Using Artificial Intelligence to Enhance Myelodysplastic Syndrome Diagnosis, Prognosis, and Treatment
Abstract
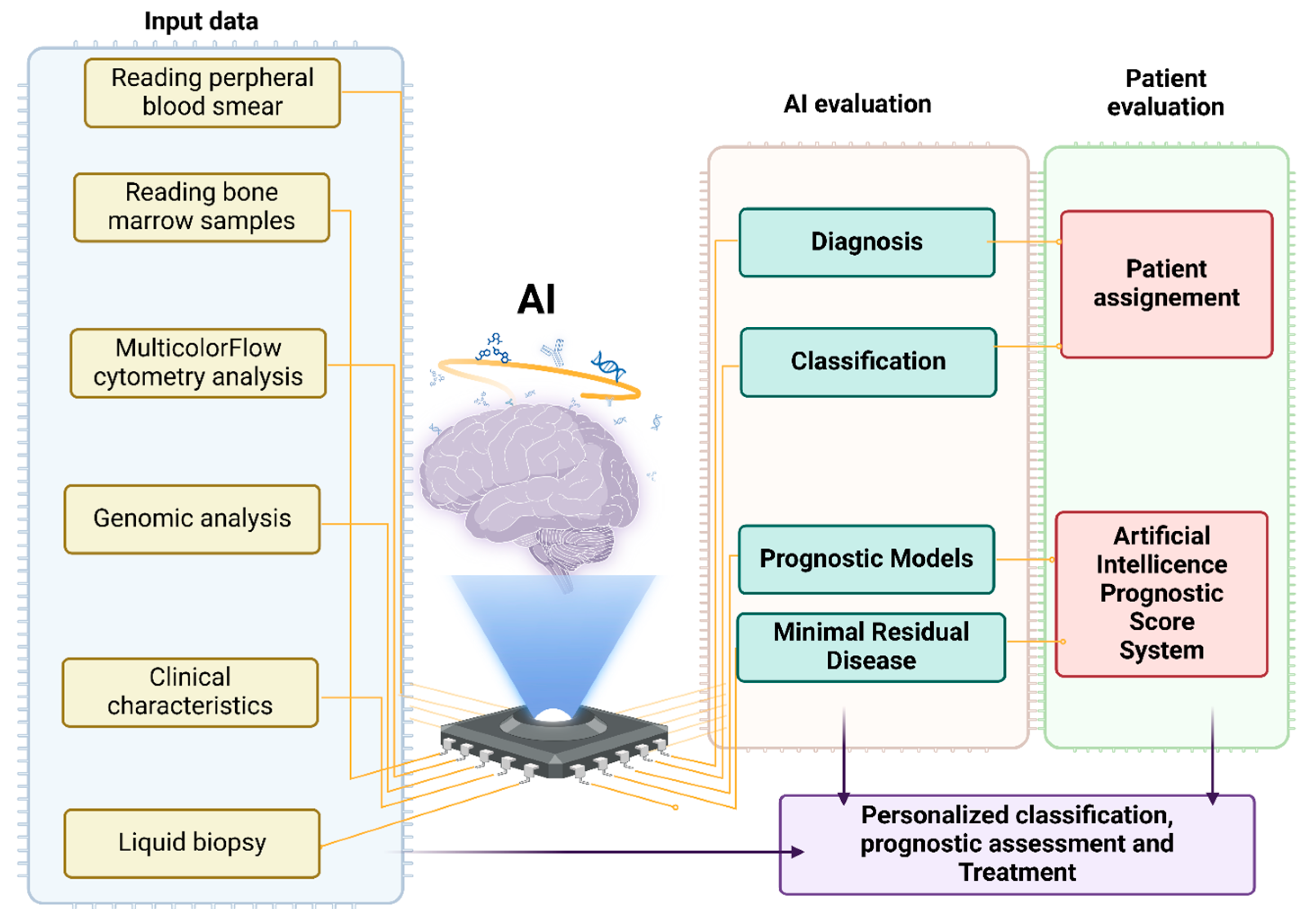
1. Introduction
General Information on Myelodysplastic Syndromes
2. Methods
2.1. Use of Artificial Intelligence in Hematology
2.2. Use of Artificial Intelligence in Myelodysplastic Syndromes
3. Results
3.1. Use of Artificial Intelligence in Reading Peripheral Blood Smears
3.2. Use of Artificial Intelligence in Reading Bone Marrow Samples
3.3. Use of Artificial Intelligence in Flow Cytometry Analysis of MDS Samples
3.4. Use of AI in the Differential Diagnosis of MDS with Aplastic Anemia and Acute Myeloid Leukemia
4. AI and Prognosis Evaluation in MDS
5. AI and Emotional Needs of MDS Patients
6. Conclusions
6.1. Future Perspectives
6.2. Challenges of Implementing AI Tools in Clinical Settings
Author Contributions
Funding
Conflicts of Interest
References
- Campo, E.; Swerdlow, S.H.; Harris, N.L.; Pileri, S.; Stein, H.; Jaffe, E.S. The 2008 WHO classification of lymphoid neoplasms and beyond, evolving concepts and practical applications. Blood 2011, 117, 5019–5032. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Komrokji, R.S.; Lanino, L.; Ball, S.; Bewersdorf, J.P.; Marchetti, M.; Maggioni, G.; Travaglino, E.; Al Ali, N.-H.; Fenaux, P.; Platzbecker, U.; et al. International Consortium on Myelodysplastic Syndromes. Data-driven, harmonised classification system for myelodysplastic syndromes: A consensus paper from the International Consortium for Myelodysplastic Syndromes. Lancet Haematol. 2021, 11, e862–e872. [Google Scholar] [CrossRef] [PubMed]
- Malcovati, L.; Hellström-Lindberg, E.; Bowen, D.; Adès, L.; Cermak, J.; del Cañizo, C.; Della Porta, M.G.; Fenaux, P.; Gattermann, N.; Germing, U.; et al. Diagnosis and treatment of primary myelodysplastic syndromes in adults, recommendations from the European LeukemiaNet. Blood 2013, 122, 2943–2964. [Google Scholar] [CrossRef]
- Haferlach, T. The molecular pathology of myelodysplastic syndrome. Pathobiology 2019, 86, 24–29. [Google Scholar]
- Tefferi, A.; Vardiman, J.W. Myelodysplastic syndromes: Mechanisms of disease. Hematol. Cell Ther. 2009, 38, 363–380. [Google Scholar]
- Greenberg, P.L.; Stone, R.M.; Al-Kali, A.; Barta, S.K.; Bejar, R.; Bennett, J.M.; Carraway, H.; De Castro, C.M.; Deeg, H.J.; DeZern, A.E.; et al. Myelodysplastic syndromes, version 2.2017, clinical practice guidelines in oncology. JNCCN 2017, 15, 60–87. [Google Scholar]
- Sekeres, M.A.; Cutler, C. How we treat higher-risk myelodysplastic syndromes. Blood 2014, 123, 829–836. [Google Scholar] [PubMed]
- Dodig, S.; Čepelak, I.; Dodig, M. Are we ready to integrate advanced artificial intelligence models in clinical laboratory? Biochem. Med. 2025, 35, 010501. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Allegra, A.; Tonacci, A.; Sciaccotta, R.; Genovese, S.; Musolino, C.; Pioggia, G.; Gangemi, S. Machine Learning and Deep Learning Applications in Multiple Myeloma Diagnosis, Prognosis, and Treatment Selection. Cancers 2022, 14, 606. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Allegra, A.; Mirabile, G.; Tonacci, A.; Genovese, S.; Pioggia, G.; Gangemi, S. Machine Learning Approaches in Diagnosis, Prognosis and Treatment Selection of Cardiac Amyloidosis. Int. J. Mol. Sci. 2023, 24, 5680. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Della Porta, M.G.; Bewersdorf, J.P.; Wang, Y.H.; Hasserjian, R.P. Future directions in myelodysplastic syndromes/neoplasms and acute myeloid leukaemia classification, from blast counts to biology. Histopathology 2025, 86, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, C.M.; Madjarova, S.J.; Williams, R.J.; Ollivier, M.; Karlsson, J.; Pareek, A.; Nwachukwu, B.U. Unsupervised machine learning methods and emerging applications in healthcare. Knee Surg. Sports Traumatol. Arthrosc. 2023, 31, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; See, K.C.; Ngiam, K.Y.; Celi, L.A.; Sun, X.; Feng, M. Reinforcement Learning for Clinical Decision Support in Critical Care, Comprehensive Review. J. Med. Int. Res. 2020, 22, e18477. [Google Scholar]
