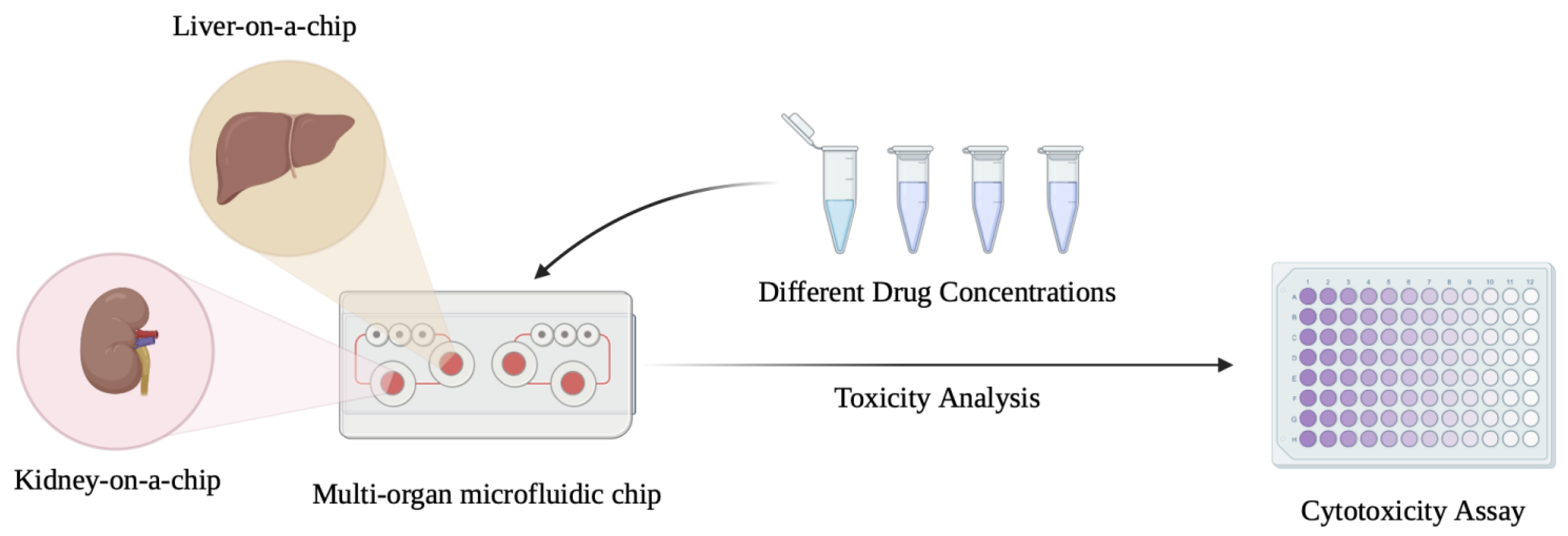
- Multi-Organ-on-chip integration: The development of multi-organ-on-chip systems that integrate the LiOC with other organs such as the kidney are yet another promising avenue that could provide a more comprehensive view of drug effects, disease progression, and organ interactions [136]. Multi-organ systems on a chip could also be vital to studying systemic diseases like metabolic syndromes, multi-organ failure, and metastasis. Advancements in microfluidics and 3D printing could potentially provide a breakthrough in this aspect [137].
- (3)
- Personalized Medicine and Precision Therapy: The medical research community is increasingly exploring the adoption of a more personalized approach to medical care, especially with regards to therapeutics. Since LiOC can be developed using patient-derived cells, individualized liver models are possible [138]. With these, researchers and clinicians could predict how an individual will respond to a given medical intervention. In the future, it is conceivable that each patient could have their own LiOC to test therapeutic options, provided that the challenges of scalability and cost are addressed.
- (4)
- Artificial Intelligence and Machine Learning: As LiOC models become more sophisticated, machine learning model and artificial intelligence (AI) integration are likely to enhance the capabilities of models. The AI algorithms could be employed to analyze the complex data generated by the LiOC systems, identifying patterns and predicting outcomes that may not be immediately apparent to researchers and clinicians [139]. For instance, AI could be utilized to optimize the conditions for organoid growth, predict drug responses, or identify biomarkers associated with specific liver diseases. They could also help accelerate the design of new LiOC platforms by predicting how different cell types, structures, and environmental conditions may affect organ phenotype and function.
- (5)
- Addressing Current Challenges in Vascularization: Currently, the vascularization of LiOCs and indeed organoids of other organs, like the lung and kidney, remain a significant challenge [140]. For LiOCs, their inability to replicate the liver’s intricate vascular network, which is essential for nutrient delivery, waste removal, drug metabolism, and plays a role in liver regeneration, is a critical limitation. Perhaps developments in the field may enable creating functional vasculatures within the organoid. Bioprinting technologies are being explored as a potential solution, but it is still early to tell.
- (6)
- Clinical Translation: Ultimately, the most exciting prospect for LiOC technology lies in its potential clinical applications, particularly in regenerative medicine. LiOC systems may be used to bioengineer liver tissue for transplantation in patients with liver failure and chronic liver disease [141]. This would help address the shortage of donated livers and would also negate the rejection of the graft. However, this remains a long-term prospect.
