MicroRNA Signatures in Endometrial Receptivity—Unlocking Their Role in Embryo Implantation and IVF Success: A Systematic Review
Abstract
1. Introduction
1.1. MicroRNAs: Biogenesis, Function, and Mechanistic Roles
- MiRNAs control the development of stromal fibroblasts into decidual cells, which produce prolactin and IGFBP1. For example, miR-21-5p, miR-193b-3p, and miR-17-5p have been linked to regulating endoplasmic reticulum stress (ERS) and the unfolded protein response (UPR) during decidualization. Disruption in these stress-adaptation mechanisms may cause poor implantation and inflammations [6].
- Immune modulation: The maternal immune system must accept the semi-allogeneic embryo. MiRNAs like miR-146a, miR-125b, and miR-124-3p impact the expression of cytokines, interleukins, and immunological checkpoint molecules, including leukemia inhibitory factor (LIF), IL-11, and SOCS1, influencing the Th1/Th2 cytokine balance, macrophage polarization, and T-regulatory cell recruitment [5,6].
- Angiogenesis and vascular remodeling: The endometrial bed must be extensively vascularized before embryo implantation can occur. RIF patients exhibit variable expression of miRNAs (miR-27a, miR-20a, and miR-126), which target angiogenic regulators such as VEGFA, HIF-1α, and FLT1 [11].
- miRNAs like miR-149 and miR-33a play a role in modulating the Wnt/β-catenin signaling pathway, which is crucial for implantation. These miRNAs control epithelial–mesenchymal transition (EMT), trophoblast invasion, and uterine receptivity by directly or indirectly targeting WNT3A and its downstream effectors [12].
- miR-146aC>G and miR-196a2T>C polymorphisms were shown to be strongly linked with increased RIF risk in Korean women, possibly because to differential maturation and expression of these miRNAs and their downstream inflammatory pathways [5].
- SNPs in miR-125a, miR-365b, and miR-149 have been linked to poor reproductive outcomes, including fetal growth limitation, due to inadequate regulation of Wnt/β-catenin signaling and angiogenesis pathways [12].
- For example, circ_0038383 was discovered to sponge miR-196b-5p, thereby upregulating HOXA9, a critical transcription factor for stromal cell development and embryo–maternal communications [6].
- LncRNAs H19 and NEAT1, which are abundant in mid-secretory endometrium, influence miR-29c, miR-20a, and other miRNAs involved in decidualization and immunological tolerance [13].
1.2. Molecular Pathways Governed by miRNAs in Endometrial Receptivity
1.3. Non-Coding RNA Crosstalk and ceRNA Networks in Endometrial Receptivity and Implant Failure
1.4. miRNAs as Biomarkers
1.5. Current Limitations and Future Directions
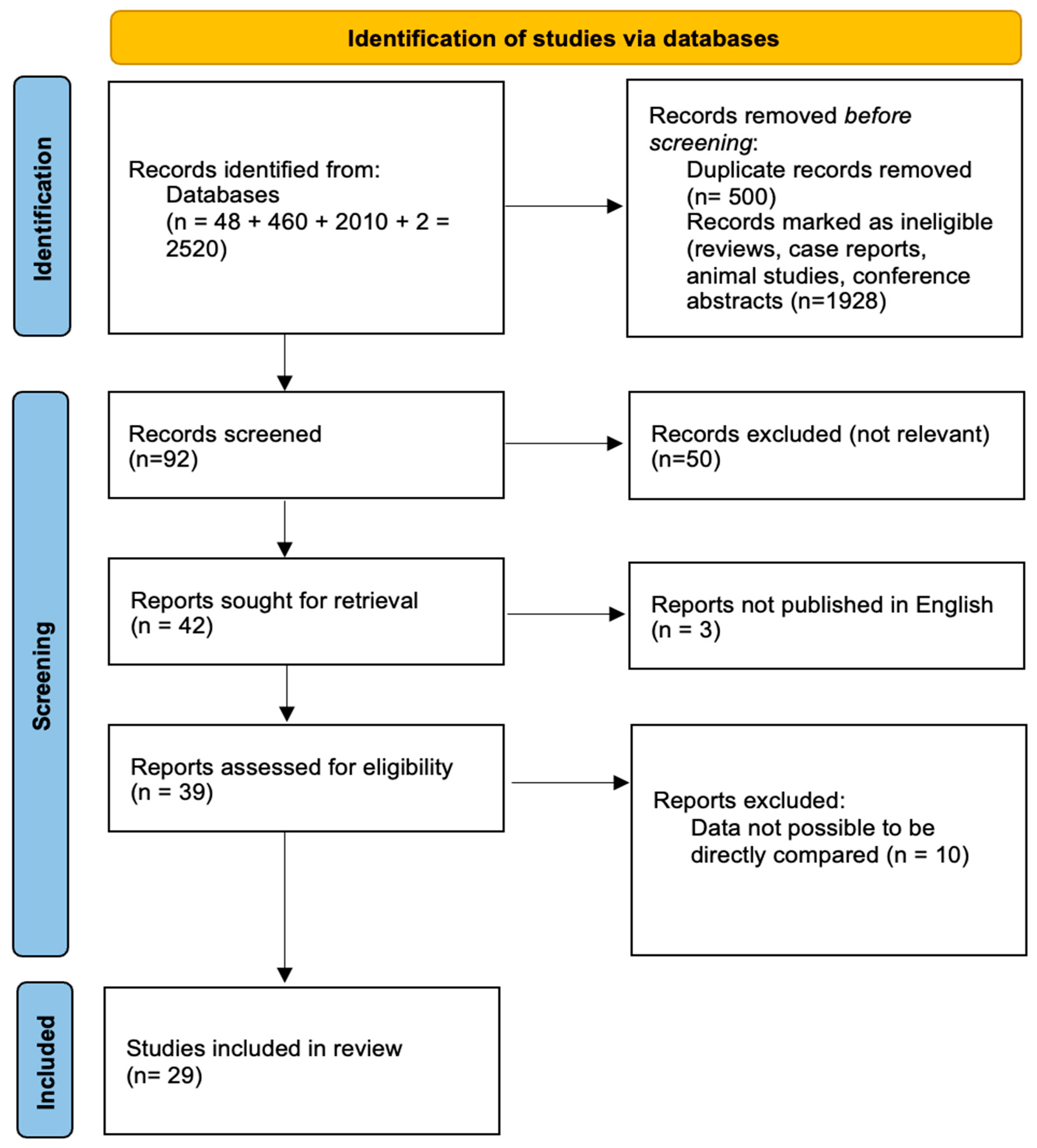
2. Materials and Methods
2.1. Search Strategy and Study Selection
2.2. Eligibility Criteria
- Study type: Only original, peer-reviewed research articles were considered. Prospective and retrospective cohort studies, case–control studies, cross-sectional studies, and randomized controlled trials (RCTs) were all considered valid designs. Basic research involving clinical human samples was also considered if the molecular endpoints were well defined. Review articles, editorials, conference abstracts, case reports, commentaries, and animal-only research were not included unless they contained human tissue validation.
