Prognostic Significance of CLDN1, INHBA, and CXCL12 in Colon Adenocarcinoma: A Multi-Omics and Single-Cell Approach
Abstract
1. Introduction
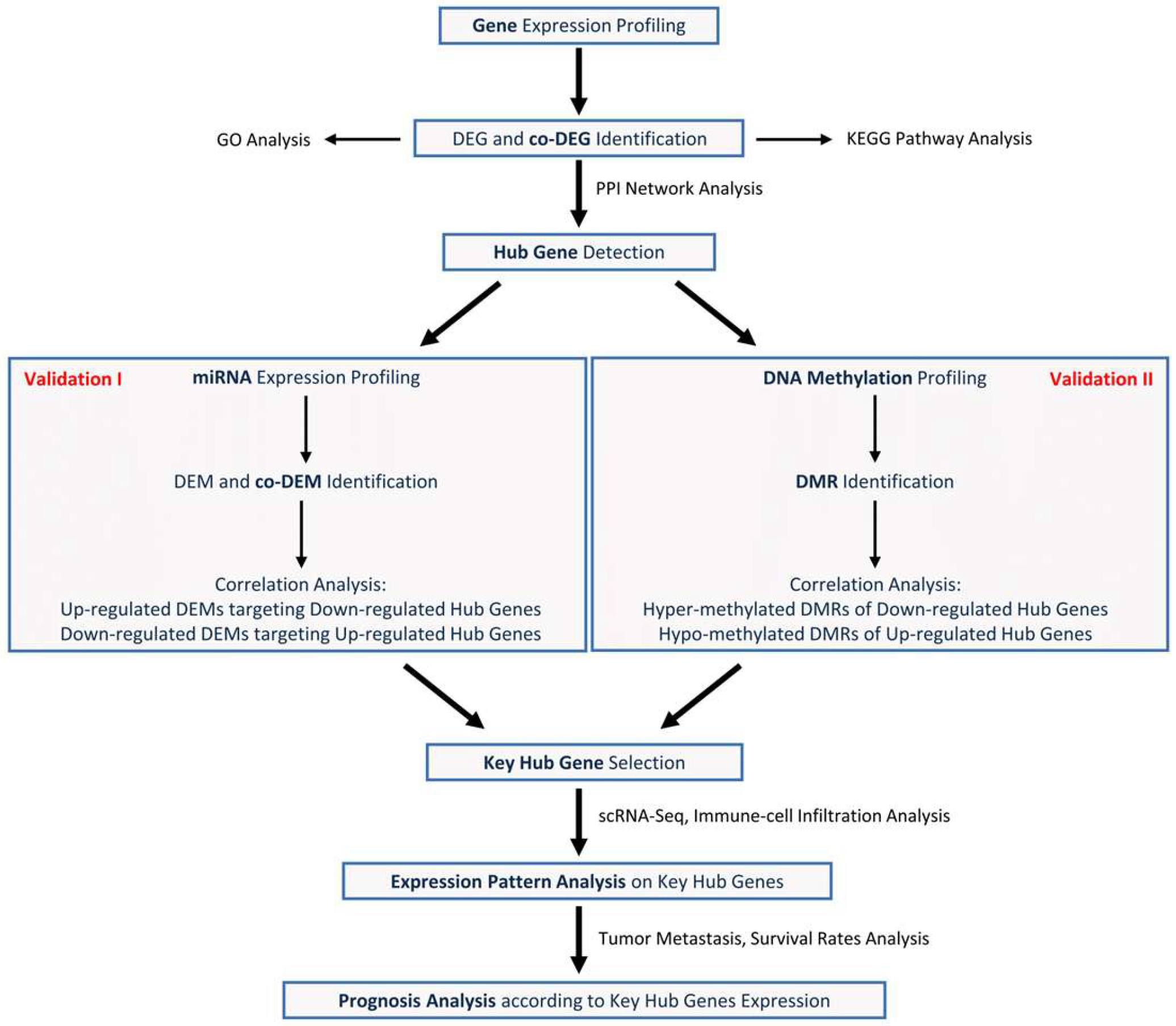
2. Materials and Methods
2.1. Microarray Data
2.2. Identification of Differentially Expressed Genes (DEGs) and Co-DEGs
2.3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis of Up- and Down-Regulated Co-DEGs
2.4. Protein–Protein Interaction (PPI) Network Buildup on Up- and Down-Regulated DEGs for Hub Genes Detection
2.5. Analysis of Differentially Expressed miRNAs (DEMs) Related to the Hub Gene Expression