- Shouval, R.; Fein, J.A.; Savani, B.; Mohty, M.; Nagler, A. Machine learning and artificial intelligence in haematology. Br. J. Haematol. 2021, 192, 239–250. [Google Scholar] [PubMed]
- Munir, K.; Elahi, H.; Ayub, A.; Frezza, F.; Rizzi, A. Cancer Diagnosis Using Deep Learning, A Bibliographic Review. Cancers 2019, 11, 1235. [Google Scholar] [CrossRef]
- Goasguen, J.E.; Bennett, J.M.; Bain, B.J.; Brunning, R.; Vallespi, M.T.; Tomonaga, M.; Zini, G.; Renault, A. (The International Working Group on Morphology of MDS). Dyserythropoiesis in the diagnosis of the myelodysplastic syndromes and other myeloid neoplasms, problem areas. Br. J. Haematol. 2018, 182, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Valent, P.; Bain, B.J.; Bennett, J.M.; Wimazal, F.; Sperr, W.R.; Mufti, G.; Horny, H.P. Idiopathic cytopenia of undetermined significance (ICUS) and idiopathic dysplasia of uncertain significance (IDUS), and their distinction from low risk MDS. Leuk Res. 2012, 36, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Kwok, B.; Hall, J.M.; Witte, J.S.; Xu, Y.; Reddy, P.; Lin, K.; Flamholz, R.; Dabbas, B.; Yung, A.; Al-Hafidh, J.; et al. MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood 2015, 126, 2355–2361. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Visconte, V.; Maciejewski, J.P.; Guarnera, L. The potential promise of machine learning in myelodysplastic syndrome. Semin Hematol. 2024, in press . [CrossRef] [PubMed]
- Khan, S.; Sajjad, M.; Hussain, T.; Ullah, A.; Imran, A.S. A review on traditional machine learning and deep learning models for WBCs classification in blood smear images. IEEE Access 2020, 9, 10657–10673. [Google Scholar]
- Xiong, W.; Ong, S.H.; Lim, J.H.; Foong, K.W.; Liu, J.; Racoceanu, D.; Chong, A.G.; Tan, K.S. Automatic area classification in peripheral blood smears. IEEE Trans. Biomed. Eng. 2010, 57, 1982–1990. [Google Scholar] [CrossRef] [PubMed]
- Jati, A.; Singh, G.; Mukherjee, R.; Ghosh, M.; Konar, A.; Chakraborty, C.; Nagar, A.K. Automatic leukocyte nucleus segmentation by intuitionistic fuzzy divergence-based thresholding. Micron 2014, 58, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Alférez, S.; Merino, A.; Bigorra, L.; Mujica, L.; Ruiz, M.; Rodellar, J. Automatic recognition of atypical lymphoid cells from peripheral blood by digital image analysis. Am. J. Clin. Pathol. 2015, 143, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Rezatofighi, S.H.; Soltanian-Zadeh, H. Automatic recognition of five types of white blood cells in peripheral blood. Comput. Med. Imaging Graph. 2011, 35, 333–343. [Google Scholar]
- Ko, B.C.; Gim, J.-W.; Nam, J.-Y. Automatic white blood cell segmentation using stepwise merging rules and gradient vector flow snake. Micron 2011, 42, 695–705. [Google Scholar]
- Habibzadeh, M.; Krzyzak, A.; Fevens, T.; Sadr, A. Counting of RBCs and WBCs in Noisy Normal Blood Smear Microscopic Images. In Medical Imaging 2011: Computer-Aided Diagnosis; Summers, R.M., van Ginneken, B., Eds.; SPIE: Philadelphia, PA, USA, 2011; Volume 7963, p. 79633i. [Google Scholar] [CrossRef]
- Pan, C.; Park, D.S.; Yoon, S.; Yang, J.C. Leukocyte image segmentation using simulated visual attention. Expert Syst. Appl. 2012, 39, 7479–7494. [Google Scholar]
- Putzu, L.; Caocci, G.; Di Ruberto, C. Leucocyte classification for leukaemia detection using image processing techniques. Artif. Intell. Med. 2014, 62, 179–191. [Google Scholar]