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kwong, A.J.; Ebel, N.H.; Kim, W.R.; Lake, J.R.; Smith, J.M.; Schladt, D.P.; Schnellinger, E.M.; Handarova, D.; Weiss, S.; Cafarella, M.; et al. OPTN/SRTR 2021 Annual Data Report: Liver. Am. J. Transplant. 2023, 23, S178–S263. [Google Scholar] [CrossRef] [PubMed]
- Devarbhavi, H.; Asrani, S.K.; Arab, J.P.; Nartey, Y.A.; Pose, E.; Kamath, P.S. Global burden of liver disease: 2023 update. J. Hepatol. 2023, 79, 516–537. [Google Scholar] [CrossRef]
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef]
- Mugaanyi, J.; Tong, J.; Lu, C.; Mao, S.; Huang, J.; Lu, C. Risk factors for acute rejection in liver transplantation and its impact on the outcomes of recipients. Transpl. Immunol. 2023, 76, 101767. [Google Scholar] [CrossRef]
- Chiang, J.Y. Bile acid metabolism and signaling. Compr. Physiol. 2013, 3, 1191–1212. [Google Scholar] [CrossRef]
- Remmer, H. The role of theliver in drug metabolism. Am. J. Med. 1970, 49, 617–629. [Google Scholar] [CrossRef]
- Kapalczynska, M.; Kolenda, T.; Przybyla, W.; Zajaczkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Blizniak, R.; Luczewski, L.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Aziz, A.U.R.; Geng, C.; Fu, M.; Yu, X.; Qin, K.; Liu, B. The Role of Microfluidics for Organ on Chip Simulations. Bioengineering 2017, 4, 39. [Google Scholar] [CrossRef]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Yoon No, D.; Lee, K.H.; Lee, J.; Lee, S.H. 3D liver models on a microplatform: Well-defined culture, engineering of liver tissue and liver-on-a-chip. Lab Chip 2015, 15, 3822–3837. [Google Scholar] [CrossRef] [PubMed]
- Moradi, E.; Jalili-Firoozinezhad, S.; Solati-Hashjin, M. Microfluidic organ-on-a-chip models of human liver tissue. Acta Biomater. 2020, 116, 67–83. [Google Scholar] [CrossRef]
- Hassan, S.; Sebastian, S.; Maharjan, S.; Lesha, A.; Carpenter, A.M.; Liu, X.; Xie, X.; Livermore, C.; Zhang, Y.S.; Zarrinpar, A. Liver-on-a-Chip Models of Fatty Liver Disease. Hepatology 2020, 71, 733–740. [Google Scholar] [CrossRef]
- Zhang, B.; Montgomery, M.; Chamberlain, M.D.; Ogawa, S.; Korolj, A.; Pahnke, A.; Wells, L.A.; Masse, S.; Kim, J.; Reis, L.; et al. Biodegradable scaffold with built-in vasculature for organ-on-a-chip engineering and direct surgical anastomosis. Nat. Mater. 2016, 15, 669–678. [Google Scholar] [CrossRef]
- van Midwoud, P.M.; Merema, M.T.; Verpoorte, E.; Groothuis, G.M. A microfluidic approach for in vitro assessment of interorgan interactions in drug metabolism using intestinal and liver slices. Lab Chip 2010, 10, 2778–2786. [Google Scholar] [CrossRef]
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-chips: Into the next decade. Nat. Rev. Drug Discov. 2021, 20, 345–361. [Google Scholar] [CrossRef]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef]
- Rensch, C.; Jackson, A.; Lindner, S.; Salvamoser, R.; Samper, V.; Riese, S.; Bartenstein, P.; Wangler, C.; Wangler, B. Microfluidics: A groundbreaking technology for PET tracer production? Molecules 2013, 18, 7930–7956. [Google Scholar] [CrossRef]
- Kimura, H.; Sakai, Y.; Fujii, T. Organ/body-on-a-chip based on microfluidic technology for drug discovery. Drug Metab. Pharmacokinet. 2018, 33, 43–48. [Google Scholar] [CrossRef]
- Cavaniol, C.; Cesar, W.; Descroix, S.; Viovy, J.L. Flowmetering for microfluidics. Lab Chip 2022, 22, 3603–3617. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.S.; Venkatesalu, S.; Dilliyappan, S.; Pasupulla, A.P.; Prathap, L.; Palaniyandi, T.; Baskar, G.; Ravi, M.; Sugumaran, A. Microfluidics as diagnostic tools. Clin. Chim. Acta 2024, 556, 117841. [Google Scholar] [CrossRef]
- Ahn, J.; Yoon, M.J.; Hong, S.H.; Cha, H.; Lee, D.; Koo, H.S.; Ko, J.E.; Lee, J.; Oh, S.; Jeon, N.L.; et al. Three-dimensional microengineered vascularised endometrium-on-a-chip. Hum. Reprod. 2021, 36, 2720–2731. [Google Scholar] [CrossRef]
- Li, M.; Gao, L.; Zhao, L.; Zou, T.; Xu, H. Toward the next generation of vascularized human neural organoids. Med. Res. Rev. 2023, 43, 31–54. [Google Scholar] [CrossRef]
- Ashammakhi, N.; Wesseling-Perry, K.; Hasan, A.; Elkhammas, E.; Zhang, Y.S. Kidney-on-a-chip: Untapped opportunities. Kidney Int. 2018, 94, 1073–1086. [Google Scholar] [CrossRef]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef]
- Huh, D.; Leslie, D.C.; Matthews, B.D.; Fraser, J.P.; Jurek, S.; Hamilton, G.A.; Thorneloe, K.S.; McAlexander, M.A.; Ingber, D.E. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 2012, 4, 159ra147. [Google Scholar] [CrossRef]
- Tony, A.; Badea, I.; Yang, C.; Liu, Y.; Wells, G.; Wang, K.; Yin, R.; Zhang, H.; Zhang, W. The Additive Manufacturing Approach to Polydimethylsiloxane (PDMS) Microfluidic Devices: Review and future directions. Polymers 2023, 15, 1926. [Google Scholar] [CrossRef]
- Alavi, S.E.; Alharthi, S.; Alavi, S.F.; Alavi, S.Z.; Zahra, G.E.; Raza, A.; Shahmabadi, H.E. Microfluidics for personalized drug delivery. Drug Discov. Today 2024, 29, 103936. [Google Scholar] [CrossRef]
- Zhu, Y.; Guo, S.; Ravichandran, D.; Ramanathan, A.; Sobczak, M.T.; Sacco, A.F.; Patil, D.; Thummalapalli, S.V.; Pulido, T.V.; Lancaster, J.N.; et al. 3D-Printed Polymeric biomaterials for health applications. Adv. Healthc. Mater. 2024, 14, 2402571. [Google Scholar] [CrossRef]
