Exploring the Molecular Mechanism and Role of Glutathione S-Transferase P in Prostate Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Download from GEO Database and Data Preprocessing
2.2. Download of Data from the GWAS Public Database
2.3. Instrumental Variables (IVs)
2.4. MR Analysis Methods
2.5. Construct Drug-Gene Regulatory Network and ceRNA Regulatory Network of Pathway Genes
2.6. Analysis of Immune Infiltration of Pathway Genes
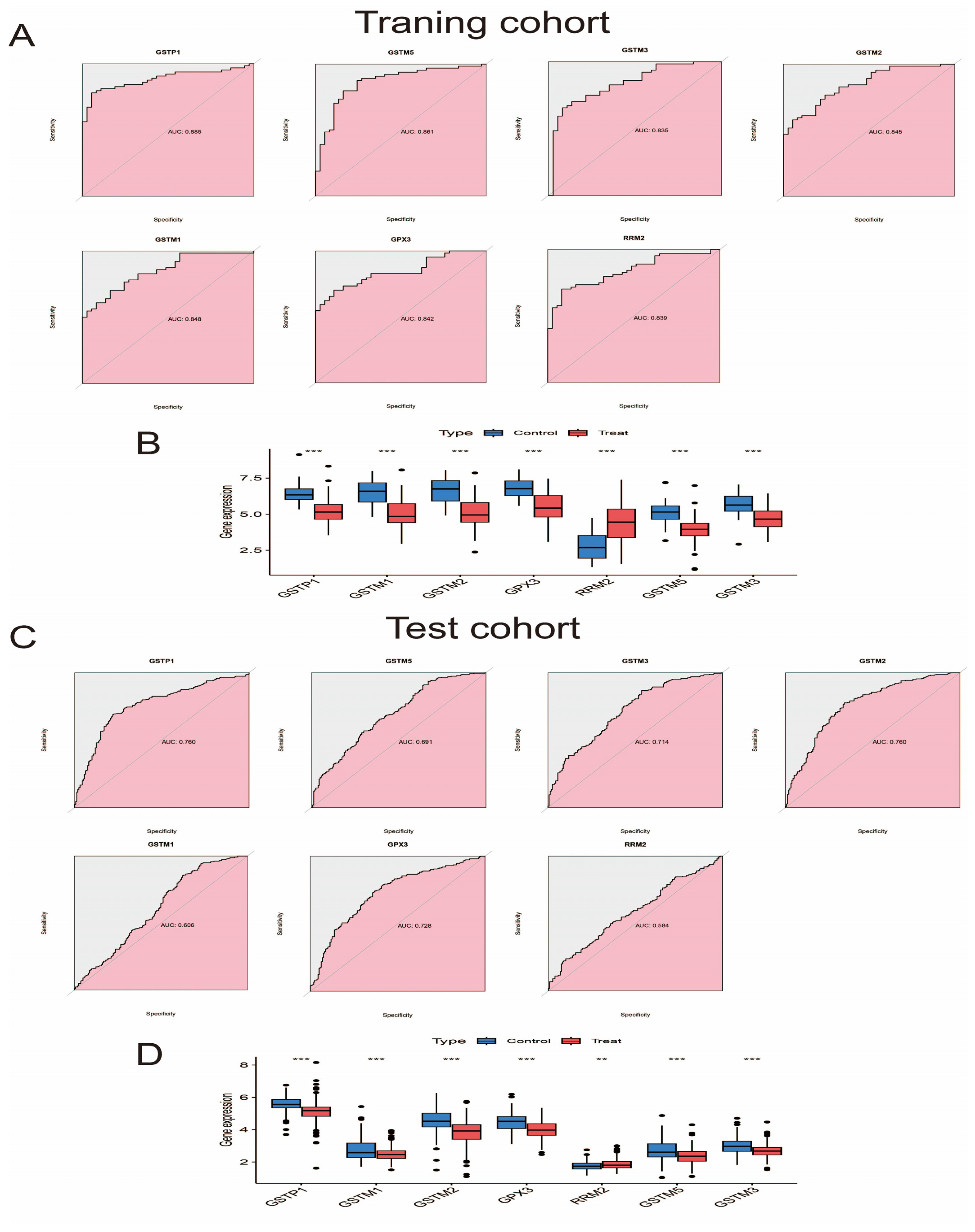
2.7. Diagnostic Ability, Expression Level, and Validation of Pathway Genes
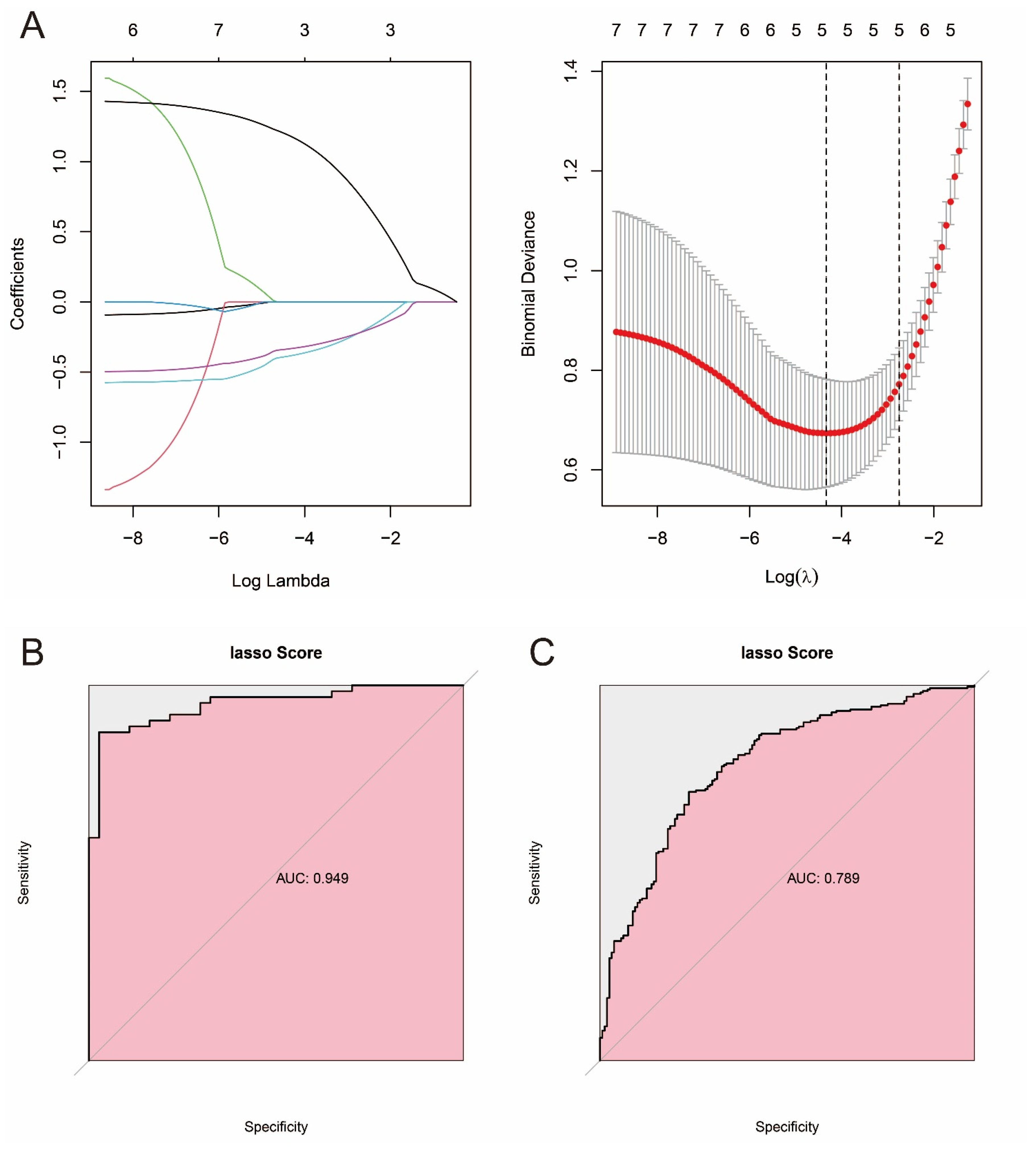
2.8. Construction of Prostate Cancer Prediction Model Based on Machine Learning and Pathway Genes
2.9. Processing of Single-Cell RNA-Seq Data
2.10. Statistical Analysis
3. Results
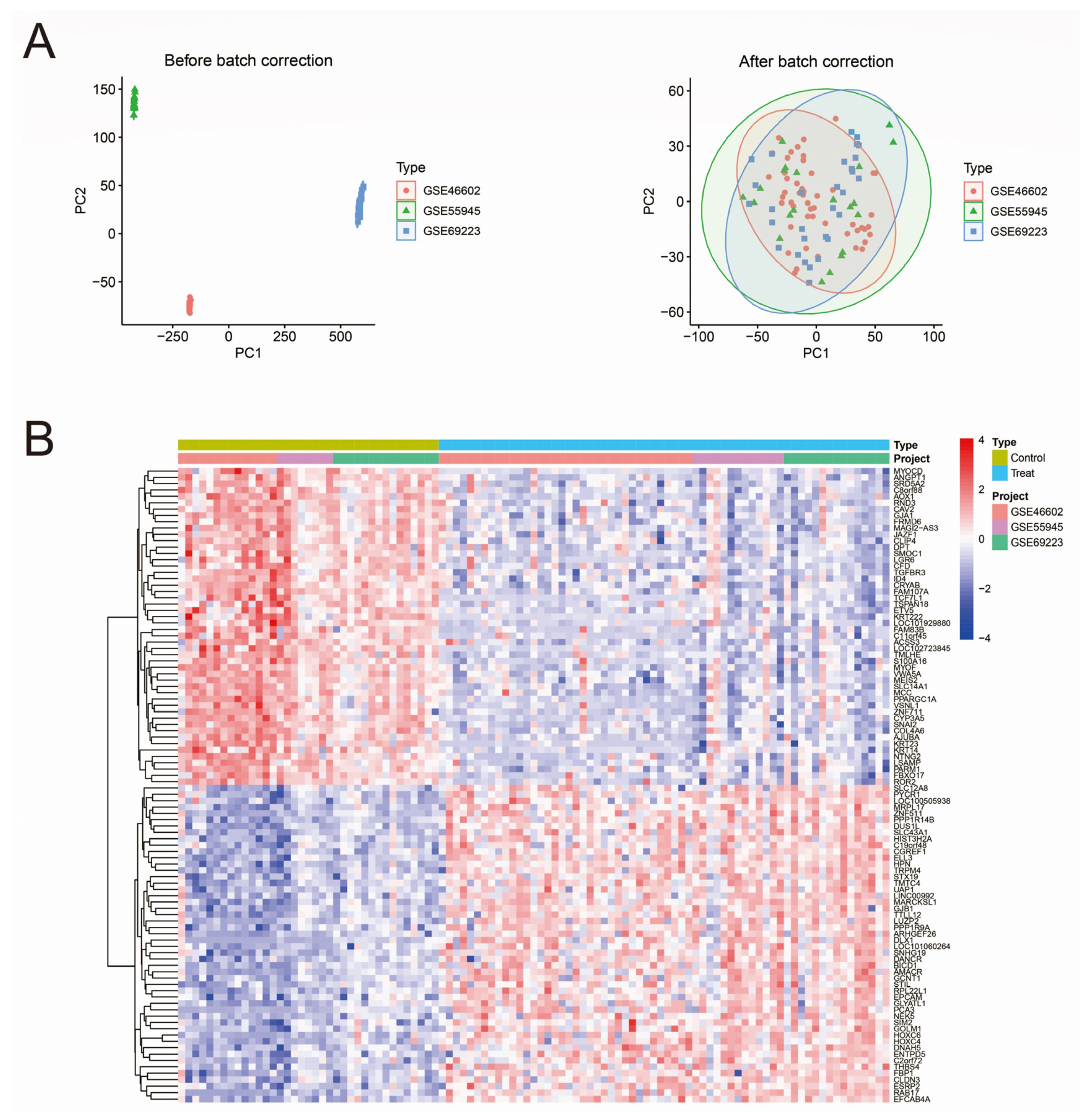
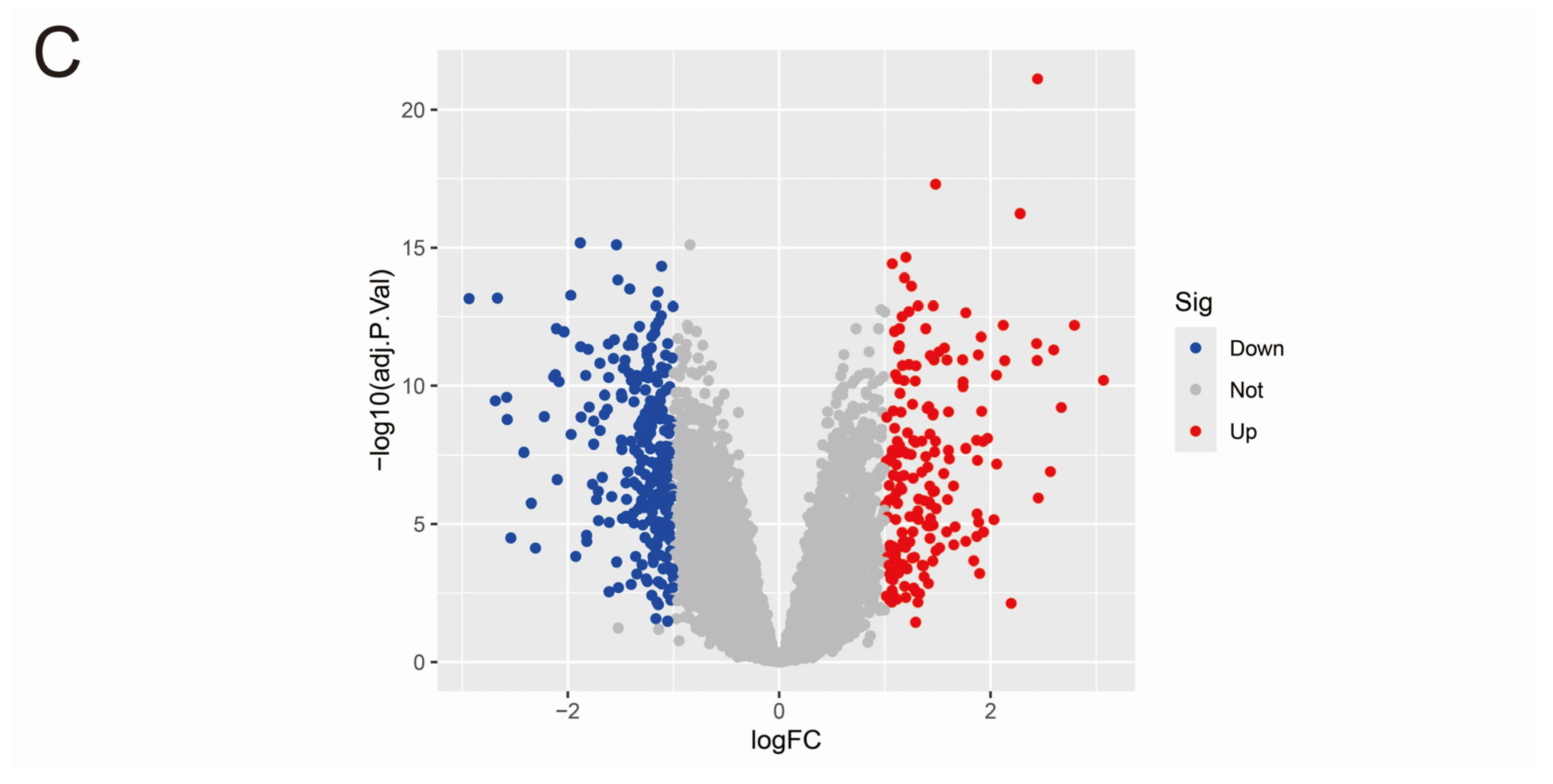
3.1. Data Merging and Gene Differential Expression Analysis
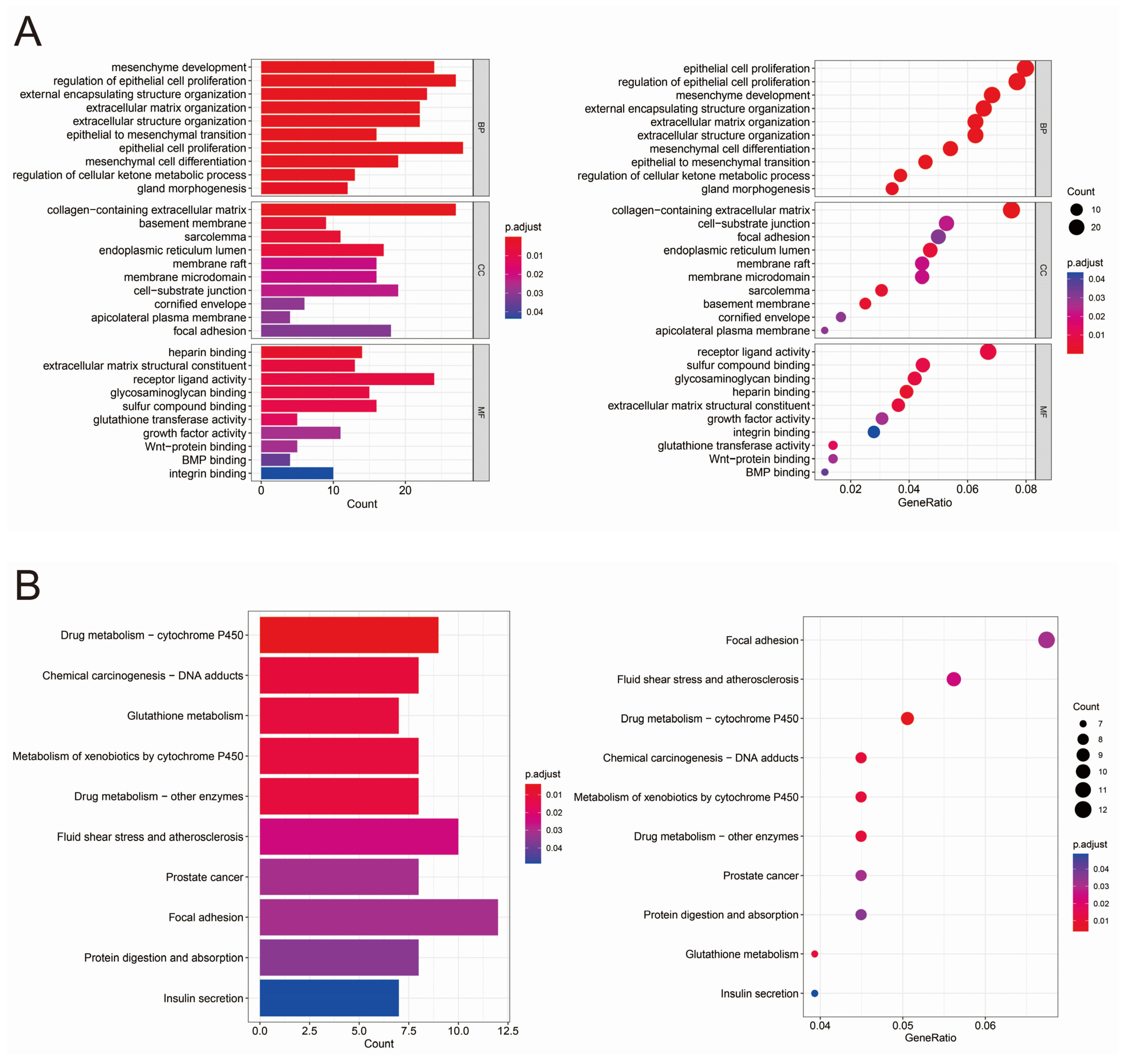
3.2. GO and KEGG Analysis of Differentially Expressed Genes
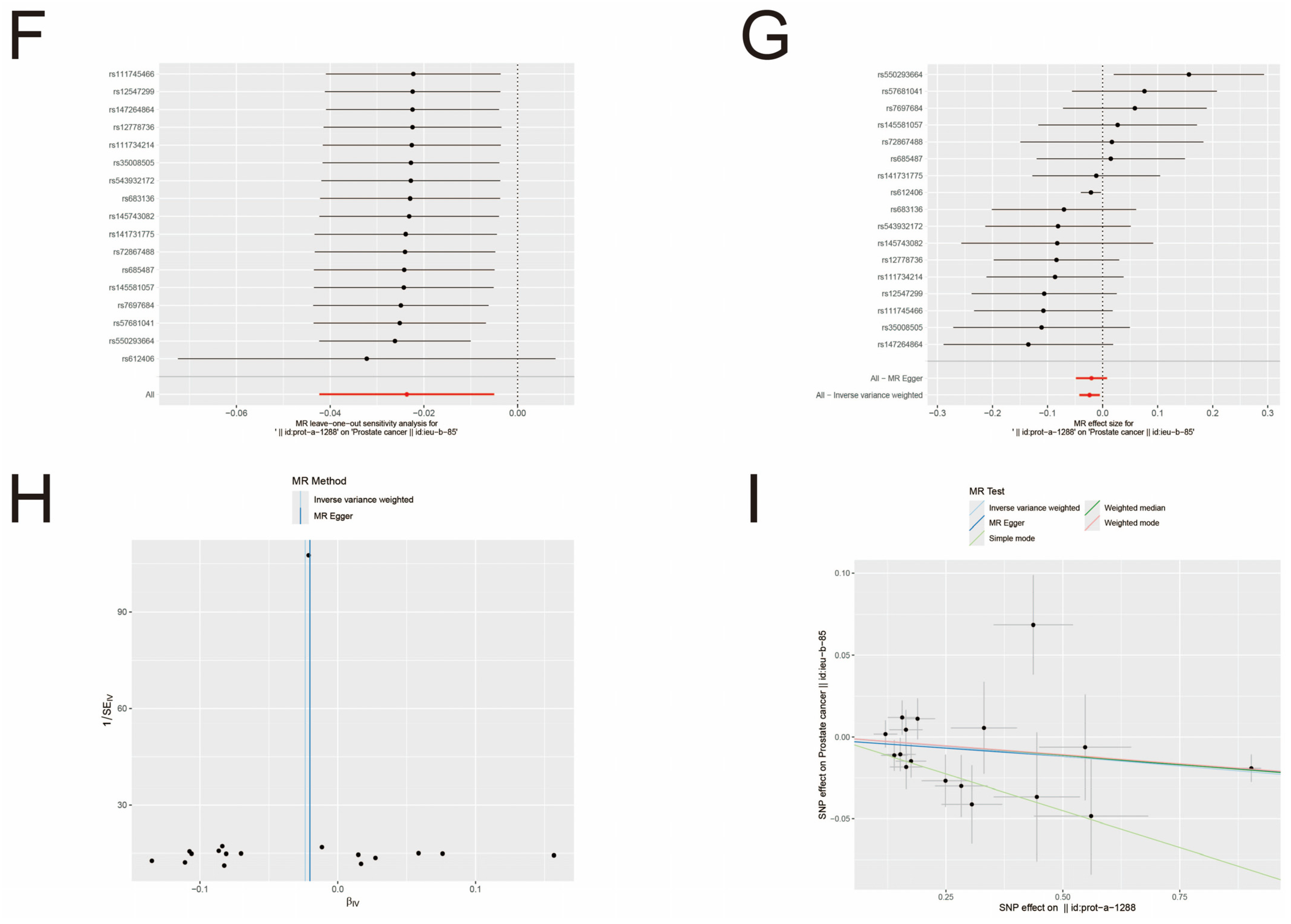
3.3. MR Analysis
3.4. Construct Drug-Gene Regulatory Network and ceRNA Regulatory Network
3.5. Immune Infiltration Analysis of Pathway Genes
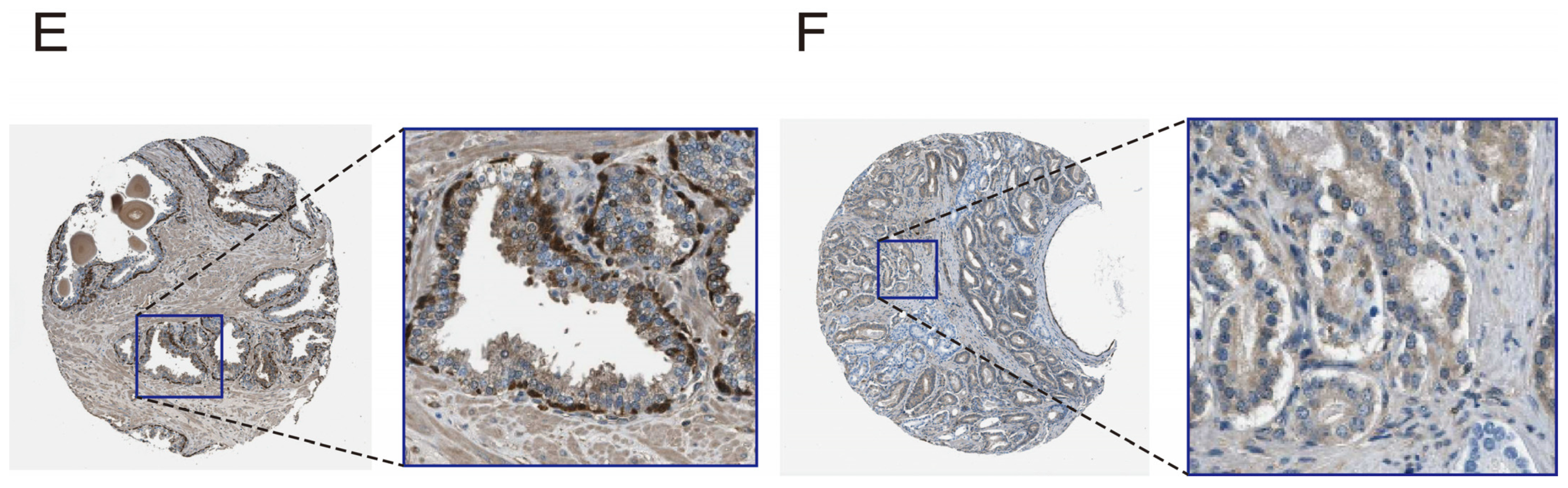
3.6. Evaluate and Verify the Diagnostic Ability and Expression Level of Pathway Genes
3.7. Constructing Prostate Cancer Prediction Models Based on Machine Learning and Pathway Genes
3.8. Single-Cell Analysis of Cell-Specific Expression Patterns of Pathway Genes in Prostate Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cornford, P.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Brunckhorst, O.; Darraugh, J.; Eberli, D.; De Meerleer, G.; De Santis, M.; Farolfi, A.; et al. EAU-EANM-ESTRO-ESUR-ISUP-SIOG Guidelines on Prostate Cancer-2024 Update. Part I: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2024, 86, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, E.M.; Srinivas, S.; Adra, N.; An, Y.; Bitting, R.; Chapin, B.; Cheng, H.H.; D’Amico, A.V.; Desai, N.; Dorff, T.; et al. NCCN Guidelines® Insights: Prostate Cancer, V 3.2024. J. Natl. Compr. Cancer Netw. 2024, 22, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Culp, M.B.; Soerjomataram, I.; Efstathiou, J.A.; Bray, F.; Jemal, A. Recent Global Patterns in Prostate Cancer Incidence and Mortality Rates. Eur. Urol. 2020, 77, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xu, Y.