- Population: Studies had to include human participants who were being evaluated or treated for infertility, in vitro fertilization (IVF), embryo transfer, RIF, or conditions known to affect endometrial receptivity. Studies on healthy endometrial controls were also considered if they were compared to non-receptive or infertile populations.
- Biological samples: Eligible research must disclose molecular analysis of human-derived samples, including the following:
- Endometrial tissue (biopsies and tissue explants);
- Uterine fluid: plasma or serum saliva;
- Embryo wasted its culture medium. These sample types were approved as long as they were collected during relevant times of the menstrual cycle (particularly the mid-secretory or luteal phase) during IVF preparation.
- Molecular biomarkers: Although miRNAs were the primary focus of the study, studies involving other non-coding RNAs, such as long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs), were also included if they provided functional or correlative evidence relevant to endometrial receptivity, WOI, implantation failure, or embryo–maternal interactions.
- Analytical methods: The studies had to use validated molecular techniques for RNA profiling and quantification, including but not limited to the following:
- Quantitative real-time polymerase chain reaction (qRT-PCR);
- Microarray hybridization;
- Next-generation sequencing (NGS);
- Bioinformatic pathway enrichment and target prediction (when combined with experimental validation).
- Outcomes: Studies needed to report on at least one of the following outcome domains to be included:
- miRNA or ncRNA expression in receptive and non-receptive endometrium;
- Diagnostic or predictive performance in terms of implantation outcomes;
- MiRNAs play a functional role in implantation-related signaling, including HOXA10, LIF-STAT3, and Wnt/β-catenin;
- Differences in miRNA expression between fertile and infertile populations;
- Association between miRNAs and IVF success, embryo transfer result, or RIF diagnosis
- Studies were excluded if they matched any of the following criteria:
- Non-human animal models were used exclusively, with no equivalent human validation;
- MiRNAs and other ncRNAs were not assessed, and no molecular data relevant to endometrial function were reported;
- Concentrated primarily on embryo development or oocyte competency, without looking at the endometrial or implantation environment;
- Sample-collection timing was unclear or there was no information on the cycle phase.
- Did not apply a proven molecular quantification approach, and there were no adequate control groups or normalization strategies;
- Were not published in English or were not available in full-text format.
2.3. Data Extraction
2.4. Quality Assessment and Risk of Bias
3. Results
Overview of Included Studies
4. Discussion
4.1. Molecular Disruption of Implantation Pathways
4.2. Clinical Relevance of Endometrial miRNA Signatures
4.3. The Central Role of HOX Genes and Decidualization Regulation
4.4. Systems Biology and Single-Cell Insights
4.5. Angiogenesis, Immune Crosstalk, and ceRNA Networks
4.6. Non-Invasive miRNA Biomarkers and Diagnostic Potential
4.7. Clinical Implications
5. Strengths and Limitations
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Diedrich, K.; Fauser, B.C.J.M.; Devroey, P.; Griesinger, G. The role of the endometrium and embryo in human implantation. Hum. Reprod. Update 2007, 13, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Pirtea, P.; De Ziegler, D.; Tao, X.; Sun, L.; Zhan, Y.; Ayoubi, J.M.; Seli, E.; Franasiak, J.M.; Scott, R.T. Rate of true recurrent implantation failure is low: Results of three successive frozen euploid single embryo transfers. Fertil. Steril. 2021, 115, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Opuchlik, K.; Pankiewicz, K.; Pierzyński, P.; Sierdziński, J.; Aleksejeva, E.; Salumets, A.; Issat, T.; Laudański, P. Factors influencing endometrial receptivity in women with recurrent implantation failure. BMC Women’s Health 2025, 25, 15. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Gao, W.; Li, D. Recurrent implantation failure: A comprehensive summary from etiology to treatment. Front. Endocrinol. 2023, 13, 1061766. [Google Scholar] [CrossRef]
- Cho, S.H.; Chung, K.W.; Kim, J.O.; Jang, H.; Yoo, J.K.; Choi, Y.; Ko, J.J.; Kim, J.H.; Nishi, Y.; Yanase, T.; et al. Association of miR-146aC>G, miR-149C>T, miR-196a2T>C, and miR-499A>G polymorphisms with risk of recurrent implantation failure in Korean women. Eur. J. Obstet. Gynecol. Reprod. Biol. 2016, 202, 14–19. [Google Scholar] [CrossRef]
- Soczewski, E.; Murrieta-Coxca, J.M.; Miranda, L.; Fuentes-Zacarías, P.; Gutiérrez-Samudio, R.; Grasso, E.; Marti, M.; PérezLeirós, C.; Morales-Prieto, D.; Markert, U.R.; et al. miRNAs associated with endoplasmic reticulum stress and unfolded protein response during decidualization. Reprod. Biomed. Online 2023, 47, 103289. [Google Scholar] [CrossRef]
- Dabi, Y.; Suisse, S.; Puchar, A.; Delbos, L.; Poilblanc, M.; Descamps, P.; Haury, J.; Golfier, F.; Jornea, L.; Bouteiller, D.; et al. Endometriosis-associated infertility diagnosis based on saliva microRNA signatures. Reprod. Biomed. Online 2023, 46, 138–149. [Google Scholar] [CrossRef]
- Bendifallah, S.; Dabi, Y.; Suisse, S.; Ilic, J.; Delbos, L.; Poilblanc, M.; Descamps, P.; Golfier, F.; Jornea, L.; Bouteiller, D.; et al. Saliva-based microRNA diagnostic signature for the superficial peritoneal endometriosis phenotype. Eur. J. Obstet. Gynecol. Reprod. Biol. 2024, 297, 187–196. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Guo, L.; Zhao, Y.; Yang, S.; Zhang, H.; Chen, F. Integrative Analysis of miRNA-mRNA and miRNA-miRNA Interactions. BioMed Res. Int. 2014, 2014, 907420. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Orekhov, A.N.; Bobryshev, Y.V. The role of miR-126 in embryonic angiogenesis, adult vascular homeostasis, and vascular repair and its alterations in atherosclerotic disease. J. Mol. Cell. Cardiol. 2016, 97, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Alset, D.; Butenko, E.V.; Pokudina, I.O.; Shkurat, T.P.; Kuznetsova, N.B.; Bushtyreva, I.O. Genetic variations of Wnt/β-catenin signaling pathway microRNA regulators as novel prenatal biomarkers of fetal growth restriction syndrome. Gene Rep. 2024, 35, 101914. [Google Scholar] [CrossRef]