2.6. Analysis of Differentially Methylated Regions (DMRs) of Hub Genes
2.7. Hub Gene Expression Pattern Analysis in the COAD scRNA-Seq Dataset
2.8. Analysis of Immune Cell Infiltration Level
2.9. Tumor Metastasis Analysis According to Hub Gene Expression
2.10. Analysis of Survival Rates According to Hub Gene Expression
2.11. Data Visualization
3. Results
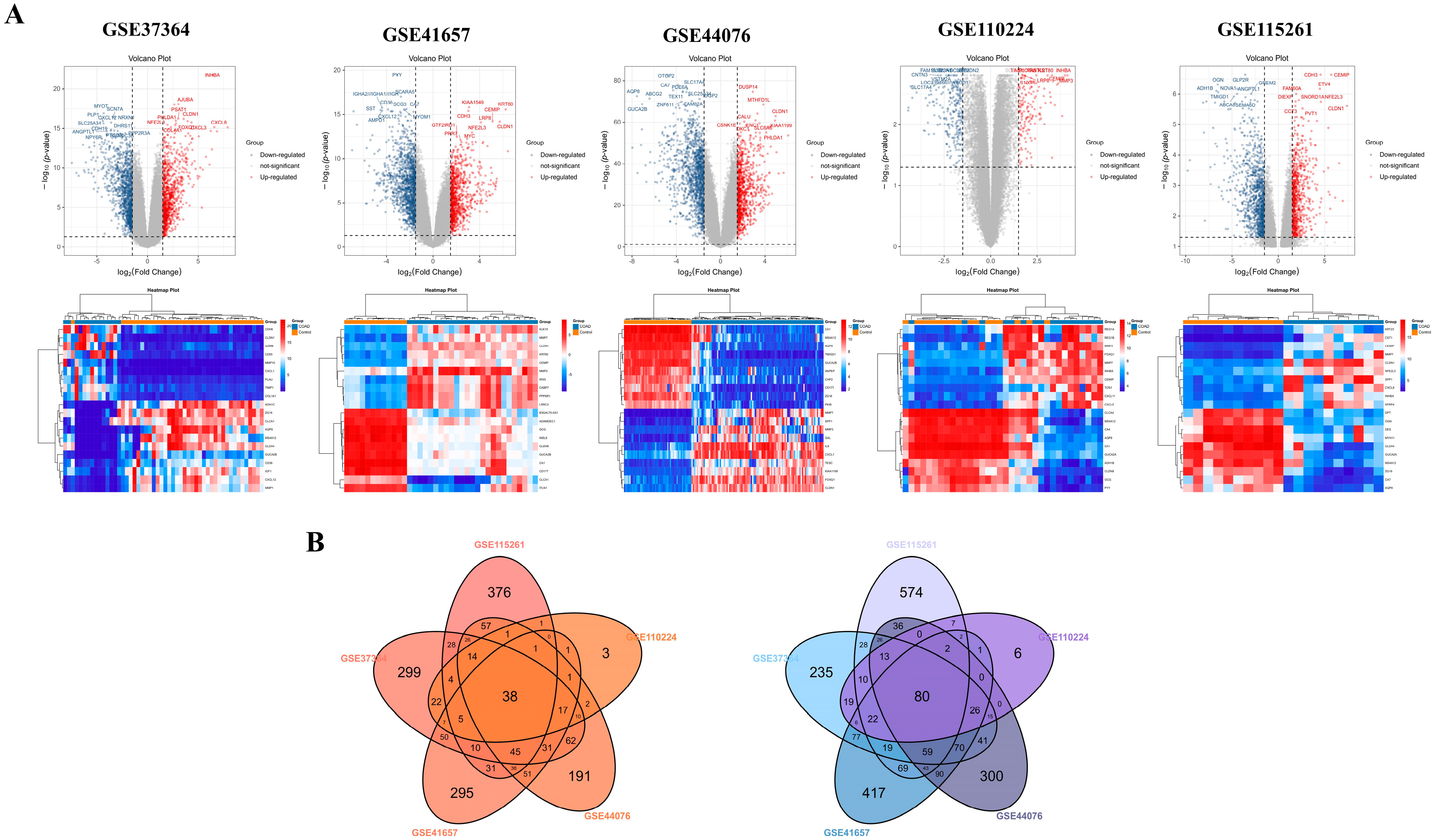
3.1. Identification of DEGs and Co-DEGs in Five COAD Gene Datasets
3.2. GO and KEGG Pathway Analysis on Up- and Down-Regulated Co-DEGs
3.3. PPI Network Construction of Co-DEGs and Detection of Hub Genes