-X.; Wang, Y.-S.; Zhou, J.-L. Lipid metabolism, amino acid metabolism, and prostate cancer: A crucial metabolic journey. Asian J. Androl. 2024, 26, 123–134. [Google Scholar] [CrossRef]
- Kennedy, L.; Sandhu, J.K.; Harper, M.-E.; Cuperlovic-Culf, M. Role of Glutathione in Cancer: From Mechanisms to Therapies. Biomolecules 2020, 10, 1429. [Google Scholar] [CrossRef]
- Silva, M.M.; Rocha, C.R.R.; Kinker, G.S.; Pelegrini, A.L.; Menck, C.F.M. The balance between NRF2/GSH antioxidant mediated pathway and DNA repair modulates cisplatin resistance in lung cancer cells. Sci. Rep. 2019, 9, 17639. [Google Scholar] [CrossRef]
- Krajka-Kuźniak, V.; Paluszczak, J.; Baer-Dubowska, W. The Nrf2-ARE signaling pathway: An update on its regulation and possible role in cancer prevention and treatment. Pharmacol. Rep. 2017, 69, 393–402. [Google Scholar] [CrossRef]
- Song, K.; Yi, J.; Shen, X.; Cai, Y. Genetic polymorphisms of glutathione S-transferase genes GSTM1, GSTT1 and risk of hepatocellular carcinoma. PLoS ONE 2012, 7, e48924. [Google Scholar] [CrossRef]
- Zendehdel, K.; Bahmanyar, S.; McCarthy, S.; Nyren, O.; Andersson, B.; Ye, W. Genetic polymorphisms of glutathione S-transferase genes GSTP1, GSTM1, and GSTT1 and risk of esophageal and gastric cardia cancers. Cancer Causes Control 2009, 20, 2031–2038. [Google Scholar] [CrossRef][Green Version]
- Harries, L.W.; Stubbins, M.J.; Forman, D.; Howard, G.C.; Wolf, C.R. Identification of genetic polymorphisms at the glutathione S-transferase Pi locus and association with susceptibility to bladder, testicular and prostate cancer. Carcinogenesis 1997, 18, 641–644. [Google Scholar] [CrossRef]