- Elsabagh, A.A.; Elhadary, M.; Elsayed, B.; Elshoeibi, A.M.; Ferih, K.; Kaddoura, R.; Alkindi, S.; Alshurafa, A.; Alrasheed, M.; Alzayed, A.; et al. Artificial intelligence in sickle disease. Blood Rev. 2023, 61, 101102. [Google Scholar] [CrossRef] [PubMed]
- Walter, W.; Pohlkamp, C.; Meggendorfer, M.; Nadarajah, N.; Kern, W.; Haferlach, C.; Haferlach, T. Artificial intelligence in hematological diagnostics, Game changer or gadget? Blood Rev. 2023, 58, 101019. [Google Scholar] [CrossRef] [PubMed]
- El Alaoui, Y.; Elomri, A.; Qaraqe, M.; Padmanabhan, R.; Yasin Taha, R.; El Omri, H.; El Omri, A.; Aboumarzouk, O. A Review of Artificial Intelligence Applications in Hematology Management, Current Practices and Future Prospects. J. Med. Int. Res. 2022, 24, e36490. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kratz, A.; Lee, S.H.; Zini, G.; Riedl, J.A.; Hur, M.; Machin, S. International Council for Standardization in Haematology. Digital morphology analyzers in hematology, ICSH review and recommendations. Int. J. Lab. Hematol. 2019, 41, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Niazi, M.K.K.; Parwani, A.V.; Gurcan, M.N. Digital pathology and artificial intelligence. Lancet. Oncol. 2019, 20, e253–e261. [Google Scholar] [CrossRef]
- Diaz, O.; Kushibar, K.; Osuala, R.; Linardos, A.; Garrucho, L.; Igual, L.; Radeva, P.; Prior, F.; Gkontra, P.; Lekadir, K. Data preparation for artificial intelligence in medical imaging, A comprehensive guide to open-access platforms and tools. Phys Med. 2021, 83, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Bengio, Y.; Courville, A.; Bengio, Y. Deep Learning; MIT Press: Cambridge, UK, 2016. [Google Scholar]
- Zhang, S.; He, Y.; Wu, W.; Tan, H.; Xie, S.; Liu, M.; Chen, W.; Sun, D. Comparison of the performance of two automatic cell morphology analyzers for peripheral-blood leukocyte morphology analysis, Mindray MC-100i and Sysmex DI-60. Int. J. Lab. Hematol. 2023, 45, 860–868. [Google Scholar] [CrossRef] [PubMed]
- Eilertsen, H.; Henriksson, C.; Hagve, T.A. The use of CellaVisionTM DM 96 in the verification of the presence of blasts in samples flagged by the Sysmex XE-5000. Int. J. Lab. Hematol. 2017, 39, 423–428. [Google Scholar]
- van der Vorm, L.N.; Hendriks, H.A.; Smits, S.M. Performance of the CellaVision DC-1 digital cell imaging analyser for differential counting and morphological classification of blood cells. J. Clin. Pathol. 2023, 76, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Katz, B.Z.; Feldman, M.D.; Tessema, M.; Benisty, D.; Toles, G.S.; Andre, A.; Shtreker, B.; Paz, F.M.; Edwards, J.; Jengehino, D.; et al. Evaluation of Scopio Labs X100 Full Field PBS, The first high-resolution full field viewing of peripheral blood specimens combined with artificial intelligence-based morphological analysis. Int. J. Lab. Hematol. 2021, 43, 1408–1416. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hematology Imaging System. EasyCell Assistant. Available online: https://www.medicacorp.com/products/hematology-imaging-analyzers/ (accessed on 21 August 2023).
- Artificial Intelligence and Machine Learning (AI/ML)-Enabled Medical Devices. Available online: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices (accessed on 21 August 2023).
- Kimura, K.; Tabe, Y.; Ai, T.; Takehara, I.; Fukuda, H.; Takahashi, H.; Naito, T.; Komatsu, N.; Uchihashi, K.; Ohsaka, A.; et al. A novel automated image analysis system using deep convolutional neural networks can assist to differentiate MDS and AA. Sci. Rep. 2019, 9, 13385. [Google Scholar]
- Dao, K.T. Myelodysplastic Syndromes, Updates and Nuances. Med. Clin. N. Am. 2017, 101, 333–350. [Google Scholar]