- Zhou, Y. The recent development and applications of fluidic channels by 3D printing. J. Biomed. Sci. 2017, 24, 80. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Wang, Y.; Liu, N.; Zhong, S.; Li, L.; Zhang, Q.; Liu, Z.; Yue, T. Advances in the model structure of in vitro Vascularized Organ-on-a-Chip. Cyborg Bionic Syst. 2024, 5, 107. [Google Scholar] [CrossRef]
- Corral-Nájera, K.; Chauhan, G.; Serna-Saldívar, S.O.; Martínez-Chapa, S.O.; Aeinehvand, M.M. Polymeric and biological membranes for organ-on-a-chip devices. Microsyst. Nanoeng. 2023, 9, 107. [Google Scholar] [CrossRef]
- Hull, S.M.; Brunel, L.G.; Heilshorn, S.C. 3D bioprinting of Cell-Laden hydrogels for improved biological functionality. Adv. Mater. 2021, 34, 2103691. [Google Scholar] [CrossRef]
- Mirshafiei, M.; Rashedi, H.; Yazdian, F.; Rahdar, A.; Baino, F. Advancements in tissue and organ 3D bioprinting: Current techniques, applications, and future perspectives. Mater. Des. 2024, 240, 112853. [Google Scholar] [CrossRef]
- Chen, X.; Anvari-Yazdi, A.F.; Duan, X.; Zimmerling, A.; Gharraei, R.; Sharma, N.; Sweilem, S.; Ning, L. Biomaterials/bioinks and extrusion bioprinting. Bioact. Mater. 2023, 28, 511–536. [Google Scholar] [CrossRef]
- Zhang, B.; Gao, L.; Ma, L.; Luo, Y.; Yang, H.; Cui, Z. 3D bioprinting: A novel avenue for manufacturing tissues and organs. Engineering 2019, 5, 777–794. [Google Scholar] [CrossRef]
- Aizarani, N.; Saviano, A.; Sagar; Mailly, L.; Durand, S.; Herman, J.S.; Pessaux, P.; Baumert, T.F.; Grun, D. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 2019, 572, 199–204. [Google Scholar] [CrossRef]
- Sasse, D.; Spornitz, U.M.; Maly, I.P. Liver architecture. Enzyme 1992, 46, 8–32. [Google Scholar] [CrossRef]
- Vernon, H.; Wehrle, C.J.; Alia, V.S.K.; Kasi, A.; Abdomen, A.; Liver, P. Study Guide Book Chapter. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Pleguezuelos-Manzano, C.; Puschhof, J.; van den Brink, S.; Geurts, V.; Beumer, J.; Clevers, H. Establishment and Culture of Human Intestinal Organoids Derived from Adult Stem Cells. Curr. Protoc. Immunol. 2020, 130, e106. [Google Scholar] [CrossRef]
- Mun, S.J.; Ryu, J.S.; Lee, M.O.; Son, Y.S.; Oh, S.J.; Cho, H.S.; Son, M.Y.; Kim, D.S.; Kim, S.J.; Yoo, H.J.; et al. Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J. Hepatol. 2019, 71, 970–985. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zhao, S.; Yu, F.; Rong, Z.; Lin, Y.; Chen, Y. Generation of human embryonic stem cell-derived lung organoids. STAR Protoc. 2022, 3, 101270. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Wu, D.; Ren, Y.; Huang, Y.; Feng, B.; Zhao, N.; Zhang, T.; Chen, X.; Chen, S.; Xu, A. Generation of hepatobiliary organoids from human induced pluripotent stem cells. J. Hepatol. 2019, 70, 1145–1158. [Google Scholar] [CrossRef]
- Lee, J.; Böscke, R.; Tang, P.C.; Hartman, B.H.; Heller, S.; Koehler, K.R. Hair Follicle Development in Mouse Pluripotent Stem Cell-Derived Skin Organoids. Cell Rep. 2018, 22, 242–254. [Google Scholar] [CrossRef]
- Frenz-Wiessner, S.; Fairley, S.D.; Buser, M.; Goek, I.; Salewskij, K.; Jonsson, G.; Illig, D.; Zu Putlitz, B.; Petersheim, D.; Li, Y.; et al. Generation of complex bone marrow organoids from human induced pluripotent stem cells. Nat. Methods 2024, 21, 868–881. [Google Scholar] [CrossRef]
- Golchin, A.; Chatziparasidou, A.; Ranjbarvan, P.; Niknam, Z.; Ardeshirylajimi, A. Embryonic Stem Cells in Clinical Trials: Current Overview of Developments and Challenges. Adv. Exp. Med. Biol. 2021, 1312, 19–37. [Google Scholar] [CrossRef]
- Tian, Z.; Yu, T.; Liu, J.; Wang, T.; Higuchi, A. Introduction to stem cells. Prog. Mol. Biol. Transl. Sci. 2023, 199, 3–32. [Google Scholar] [CrossRef]
- Clevers, H.; Watt, F.M. Defining Adult Stem Cells by Function, not by Phenotype. Annu. Rev. Biochem. 2018, 87, 1015–1027. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Cerneckis, J.; Cai, H.; Shi, Y. Induced pluripotent stem cells (iPSCs): Molecular mechanisms of induction and applications. Signal Transduct. Target. Ther. 2024, 9, 112. [Google Scholar] [CrossRef]
- Qin, D.; Liu, S.; Lu, Y.; Yan, Y.; Zhang, J.; Cao, S.; Chen, M.; Chen, N.; Huang, W.; Wang, L.; et al. Lgr5 (+) cell fate regulation by coordination of metabolic nuclear receptors during liver repair. Theranostics 2022, 12, 6130–6142. [Google Scholar] [CrossRef] [PubMed]
- Huch, M.; Dorrell, C.; Boj, S.F.; van Es, J.H.; Li, V.S.; van de Wetering, M.; Sato, T.; Hamer, K.; Sasaki, N.; Finegold, M.J.; et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 2013, 494, 247–250. [Google Scholar] [CrossRef]
- Huch, M.; Gehart, H.; van Boxtel, R.; Hamer, K.; Blokzijl, F.; Verstegen, M.M.; Ellis, E.; van Wenum, M.; Fuchs, S.A.; de Ligt, J.; et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015, 160, 299–312. [Google Scholar] [CrossRef]
- Frank, E.; Cailleret, M.; Nelep, C.; Fragner, P.; Polentes, J.; Herardot, E.; El Kassar, L.; Giraud-Triboult, K.; Monville, C.; Ben M’Barek, K. Semi-automated optimized method to isolate CRISPR/Cas9 edited human pluripotent stem cell clones. Stem. Cell Res. Ther. 2023, 14, 110. [Google Scholar] [CrossRef]
- Wang, J.; Singh, M.; Sun, C.; Besser, D.; Prigione, A.; Ivics, Z.; Hurst, L.D.; Izsvak, Z. Isolation and cultivation of naive-like human pluripotent stem cells based on HERVH expression. Nat. Protoc. 2016, 11, 327–346. [Google Scholar] [CrossRef]