- Bowden, J.; Holmes, M.V. Meta-analysis and Mendelian randomization: A review. Res. Synth. Methods 2019, 10, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Asantewaa, G.; Harris, I.S. Glutathione and its precursors in cancer. Curr. Opin. Biotechnol. 2021, 68, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Townsend, D.; Tew, K. Cancer drugs, genetic variation and the glutathione-S-transferase gene family. Am. J. Pharmacogenom. 2003, 3, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.R.; Reindl, K.M. Glutathione S-Transferases in Cancer. Antioxidants 2021, 10, 701. [Google Scholar] [CrossRef]
- Schnekenburger, M.; Karius, T.; Diederich, M. Regulation of epigenetic traits of the glutathione S-transferase P1 gene: From detoxification toward cancer prevention and diagnosis. Front. Pharmacol. 2014, 5, 170. [Google Scholar] [CrossRef]
- Hauptstock, V.; Kuriakose, S.; Schmidt, D.; Düster, R.; Müller, S.C.; von Ruecker, A.; Ellinger, J. Glutathione-S-transferase pi 1 (GSTP1) gene silencing in prostate cancer cells is reversed by the histone deacetylase inhibitor depsipeptide. Biochem. Biophys. Res. Commun. 2011, 412, 606–611. [Google Scholar] [CrossRef]
- Okino, S.T.; Pookot, D.; Majid, S.; Zhao, H.; Li, L.-C.; Place, R.F.; Dahiya, R. Chromatin changes on the GSTP1 promoter associated with its inactivation in prostate cancer. Mol. Carcinog. 2007, 46, 839–846. [Google Scholar] [CrossRef]
- Stirzaker, C.; Song, J.Z.; Davidson, B.; Clark, S.J. Transcriptional gene silencing promotes DNA hypermethylation through a sequential change in chromatin modifications in cancer cells. Cancer Res. 2004, 64, 3871–3877. [Google Scholar] [CrossRef]
- Strunnikova, M.; Schagdarsurengin, U.; Kehlen, A.; Garbe, J.C.; Stampfer, M.R.; Dammann, R. Chromatin inactivation precedes de novo DNA methylation during the progressive epigenetic silencing of the RASSF1A promoter. Mol. Cell. Biol. 2005, 25, 3923–3933. [Google Scholar] [CrossRef]
- Kanwal, R.; Pandey, M.; Bhaskaran, N.; Maclennan, G.T.; Fu, P.; Ponsky, L.E.; Gupta, S. Protection against oxidative DNA damage and stress in human prostate by glutathione S-transferase P1. Mol. Carcinog. 2014, 53, 8–18. [Google Scholar] [CrossRef]