3.4. Identification of DEMs Regulating Hub Gene Expression in Three COAD Datasets
3.5. Identification of DMRs Modulating the Expression of Hub Genes
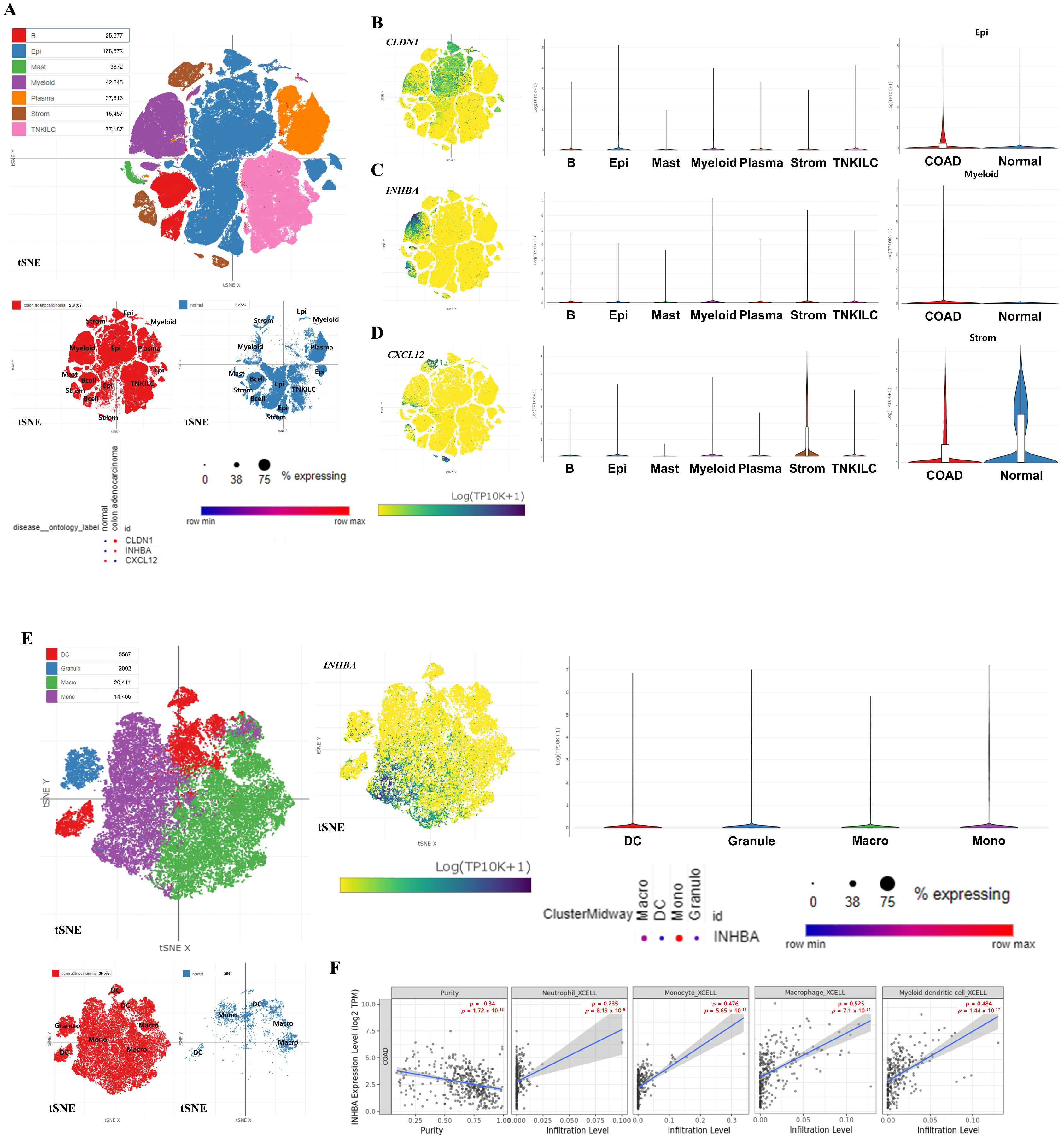
3.6. Expression Pattern Analysis of Hub Genes Using COAD scRNA-Seq Dataset
3.7. Correlation Analysis Between Immune Cell Infiltration and INHBA Expression Levels
3.8. Tumor Progress Analysis Based on the Expression of Hub Genes
4. Discussion
- CLDN1 and INHBA are consistently over-expressed in COAD and are associated with poor prognosis and tumor progression, suggesting their potential role as negative prognostic biomarkers.