- Begum, M.I.A.; Chuan, L.; Hong, S.-T.; Chae, H.-S. The Pathological Role of miRNAs in Endometriosis. Biomedicines 2023, 11, 3087. [Google Scholar] [CrossRef]
- Maenhoudt, N.; De Moor, A.; Vankelecom, H. Modeling Endometrium Biology and Disease. J. Pers. Med. 2022, 12, 1048. [Google Scholar] [CrossRef]
- Makieva, S.; Giacomini, E.; Ottolina, J.; Sanchez, A.M.; Papaleo, E.; Viganò, P. Inside the Endometrial Cell Signaling Subway: Mind the Gap(s). Int. J. Mol. Sci. 2018, 19, 2477. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef]
- Salmasi, S.; Heidar, M.S.; Khaksary Mahabady, M.; Rashidi, B.; Mirzaei, H. MicroRNAs, endometrial receptivity and molecular pathways. Reprod. Biol. Endocrinol. 2024, 22, 139. [Google Scholar] [CrossRef]
- Ashary, N.; Laheri, S.; Modi, D. Homeobox genes in endometrium: From development to decidualization. Int. J. Dev. Biol. 2020, 64, 227–237. [Google Scholar] [CrossRef]
- Bahmyari, S.; Jamali, Z.; Khatami, S.H.; Vakili, O.; Roozitalab, M.; Savardashtaki, A.; Solati, A.; Mousavi, P.; Shabaninejad, Z.; Vakili, S.; et al. microRNAs in female infertility: An overview. Cell Biochem. Funct. 2021, 39, 955–969. [Google Scholar] [CrossRef]
- Matsuyama, S.; Whiteside, S.; Li, S.-Y. Implantation and Decidualization in PCOS: Unraveling the Complexities of Pregnancy. Int. J. Mol. Sci. 2024, 25, 1203. [Google Scholar] [CrossRef]
- Peng, Y.; Guo, R.; Shi, B.; Li, D. The role of long non-coding RNA H19 in infertility. Cell Death Discov. 2023, 9, 268. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yan, J. Update of Wnt signaling in implantation and decidualization. Reprod. Med. Biol. 2016, 15, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Tabnak, P.; Masrouri, S.; Geraylow, K.R.; Zarei, M.; Esmailpoor, Z.H. Targeting miRNAs with anesthetics in cancer: Current understanding and future perspectives. Biomed. Pharmacother. 2021, 144, 112309. [Google Scholar] [CrossRef] [PubMed]
- Gurugubelli Krishna, R.; Vishnu Bhat, B. Molecular mechanisms of intrauterine growth restriction. J. Matern.-Fetal Neonatal Med. 2018, 31, 2634–2640. [Google Scholar] [CrossRef]
- Guzeloglu-Kayisli, O.; Kayisli, U.; Taylor, H. The Role of Growth Factors and Cytokines during Implantation: Endocrine and Paracrine Interactions. Semin. Reprod. Med. 2009, 27, 062–079. [Google Scholar] [CrossRef]
- Gonzalez-Avila, G.; Sommer, B.; Mendoza-Posada, D.A.; Ramos, C.; Garcia-Hernandez, A.A.; Falfan-Valencia, R. Matrix metalloproteinases participation in the metastatic process and their diagnostic and therapeutic applications in cancer. Crit. Rev. Oncol./Hematol. 2019, 137, 57–83. [Google Scholar] [CrossRef]
- Von Grothusen, C.; Frisendahl, C.; Modhukur, V.; Lalitkumar, P.G.; Peters, M.; Faridani, O.R.; Salumets, A.; Boggavarapu, N.R.; Gemzell-Danielsson, K. Uterine fluid microRNAs are dysregulated in women with recurrent implantation failure. Hum. Reprod. 2022, 37, 734–746. [Google Scholar] [CrossRef]
- Testa, U.; Pelosi, E.; Castelli, G.; Labbaye, C. miR-146 and miR-155: Two Key Modulators of Immune Response and Tumor Development. ncRNA 2017, 3, 22. [Google Scholar] [CrossRef]
- Dong, X.; Zhou, M.; Li, X.; Huang, H.; Sun, Y. Gene profiling reveals the role of inflammation, abnormal uterine muscle contraction and vascularity in recurrent implantation failure. Front. Genet. 2023, 14, 1108805. [Google Scholar] [CrossRef]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N.; Hahne, J.C. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target. Oncol. 2020, 15, 261–278. [Google Scholar] [CrossRef]
- Ha, T.-Y. The Role of MicroRNAs in Regulatory T Cells and in the Immune Response. Immune Netw. 2011, 11, 11–41. [Google Scholar] [CrossRef] [PubMed]
- Mortlock, S.; Restuadi, R.; Levien, R.; Girling, J.E.; Holdsworth-Carson, S.J.; Healey, M.; Zhu, Z.; Qi, T.; Wu, Y.; Lukowski, S.W.; et al. Genetic regulation of methylation in human endometrium and blood and gene targets for reproductive diseases. Clin. Epigenetics 2019, 11, 49. [Google Scholar] [CrossRef] [PubMed]
- Weiss, C.N.; Ito, K. A Macro View of MicroRNAs: The Discovery of MicroRNAs and Their Role in Hematopoiesis and Hematologic Disease. In International Review of Cell and Molecular Biology; Elsevier: Amsterdam, The Netherlands, 2017; Volume 334, pp. 99–175. ISBN 978-0-12-811868-9. [Google Scholar]
- Kolanska, K.; Bendifallah, S.; Canlorbe, G.; Mekinian, A.; Touboul, C.; Aractingi, S.; Chabbert-Buffet, N.; Daraï, E. Role of miRNAs in Normal Endometrium and in Endometrial Disorders: Comprehensive Review. J. Clin. Med. 2021, 10, 3457. [Google Scholar] [CrossRef] [PubMed]
- Canan, S.; İnan, M.A.; Erdem, A.; Demirdağ, E.; Gündüz, M.İ.; Erdem, Ö.; Erdem, M. Evaluation of endometrial receptivity in recurrent pregnancy loss and recurrent implantation failure. Turk. J. Obstet. Gynecol. 2024, 21, 22–27. [Google Scholar] [CrossRef]
- Yang, J.; Qi, M.; Fei, X.; Wang, X.; Wang, K. LncRNA H19: A novel oncogene in multiple cancers. Int. J. Biol. Sci. 2021, 17, 3188–3208. [Google Scholar] [CrossRef]
- Ye, L.; Dimitriadis, E. Endometrial Receptivity–Lessons from “Omics”. Biomolecules 2025, 15, 106. [Google Scholar] [CrossRef]
- Huang, X.; Zhong, R.; He, X.; Deng, Q.; Peng, X.; Li, J.; Luo, X. Investigations on the mechanism of progesterone in inhibiting endometrial cancer cell cycle and viability via regulation of long noncoding RNA NEAT1/microRNA-146b-5p mediated Wnt/β-catenin signaling. IUBMB Life 2019, 71, 223–234. [Google Scholar] [CrossRef]
- Treeck, O.; Haerteis, S.; Ortmann, O. Non-Coding RNAs Modulating Estrogen Signaling and Response to Endocrine Therapy in Breast Cancer. Cancers 2023, 15, 1632. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Gahan, P.B. Interplay between LncRNAs and microRNAs in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 8095. [Google Scholar] [CrossRef]