- Choi, J.W.; Ku, Y.; Yoo, B.W.; Kim, J.A.; Lee, D.S.; Chai, Y.J.; Kong, H.J.; Kim, H.C. White blood cell differential count of maturation stages in bone marrow smear using dual-stage convolutional neural networks. PLoS ONE 2017, 12, e0189259. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kainz, P.; Burgsteiner, H.; Asslaber, M.; Ahammer, H. Training echo state networks for rotation-invariant bone marrow cell classification. Neural. Comput. Appl. 2017, 28, 1277–1292. [Google Scholar] [CrossRef]
- Allam, S.; Nasr, K.; Khalid, F.; Shah, Z.; Khan Suheb, M.Z.; Mulla, S.; Vikash, S.; Bou Zerdan, M.; Anwer, F.; Chaulagain, C.P. Liquid biopsies and minimal residual disease in myeloid malignancies. Front Oncol. 2023, 13, 1164017. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, N.; Jeong, S.; Park, M.J.; Song, W. Deep learning application of the discrimination of bone marrow aspiration cells in patients with myelodysplastic syndromes. Sci. Rep. 2022, 12, 18677. [Google Scholar] [CrossRef]
- Mori, J.; Kaji, S.; Kawai, H.; Kida, S.; Tsubokura, M.; Fukatsu, M.; Harada, K.; Noji, H.; Ikezoe, T.; Maeda, T.; et al. Assessment of dysplasia in bone marrow smear with convolutional neural network. Sci. Rep. 2020, 10, 14734. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, Y.Y.; Huang, T.C.; Ye, R.H.; Fang, W.H.; Lai, S.W.; Chang, P.Y.; Liu, W.N.; Kuo, T.Y.; Lee, C.H.; Tsai, W.C.; et al. A Hematologist-Level Deep Learning Algorithm (BMSNet) for Assessing the Morphologies of Single Nuclear Balls in Bone Marrow Smears, Algorithm Development. JMIR Med. Inform. 2020, 8, e15963. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Brück, O.E.; Lallukka-Brück, S.E.; Hohtari, H.R.; Ianevski, A.; Ebeling, F.T.; Kovanen, P.E.; Kytölä, S.I.; Aittokallio, T.A.; Ramos, P.M.; Porkka, K.V.; et al. Machine learning of bone marrow histopathology identifies genetic and clinical determinants in patients with MDS. Blood Cancer Discov. 2021, 2, 238–249. [Google Scholar]
- Ogata, K.; Della Porta, M.G.; Malcovati, L.; Picone, C.; Yokose, N.; Matsuda, A.; Yamashita, T.; Tamura, H.; Tsukada, J.; Dan, K. Diagnostic utility of flow cytometry in low-grade myelodysplastic syn dromes, A prospective validation study. Haematologica 2009, 94, 1066–1074. [Google Scholar]
- Dhingra, G.; Dass, J.; Arya, V.; Gupta, N.; Saraf, A.; Langer, S.; Aggarwal, S.; Kotwal, J.; Bhargava, M. Evaluation of multiparametric flow cytometry in diagnosis & prognosis of myelodysplastic syndrome in India. Indian J. Med. Res. 2020, 152, 254–262. [Google Scholar]
- Herbig, M.; Jacobi, A.; Wobus, M.; Weidner, H.; Mies, A.; Kräter, M.; Otto, O.; Thiede, C.; Weickert, M.T.; Götze, K.S.; et al. Machine learning assisted real-time deformability cytometry of CD34+ cells allow to identify patients with myelodysplastic syndromes. Sci. Rep. 2022, 12, 870. [Google Scholar] [CrossRef]
- Probst, C.; Zeng, Y.; Zhu, R.R. Characterization of protein particles in therapeutic formulations using imaging flow cytometry. J. Pharm. Sci. 2017, 106, 1952–1960. [Google Scholar]
- Rodrigues, M.A.; Probst, C.E.; Zayats, A.; Davidson, B.; Riedel, M.; Li, Y.; Venkatachalam, V. The in vitro micronucleus assay using imaging flow cytometry and deep learning. NPJ Syst. Biol. Appl. 2021, 7, 20. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rodrigues, M.A. Automation of the in vitro micronucleus assay using the Imagestream® imaging flow cytometer. Cytom. Part A 2018, 93, 706–726. [Google Scholar]
- Clichet, V.; Lebon, D.; Chapuis, N.; Zhu, J.; Bardet, V.; Marolleau, J.P.; Garcon, L.; Caulier, A.; Boyer, T. Artificial intelligence to empower diagnosis of myelodysplastic syndromes by multiparametric flow cytometry. Haematologica 2023, 108, 2435–2443. [Google Scholar]