- Teo, A.K.; Valdez, I.A.; Dirice, E.; Kulkarni, R.N. Comparable generation of activin-induced definitive endoderm via additive Wnt or BMP signaling in absence of serum. Stem Cell Rep. 2014, 3, 5–14. [Google Scholar] [CrossRef]
- Diekmann, U.; Wolling, H.; Dettmer, R.; Niwolik, I.; Naujok, O.; Buettner, F.F.R. Chemically defined and xenogeneic-free differentiation of human pluripotent stem cells into definitive endoderm in 3D culture. Sci. Rep. 2019, 9, 996. [Google Scholar] [CrossRef]
- Twaroski, K.; Mallanna, S.K.; Jing, R.; DiFurio, F.; Urick, A.; Duncan, S.A. FGF2 mediates hepatic progenitor cell formation during human pluripotent stem cell differentiation by inducing the WNT antagonist NKD1. Genes Dev. 2015, 29, 2463–2474. [Google Scholar] [CrossRef]
- Kamiya, A.; Kinoshita, T.; Miyajima, A. Oncostatin M and hepatocyte growth factor induce hepatic maturation via distinct signaling pathways. FEBS Lett. 2001, 492, 90–94. [Google Scholar] [CrossRef]
- Yang, X.; Shao, C.; Duan, L.; Hou, X.; Huang, Y.; Gao, L.; Zong, C.; Liu, W.; Jiang, J.; Ye, F.; et al. Oncostatin M promotes hepatic progenitor cell activation and hepatocarcinogenesis via macrophage-derived tumor necrosis factor-alpha. Cancer Lett. 2021, 517, 46–54. [Google Scholar] [CrossRef]
- Ma, Y.; Hu, L.; Tang, J.; Guo, W.; Feng, Y.; Liu, Y.; Tang, F. Three-Dimensional Cell Co-Culture Liver Models and Their Applications in Pharmaceutical Research. Int. J. Mol. Sci. 2023, 24, 6248. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, J.T.; Zhang, W.; Atlas, S.I.; Bryan, R.A.; Inman, S.W.; Vukasinovic, J. A 3D Cell Culture Organ-on-a-Chip Platform With a Breathable Hemoglobin Analogue Augments and Extends Primary Human Hepatocyte Functions in vitro. Front. Mol. Biosci. 2020, 7, 568777. [Google Scholar] [CrossRef] [PubMed]
- Habanjar, O.; Diab-Assaf, M.; Caldefie-Chezet, F.; Delort, L. 3D Cell Culture Systems: Tumor Application, Advantages, and Disadvantages. Int. J. Mol. Sci. 2021, 22, 12200. [Google Scholar] [CrossRef]
- Yi, S.A.; Zhang, Y.; Rathnam, C.; Pongkulapa, T.; Lee, K.B. Bioengineering Approaches for the Advanced Organoid Research. Adv. Mater. 2021, 33, e2007949. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Koo, I.S.; Hwang, H.J.; Lee, D.W. In Vitro three-dimensional (3D) cell culture tools for spheroid and organoid models. SLAS Discov. 2023, 28, 119–137. [Google Scholar] [CrossRef]
- Decaens, C.; Durand, M.; Grosse, B.; Cassio, D. Which in vitro models could be best used to study hepatocyte polarity? Biol. Cell 2008, 100, 387–398. [Google Scholar] [CrossRef]
- Calabrese, D.; Roma, G.; Bergling, S.; Carbone, W.; Mele, V.; Nuciforo, S.; Fofana, I.; Campana, B.; Szkolnicka, D.; Hay, D.C.; et al. Liver biopsy derived induced pluripotent stem cells provide unlimited supply for the generation of hepatocyte-like cells. PLoS ONE 2019, 14, e0221762. [Google Scholar] [CrossRef]
- Diehl, D.L. Top tips regarding EUS-guided liver biopsy. Gastrointest. Endosc. 2022, 95, 368–371. [Google Scholar] [CrossRef]
- Castell, J.V.; Gomez-Lechon, M.J. Liver cell culture techniques. Methods Mol. Biol. 2009, 481, 35–46. [Google Scholar] [CrossRef]
- Moon, H.R.; Mun, S.J.; Kim, T.H.; Kim, H.; Kang, D.; Kim, S.; Shin, J.H.; Choi, D.; Ahn, S.J.; Son, M.J. Guidelines for Manufacturing and Application of Organoids: Liver. Int. J. Stem Cells 2024, 17, 120–129. [Google Scholar] [CrossRef]
- Akbari, S.; Arslan, N.; Senturk, S.; Erdal, E. Next-Generation Liver Medicine Using Organoid Models. Front. Cell Dev. Biol. 2019, 7, 345. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Li, C.; Gong, W. Toward reproducible tumor organoid culture: Focusing on primary liver cancer. Front. Immunol. 2024, 15, 1290504. [Google Scholar] [CrossRef]
- Sang, C.; Lin, J.; Ji, S.; Gao, Q. Progress, application and challenges of liver organoids. Clin. Cancer Bull. 2024, 3, 7. [Google Scholar] [CrossRef]
- Hu, Y.; Geng, Q.; Wang, L.; Wang, Y.; Huang, C.; Fan, Z.; Kong, D. Research progress and application of liver organoids for disease modeling and regenerative therapy. J. Mol. Med. 2024, 102, 859–874. [Google Scholar] [CrossRef]
- Broutier, L.; Mastrogiovanni, G.; Verstegen, M.M.; Francies, H.E.; Gavarro, L.M.; Bradshaw, C.R.; Allen, G.E.; Arnes-Benito, R.; Sidorova, O.; Gaspersz, M.P.; et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017, 23, 1424–1435. [Google Scholar] [CrossRef]
- Serra-Camprubi, Q.; Verdaguer, H.; Oliveros, W.; Lupion-Garcia, N.; Llop-Guevara, A.; Molina, C.; Vila-Casadesus, M.; Turpin, A.; Neuzillet, C.; Frigola, J.; et al. Human Metastatic Cholangiocarcinoma Patient-Derived Xenografts and Tumoroids for Preclinical Drug Evaluation. Clin. Cancer Res. 2023, 29, 432–445. [Google Scholar] [CrossRef]
- Ozkan, A.; Stolley, D.L.; Cressman, E.N.K.; McMillin, M.; Yankeelov, T.E.; Rylander, M.N. Vascularized Hepatocellular Carcinoma on a Chip to Control Chemoresistance through Cirrhosis, Inflammation and Metabolic Activity. Small Struct. 2023, 4, 2200403. [Google Scholar] [CrossRef]
- Kopczynski, M.; Rumienczyk, I.; Kulecka, M.; Statkiewicz, M.; Pysniak, K.; Sandowska-Markiewicz, Z.; Wojcik-Trechcinska, U.; Goryca, K.; Pyziak, K.; Majewska, E.; et al. Selective Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibition by the SCH772984 Compound Attenuates In Vitro and In Vivo Inflammatory Responses and Prolongs Survival in Murine Sepsis Models. Int. J. Mol. Sci. 2021, 22, 10204. [Google Scholar] [CrossRef]