- CXCL12 is down-regulated and epigenetically silenced in COAD and may be in-volved in early tumor suppression, offering insight into immune micro-environment modulation.
- Epigenetic regulation (via miRNAs and DNA methylation) plays a critical role in gene dysregulation in COAD and may represent therapeutic targets or predictive markers.
- Integrated multi-omics analysis improves the identification of functionally relevant and clinically applicable biomarkers for personalized treatment strategies in COAD.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef] [PubMed]
- Marcellinaro, R.; Spoletini, D.; Grieco, M.; Avella, P.; Cappuccio, M.; Troiano, R.; Lisi, G.; Garbarino, G.M.; Carlini, M. Colorectal cancer: Current updates and future perspectives. J. Clin. Med. 2024, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Yi, K.; Wu, J.; Tang, X.; Zhang, Q.; Wang, B.; Wang, F. Identification of a novel glycolysis-related gene signature for predicting the survival of patients with colon adenocarcinoma. Scand. J. Gastroenterol. 2022, 57, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, J.; Gao, J.; Ding, M.; Li, H. The expression of SERPINE1 in colon cancer and its regulatory network and prognostic value. BMC Gastroenterol. 2023, 23, 33. [Google Scholar] [CrossRef]
- Huang, H.; Xu, S.; Chen, A.; Li, F.; Wu, J.; Tu, X.; Hu, K. Identification of a 5-gene-based scoring system by WGCNA and LASSO to predict prognosis for rectal cancer patients. Anal. Cell Pathol. 2021, 2021, 6697407. [Google Scholar] [CrossRef]
- Dayde, D.; Tanaka, I.; Jain, R.; Tai, M.C.; Taguchi, A. Predictive and prognostic molecular biomarkers for response to neoadjuvant chemoradiation in rectal cancer. Int. J. Mol. Sci. 2017, 18, 573. [Google Scholar] [CrossRef]
- De Felice, F.; Crocetti, D.; Maiuri, V.; Parisi, M.; Marampon, F.; Izzo, L.; De Toma, G.; Musio, D.; Tombolini, V. Locally advanced rectal cancer: Treatment approach in elderly patients. Curr. Treat. Options Oncol. 2020, 21, 1. [Google Scholar] [CrossRef]
- Ciardiello, F.; Ciardiello, D.; Martini, G.; Napolitano, S.; Tabernero, J.; Cervantes, A. Clinical management of metastatic colorectal cancer in the era of precision medicine. CA Cancer J. Clin. 2022, 72, 372–401. [Google Scholar] [CrossRef]
- Dariya, B.; Aliya, S.; Merchant, N.; Alam, A.; Nagaraju, G.P. Colorectal cancer biology, diagnosis, and therapeutic approaches. Crit. Rev. Oncog. 2020, 25, 71–94. [Google Scholar] [CrossRef]
- Ding, S.; Sun, X.; Zhu, L.; Li, Y.; Chen, W.; Shen, K. Identification of a novel immune-related prognostic signature associated with tumor microenvironment for breast cancer. Int. Immunopharmacol. 2021, 100, 108122. [Google Scholar] [CrossRef]
- Luo, R.; Guo, W.; Wang, H. A comprehensive analysis of tumor microenvironment-related genes in colon cancer. Clin. Transl. Oncol. 2021, 23, 1769–1781. [Google Scholar] [CrossRef] [PubMed]
- Babar, Q.; Saeed, A.; Tabish, T.A.; Sarwar, M.; Thorat, N.D. Targeting the tumor microenvironment: Potential strategy for cancer therapeutics. Biochim. Biophys. Acta Mol. Basis Dis. 2023, 1869, 166746. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Xiao, F.; Chen, M.; Gao, H. Tumor-microenvironment-responsive nanomedicine for enhanced cancer immunotherapy. Adv. Sci. 2022, 9, 2103836. [Google Scholar] [CrossRef] [PubMed]