- Duetz, C.; Van Gassen, S.; Westers, T.M.; van Spronsen, M.F.; Bachas, C.; Saeys, Y.; van de Loosdrecht, A.A. Computational flow cytometry as a diagnostic tool in suspected-myelodysplastic syndromes. Cytom. A 2021, 99, 814–824. [Google Scholar]
- Porwit, A.; Violidaki, D.; Axler, O.; Lacombe, F.; Ehinger, M.; Béné, M.C. Unsupervised cluster analysis and subset characterization of abnormal erythropoiesis using the bioinformatic Flow-Self Organizing Maps algorithm. Cytom. B Clin. Cytom. 2022, 102, 134–142. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rosenberg, C.A.; Rodrigues, M.A.; Bill, M.; Ludvigsen, M. Comparative analysis of feature-based ML and CNN for binucleated erythroblast quantification in myelodysplastic syndrome patients using imaging flow cytometry data. Sci. Rep. 2024, 14, 9349. [Google Scholar] [CrossRef]
- Wang, M.; Dong, C.; Gao, Y.; Li, J.; Han, M.; Wang, L. A Deep Learning Model for the Automatic Recognition of Aplastic Anemia, Myelodysplastic Syndromes, and Acute Myeloid Leukemia Based on Bone Marrow Smear. Front. Oncol. 2022, 12, 844978. [Google Scholar]
- Elshoeibi, A.M.; Badr, A.; Elsayed, B.; Metwally, O.; Elshoeibi, R.; Elhadary, M.R.; Elshoeibi, A.; Attya, M.A.; Khadadah, F.; Alshurafa, A.; et al. Integrating AI and ML in Myelodysplastic Syndrome Diagnosis, State-of-the-Art and Future Prospects. Cancers 2023, 16, 65. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Greenberg, P.L.; Tuechler, H.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Solé, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; Dreyfus, F.; et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood 2012, 120, 2454–2465. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Della Porta, M.G.; Tuechler, H.; Malcovati, L.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Solé, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; et al. Validation of WHO classification-based Prognostic Scoring System (WPSS) for myelodysplastic syndromes and comparison with the revised International Prognostic Scoring System (IPSS-R). A study of the International Working Group for Prognosis in Myelodysplasia (IWG-PM). Leukemia 2015, 29, 1502–1513. [Google Scholar] [CrossRef] [PubMed]
- Bersanelli, M.; Travaglino, E.; Meggendorfer, M.; Matteuzzi, T.; Sala, C.; Mosca, E.; Chiereghin, C.; Di Nanni, N.; Gnocchi, M.; Zampini, M.; et al. Classification and Personalized Prognostic Assessment on the Basis of Clinical and Genomic Features in Myelodysplastic Syndromes. J. Clin. Oncol. 2021, 39, 1223–1233. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Khanna, V.; Lu, R.; Kumar, J.; Molina, A.; Stehr, H.; Spiteri, E.; Spinner, M.; Silva, O.; Fernandez-Pol, S.; Tan, B.; et al. The clinical, molecular, and prognostic features of the 2022 WHO and ICC classification systems for myelodysplastic neoplasms. Leuk Res. 2024, 136, 107433. [Google Scholar] [CrossRef] [PubMed]
- Huber, S.; Haferlach, T.; Müller, H.; Meggendorfer, M.; Hutter, S.; Hoermann, G.; Baer, C.; Kern, W.; Haferlach, C. MDS subclassification-do we still have to count blasts? Leukemia 2023, 37, 942–945. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bernard, E.; Tuechler, H.; Greenberg, P.L.; Hasserjian, R.P.; Arango Ossa, J.E.; Nannya, Y.; Devlin, S.M.; Creignou, M.; Pinel, P.; Monnier, L.; et al. Molecular International Prognostic Scoring System for Myelodysplastic Syndromes. NEJM Evid. 2022, 1, EVIDoa2200008. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, Y.; Qin, T.; Xu, Z.; Qu, S.; Pan, L.; Li, B.; Jia, Y.; Li, C.; Wang, H.; et al. IPSS-M has greater survival predictive accuracy compared with IPSS-R in persons ≥ 60 years with myelodysplastic syndromes. Exp. Hematol. Oncol. 2022, 11, 73. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, W.H.; Tsai, M.T.; Tsai, C.H.; Tien, F.M.; Lo, M.Y.; Tseng, M.H.; Kuo, Y.Y.; Liu, M.C.; Yang, Y.T.; Chen, J.C.; et al. Validation of the molecular international prognostic scoring system in patients with myelodysplastic syndromes defined by international consensus classification. Blood Cancer J. 2023, 13, 120. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ma, J.; Gu, Y.; Wei, Y.; Wang, X.; Wang, P.; Song, C.; Ge, Z. Evaluation of new IPSS-Molecular model and comparison of different prognostic systems in patients with myelodysplastic syndrome. Blood Sci. 2023, 5, 187–195. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sauta, E.; Robin, M.; Bersanelli, M.; Travaglino, E.; Meggendorfer, M.; Zhao, L.P.; Caballero Berrocal, J.C.; Sala, C.; Maggioni, G.; Bernardi, M. Real-World Validation of Molecular International Prognostic Scoring System for Myelodysplastic Syndromes. J. Clin. Oncol. 2023, 41, 2827–2842. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, T.; Jiang, B.; Luo, Y.; Zhao, Y.; Ouyang, G.; Yu, J.; Lan, J.; Lu, Y.; Lai, X.; Ye, B.; et al. Comparison of the prognostic predictive value of Molecular International Prognostic Scoring System and Revised International Prognostic Scoring System in patients undergoing allogeneic hematopoietic stem cell transplantation for myelodysplastic neoplasms. Am. J. Hematol. 2023, 98, E391–E394. [Google Scholar] [CrossRef] [PubMed]
- Lincango, M.; Andreoli, V.; Rivello, H.G.; Bender, A.; Catalán, A.I.; Rahhal, M.; Delamer, R.; Asinari, M.; Orgueira, A.M.; Castro, M.B.; et al. Assessing the Relevance of Non-molecular Prognostic Systems for Myelodysplastic Syndrome in the Era of Next-Generation Sequencing. Ann Lab. Med. 2025, 45, 44–52. [Google Scholar] [CrossRef]
- D’Amico, S.; Dall’Olio, L.; Rollo, C.; Alonso, P.; Prada-Luengo, I.; Dall’Olio, D.; Sala, C.; Sauta, E.; Asti, G.; Lanino, L.; et al. MOSAIC, An Artificial Intelligence-Based Framework for Multimodal Analysis, Classification, and Personalized Prognostic Assessment in Rare Cancers. JCO Clin. Cancer Inform. 2024, 8, e2400008. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kewan, T.; Durmaz, A.; Bahaj, W.; Gurnari, C.; Terkawi, L.; Awada, H.; Ogbue, O.D.; Ahmed, R.; Pagliuca, S.; Awada, H.; et al. Molecular patterns identify distinct subclasses of myeloid neoplasia. Nat. Commun. 2023, 14, 3136. [Google Scholar] [CrossRef]
- Stomper, J.; Lubbert, M. Can we predict responsiveness to hypomethylating agents in AML? Semin. Hematol. 2019, 56, 118–124. [Google Scholar] [CrossRef]
- Musolino, C.; Sant’antonio, E.; Penna, G.; Alonci, A.; Russo, S.; Granata, A.; Allegra, A. Epigenetic therapy in myelodysplastic syndromes. Eur. J. Haematol. 2010, 84, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Nazha, A.; Komrokji, R.; Meggendorfer, M.; Jia, X.; Radakovich, N.; Shreve, J.; Hilton, C.B.; Nagata, Y.; Hamilton, B.K.; Mukherjee, S.; et al. Personalized prediction model to risk stratify patients with myelodysplastic syndromes. J. Clin. Oncol. Off J. Am. Soc. Clin. Oncol. 2021, 39, 3737–3746. [Google Scholar]
- Radakovich, N.; Sallman, D.A.; Buckstein, R.; Brunner, A.; Dezern, A.; Mukerjee, S.; Komrokji, R.; Al-Ali, N.; Shreve, J.; Rouphail, Y.; et al. A machine learning model of response to hypomethylating agents in myelodysplastic syndromes. IScience 2022, 25, 104931. [Google Scholar]