- Ewart, L.; Apostolou, A.; Briggs, S.A.; Carman, C.V.; Chaff, J.T.; Heng, A.R.; Jadalannagari, S.; Janardhanan, J.; Jang, K.J.; Joshipura, S.R.; et al. Performance assessment and economic analysis of a human Liver-Chip for predictive toxicology. Commun. Med. 2022, 2, 154. [Google Scholar] [CrossRef]
- Liu, J.; Feng, C.; Zhang, M.; Song, F.; Liu, H. Design and Fabrication of a Liver-on-a-chip Reconstructing Tissue-tissue Interfaces. Front. Oncol. 2022, 12, 959299. [Google Scholar] [CrossRef]
- Gough, A.; Soto-Gutierrez, A.; Vernetti, L.; Ebrahimkhani, M.R.; Stern, A.M.; Taylor, D.L. Human biomimetic liver microphysiology systems in drug development and precision medicine. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 252–268. [Google Scholar] [CrossRef] [PubMed]
- Ronaldson-Bouchard, K.; Teles, D.; Yeager, K.; Tavakol, D.N.; Zhao, Y.; Chramiec, A.; Tagore, S.; Summers, M.; Stylianos, S.; Tamargo, M.; et al. A multi-organ chip with matured tissue niches linked by vascular flow. Nat. Biomed. Eng. 2022, 6, 351–371. [Google Scholar] [CrossRef]
- Kennedy, J.I.; Davies, S.P.; Hewett, P.W.; Wilkinson, A.L.; Oo, Y.H.; Lu, W.Y.; El Haj, A.J.; Shetty, S. Organ-on-a-chip for studying immune cell adhesion to liver sinusoidal endothelial cells: The potential for testing immunotherapies and cell therapy trafficking. Front. Cell Dev. Biol. 2024, 12, 1359451. [Google Scholar] [CrossRef]
- Chhabra, A.; Song, H.G.; Grzelak, K.A.; Polacheck, W.J.; Fleming, H.E.; Chen, C.S.; Bhatia, S.N. A vascularized model of the human liver mimics regenerative responses. Proc. Natl. Acad. Sci. USA 2022, 119, e2115867119. [Google Scholar] [CrossRef]
- McCarty, W.J.; Usta, O.B.; Yarmush, M.L. A Microfabricated Platform for Generating Physiologically-Relevant Hepatocyte Zonation. Sci. Rep. 2016, 6, 26868. [Google Scholar] [CrossRef]
- Deguchi, S.; Takayama, K. State-of-the-art liver disease research using liver-on-a-chip. Inflamm. Regen. 2022, 42, 62. [Google Scholar] [CrossRef]
- Li, X.; George, S.M.; Vernetti, L.; Gough, A.H.; Taylor, D.L. A glass-based, continuously zonated and vascularized human liver acinus microphysiological system (vLAMPS) designed for experimental modeling of diseases and ADME/TOX. Lab Chip 2018, 18, 2614–2631. [Google Scholar] [CrossRef]
- Park, S.E.; Georgescu, A.; Huh, D. Organoids-on-a-chip. Science 2019, 364, 960–965. [Google Scholar] [CrossRef]
- Shao, W.; Xu, H.; Zeng, K.; Ye, M.; Pei, R.; Wang, K. Advances in liver organoids: Replicating hepatic complexity for toxicity assessment and disease modeling. Stem Cell Res. Ther. 2025, 16, 27. [Google Scholar] [CrossRef] [PubMed]
- Persaud, A.; Maus, A.; Strait, L.; Zhu, D. 3D Bioprinting with Live Cells. Eng. Regen. 2022, 3, 292–309. [Google Scholar] [CrossRef]
- Ma, X.; Qu, X.; Zhu, W.; Li, Y.S.; Yuan, S.; Zhang, H.; Liu, J.; Wang, P.; Lai, C.S.; Zanella, F.; et al. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc. Natl. Acad. Sci. USA 2016, 113, 2206–2211. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Zhang, Z.; Wang, X. The Prospect of Hepatic Decellularized Extracellular Matrix as a Bioink for Liver 3D Bioprinting. Biomolecules 2024, 14, 1019. [Google Scholar] [CrossRef]
- Zhang, X.; Karim, M.; Hasan, M.M.; Hooper, J.; Wahab, R.; Roy, S.; Al-Hilal, T.A. Cancer-on-a-Chip: Models for Studying Metastasis. Cancers 2022, 14, 648. [Google Scholar] [CrossRef]
- Fang, L.; Liu, Y.; Qiu, J.; Wan, W. Bioprinting and its Use in Tumor-On-A-Chip Technology for Cancer Drug Screening: A Review. Int. J. Bioprint. 2022, 8, 603. [Google Scholar] [CrossRef]
- Chen, L.; Wei, X.; Gu, D.; Xu, Y.; Zhou, H. Human liver cancer organoids: Biological applications, current challenges, and prospects in hepatoma therapy. Cancer Lett. 2023, 555, 216048. [Google Scholar] [CrossRef]
- Yang, H.; Cheng, J.; Zhuang, H.; Xu, H.; Wang, Y.; Zhang, T.; Yang, Y.; Qian, H.; Lu, Y.; Han, F.; et al. Pharmacogenomic profiling of intra-tumor heterogeneity using a large organoid biobank of liver cancer. Cancer Cell 2024, 42, 535–551.e8. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Chen, S.; Zhang, L.; Rao, J.; Lu, X.; Ma, Y. Liver organoids: A promising three-dimensional model for insights and innovations in tumor progression and precision medicine of liver cancer. Front. Immunol. 2023, 14, 1180184. [Google Scholar] [CrossRef]
- Kacar, Z.; Slud, E.; Levy, D.; Candia, J.; Budhu, A.; Forgues, M.; Wu, X.; Raziuddin, A.; Tran, B.; Shetty, J.; et al. Characterization of tumor evolution by functional clonality and phylogenetics in hepatocellular carcinoma. Commun. Biol. 2024, 7, 383. [Google Scholar] [CrossRef]
- Bresnahan, E.; Ramadori, P.; Heikenwalder, M.; Zender, L.; Lujambio, A. Novel patient-derived preclinical models of liver cancer. J. Hepatol. 2020, 72, 239–249. [Google Scholar] [CrossRef]
- Geurts, M.H.; Clevers, H. CRISPR engineering in organoids for gene repair and disease modelling. Nat. Rev. Bioeng. 2023, 1, 32–45. [Google Scholar] [CrossRef]
- Liang, J.; Wei, J.; Cao, J.; Qian, J.; Gao, R.; Li, X.; Wang, D.; Gu, Y.; Dong, L.; Yu, J.; et al. In-organoid single-cell CRISPR screening reveals determinants of hepatocyte differentiation and maturation. Genome Biol. 2023, 24, 251. [Google Scholar] [CrossRef] [PubMed]