- Noto, C.N.; Hoft, S.G.; DiPaolo, R.J. Mast Cells as Important Regulators in Autoimmunity and Cancer Development. Front. Cell Dev. Biol. 2021, 9, 752350. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, X.; Wei, H.; Liu, Y.; Li, N. Mast cells in colorectal cancer tumour progression, angiogenesis, and lymphangiogenesis. Front. Immunol. 2023, 14, 1209056. [Google Scholar] [CrossRef] [PubMed]
- Zudaire, E.; Martínez, A.; Garayoa, M.; Pío, R.; Kaur, G.; Woolhiser, M.R.; Metcalfe, D.D.; Hook, W.A.; Siraganian, R.P.; Guise, T.A.; et al. Adrenomedullin is a cross-talk molecule that regulates tumor and mast cell function during human carcinogenesis. Am. J. Pathol. 2006, 168, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Bresnick, A.R.; Weber, D.J.; Zimmer, D.B. S100 proteins in cancer. Nat. Rev. Cancer 2015, 15, 96–109. [Google Scholar] [CrossRef]
- Nasser, M.W.; Elbaz, M.; Ahirwar, D.K.; Ganju, R.K. Conditioning solid tumor microenvironment through inflammatory chemokines and S100 family proteins. Cancer Lett. 2015, 365, 11–22. [Google Scholar] [CrossRef]
- Delangre, E.; Oppliger, E.; Berkcan, S.; Gjorgjieva, M.; Correia de Sousa, M.; Foti, M. S100 Proteins in Fatty Liver Disease and Hepatocellular Carcinoma. Int. J. Mol. Sci. 2022, 23, 11030. [Google Scholar] [CrossRef]
- Grebhardt, S.; Veltkamp, C.; Ströbel, P.; Mayer, D. Hypoxia and HIF-1 increase S100A8 and S100A9 expression in prostate cancer. Int. J. Cancer 2012, 131, 2785–2794. [Google Scholar] [CrossRef]
- Diederichs, S.; Bulk, E.; Steffen, B.; Ji, P.; Tickenbrock, L.; Lang, K.; Zänker, K.S.; Metzger, R.; Schneider, P.M.; Gerke, V.; et al. S100 family members and trypsinogens are predictors of distant metastasis and survival in early-stage non-small cell lung cancer. Cancer Res. 2004, 64, 5564–5569. [Google Scholar] [CrossRef]
- Shafi, A.A.; Yen, A.E.; Weigel, N.L. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol. Ther. 2013, 140, 223–238. [Google Scholar] [CrossRef]
- Desai, K.; McManus, J.M.; Sharifi, N. Hormonal Therapy for Prostate Cancer. Endocr. Rev. 2021, 42, 354–373. [Google Scholar] [CrossRef]