- Zhang, H.; Li, C.; Li, W.; Xin, W.; Qin, T. Research Advances in Adenomyosis-Related Signaling Pathways and Promising Targets. Biomolecules 2024, 14, 1402. [Google Scholar] [CrossRef]
- Ansari, S.K.; Gokalp, E.E.; Ozkara, E.; Aykac, O.; Cilingir, O.; Colak, E.; Ozdemir, A.O.; Artan, S. The Role of miRNA Expression Profiles in Different Biofluids İn Aneurysm Rupture. J. Korean Neurosurg. Soc. 2025. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-J.; Hsu, A.; Lin, P.-Y.; Chen, Y.-L.; Wu, K.-W.; Chen, K.-C.; Wang, T.; Yi, Y.-C.; Kung, H.-F.; Chang, J.-C.; et al. Development of a Predictive Model for Optimization of Embryo Transfer Timing Using Blood-Based microRNA Expression Profile. Int. J. Mol. Sci. 2023, 25, 76. [Google Scholar] [CrossRef] [PubMed]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- DaCunza, J.T.; Wickman, J.R.; Ajit, S.K. miRNA packaging into small extracellular vesicles and implications in pain. Pain Rep. 2024, 9, e1198. [Google Scholar] [CrossRef]
- Chen, C.-H.; Lu, F.; Yang, W.-J.; Chen, W.-M.; Yang, P.E.; Kang, S.-T.; Wang, T.; Chang, P.-C.; Feng, C.-T.; Yang, J.-H.; et al. Development of a Novel Endometrial Signature Based on Endometrial microRNA for Determining the Optimal Timing for Embryo Transfer. Biomedicines 2024, 12, 700. [Google Scholar] [CrossRef]
- Omes, C.; Conti, A.; Benedetti, L.; Tomasoni, V.; De Marchi, D.; Nappi, R.E.; Cusella De Angelis, M.G.; Ceccarelli, G. Expression of miRNA from spent pre-implantation embryos culture media. Reprod. Biol. 2024, 24, 100847. [Google Scholar] [CrossRef]
- Balaguer, N.; Moreno, I.; Herrero, M.; Gonzáléz-Monfort, M.; Vilella, F.; Simón, C. MicroRNA-30d deficiency during preconception affects endometrial receptivity by decreasing implantation rates and impairing fetal growth. Am. J. Obstet. Gynecol. 2019, 221, 46.e1–46.e16. [Google Scholar] [CrossRef]
- Witwer, K.W. Circulating MicroRNA Biomarker Studies: Pitfalls and Potential Solutions. Clin. Chem. 2015, 61, 56–63. [Google Scholar] [CrossRef]
- Xiang, M.; Zeng, Y.; Yang, R.; Xu, H.; Chen, Z.; Zhong, J.; Xie, H.; Xu, Y.; Zeng, X. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem. Biophys. Res. Commun. 2014, 454, 210–214. [Google Scholar] [CrossRef]
- Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. miRNA: A Promising Therapeutic Target in Cancer. Int. J. Mol. Sci. 2022, 23, 11502. [Google Scholar] [CrossRef]
- Revel, A.; Achache, H.; Stevens, J.; Smith, Y.; Reich, R. MicroRNAs are associated with human embryo implantation defects. Hum. Reprod. 2011, 26, 2830–2840. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhao, Y.; Yu, Y.; Li, R.; Qiao, J. MiR-125b regulates endometrial receptivity by targeting MMP26 in women undergoing IVF-ET with elevated progesterone on HCG priming day. Sci. Rep. 2016, 6, 25302. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, L.; Ma, X.; Yue, F.; Wang, Y.; Wang, L.; Jin, P.; Zhang, X. Altered Circular RNA Expression in Patients with Repeated Implantation Failure. Cell Physiol. Biochem. 2017, 44, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Di Pietro, C.; Caruso, S.; Battaglia, R.; Iraci Sareri, M.; La Ferlita, A.; Strino, F.; Bonaventura, G.; Di Mauro, M.; Barcellona, M.L.; Perciavalle, V.; et al. MiR-27a-3p and miR-124-3p, upregulated in endometrium and serum from women affected by Chronic Endometritis, are new potential molecular markers of endometrial receptivity. Am. J. Rep. Immunol. 2018, 80, e12858. [Google Scholar] [CrossRef]
- Xu, H.; Zhou, M.; Cao, Y.; Zhang, D.; Han, M.; Gao, X.; Xu, B.; Zhang, A. Genome-wide analysis of long noncoding RNAs, microRNAs, and mRNAs forming a competing endogenous RNA network in repeated implantation failure. Gene 2019, 720, 144056. [Google Scholar] [CrossRef]
- Zhai, J.; Yao, G.-D.; Wang, J.-Y.; Yang, Q.-L.; Wu, L.; Chang, Z.-Y.; Sun, Y.-P. Metformin Regulates Key MicroRNAs to Improve Endometrial Receptivity Through Increasing Implantation Marker Gene Expression in Patients with PCOS Undergoing IVF/ICSI. Reprod. Sci. 2019, 26, 1439–1448. [Google Scholar] [CrossRef]
- Drissennek, L.; Baron, C.; Brouillet, S.; Entezami, F.; Hamamah, S.; Haouzi, D. Endometrial miRNome profile according to the receptivity status and implantation failure. Hum. Fertil. 2022, 25, 356–368. [Google Scholar] [CrossRef]
- Grasso, A.; Navarro, R.; Balaguer, N.; Moreno, I.; Alama, P.; Jimenez, J.; Simón, C.; Vilella, F. Endometrial Liquid Biopsy Provides a miRNA Roadmap of the Secretory Phase of the Human Endometrium. J. Clin. Endocrinol. Metab. 2020, 105, 877–889. [Google Scholar] [CrossRef]
- Riyanti, A.; Febri, R.R.; Zakirah, S.C.; Harzif, A.K.; Rajuddin, R.; Muharam, R.; Asmarinah, A.; Wiweko, B. Suppressing HOXA-10 Gene Expression by MicroRNA 135b During the Window of Implantation in Infertile Women. J. Reprod. Infertil. 2020, 21, 217–221. [Google Scholar]
- Li, T.; Greenblatt, E.M.; Shin, M.E.; Brown, T.J.; Chan, C. Cargo small non-coding RNAs of extracellular vesicles isolated from uterine fluid associate with endometrial receptivity and implantation success. Fertil. Steril. 2021, 115, 1327–1336. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, L.; Shan, Y.; Chen, G.; Chu, Y.; Dai, H.; Liu, X.; Bao, H. Hsa_circ_0038383-mediated competitive endogenous RNA network in recurrent implantation failure. Aging 2021, 13, 6076–6090. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; He, D.; Zeng, H.; Luo, J.; Yang, S.; Chen, J.; Abdullah, R.K.; Liu, N. Expression and significance of miR-30d-5p and SOCS1 in patients with recurrent implantation failure during implantation window. Reprod. Biol. Endocrinol. 2021, 19, 138. [Google Scholar] [CrossRef] [PubMed]
- Azarpoor, A.; Ardeshirylajimi, A.; Yeganeh, S.M.; Pourmatrood, E.; Dehghan, Z.; Farifteh Nobijari, F.; Salehi, M. Expressions of miR-199a-5p and miR-125b-5p and their target genes in the endometrium of recurrent implantation failure patients following in uterus infusion of autologous peripheral blood mononuclear cells. Trends Immunother. 2022, 6, 50. [Google Scholar] [CrossRef]