- Ko, B.S.; Wang, Y.F.; Li, J.L.; Li, C.C.; Weng, P.F.; Hsu, S.C.; Hou, H.A.; Huang, H.H.; Yao, M.; Lin, C.T.; et al. Clinically validated machine learning algorithm for detecting residual diseases with multicolor flow cytometry analysis in acute myeloid leukemia and myelodysplastic syndrome. EBioMedicine 2018, 37, 91–100. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Booth, A.; Bell, T.; Halhol, S.; Pan, S.; Welch, V.; Merinopoulou, E.; Lambrelli, D.; Cox, A. Using Social Media to Uncover Treatment Experiences and Decisions in Patients With Acute Myeloid Leukemia or Myelodysplastic Syndrome Who Are Ineligible for Intensive Chemotherapy, Patient-Centric Qualitative Data Analysis. J. Med. Int. Res. 2019, 21, e14285. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Frank, P.P.; Lu, M.X.E.; Sasse, E.C. Educational and Emotional Needs of Patients with Myelodysplastic Syndromes, An AI Analysis of Multi-Country Social Media. Adv. Ther. 2023, 40, 159–173. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Billingham, L.; Malottki, K.; Steven, N. Research methods to change clinical practice for patients with rare cancers. Lancet. Oncol. 2016, 17, e70–e80. [Google Scholar] [CrossRef]
- Blay, J.Y.; Coindre, J.M.; Ducimetière, F.; Ray-Coquard, I. The value of research collaborations and consortia in rare cancers. Lancet. Oncol. 2016, 17, e62–e69. [Google Scholar] [CrossRef] [PubMed]
- Rajpurkar, P.; Chen, E.; Banerjee, O.; Topol, E.J. AI in health and medicine. Nat. Med. 2022, 28, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Kamrani, S.M.E.; Hadizadeh, F. A coarse-grain MD (molecular dynamic) simulation of PCL-PEG and PLA-PEG aggregation as a computational model for prediction of the drug-loading efficacy of doxorubicin. J Biomol Struct Dyn. 2019, 37(16), 4215–4221. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Gilbert, J.A.; Zhu, H.; Huang, S.-M.; Kunkoski, E.; Das, P.; Bergman, K.; Buschmann, M.; ElZarrad, M.K. Emerging clinical pharmacology topics in drug development and precision medicine. In Atkinson’s Principles of Clinical Pharmacology; Elsevier: Amsterdam, The Netherlands, 2022; pp. 691–708. [Google Scholar]
- Docherty, J.R.; Alsufyani, H.A. Pharmacology of drugs used as stimulants. J. Clin. Pharmacol. 2021, 61, S53–S69. [Google Scholar] [PubMed]
- Abdelbasset, W.K.; Elsayed, S.H.; Alshehri, S.; Huwaimel, B.; Alobaida, A.; Alsubaiyel, A.M.; Alqahtani, A.A.; El Hamd, M.A.; Venkatesan, K.; AboRas, K.M.; et al. Development of GBRT Model as a Novel and Robust Mathematical Model to Predict and Optimize the Solubility of Decitabine as an Anti-Cancer Drug. Molecules 2022, 27, 5676. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rabbani, N.; Kim, G.Y.E.; Suarez, C.J.; Chen, J.H. Applications of machine learning in routine laboratory medicine, Current state and future directions. Clin. Biochem. 2022, 103, 1–7. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Baxi, V.; Edwards, R.; Montalto, M.; Saha, S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod. Pathol. 2022, 35, 23–32. [Google Scholar] [PubMed]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The5th edition of the World Health Organization Classification of Haematolymphoid Tumours, Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar]
- Li, S.; Yi, H.; Leng, Q.; Wu, Y.; Mao, Y. New perspectives on cancer clinical research in the era of big data and machine learning. Surg. Oncol. 2024, 52, 102009. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.R.; Ye, Y.; Crawford, J.M.; Becich, M.J.; Roy, S.; Botkin, J.R.; de Baca, M.E.; Pantanowitz, L. The Ethics of Artificial Intelligence in Pathology and Laboratory Medicine, Principles and Practice. Acad. Pathol. 2021, 8, 2374289521990784. [Google Scholar]
- Fan, B.E.; Yong, B.S.J.; Li, R.; Wang, S.S.Y.; Aw, M.Y.N.; Chia, M.F.; Chen, D.T.Y.; Neo, Y.S.; Occhipinti, B.; Ling, R.R.; et al. From microscope to micropixels, A rapid review of artificial intelligence for the peripheral blood film. Blood Rev. 2024, 64, 101144. [Google Scholar] [CrossRef] [PubMed]