- Lafront, C.; Germain, L.; Campolina-Silva, G.H.; Weidmann, C.; Berthiaume, L.; Hovington, H.; Brisson, H.; Jobin, C.; Frégeau-Proulx, L.; Cotau, R.; et al. The estrogen signaling pathway reprograms prostate cancer cell metabolism and supports proliferation and disease progression. J. Clin. Investig. 2024, 134, e170809. [Google Scholar] [CrossRef]
- Joerger, A.C.; Fersht, A.R. The p53 Pathway: Origins, Inactivation in Cancer, and Emerging Therapeutic Approaches. Annu. Rev. Biochem. 2016, 85, 375–404. [Google Scholar] [CrossRef] [PubMed]
- Hernández Borrero, L.J.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Et Biophys. Acta (BBA) Rev. Cancer 2021, 1876, 188556. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.; Levine, A. p53 Research: The past thirty years and the next thirty years. Cold Spring Harb. Perspect. Biol. 2010, 2, a000893. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Su, Z.; Tavana, O.; Gu, W. Understanding the complexity of p53 in a new era of tumor suppression. Cancer Cell 2024, 42, 946–967. [Google Scholar] [CrossRef]
- Ventura, A.; Kirsch, D.G.; McLaughlin, M.E.; Tuveson, D.A.; Grimm, J.; Lintault, L.; Newman, J.; Reczek, E.E.; Weissleder, R.; Jacks, T. Restoration of p53 function leads to tumour regression in vivo. Nature 2007, 445, 661–665. [Google Scholar] [CrossRef]
- Martins, C.P.; Brown-Swigart, L.; Evan, G.I. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 2006, 127, 1323–1334. [Google Scholar] [CrossRef]
- Xue, W.; Zender, L.; Miething, C.; Dickins, R.A.; Hernando, E.; Krizhanovsky, V.; Cordon-Cardo, C.; Lowe, S.W. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007, 445, 656–660. [Google Scholar] [CrossRef]
- Morana, O.; Wood, W.; Gregory, C.D. The Apoptosis Paradox in Cancer. Int. J. Mol. Sci. 2022, 23, 1328. [Google Scholar] [CrossRef]
- Xu, X.; Lai, Y.; Hua, Z.-C. Apoptosis and apoptotic body: Disease message and therapeutic target potentials. Biosci. Rep. 2019, 39, BSR20180992. [Google Scholar] [CrossRef]