- Driehuis, E.; Clevers, H. CRISPR/Cas 9 genome editing and its applications in organoids. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G257–G265. [Google Scholar] [CrossRef] [PubMed]
- Van Norman, G.A. Limitations of Animal Studies for Predicting Toxicity in Clinical Trials: Part 2: Potential Alternatives to the Use of Animals in Preclinical Trials. JACC Basic Transl. Sci. 2020, 5, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.J.; Otieno, M.A.; Ronxhi, J.; Lim, H.K.; Ewart, L.; Kodella, K.R.; Petropolis, D.B.; Kulkarni, G.; Rubins, J.E.; Conegliano, D.; et al. Reproducing human and cross-species drug toxicities using a Liver-Chip. Sci. Transl. Med. 2019, 11, 517. [Google Scholar] [CrossRef]
- Yu, Y.; Sun, B.; Ye, X.; Wang, Y.; Zhao, M.; Song, J.; Geng, X.; Marx, U.; Li, B.; Zhou, X. Hepatotoxic assessment in a microphysiological system: Simulation of the drug absorption and toxic process after an overdosed acetaminophen on intestinal-liver-on-chip. Food Chem. Toxicol. 2024, 193, 115016. [Google Scholar] [CrossRef]
- Jiao, D.; Xie, L.; Xing, W. A pumpless liver-on-a-chip for drug hepatotoxicity analysis. Analyst 2024, 149, 4675–4686. [Google Scholar] [CrossRef]
- Deng, Q.; Yang, Y.; Liu, Y.; Zou, M.; Huang, G.; Yang, S.; Li, L.; Qu, Y.; Luo, Y.; Zhang, X. Assessing immune hepatotoxicity of troglitazone with a versatile liver-immune-microphysiological-system. Front. Pharmacol. 2024, 15, 1335836. [Google Scholar] [CrossRef]
- Shinozawa, T.; Kimura, M.; Cai, Y.; Saiki, N.; Yoneyama, Y.; Ouchi, R.; Koike, H.; Maezawa, M.; Zhang, R.R.; Dunn, A.; et al. High-Fidelity Drug-Induced Liver Injury Screen Using Human Pluripotent Stem Cell-Derived Organoids. Gastroenterology 2021, 160, 831–846.e10. [Google Scholar] [CrossRef]
- Segovia-Zafra, A.; Di Zeo-Sanchez, D.E.; Lopez-Gomez, C.; Perez-Valdes, Z.; Garcia-Fuentes, E.; Andrade, R.J.; Lucena, M.I.; Villanueva-Paz, M. Preclinical models of idiosyncratic drug-induced liver injury (iDILI): Moving towards prediction. Acta Pharm. Sin. B 2021, 11, 3685–3726. [Google Scholar] [CrossRef]
- Tanataweethum, N.; Trang, A.; Lee, C.; Mehta, J.; Patel, N.; Cohen, R.N.; Bhushan, A. Investigation of insulin resistance through a multiorgan microfluidic organ-on-chip. Biomed. Mater. 2022, 17, 025002. [Google Scholar] [CrossRef]
- Whiteford, J.; Arokiasamy, S.; Thompson, C.L.; Dufton, N.P. Novel application of live imaging to determine the functional cell biology of endothelial-to-mesenchymal transition (EndMT) within a liver-on-a-chip platform. Vitr. Model 2022, 1, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zheng, X.; Xie, M.; Lin, Z.; Du, F.; Shi, X.; Li, R. Mice Hepatic Organoids for Modeling Nonalcoholic Fatty Liver Disease and Drug Response. Stem Cells Dev. 2024, 33, 387–398. [Google Scholar] [CrossRef]
- Nuciforo, S.; Heim, M.H. Organoids to model liver disease. JHEP Rep. 2021, 3, 100198. [Google Scholar] [CrossRef]
- Ramli, M.N.B.; Lim, Y.S.; Koe, C.T.; Demircioglu, D.; Tng, W.; Gonzales, K.A.U.; Tan, C.P.; Szczerbinska, I.; Liang, H.; Soe, E.L.; et al. Human Pluripotent Stem Cell-Derived Organoids as Models of Liver Disease. Gastroenterology 2020, 159, 1471–1486.e12. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, W.; Zhao, B. Human liver organoid: Modeling liver steatosis and beyond. Cell Regen. 2023, 12, 17. [Google Scholar] [CrossRef]
- Huang, R.; Zhang, X.; Gracia-Sancho, J.; Xie, W.F. Liver regeneration: Cellular origin and molecular mechanisms. Liver Int. 2022, 42, 1486–1495. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, S.; Fu, J.; Chen, Y.; Xu, J.; Wei, W.; Song, H.; Zhao, X.; Wang, H. Liver regeneration after injury: Mechanisms, cellular interactions and therapeutic innovations. Clin. Transl. Med. 2024, 14, e1812. [Google Scholar] [CrossRef]
- Reza, H.A.; Okabe, R.; Takebe, T. Organoid transplant approaches for the liver. Transpl. Int. 2021, 34, 2031–2045. [Google Scholar] [CrossRef]
- Tadokoro, T.; Murata, S.; Kato, M.; Ueno, Y.; Tsuchida, T.; Okumura, A.; Kuse, Y.; Konno, T.; Uchida, Y.; Yamakawa, Y.; et al. Human iPSC-liver organoid transplantation reduces fibrosis through immunomodulation. Sci. Transl. Med. 2024, 16, eadg0338. [Google Scholar] [CrossRef]
- Peng, W.C.; Kraaier, L.J.; Kluiver, T.A. Hepatocyte organoids and cell transplantation: What the future holds. Exp. Mol. Med. 2021, 53, 1512–1528. [Google Scholar] [CrossRef]
- Mehta, V.; Karnam, G.; Madgula, V. Liver-on-chips for drug discovery and development. Mater. Today Bio 2024, 27, 101143. [Google Scholar] [CrossRef] [PubMed]
- Berger, D.R.; Ware, B.R.; Davidson, M.D.; Allsup, S.R.; Khetani, S.R. Enhancing the functional maturity of induced pluripotent stem cell-derived human hepatocytes by controlled presentation of cell-cell interactions in vitro. Hepatology 2015, 61, 1370–1381. [Google Scholar] [CrossRef] [PubMed]
- Bonanini, F.; Kurek, D.; Previdi, S.; Nicolas, A.; Hendriks, D.; de Ruiter, S.; Meyer, M.; Clapes Cabrer, M.; Dinkelberg, R.; Garcia, S.B.; et al. In vitro grafting of hepatic spheroids and organoids on a microfluidic vascular bed. Angiogenesis 2022, 25, 455–470. [Google Scholar] [CrossRef]