- Babian, S.; Salehpour, S.; Nazari, L.; Ghorbanmehr, N. The expression level of mir-21-3p in platelet-rich plasma: A potential effective factor and predictive biomarker in recurrent implantation failure. Mol. Reprod. Dev. 2022, 89, 498–505. [Google Scholar] [CrossRef]
- Li, L.; Liu, L.; Kou, Z.; Huo, M.; An, J.; Zhang, X. GnRH agonist treatment regulates IL-6 and IL-11 expression in endometrial stromal cells for patients with HRT regiment in frozen embryo transfer cycles. Reprod. Biol. 2022, 22, 100608. [Google Scholar] [CrossRef]
- Juárez-Barber, E.; Segura-Benítez, M.; Carbajo-García, M.C.; Bas-Rivas, A.; Faus, A.; Vidal, C.; Giles, J.; Labarta, E.; Pellicer, A.; Cervelló, I.; et al. Extracellular vesicles secreted by adenomyosis endometrial organoids contain miRNAs involved in embryo implantation and pregnancy. Reprod. Biomed. Online 2023, 46, 470–481. [Google Scholar] [CrossRef]
- He, Y.; Ju, Y.; Lei, H.; Dong, J.; Jin, N.; Lu, J.; Chen, S.; Wang, X. MiR-135a-5p regulates window of implantation by suppressing pinopodes development and decidualization of endometrial stromal cells. J. Assist. Reprod. Genet. 2024, 41, 1645–1659. [Google Scholar] [CrossRef]
- Yang, W.-J.; Lu, F.; Wang, C.-Y.; Hong, J.-J.; Wang, T.; Yang, P.E.; Huang, J.Y.-J. Different Dosages of Progesterone in Luteal Phase Support Reflect Varying Endometrial microRNA Expression in Frozen Embryo Transfer Cycles. Int. J. Mol. Sci. 2024, 25, 3670. [Google Scholar] [CrossRef]
- Tan, Y.; Du, B.; Chen, X.; Chen, M. Correlation of MicroRNA-31 with Endometrial Receptivity in Patients with Repeated Implantation Failure of In Vitro Fertilization and Embryo Transfer. Organogenesis 2025, 21, 2460263. [Google Scholar] [CrossRef]
- Chettiar, V.; Patel, A.; Chettiar, S.S.; Jhala, D.D. Meta-analysis of endometrial transcriptome data reveals novel molecular targets for recurrent implantation failure. J. Assist. Reprod. Genet. 2024, 41, 1417–1431. [Google Scholar] [CrossRef]
- Xu, S.; Diao, H.; Xiong, Y.; Zhang, C.; Zhang, Y.; Zhang, Y. The study on the clinical efficacy of endometrial receptivity analysis and influence factors of displaced window of implantation. Sci. Rep. 2025, 15, 7326. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.; Qian, W.; Zhang, C.; Zhou, L.; Hou, Z. Study on microRNA expression in endometrium of luteal phase and its relationship with infertility of endometriosis. Zhonghua Fu Chan Ke Za Zhi 2013, 48, 907–910. [Google Scholar] [PubMed]
- Cheng, F.; Lu, L.; Wang, H.; Cheng, H.; Zhang, D. Expression and Significance of miR-126 and miR-145 in Infertility due to Endometriosis. J. Coll. Physicians Surg. Pak. 2019, 29, 585–587. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Broaddus, R.; Cheng, W.; Xie, S.; Naora, H. Deregulation of the HOXA10 Homeobox Gene in Endometrial Carcinoma: Role in Epithelial-Mesenchymal Transition. Cancer Res. 2006, 66, 889–897. [Google Scholar] [CrossRef]
- Zanatta, A.; Rocha, A.M.; Carvalho, F.M.; Pereira, R.M.A.; Taylor, H.S.; Motta, E.L.A.; Baracat, E.C.; Serafini, P.C. The role of the Hoxa10/HOXA10 gene in the etiology of endometriosis and its related infertility: A review. J. Assist. Reprod. Genet. 2010, 27, 701–710. [Google Scholar] [CrossRef]
- Marwood, M.; Visser, K.; Salamonsen, L.A.; Dimitriadis, E. Interleukin-11 and Leukemia Inhibitory Factor Regulate the Adhesion of Endometrial Epithelial Cells: Implications in Fertility Regulation. Endocrinology 2009, 150, 2915–2923. [Google Scholar] [CrossRef]
- Wang, H.-Q.; Zhang, W.-D.; Yuan, B.; Zhang, J.-B. Advances in the Regulation of Mammalian Follicle-Stimulating Hormone Secretion. Animals 2021, 11, 1134. [Google Scholar] [CrossRef]
- Menkhorst, E.; So, T.; Rainczuk, K.; Barton, S.; Zhou, W.; Edgell, T.; Dimitriadis, E. Endometrial stromal cell miR-19b-3p release is reduced during decidualization implying a role in decidual-trophoblast cross-talk. Front. Endocrinol. 2023, 14, 1149786. [Google Scholar] [CrossRef]
- Fu, X.; Guo, X.; Xu, H.; Li, Y.; Jin, B.; Zhang, X.; Shu, C.; Fan, Y.; Yu, Y.; Tian, Y.; et al. Varied cellular abnormalities in thin vs. normal endometrium in recurrent implantation failure by single-cell transcriptomics. Reprod. Biol. Endocrinol. 2024, 22, 90. [Google Scholar] [CrossRef]
| Author, Year | Study Design | Sample Size | Sample Source | Molecular Method | Normalization Strategy | Validation Cohort | Risk of Bias |
|---|---|---|---|---|---|---|---|
| Revel et al., 2011 [52] | Retrospective | 11 RIF-IVF patients vs. 5 fertile women | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Chen et al., 2016 [53] | Prospective | Human: 16 women with elevated P vs. 9 with normal P; Mouse: groups treated with miR-125b agomir vs. NC/water | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Cho et al., 2016 [5] | Retrospective | 120 RIF patients vs. 234 fertile controls | Endometrial tissue | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Liu et al., 2017 [54] | Prospective | 6 RIF patients vs. 6 fertile controls (3 each for microarray; 3 each for qRT-PCR validation) | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Di Pietro et al., 2018 [55] | Prospective | 15 CE patients vs. 15 healthy controls (serum and endometrial samples) | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Xu et al., 2019 [56] | Prospective | RNA-Seq: 5 RIF patients vs. 5 controls Validation: 14 RIF patients vs. 17 controls | Endometrial tissue | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Zhai et al., 2019 [57] | RCT | 60 PCOS patients with metformin vs. 60 PCOS patients without metformin | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Drissennek et al., 2020 [58] | Retrospective | Discovery cohort: 5 non-receptive vs. 15 receptive patients Validation cohort: 27 non-receptive vs. 25 receptive patients (total 123 RIF patients analyzed) | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Grasso et al., 2020 [59] | Prospective | 58 ELB samples from 40 healthy ovum donors (each in natural and HRT cycles) | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Riyanti et al., 2020 [60] | Prospective | 14 infertile women vs. 9 fertile controls | Endometrial tissue | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Li et al., 2021 [61] | Prospective | Group A: 22 fertile women (LH+2 vs. LH+7) Group B: 51 IVF patients (hCG+2 vs. hCG+7; 18 conceived, 33 not) | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Zhao et al., 2021, aging [62] | Prospective | 18 RIF patients vs. 16 fertile controls (validation cohort) GEO datasets: circRNA (8 vs. 8), miRNA (7 vs. 5), mRNA (43 vs. 42) | Endometrial tissue | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Zhao et al., 2021 (Reprod. Biol. Endocrinol.) [63] | Prospective | 20 RIF patients vs. 10 fertile controls | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Azarpoor et al., 2022 [64] | Prospective | 10 RIF+PBMCs vs. 10 RIF vs. 10 fertile patients | Endometrial tissue | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Babian et al., 2022 [65] | Prospective | 30 pregnant vs. 30 non-pregnant RIF patients (PRP-treated) | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Li et al., 2022 [66] | Retrospective | 1143 FET cycles (Group A: 290 HT only; Group B-D: 293 + 310 + 250 = 853 with GnRHa pretreatment) | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Von Grothusen et al., 2022 [27] | Prospective | 33 RIF patients vs. 15 fertile controls | Endometrial tissue | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Chen et al., 2023 [43] | Retrospective | 111 in model training; 73 in validation | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Dabi et al., 2023 [7] | Prospective | 153 women with endometriosis (79 infertile, 74 fertile) vs. 31 healthy controls | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Juarez-Barber et al., 2023 [67] | Prospective | 4 adenomyosis patients vs. 4 fertile controls | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Omes et al., 2023 [47] | Prospective | 53 embryos from 16 IVF patients (BLOK vs. NE/DG; 5 pools each group) | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Soczewski et al., 2023 [6] | Retrospective | n = 20 (10 RPL vs. 10 RIF patients) | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Alset et al., 2024 [12] | Prospective | 65 total (30 FGR patients vs. 35 healthy controls) | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Bendifallah et al., 2024 [8] | Prospective | 153 total (16 SPE vs. 137 non-SPE endometriosis phenotypes) | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Chen et al., 2024 [46] | Retrospective | 200 total: 150 successful implantations vs. 50 failed implantations | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| He et al., 2024 [68] | Prospective | 54 women (FET with clinical pregnancy); human and mouse models used | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Yang et al., 2024 [69] | Retrospective | 22 IVF patients (no separate control group; all underwent mock FET cycles) | Serum/saliva/plasma | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Tan et al., 2025 [70] | Retrospective | 80 total (40 RIF patients vs. 40 controls) | Endometrial tissue | qRT-PCR | U6/SNORD/endogenous control | Not reported | Moderate |
| Author, Year | Type of Study | Sample Size | Inclusion Criteria | Exclusion Criteria | Type of microRNA | Target Gene/Protein | Receptivity |
|---|---|---|---|---|---|---|---|
| Revel et al., 2011 [52] | retrospective | 11 RIF-IVF patients vs. 5 fertile women | Women < 41 years, ≥4 IVF-ET failures, good ovarian reserve (FSH < 8 mIU/mL), >8 oocytes per retrieval, normal uterine cavity, endometrial thickness ≥ 8 mm | Infertility from severe male factor, oral contraceptives or IUD use, breastfeeding, hormonal therapy during study cycle | 13 miRNAs (↑ miR-23b, miR-145, miR-99a, miR-27b, miR-652, miR-139-5p, miR-195, miR-342-3p, miR-150, miR-374b; ↓ miR-32, miR-628-5p, miR-874) | miR-145 → N-cadherin, H2AFX, netrin-4; miR-23b → sFRP-4 | Impaired |
| Chen et al., 2016 [53] | prospective | Human: 16 women with elevated P vs. 9 with normal P; Mouse: groups treated with miR-125b agomir vs. NC/water | Women aged 23–38 years, regular cycles, undergoing IVF-ET for male factor infertility, no uterine pathology | PCOS, endometriosis, ovarian tumor, hydrosalpinx, steroid use within 3 months | miR-125b | MMP26 | Impaired |
| Cho et al., 2016 [5] | retrospective | 120 RIF patients vs. 234 fertile controls | Women with RIF: failure after ≥2 fresh IVF-ET cycles, >10 cleaved embryos, high-quality embryos; <40 years old; Korean ethnicity | Anatomical/chromosomal/hormonal/infectious/autoimmune/thrombotic causes of implantation failure; Müllerian anomaly, hypothyroidism, chromosomal abnormality, antiphospholipid syndrome; male partners with abnormal semen/karyotype/hormones | miR-146aC>G, miR-149C>T, miR-196a2T>C, miR-499A>G | CCND2 (Cyclin D2), COL8A2, NF2, FZD4 (via miR-146a-3p alleles) | Impaired |
| Liu et al., 2017 [54] | Prospective | 6 RIF patients vs. 6 fertile controls (3 each for microarray; 3 each for qRT-PCR validation) | Women with ≥3 IVF-ET failures with high-grade embryos transferred; fertile women with ≥1 live birth for controls | N/A | circRNA sponges: hsa_circRNA_070616, _103716, etc. → miR-574-5p | miR-574-5p (indirect target: MACC-1, others via sponge activity) | Impaired |
| Di Pietro et al., 2018 [55] | Prospective | 15 CE patients vs. 15 healthy controls (serum and endometrial samples) | Infertile women undergoing hysteroscopy with confirmed histological CE (study group); healthy women (controls) | N/A | miR-27a-3p, miR-124-3p | IGF1 (miR-27a-3p), IL11 (miR-124-3p) | Impaired |
| Xu et al., 2019 [56] | Prospective | RNA-Seq: 5 RIF patients vs. 5 controls Validation: 14 RIF patients vs. 17 controls | Women aged 25–35 years RIF: ≥3 embryo transfers with ≥4 good-quality embryos Controls: peritubal infertility, pregnant within 1–2 ETs | uterine anomaly, impaired glucose tolerance, abnormal thyroid function, antiphospholipid antibodies, or other diseases | 86 differentially expressed miRNAs (e.g., miR-424-5p, miR-488-3p, miR-548o-3p, miR-30d-5p, miR-124-5p, etc.) | Multiple targets via ceRNA network (e.g., DOCK8, MYH11, IL6 pathways; miRNAs target via lncRNAs like NEAT1, TRG-AS1, SMIM25, H19, etc.) | Impaired |