- Zhu, M.; Liu, D.; Liu, G.; Zhang, M.; Pan, F. Caspase-Linked Programmed Cell Death in Prostate Cancer: From Apoptosis, Necroptosis, and Pyroptosis to PANoptosis. Biomolecules 2023, 13, 1715. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Kulik, G. Signaling Pathways That Control Apoptosis in Prostate Cancer. Cancers 2021, 13, 937. [Google Scholar] [CrossRef] [PubMed]
- Wosik, K.; Biernacki, K.; Khouzam, M.-P.; Prat, A. Death receptor expression and function at the human blood brain barrier. J. Neurol. Sci. 2007, 259, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Cueto, A.; Hernandez-Cueto, D.; Antonio-Andres, G.; Mendoza-Marin, M.; Jimenez-Gutierrez, C.; Sandoval-Mejia, A.L.; Mora-Campos, R.; Gonzalez-Bonilla, C.; Vega, M.I.; Bonavida, B.; et al. Death receptor 5 expression is inversely correlated with prostate cancer progression. Mol. Med. Rep. 2014, 10, 2279–2286. [Google Scholar] [CrossRef]
- Carneiro, B.A.; El-Deiry, W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef]




| GWAS Database | |||
|---|---|---|---|
| Phenotype | Consortium | Sample Size | GWAS ID |
| Exposure | |||
| Glutathione peroxidase 1 measurement | NA | 10,708 | ebi-a-GCST90019397 |
| Glutathione peroxidase 7 | NA | 3301 | prot-a-1265 |
| Glutathione S-transferase A1 | NA | 3301 | prot-a-1283 |
| Glutathione S-transferase A3 | NA | 3301 | prot-a-1284 |
| Glutathione S-transferase A4 | NA | 3301 | prot-a-1285 |
| Glutathione S-transferase kappa 1 | NA | 3301 | prot-a-1286 |
| Glutathione S-transferase Mu 1 | NA | 3301 | prot-a-1287 |
| Glutathione S-transferase omega-1 | NA | 3301 | prot-a-1288 |
| Glutathione S-transferase P | NA | 3301 | prot-a-1289 |
| Glutathione S-transferase theta-2B | NA | 3301 | prot-a-1291 |
| Lactoylglutathione lyase | NA | 3301 | prot-a-1218 |
| S-Formylglutathione hydrolase | NA | 3301 | prot-a-989 |
| Cysteine-glutathione disulfide | NA | 1997 | met-a-680 |
| Glutathione S-transferase Pi | NA | NA | prot-c-4911_49_2 |
| Outcome | |||
| Prostate cancer | PRACTICAL | 140,254 | ieu-b-85 |
| GEO Database | |||
| Accession | Sample Source | Sequencing Type | Dataset Usage |
| GSE46602 | Prostate cancer | Array | Training dataset |
| GSE55945 | Prostate cancer | Array | Training dataset |