- Arneth, B. MicroRNAs in the tumor microenvironment. Medicina 2019, 56, 15. [Google Scholar] [CrossRef]
- Yang, J.; Xu, J.; Wang, W.; Zhang, B.; Yu, X.; Shi, S. Epigenetic regulation in the tumor microenvironment: Molecular mechanisms and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 210. [Google Scholar] [CrossRef]
- Chen, S.; Shen, X. Long non-coding RNAs: Functions and mechanisms in colon cancer. Mol. Cancer 2020, 19, 167. [Google Scholar] [CrossRef]
- Sean, D.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar]
- Sherman, B.T.; Huang, D.W.; Tan, Q.; Guo, Y.; Bour, S.; Liu, D.; Stephens, R.; Baseler, M.W.; Lane, C.H.; Lempicki, R.A. DAVID Knowledgebase: A gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinform. 2007, 8, 426. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA, and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.Y.; Kingston, E.R.; Kleaveland, B.; Lin, D.H.; Stubna, M.W.; Bartel, D.P. The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science 2020, 370, 6523. [Google Scholar] [CrossRef]
- Egger, G.; Aparicio, A.; Jones, P.A.; Liang, G. Epigenetics in human diseases and prospects of epigenetic therapy. Nature 2004, 429, 457–463. [Google Scholar] [CrossRef]
- Tarhan, L.; Bistline, J.; Chang, J.; Galloway, B.; Hanna, E.; Weitz, E. Single cell portal: An interactive home for single-cell genomics data. bioRxiv 2023. bioRxiv: 2023.07.13.548886. [Google Scholar]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Bartha, Á.; Győrffy, B. Tnmplot.Com: A web tool for the comparison of gene expression in normal, tumor and metastatic tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Győrffy, B. Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. Br. J. Pharmacol. 2024, 181, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Miao, B.; Wang, S.; Dong, W.; Xu, H.; Si, C.; Wang, W.; Duan, S.; Lou, J.; Bao, Z.; et al. Hiplot: A comprehensive and easy-to-use web service for boosting publication-ready biomedical data visualization. Brief. Bioinform. 2022, 23, bbac261. [Google Scholar] [CrossRef]
- Wang, X.; Duanmu, J.; Fu, X.; Li, T.; Jiang, Q. Analyzing and validating the prognostic value and mechanism of colon cancer immune microenvironment. J. Transl. Med. 2020, 18, 324. [Google Scholar] [CrossRef]
- Neophytou, C.M.; Panagi, M.; Stylianopoulos, T.; Papageorgis, P. The role of tumor microenvironment in cancer metastasis: Molecular mechanisms and therapeutic opportunities. Cancers 2021, 13, 2053. [Google Scholar] [CrossRef]
- Pajares, M.J.; Alemany-cosme, E.; Goñi, S.; Bandres, E.; Palanca-Ballester, C.; Sandoval, J. Epigenetic regulation of microRNAs in cancer: Shortening the distance from bench to bedside. Int. J. Mol. Sci. 2021, 22, 7350. [Google Scholar] [CrossRef] [PubMed]
- Aure, M.R.; Fleischer, T.; Bjørklund, S.; Ankill, J.; Castro-Mondragon, J.A.; Bathen, T.F.; Borgen, E.; Engebråten, O.; Hartman-Johnsen, O.J.; Garred, Ø.; et al. Crosstalk between microRNA expression and DNA methylation drives the hormone-dependent phenotype of breast cancer. Genome Med. 2021, 13, 72. [Google Scholar] [CrossRef]
- Okayama, H.; Schetter, A.J.; Harris, C.C. MicroRNAs and inflammation in the pathogenesis and progression of colon cancer. Dig. Dis. 2012, 30, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Fan, X.M. The pathological role of microRNAs and inflammation in colon carcinogenesis. Clin. Res. Hepatol. Gastroenterol. 2015, 39, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Okano, M.; Yamamoto, H.; Ohkuma, H.; Kano, Y.; Kim, H.; Nishikawa, S.; Konno, M.; Kawamoto, K.; Haraguchi, N.; Takemasa, I.; et al. Significance of INHBA expression in human colorectal cancer. Oncol. Rep. 2013, 30, 2903–2908. [Google Scholar] [CrossRef]