| Zhai et al., 2019 [57] | RCT | 60 PCOS patients with metformin vs. 60 PCOS patients without metformin | Women diagnosed with PCOS (Rotterdam criteria), undergoing IVF/ICSI, with insulin resistance | N/A | miR-491-3p, miR-1910-3p (↓ with metformin) | HOXA10, ITGB3 | Improved |
| Drissennek et al., 2020 [58] | Retrospective | Discovery cohort: 5 non-receptive vs. 15 receptive patients Validation cohort: 27 non-receptive vs. 25 receptive patients (total 123 RIF patients analyzed) | Women with ≥3 failed embryo transfers (mean 9.1 failed attempts; 9.8 non-implanted embryos), undergoing hormone replacement and endometrial biopsy in implantation window | N/A | miR-455-3p, miR-4423-3p (↓ in receptive); miR-152-3p, miR-155-5p (↑ in implantation failure) | miR-155-5p → TGFβ signaling, leukocyte extravasation; miR-152-3p → cell migration and adhesion genes | Impaired |
| Grasso et al., 2020 [59] | Prospective | 58 ELB samples from 40 healthy ovum donors (each in natural and HRT cycles) | Healthy women (18–34 years), regular cycles, normal BMI (19–29), normal karyotype, participation in ovum donation program | N/A | Receptivity-associated ↑: miR-30d-5p, miR-873-3p, miR-345-5p, miR-30d-3p, miR-141-3p, miR-30b-3p, miR-223-3p, miR-582-5p; ↓: various others in non-receptive phases | MAPK8, FOXO1, HOXA11, ITGB3, RASSF2, IGF2, KIF11, ATP5B, VCAM1, MMP2, MMP9, SLC1A1, SLC7A1 (differ by cycle and miRNA) | Improved |
| Riyanti et al., 2020 [60] | Prospective | 14 infertile women vs. 9 fertile controls | Infertile: women undergoing IVF with normal hormonal profiles, sperm analysis, mid-secretory phase endometrium (≥8 mm), regular cycles, normal uterus imaging. Fertile: ≥1 normal pregnancy/delivery, attending for Pap smear/screening | N/A | miR-135b | HOXA10 | Impaired |
| Li et al., 2021 [61] | Prospective | Group A: 22 fertile women (LH+2 vs. LH+7) Group B: 51 IVF patients (hCG+2 vs. hCG+7; 18 conceived, 33 not) | Age < 40, regular menstrual cycles, normal uterine cavity, no history of infertility (Group A); IVF patients on first/second cycle, no uterine abnormalities (Group B) | IVF patients with no embryo transfer or missing data excluded | 12 conserved EV-sncRNAs: 11 miRNAs (e.g., miR-503-5p, miR-196a-5p, and miR-18a-5p) and 1 piRNA (piR-hsa-456); hsa-miR-362-3p ↑ in non-pregnant IVF patients | TGF-β pathway, Hippo signaling, immune response, ECM, cell junction pathways | Improved (in fertile and pregnant cycles); Impaired (↑ miR-362-3p in non-pregnant) |
| Zhao et al., 2021 (Aging) [62] | Prospective | 18 RIF patients vs. 16 fertile controls (validation cohort) GEO datasets: circRNA (8 vs. 8), miRNA (7 vs. 5), mRNA (43 vs. 42) | Women undergoing IVF-ET RIF: ≥3 IVF-ET failures Controls: clinical pregnancy after 1–2 ETs for tubal infertility | endometrial disease, hydrosalpinx, PCOS; all with normal cycles, hormone levels, endometrial thickness | miR-196b-5p, miR-424-5p (↓) | HOXA9 (via miR-196b-5p), PBX1, HOXA3 (via miR-424-5p) | Impaired |
| Zhao et al., 2021 (Reprod Biol Endocrinol) [63] | Prospective | 20 RIF patients vs. 10 fertile controls | Women aged 25–40; regular menstrual cycles (25–35 days); BMI 18.5–24.9; normal hormone profile (FSH, LH, and E2); infertility due to male or tubal factors | Endometrial/uterine pathology (fibroids, adenomyosis, endometritis, etc.); PCOS; hydrosalpinx; abnormal karyotypes; autoimmunity; endocrine disorders; recent contraceptive use or IUD (within 6 months) | miR-30d-5p | SOCS1 (↑ in RIF), LIF, p-STAT3 (↓ in RIF) | Impaired |
| Azarpoor et al., 2022 [64] | Prospective | 10 RIF+PBMCs vs. 10 RIF patients vs. 10 fertile patients | 30–35 y, ≥2 RIF, normal endometrium in controls | Tubal/male/unexplained infertility, recurrent miscarriage, infections, systemic/uterine pathologies | miR-199a-5p, miR-125b-5p | LIF, FGFR-2 | Improved |
| Babian et al., 2022 [65] | Prospective | 30 pregnant vs. 30 non-pregnant RIF patients (PRP-treated) | Women with ≥3 IVF failures and ≥1–2 high-quality embryos per transfer; frozen–thawed embryo transfer cycles; PRP treatment | Anatomical, chromosomal, and hormonal causes of RIF (e.g., hyperprolactinemia, luteal insufficiency, and thyroid disease); systemic diseases; infections | miR-21-3p (↑ in pregnant), miR-21-5p, miR-494-3p, miR-145-5p | RBPMS → SMAD4 inhibition; CCAR1 → reduced apoptosis/proliferation regulation | Improved |
| Li et al., 2022 [66] | Retrospective | 1143 FET cycles (Group A: 290 HT only; Group B-D: 293 + 310 + 250 = 853 with GnRHa pretreatment) | Women aged 18–40 with regular cycles (26–35 days), ovulatory, undergoing HRT-FET cycles | Contraindications to estrogen/progesterone, intrauterine adhesions, congenital uterine anomalies, donor oocyte use | miR-124-3p | IL-6, IL-11 | Improved |
| Von Grothusen et al., 2022 [27] | Prospective | 33 RIF patients vs. 15 fertile controls | Women aged 18–42; RIF: ≥3 failed IVF/ICSI cycles with high-quality embryos Fertile: ≥1 spontaneous pregnancy, regular cycles (21–35 days), no hormonal contraception for ≥3 months prior to sampling | Systemic, endocrinological, or gynecological diseases; infertility; ongoing pregnancy; breastfeeding; >2 miscarriages; regular NSAID use | 61 dysregulated miRNAs in UF: Downregulated: miR-486-5p, miR-92b-3p (validated) Upregulated: miR-320a, miR-127-3p, miR-224-5p, etc. | VEGFA, CDH1 (E-cadherin), PI3K/AKT, JAK/STAT, TGF-β, Wnt, estrogen signaling pathways (predicted targets) | Impaired |
| Chen et al., 2023 [43] | Retrospective | 111 in model training; 73 in validation | Women aged 21–45; BMI >18.5; ≥1 good-quality frozen blastocyst; no ovulatory disorders, endometriosis, myomas, polyps, or hydrosalpinx | N/A | let-7g-5p, let-7b-5p, miR-423-5p, miR-122-5p, miR-143-3p, miR-375-3p, miR-191-5p | IGF2R (via let-7g-5p), CCND2 (via miR-143-3p), cell cycle pathways | Improved |