| GSE69223 | Prostate cancer | Array | Training dataset |
| GSE62872 | Prostate cancer | Array | Testing dataset |
| GSE193337 | Prostate cancer | RNA-seq | Single cell analysis |
| Gene | Drug | Interaction_Types | Sources |
|---|---|---|---|
| GSTP1 | EZATIOSTAT HYDROCHLORIDE | inhibitor | ChemblInteractions |
| GSTP1 | CARBOPLATIN | NCI|CIViC | |
| GSTP1 | CURCUMIN | NCI | |
| GSTP1 | LEUCOVORIN | PharmGKB | |
| GSTP1 | BLEOMYCIN | PharmGKB | |
| GSTP1 | ISONIAZID | PharmGKB | |
| GSTP1 | OXALIPLATIN | NCI|PharmGKB | |
| GSTP1 | AZACITIDINE | NCI | |
| GSTP1 | IFOSFAMIDE | NCI | |
| GSTP1 | GARLIC | NCI | |
| GSTP1 | EZATIOSTAT | TdgClinicalTrial|TTD | |
| GSTP1 | SULFORAPHANE | NCI | |
| GSTP1 | PYRIMETHAMINE | PharmGKB | |
| GSTP1 | PLATINUM | PharmGKB | |
| GSTP1 | CYCLOPHOSPHAMIDE | NCI|PharmGKB | |
| GSTP1 | DROLOXIFENE | NCI | |
| GSTP1 | RESVERATROL | NCI | |
| GSTP1 | OMEPRAZOLE | NCI | |
| GSTP1 | DOXORUBICIN | PharmGKB | |
| GSTP1 | EPIRUBICIN | PharmGKB | |
| GSTP1 | IRINOTECAN HYDROCHLORIDE | NCI | |
| GSTP1 | CYTARABINE | NCI | |
| GSTP1 | VITAMIN E | NCI | |
| GSTP1 | MELPHALAN | NCI | |
| GSTP1 | ALCOHOL | NCI | |
| GSTP1 | ADRIAMYCIN | NCI | |
| GSTP1 | PREDNISONE | NCI | |
| GSTP1 | EXATECAN MESYLATE | NCI | |
| GSTP1 | THIOTEPA | PharmGKB | |
| GSTP1 | CAMPTOTHECIN | NCI | |
| GSTP1 | DOCETAXEL | NCI | |
| GSTP1 | FLUOROURACIL | PharmGKB | |
| GSTP1 | DECITABINE | NCI | |
| GSTP1 | PACLITAXEL | CIViC | |
| GSTP1 | BUSULFAN | NCI|PharmGKB | |
| GSTP1 | DEXAMETHASONE | NCI | |
| GSTP1 | SELENOMETHIONINE | NCI | |
| GSTP1 | DITIOCARB | NCI | |
| GSTP1 | MISONIDAZOLE | NCI | |
| GSTP1 | LYCOPENE | NCI | |
| GSTP1 | DAUNORUBICIN | PharmGKB | |
| GSTP1 | RIFAMPIN | PharmGKB | |
| GSTP1 | ETOPOSIDE | PharmGKB | |
| GSTP1 | CANFOSFAMIDE | TdgClinicalTrial|TTD | |
| GSTP1 | CISPLATIN | NCI|CIViC|PharmGKB | |
| GSTP1 | HYDROQUINONE | NCI | |
| GSTP1 | PERFOSFAMIDE | NCI | |
| GSTP1 | VERAPAMIL | NCI | |
| GSTM1 | CYCLOPHOSPHAMIDE | PharmGKB | |
| GSTM1 | PACLITAXEL | PharmGKB | |
| GSTM1 | CURCUMIN | NCI | |
| GSTM1 | DOXORUBICIN | PharmGKB | |
| GSTM1 | DICUMAROL | NCI | |