- Seder, C.W.; Hnrtojo, W.; Lin, L.; Silvers, A.L.; Wang, Z.; Thomas, D.G.; Giordano, T.J.; Chen, G.; Chang, A.C.; Orringer, M.B.; et al. Upregulated INHBA expression may promote cell proliferation and is associated with poor survival in lung adenocarcinoma. Neoplasia 2009, 11, 388–396. [Google Scholar] [CrossRef]
- Katayama, Y.; Oshima, T.; Sakamaki, K.; Aoyama, T.; Sato, T.; Masudo, K.; Shiozawa, M.; Yoshikawa, T.; Rino, Y.; Imada, T.; et al. Clinical significance of INHBA gene expression in patients with gastric cancer who receive curative resection followed by adjuvant s-1 chemotherapy. In Vivo 2017, 31, 565–571. [Google Scholar]
- Seder, C.W.; Hartojo, W.; Lin, L.; Silvers, A.L.; Wang, Z.; Thomas, D.G.; Giordano, T.J.; Chen, G.; Chang, A.C.; Orringer, M.B.; et al. INHBA overexpression promotes cell proliferation and may be epigenetically regulated in esophageal adenocarcinoma. J. Thorac. Oncol. 2009, 4, 455–462. [Google Scholar] [CrossRef]
- Wildi, S.; Kleeff, J.; Maruyama, H.; Maurer, C.A.; Büchler, M.W.; Korc, M. Overexpression of activin A in stage IV colorectal cancer. Gut 2001, 49, 409–417. [Google Scholar] [CrossRef]
- Dean, M.; Davis, D.A.; Burdette, J.E. Activin A stimulates migration of the fallopian tube epithelium, an origin of high-grade serous ovarian cancer, through non-canonical signaling. Cancer Lett. 2017, 391, 114–124. [Google Scholar] [CrossRef]
- Lee, H.Y.; Li, C.C.; Huang, C.N.; Li, W.M.; Yeh, H.C.; Ke, H.L.; Yang, K.F.; Liang, P.I.; Li, C.F.; Wu, W.J. INHBA overexpression indicates poor prognosis in urothelial carcinoma of the urinary bladder and upper tract. J. Surg. Oncol. 2015, 111, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Liu, Y. INHBA promotes the proliferation, migration, and invasion of colon cancer cells through the upregulation of VCAN. J. Int. Med. Res. 2021, 49, 03000605211014998. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, Q.; Li, M.; Jiang, S.; Wang, X. MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA. Biochem. Biophys. Res. Commun. 2014, 444, 199–204. [Google Scholar] [CrossRef]
- Xu, L.; Wen, T.; Liu, Z.; Xu, F.; Yang, L.; Liu, J.; Feng, G.; An, G. MicroRNA-375 suppresses human colorectal cancer metastasis by targeting Frizzled 8. Oncotarget 2016, 7, 40644–40656. [Google Scholar] [CrossRef]
- Han, K.H.; Cho, H.; Han, K.R.; Mun, S.K.; Kim, Y.K.; Park, I.; Chang, M. Role of microRNA-375-3p-mediated regulation in tinnitus development. Int. J. Mol. Med. 2021, 48, 136. [Google Scholar] [CrossRef]
- Kang, W.; Huang, T.; Zhou, Y.; Zhang, J.; Lung, R.W.M.; Tong, J.H.M.; Chan, A.W.H.; Zhang, B.; Wong, C.C.; Wu, F.; et al. MiR-375 is involved in the Hippo pathway by targeting the YAP1/TEAD4-CTGF axis in the gastric carcinogenesis article. Cell Death Dis. 2018, 9, 92. [Google Scholar] [CrossRef]
- Łukaszewicz-Zając, M.; Mroczko, B. Claudins-promising biomarkers for selected gastrointestinal (GI) malignancies? Cancers 2023, 16, 152. [Google Scholar] [CrossRef]
- Morin, P.J. Claudin proteins in human cancer: Promising new targets for diagnosis and therapy. Cancer Res. 2005, 65, 9603–9606. [Google Scholar] [CrossRef] [PubMed]