- Hu, H.; Gehart, H.; Artegiani, B.; C, L.O.I.; Dekkers, F.; Basak, O.; van Es, J.; Chuva de Sousa Lopes, S.M.; Begthel, H.; Korving, J.; et al. Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell 2018, 175, 1591–1606.e19. [Google Scholar] [CrossRef]
- Olgasi, C.; Cucci, A.; Follenzi, A. iPSC-Derived Liver Organoids: A Journey from Drug Screening, to Disease Modeling, Arriving to Regenerative Medicine. Int. J. Mol. Sci. 2020, 21, 6215. [Google Scholar] [CrossRef]
- Lim, A.Y.; Kato, Y.; Sakolish, C.; Valdiviezo, A.; Han, G.; Bajaj, P.; Stanko, J.; Ferguson, S.S.; Villenave, R.; Hewitt, P.; et al. Reproducibility and Robustness of a Liver Microphysiological System PhysioMimix LC12 under Varying Culture Conditions and Cell Type Combinations. Bioengineering 2023, 10, 1195. [Google Scholar] [CrossRef]
- Bircsak, K.M.; DeBiasio, R.; Miedel, M.; Alsebahi, A.; Reddinger, R.; Saleh, A.; Shun, T.; Vernetti, L.A.; Gough, A. A 3D microfluidic liver model for high throughput compound toxicity screening in the OrganoPlate(R). Toxicology 2021, 450, 152667. [Google Scholar] [CrossRef]
- Jiang, F.; Zhou, C.; Li, Y.; Deng, H.; Gong, T.; Chen, J.; Chen, T.; Yang, J.; Zhu, P. Metabolic engineering of yeasts for green and sustainable production of bioactive ginsenosides F2 and 3beta,20S-Di-O-Glc-DM. Acta Pharm. Sin. B 2022, 12, 3167–3176. [Google Scholar] [CrossRef]
- Tamargo-Rubio, I.; Simpson, A.B.; Hoogerland, J.A.; Fu, J. Human induced pluripotent stem cell-derived liver-on-a-chip for studying drug metabolism: The challenge of the cytochrome P450 family. Front. Pharmacol. 2023, 14, 1223108. [Google Scholar] [CrossRef]
- Liu, M.; Xiang, Y.; Yang, Y.; Long, X.; Xiao, Z.; Nan, Y.; Jiang, Y.; Qiu, Y.; Huang, Q.; Ai, K. State-of-the-art advancements in Liver-on-a-chip (LOC): Integrated biosensors for LOC. Biosens. Bioelectron. 2022, 218, 114758. [Google Scholar] [CrossRef]
- Bavli, D.; Prill, S.; Ezra, E.; Levy, G.; Cohen, M.; Vinken, M.; Vanfleteren, J.; Jaeger, M.; Nahmias, Y. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 2016, 113, E2231–E2240. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, N.; Wills, L.; Akers, K.; Su, Y.; Niccum, P.; Murali, T.M.; Rajagopalan, P. Comparative transcriptomic and phenotypic analysis of induced pluripotent stem cell hepatocyte-like cells and primary human hepatocytes. Cell Tissue Res. 2024, 396, 119–139. [Google Scholar] [CrossRef]
- Damiati, S.; Kompella, U.B.; Damiati, S.A.; Kodzius, R. Microfluidic Devices for Drug Delivery Systems and Drug Screening. Genes 2018, 9, 103. [Google Scholar] [CrossRef]
- Luo, Y.; Li, X.; Zhao, Y.; Zhong, W.; Xing, M.; Lyu, G. Development of Organs-on-Chips and Their Impact on Precision Medicine and Advanced System Simulation. Pharmaceutics 2023, 15, 2094. [Google Scholar] [CrossRef]
- Picollet-D’hahan, N.; Zuchowska, A.; Lemeunier, I.; Le Gac, S. Multiorgan-on-a-Chip: A Systemic Approach To Model and Decipher Inter-Organ Communication. Trends Biotechnol. 2021, 39, 788–810. [Google Scholar] [CrossRef]
- Wu, X.; Shi, W.; Liu, X.; Gu, Z. Recent advances in 3D-printing-based organ-on-a-chip. Deleted J. 2024, 1, 100003. [Google Scholar] [CrossRef]
- Ingber, D.E. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat. Rev. Genet. 2022, 23, 467–491. [Google Scholar] [CrossRef]
- Dai, M.; Xiao, G.; Shao, M.; Zhang, Y.S. The Synergy between Deep Learning and Organs-on-Chips for High-Throughput Drug Screening: A Review. Biosensors 2023, 13, 389. [Google Scholar] [CrossRef]
- Novosel, E.C.; Kleinhans, C.; Kluger, P.J. Vascularization is the key challenge in tissue engineering. Adv. Drug Deliv. Rev. 2011, 63, 300–311. [Google Scholar] [CrossRef]
- Azizipour, N.; Avazpour, R.; Rosenzweig, D.H.; Sawan, M.; Ajji, A. Evolution of Biochip Technology: A Review from Lab-on-a-Chip to Organ-on-a-Chip. Micromachines 2020, 11, 599. [Google Scholar] [CrossRef]

| Aspect | Source of Culture Cells | |
|---|---|---|
| hPSCs | Biopsy Sample | |
| Cell Source | Derived from human pluripotent cells (hPSCs), including ESCs and iPSCs [71] | Derived from PLCs obtained from biopsy samples [68] |
| Availability | Unlimited supply as hPSCs can be expanded indefinitely [71,72] | Limited supply due to the small size of biopsy samples, which restricts the number of cells available for culture [73] |
| Differentiation Process | Requires specific differentiation protocol to mimic liver development stages, involving growth factors and specific media [71] | Involves spontaneous outgrowth of PLCs from biopsy tissue which may not allow for selective cell type enrichment [68] |
| Time to Generate Organoids | Longer, as it involves several stages of differentiation and maturation (weeks) | Shorter, as liver cells are already committed to a liver lineage (days to a few weeks) |
| Growth Factors Required | Sequential use of Activin A, FGF2, BMP4, HGF, and oncostatin M to drive differentiation | Limited growth factors needed; typically used factors like HGF and Wnt for cell expansion and maturation |
| Culture Conditions | Maintained in a defined growth medium supplemented with various growth factors and hormones [71] | Cultures in a medium that supports the outgrowth of PLCs, typically using a reduced growth factor matrix |