| GSTM1 | VINBLASTINE | PharmGKB | |
| GSTM1 | SULFORAPHANE | NCI | |
| GSTM1 | CHLORAMBUCIL | NCI | |
| GSTM1 | CLOZAPINE | PharmGKB | |
| GSTM1 | AZATHIOPRINE | PharmGKB | |
| GSTM1 | TROGLITAZONE | NCI | |
| GSTM1 | NITROGLYCERIN | NCI | |
| GSTM1 | IRINOTECAN HYDROCHLORIDE | NCI | |
| GSTM1 | METHOTREXATE | NCI | |
| GSTM1 | BLEOMYCIN | PharmGKB | |
| GSTM1 | PREDNISONE | NCI | |
| GSTM1 | SULFAMETHOXAZOLE | PharmGKB | |
| GSTM1 | DOCETAXEL | NCI | |
| GSTM1 | DACARBAZINE | PharmGKB | |
| GSTM1 | TRIMETHOPRIM | PharmGKB | |
| GSTM1 | BUSULFAN | PharmGKB | |
| RRM2 | HYDROXYUREA | inhibitor | ChemblInteractions|TTD |
| RRM2 | FLUDARABINE PHOSPHATE | inhibitor | ChemblInteractions |
| RRM2 | GALLIUM NITRATE | inhibitor | ChemblInteractions |
| RRM2 | CLOFARABINE | inhibitor | ChemblInteractions |
| RRM2 | GEMCITABINE HYDROCHLORIDE | inhibitor | ChemblInteractions |
| RRM2 | GEMCITABINE | inhibitor | ClearityFoundationClinicalTrial|TTD |
| RRM2 | TEZACITABINE | inhibitor | ChemblInteractions |
| RRM2 | CLADRIBINE | inhibitor | PharmGKB |
| RRM2 | LY-2334737 | TTD | |
| RRM2 | CYTARABINE | PharmGKB | |
| RRM2 | TRIAPINE | TdgClinicalTrial|TTD | |
| RRM2 | GEMCITABINE ELAIDATE | TTD | |
| GSTM3 | OLANZAPINE | PharmGKB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.; Yin, H. Exploring the Molecular Mechanism and Role of Glutathione S-Transferase P in Prostate Cancer. Biomedicines 2025, 13, 1051. https://doi.org/10.3390/biomedicines13051051
Huang S, Yin H. Exploring the Molecular Mechanism and Role of Glutathione S-Transferase P in Prostate Cancer. Biomedicines. 2025; 13(5):1051. https://doi.org/10.3390/biomedicines13051051
Chicago/Turabian StyleHuang, Shan, and Hang Yin. 2025. "Exploring the Molecular Mechanism and Role of Glutathione S-Transferase P in Prostate Cancer" Biomedicines 13, no. 5: 1051. https://doi.org/10.3390/biomedicines13051051
APA StyleHuang, S., & Yin, H. (2025). Exploring the Molecular Mechanism and Role of Glutathione S-Transferase P in Prostate Cancer. Biomedicines, 13(5), 1051. https://doi.org/10.3390/biomedicines13051051