| Dabi et al., 2023 [7] | Prospective | 153 women with endometriosis (79 infertile; 74 fertile) vs. 31 healthy controls | Women with pelvic pain suggestive of endometriosis; diagnosis confirmed by MRI or laparoscopy; fertile or infertile status assessed | Prior surgery for endometriosis, unclear fertility status, male factor infertility | 34 dysregulated salivary miRNAs (e.g., miR-6818-5p, miR-498, miR-1910-3p, miR-3119, miR-501-5p, etc.) | Predicted: PI3K/Akt, JAK/STAT, EGFR, MAPK, Wnt/β-catenin signaling pathways | Impaired |
| Juarez-Barber et al., 2023 [67] | Prospective | 4 adenomyosis patients vs. 4 fertile controls | Women aged 18–45; secretory endometrial biopsies; validated adenomyosis diagnosis; fertile controls from oocyte donation program | No hormonal treatments within 3 months; absence of other gynecological pathology | Secretory EV-miRNAs: miR-21-5p, miR-24-3p, miR-26a-5p, miR-92a-3p, miR-92b-3p, miR-200c-3p, miR-423a-5p Gestational EV-miRNAs: miR-21-5p, miR-26a-5p, miR-30a-5p, miR-30c-5p, miR-222-3p, miR-423a-5p | PTEN, PLAGL2, MDM4, CELF1 (downregulated in adenomyosis organoids) | Impaired |
| Omes et al., 2023 [47] | Prospective | 53 embryos from 16 IVF patients (BLOK vs. NE/DG; 5 pools each group) | Women undergoing IVF (mean age 37.3 ± 3.4 y); embryos cultured up until day 5/6; standard grading per Istanbul Consensus | N/A | hsa-miR-661 hsa-miR-21–5p hsa-miR-372–5p | hsa-miR-661: associated with epithelial morphogenesis, apoptosis (MDM2/p53 pathway), cell structure integrity; potential role in impairing implantation | Impaired |
| Soczewski et al., 2023 [6] | Retrospective | n = 20 (10 RPL vs. 10 RIF patients) | Age 28–40, regular cycles (28–31 days), no infectious, endocrine or anatomic disease | Infectious, endocrine, anatomic disease | miR-17-5p miR-21-5p miR-193b-3p | miR-17-5p → TXNIP miR-21-5p → TXNIP, IL-1β miR-193b-3p → NLRP3 | Impaired in RPL group; Improved in RIF group |
| Alset et al., 2024 [12] | Prospective | 65 total (30 FGR vs. 35 healthy controls) | Pregnant women aged 19–39, recruited 2018–2021, FGR diagnosed by Doppler ultrasound | IVF, fetal chromosomal/congenital anomalies, previous pregnancies | miR-125a (rs12976445) miR-365b (rs121224) miR-33a (rs9620000) miR-149 (rs2292832) | miR-33a → β-catenin, GSK3 miR-125a → WNT2 miR-365b → PPP5C, Dvl2, VGLL4 miR-149 → WNT3a, TNKS, WNT1 | Impaired |
| Bendifallah et al., 2024 [8] | Prospective | 153 total (16 SPE vs. 137 non-SPE endometriosis phenotypes) | Women aged 18–43 with pelvic pain suggestive of endometriosis; confirmed endometriosis by laparoscopy and/or MRI | Suspected adenomyosis on MRI | 89 miRNAs total signature; Key miRNAs: miR-4421, miR-3153, miR-3974, miR-4632-3p, miR-4674, miR-6511a-5p, miR-190a-5p | PI3K/Akt, Wnt/β-catenin, VEGF, MAPK, NF-κB pathways | Impaired |
| Chen et al., 2024 [46] | Retrospective | 200 total: 150 successful implantations vs. 50 failed implantations | Infertile women undergoing IVF with HRT cycles and personalized embryo transfer; endometrial biopsy performed | N/A | 143 miRNAs used to build classifiers (e.g., miR-145, miR-155-5p, miR-20b-5p, and miR-718) | IGF1R, ZEB1/2, genes involved in embryo–endometrium crosstalk, preeclampsia, and tissue remodeling | Improved |
| He et al., 2024 [68] | Prospective | 54 women (FET with clinical pregnancy); human and mouse models used | Women aged 20–39; natural cycle FET; BMI 18–25; infertility due to tubal/male factors; regular cycles | Hormonal treatment within 3 months; endometriosis, adenomyosis, hydrosalpinx, endometrial lesions; endometrial thickness < 7 mm on hCG day | miR-135a-5p | HOXA10, BMPR2(also influenced by CEBPD) | Impaired |
| Yang et al., 2024 [69] | Retrospective | 22 IVF patients (no separate control group; all underwent mock FET cycles) | Women undergoing hormone-replacement therapy (HRT) FET; assessed at 120 ± 4 h post-progesterone | N/A | ~100 miRNAs analyzed via MIRA; specific miRNAs include miR-30b, miR-181, miR-223-3p, miR-21, miR-22 | Not gene-specific; platform evaluates miRNAs reflecting receptivity states and WOI displacement | Impaired in all initial cycles; Improved after personalized progesterone adjustment in 91% (20/22) |
| Tan et al., 2025 [70] | Retrospective | 80 total (40 RIF patients vs. 40 controls) | Women <40 y/o undergoing IVF-ET; ≥3 failed transfers with high-quality embryos (RIF group); vs. pregnancy in one transfer (control) | Chromosomal abnormalities, genital malformations, mental illness, endometritis, polyps, PCOS, thyroid/pituitary dysfunction, major organ diseases, immune disorders, hepatitis, malignancy | miR-31 | N/A | Impaired |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Voros, C.; Varthaliti, A.; Athanasiou, D.; Mavrogianni, D.; Bananis, K.; Athanasiou, A.; Athanasiou, A.; Papahliou, A.-M.; Zografos, C.G.; Kondili, P.; et al. MicroRNA Signatures in Endometrial Receptivity—Unlocking Their Role in Embryo Implantation and IVF Success: A Systematic Review. Biomedicines 2025, 13, 1189. https://doi.org/10.3390/biomedicines13051189
Voros C, Varthaliti A, Athanasiou D, Mavrogianni D, Bananis K, Athanasiou A, Athanasiou A, Papahliou A-M, Zografos CG, Kondili P, et al. MicroRNA Signatures in Endometrial Receptivity—Unlocking Their Role in Embryo Implantation and IVF Success: A Systematic Review. Biomedicines. 2025; 13(5):1189. https://doi.org/10.3390/biomedicines13051189
Chicago/Turabian StyleVoros, Charalampos, Antonia Varthaliti, Diamantis Athanasiou, Despoina Mavrogianni, Kyriakos Bananis, Antonia Athanasiou, Aikaterini Athanasiou, Anthi-Maria Papahliou, Constantinos G. Zografos, Panagiota Kondili, and et al. 2025. "MicroRNA Signatures in Endometrial Receptivity—Unlocking Their Role in Embryo Implantation and IVF Success: A Systematic Review" Biomedicines 13, no. 5: 1189. https://doi.org/10.3390/biomedicines13051189
APA StyleVoros, C., Varthaliti, A., Athanasiou, D., Mavrogianni, D., Bananis, K., Athanasiou, A., Athanasiou, A., Papahliou, A.-M., Zografos, C. G., Kondili, P., Daskalaki, M. A., Mazis Kourakos, D., Vaitsis, D., Theodora, M., Antsaklis, P., Loutradis, D., & Daskalakis, G. (2025). MicroRNA Signatures in Endometrial Receptivity—Unlocking Their Role in Embryo Implantation and IVF Success: A Systematic Review. Biomedicines, 13(5), 1189. https://doi.org/10.3390/biomedicines13051189