| Cell Composition | Can be engineered to reflect specific cell types and functions, achieving a more controlled composition [71,72] | May not fully recapitulate the diverse cell types present in the original tissue due to outgrowth limitations [68] |
| Complexity of Organoids | Incorporate hepatocytes and non-parenchymal cells but may need additional co-culture systems for complete cell representation | Naturally contain all liver cell types, including non-parenchymal cells, leading to higher initial complexity |
| Functional Assessment | Potentially better at mimicking liver function due to controlled differentiation and composition | May exhibit variable liver functions depending on the success of PLC outgrowth and the inherent characteristics of the biopsy sample |
| Applications | Primarily used for drug screening, disease modeling, and regenerative medicine due to their versatility | Primarily utilized for studying liver pathologies and personalized medicine, but with limitations in functional modeling |
| Ethical Considerations | Ethical concerns may arise with the use of ESCs; iPSC generation avoids many of these issues | Minimal ethical concerns, as the organoids are derived from patient biopsy tissue |
| Challenges | Requires careful optimization of differentiation protocols and culture conditions to maintain functionality | Faces challenges such as low cell yield, genetic heterogeneity, and difficulty in maintaining the native microenvironment |
| Research Group/Model | Cell Types Included | Key Features |
|---|---|---|
| Ingber Lab (Wyss Institute) [26] | Human Hepatocytes, Endothelial Cells, Kupffer Cells, Stellate Cells | Derived from PLCs Microfluidic channels; species-specific toxicity modeling |
| University of Pittsburgh [82] | Hepatocytes, Stellate Cells, Kupffer Cells, En-dothelial Cells | Self-assembling plate-like structures; fluo-rescent biosensors |
| Columbia University [83] | iPSC-Derived Hepatocytes, Endothelial Cells | 3D biomaterial environment; integration with other tissue types |
| University of Birmingham [84] | Liver Blood Vessel Cells, Immune Cells | Real-time tracking of immune cell behavior; immunotherapy focus |
| MIT and Boston University [85] | Primary Hepatocytes, Endothelial Cells | Multi-organ platform for integrated drug screening |
| McCarty et al. [86] | Primary Human/Rat Hepatocytes | Gradient generator for zonal metabolic studies |
| Tri-Vascular Liver-on-a-Chip (TVLOC) [81] | Hepatocytes, HSCs, LSECs, KCs | Trivascular system; substance concentration gradient; PMMA microchannels |
| SQL-SAL [87] | Hepatocytes, Endothelial Cells, KCs, HSCs | Sequentially layered self-assembly liver model. |
| vLAMPS [88] | Primary Hepatocytes, LSECs, HSCs, KCs | Oxygen gradient replication, functional acinar modeling |
| Company | Platform Name | Key Features | Regulatory Partnerships | Translational Highlights |
|---|---|---|---|---|
| Emulate (Boston, MA, USA) | Human Emulation System™ (Liver-Chip S1) | PDMS-based microfluidic chip with co-culture of hepatocytes and non-parenchymal cells | (1) CRADA with FDA for toxicology research; (2) accepted into FDA’s ISTAND pilot program for DILI prediction | (1) Partnered with Janssen and Takeda; (2) demonstrated >85% DILI prediction accuracy |
| CN Bio (Cambridge, UK) | PhysioMimix® Liver MPS | (1) Perfused 3D liver model with open-well design; (2) compatible with imaging and long-term culture | (1) Extended collaboration with FDA’s CDER; (2) used in FDA preclinical workflows | (1) Supported IND-enabling studies for INI-822 (NASH therapeutic); (2) data used in regulatory documentation |
| Mimetas (Leiden, The Netherlands) | OrganoPlate® | 96-well microfluidic platform without pumps; ideal for high-throughput screening | Collaboration with HUB for disease model development | (1) Widely adopted in pharmaceutical R&D; (2) used in toxicology and fibrosis modeling |
| Organovo (San Diego, CA, USA) | ExVive™ Human Liver Tissue | Bioprinted 3D liver constructs from primary human cells | No formal regulatory partnerships reported | Applied in preclinical DILI and fibrosis studies |
| InSphero (Schlieren, Switzerland) | 3D InSight™ Human Liver Microtissues | Multicellular 3D spheroids in hanging-drop or plate format | No formal regulatory partnerships reported | Used extensively in drug discovery pipelines |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mugaanyi, J.; Huang, J.; Fang, J.; Musinguzi, A.; Lu, C.; Chen, Z. Developments and Applications of Liver-on-a-Chip Technology—Current Status and Future Prospects. Biomedicines 2025, 13, 1272. https://doi.org/10.3390/biomedicines13061272
Mugaanyi J, Huang J, Fang J, Musinguzi A, Lu C, Chen Z. Developments and Applications of Liver-on-a-Chip Technology—Current Status and Future Prospects. Biomedicines. 2025; 13(6):1272. https://doi.org/10.3390/biomedicines13061272
Chicago/Turabian StyleMugaanyi, Joseph, Jing Huang, Jiongze Fang, Arthur Musinguzi, Caide Lu, and Zaozao Chen. 2025. "Developments and Applications of Liver-on-a-Chip Technology—Current Status and Future Prospects" Biomedicines 13, no. 6: 1272. https://doi.org/10.3390/biomedicines13061272
APA StyleMugaanyi, J., Huang, J., Fang, J., Musinguzi, A., Lu, C., & Chen, Z. (2025). Developments and Applications of Liver-on-a-Chip Technology—Current Status and Future Prospects. Biomedicines, 13(6), 1272. https://doi.org/10.3390/biomedicines13061272