- Ouban, A. Claudin-1 role in colon cancer: An update and a review. Histol. Histopathol. 2018, 33, 1013–1019. [Google Scholar]
- Huang, J.; Li, J.; Qu, Y.; Zhang, J.; Zhang, L.; Chen, X.; Liu, B.; Zhu, Z. The expression of Claudin 1 correlates with β-catenin and is a prognostic factor of poor outcome in gastric cancer. Int. J. Oncol. 2014, 44, 1293–1301. [Google Scholar] [CrossRef]
- De Vicente, J.C.; Fernández-Valle, Á.; Vivanco-Allende, B.; Rodríguez Santamarta, T.; Lequerica-Fernández, P.; Hernández-Vallejo, G.; Allonca-Campa, E. The prognostic role of claudins -1 and -4 in oral squamous cell carcinoma. Anticancer Res. 2015, 35, 2949–2960. [Google Scholar]
- De Oliveira, S.S.; De Oliveira, I.M.; De Souza, W.; Morgado-Díaz, J.A. Claudins upregulation in human colorectal cancer. FEBS Lett. 2005, 579, 6179–6185. [Google Scholar] [CrossRef] [PubMed]
- Kinugasa, T.; Akagi, Y.; Shirouzu, K. Claudin-1 protein is a major factor involved in the tumorigenesis of colorectal. Gastroenterology 2009, 136, A-320. [Google Scholar] [CrossRef]
- Dhawan, P.; Singh, A.B.; Deane, N.G.; No, Y.R.; Shiou, S.R.; Schmidt, C.; Neff, J.; Washington, M.K.; Beauchamp, R.D. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J. Clin. Investig. 2005, 115, 1765–1776. [Google Scholar] [CrossRef]
- Kinugasa, T.; Akagi, Y.; Ochi, T.; Tanaka, N. Increased claudin-1 protein expression in hepatic metastatic lesions of colorectal cancer. Anticancer Res. 2012, 32, 2309–2314. [Google Scholar] [PubMed]
- Abdelzaher, E.; Rizk, A.M.; Bessa, S.S.; Omer, K.M. Predictive value of immunohistochemical expression of claudin-1 in colonic carcinoma. J. Egypt. Natl. Cancer Inst. 2011, 23, 123–131. [Google Scholar] [CrossRef][Green Version]
- Shibutani, M.; Noda, E.; Maeda, K.; Nagahara, H.; Ohtani, H.; Hirakawa, K. Low expression of Claudin-1 and presence of poorly differentiated tumor clusters correlate with poor prognosis in colorectal cancer. Anticancer Res. 2013, 33, 3301–3306. [Google Scholar]
- Nishikawa, E.; Osada, H.; Okazaki, Y.; Arima, C.; Tomida, S.; Tatematsu, Y.; Taguchi, A.; Shimada, Y.; Yanagisawa, K.; Yatabe, Y.; et al. MiR-375 is activated by ASH1 and inhibits YAP1 in a lineage-dependent manner in lung cancer. Cancer Res. 2011, 71, 6165–6173. [Google Scholar] [CrossRef]
- Hozhabri, H.; Lashkari, A.; Razavi, S.M.; Mohammadian, A. Integration of gene expression data identifies key genes and pathways in colorectal cancer. Med. Oncol. 2021, 38, 7. [Google Scholar] [CrossRef]
- Wendt, M.K.; Johanesen, P.A.; Kang-Decker, N.; Binion, D.G.; Shah, V.; Dwinell, M.B. Silencing of epithelial CXCL12 expression by DNA hypermethylation promotes colonic carcinoma metastasis. Oncogene 2006, 25, 4986–4997. [Google Scholar] [CrossRef]
- Drury, L.J.; Wendt, M.K.; Dwinell, M.B. CXCL12 chemokine expression and secretion regulates colorectal carcinoma cell anoikis through Bim-mediated intrinsic apoptosis. PLoS ONE 2010, 5, e12895. [Google Scholar] [CrossRef] [PubMed]
- Bocchi, M.; de Sousa Pereira, N.; de Oliveira, K.B.; Amarante, M.K. Involvement of CXCL12/CXCR4 axis in colorectal cancer: A mini-review. Mol. Biol. Rep. 2023, 50, 6233–6239. [Google Scholar] [CrossRef]
- Janssens, R.; Struyf, S.; Proost, P. The unique structural and functional features of CXCL12. Cell. Mol. Immunol. 2018, 15, 299–311. [Google Scholar] [CrossRef]