| Material or Technique Employed | Type of Analysis | AI Model | Features of Dataset | Sensitivity and Specificity | Strengths or Weaknesses | Ref. |
|---|---|---|---|---|---|---|
| Peripheral Blood | Morphology | Convolutional neural network-powered deep learning | A highly trained cell image recognition system | 93–99.8% 96–100% | Ability to differentiate MDS and Aplastic Anemia | [42] |
| Bone Marrow | Research of dysplasia | Convolutional neural network-derived machine learning | 6453 cell images (c.i.) for training; 806 c.i. for validation: 806 c.i. for testing Inception V3 architecture | 90% 99.9% | Difficulty to measure the proportion of dysplasia | [47] |
| Bone Marrow | Evaluation of reduced granules | Deep neural network | 1797 labeled images. Faster R-CNN was trained with ResNet-101 backbone | 85.2% 98.9% | Low sensitivity | [48] |
| Bone Marrow | Morphology | Deep learning model | 17,319 annotated cells (development cohort) | Precision 67.4% | Precision inferior to pathologist analysis | [49] |
| Bone Marrow | Different cellular features | Convolutional neural network | 236 samples from 143 MDS subjects; 87 samples from 51 MDS/MPN subjects; 11 healthy controls samples from 11 subjects | - - | WHO subtypes and MDS categories only partially overlapped | [50] |
| Real-Time Deformability Cytometry | Analysis of dyserythropoiesis through morpho-mechanical pattern | Random forest | Phenotype of BM-derived CD34+ HSCs from MDS patients and healthy donors. Seven features were extracted from the contour of each cell and a RF model was trained to distinguish between the healthy state and MDS | Accuracy of 82.9% | The efficiency of CD34 isolation is low | [53] |
| Flow Cytometry | Hematogone ratio, ratio of CD34+ progenitors | Machine learning | Cohort of 191 patients; external cohort of 89 patients | 91.8% 92.5% | Superior sensitivity to Ogata score, prediction of evolution | [57] |
| Flow Cytometry | Erythroid and myeloid progenitors | Random forest ML | Training cohort 71 MDS, 81 controls; validation cohort 30 MDS, 27 controls; validation cohort 25 MDS with excess of blasts | 90% 93% | Processing time less than two minutes | [58] |
| Flow Cytometry | Analysis of erythropoiesis | Flow-self organizing maps algorithm | Unsupervised clustering analysis; 11 MDS patients | - - | Evidence of subtle erythropoiesis changes | [59] |
| Flow Cytometry | Detection of binucleated erythroblasts | Convolutional neural network | Bone marrow samples from 14 MDS patients, six ICUS/CCUS patients, six non-MDS controls, and 11 healthy controls | 98.2% 78.2% | Increased diagnostic precision | [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stagno, F.; Mirabile, G.; Rizzotti, P.; Bottaro, A.; Pagana, A.; Gangemi, S.; Allegra, A. Using Artificial Intelligence to Enhance Myelodysplastic Syndrome Diagnosis, Prognosis, and Treatment. Biomedicines 2025, 13, 835. https://doi.org/10.3390/biomedicines13040835
Stagno F, Mirabile G, Rizzotti P, Bottaro A, Pagana A, Gangemi S, Allegra A. Using Artificial Intelligence to Enhance Myelodysplastic Syndrome Diagnosis, Prognosis, and Treatment. Biomedicines. 2025; 13(4):835. https://doi.org/10.3390/biomedicines13040835
Chicago/Turabian StyleStagno, Fabio, Giuseppe Mirabile, Patricia Rizzotti, Adele Bottaro, Antonio Pagana, Sebastiano Gangemi, and Alessandro Allegra. 2025. "Using Artificial Intelligence to Enhance Myelodysplastic Syndrome Diagnosis, Prognosis, and Treatment" Biomedicines 13, no. 4: 835. https://doi.org/10.3390/biomedicines13040835
APA StyleStagno, F., Mirabile, G., Rizzotti, P., Bottaro, A., Pagana, A., Gangemi, S., & Allegra, A. (2025). Using Artificial Intelligence to Enhance Myelodysplastic Syndrome Diagnosis, Prognosis, and Treatment. Biomedicines, 13(4), 835. https://doi.org/10.3390/biomedicines13040835