- Khare, T.; Bissonnette, M.; Khare, S. Cxcl12-cxcr4/cxcr7 axis in colorectal cancer: Therapeutic target in preclinical and clinical studies. Int. J. Mol. Sci. 2021, 22, 7371. [Google Scholar] [CrossRef] [PubMed]
- Tamaru, S.; Mizuno, Y.; Tochigi, H.; Kajihara, T.; Okazaki, Y.; Okagaki, R.; Kamei, Y.; Ishihara, O.; Itakura, A. MicroRNA-135b suppresses extravillous trophoblast-derived HTR-8/SVneo cell invasion by directly downregulating CXCL12 under low oxygen conditions. Biochem. Biophys. Res. Commun. 2015, 461, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Primeaux, M.; Liu, X.; Gowrikumar, S.; Fatima, I.; Fisher, K.W.; Bastola, D.; Vecchio, A.J.; Singh, A.B.; Dhawan, P. Claudin-1 interacts with EPHA2 to promote cancer stemness and chemoresistance in colorectal cancer. Cancer Lett. 2023, 579, 216479. [Google Scholar] [CrossRef]
- Cherradi, S.; Garambois, V.; Marines, J.; Andrade, A.F.; Fauvre, A.; Morand, O.; Fargal, M.; Mancouri, F.; Ayrolles-Torro, A.; Vezzo-Vié, N.; et al. Improving the response to oxaliplatin by targeting chemotherapy-induced CLDN1 in resistant metastatic colorectal cancer cells. Cell Biosci. 2023, 13, 72. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, T.; Qin, R.; Cai, X.; Zhou, Y.; Wang, X.; Shang, Z.; Li, G.; Yang, R.; Dong, C.; et al. The New Role of HNF1A-NAS1/miR-214/INHBA Signaling Axis in Colorectal Cancer. Front. Biosci. Landmark 2023, 28, 301. [Google Scholar] [CrossRef]
- Lin, H.; Hong, Y.G.; Zhou, J.D.; Gao, X.H.; Yuan, P.H.; Xin, C.; Huang, Z.P.; Zhang, W.; Hao, L.Q.; Hou, K.Z. LncRNA INHBA-AS1 promotes colorectal cancer cell proliferation by sponging miR-422a to increase AKT1 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 9940–9948. [Google Scholar]
- Qiu, S.; Li, B.; Xia, Y.; Xuan, Z.; Li, Z.; Xie, L.; Gu, C.; Lv, J.; Lu, C.; Jiang, T.; et al. CircTHBS1 drives gastric cancer progression by increasing INHBA mRNA expression and stability in a ceRNA- and RBP-dependent manner. Cell Death Dis. 2022, 13, 266. [Google Scholar] [CrossRef]
- Wang, D.; Wang, X.; Song, Y.; Si, M.; Sun, Y.; Liu, X.; Cui, S.; Qu, X.; Yu, X. Exosomal miR-146a-5p and miR-155-5p promote CXCL12/CXCR7-induced metastasis of colorectal cancer by crosstalk with cancer-associated fibroblasts. Cell Death Dis. 2022, 13, 380. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheon, J.; Kim, S.H.; Park, J.; Kim, T.H. Prognostic Significance of CLDN1, INHBA, and CXCL12 in Colon Adenocarcinoma: A Multi-Omics and Single-Cell Approach. Biomedicines 2025, 13, 1035. https://doi.org/10.3390/biomedicines13051035
Cheon J, Kim SH, Park J, Kim TH. Prognostic Significance of CLDN1, INHBA, and CXCL12 in Colon Adenocarcinoma: A Multi-Omics and Single-Cell Approach. Biomedicines. 2025; 13(5):1035. https://doi.org/10.3390/biomedicines13051035
Chicago/Turabian StyleCheon, Jaehwan, Sang Hyun Kim, Jaehyung Park, and Tae Hoon Kim. 2025. "Prognostic Significance of CLDN1, INHBA, and CXCL12 in Colon Adenocarcinoma: A Multi-Omics and Single-Cell Approach" Biomedicines 13, no. 5: 1035. https://doi.org/10.3390/biomedicines13051035
APA StyleCheon, J., Kim, S. H., Park, J., & Kim, T. H. (2025). Prognostic Significance of CLDN1, INHBA, and CXCL12 in Colon Adenocarcinoma: A Multi-Omics and Single-Cell Approach. Biomedicines, 13(5), 1035. https://doi.org/10.3390/biomedicines13051035






