NSD Family-Mediated H3K36 Methylation in Human Cancer: Mechanisms and Therapeutic Opportunities
Abstract
1. Introduction
2. H3K36 Methylation and NSD Family Methyltransferases
2.1. Roles and Regulators of H3K36 Methylation
2.2. NSD Family Members, Structures, and Isoforms
2.3. Biological Functions of NSD Family
2.3.1. Epigenetic Regulation of Gene Expression
2.3.2. Regulation of Development and Cell Differentiation
2.3.3. Regulation of Genomic Stability and DNA Damage Response
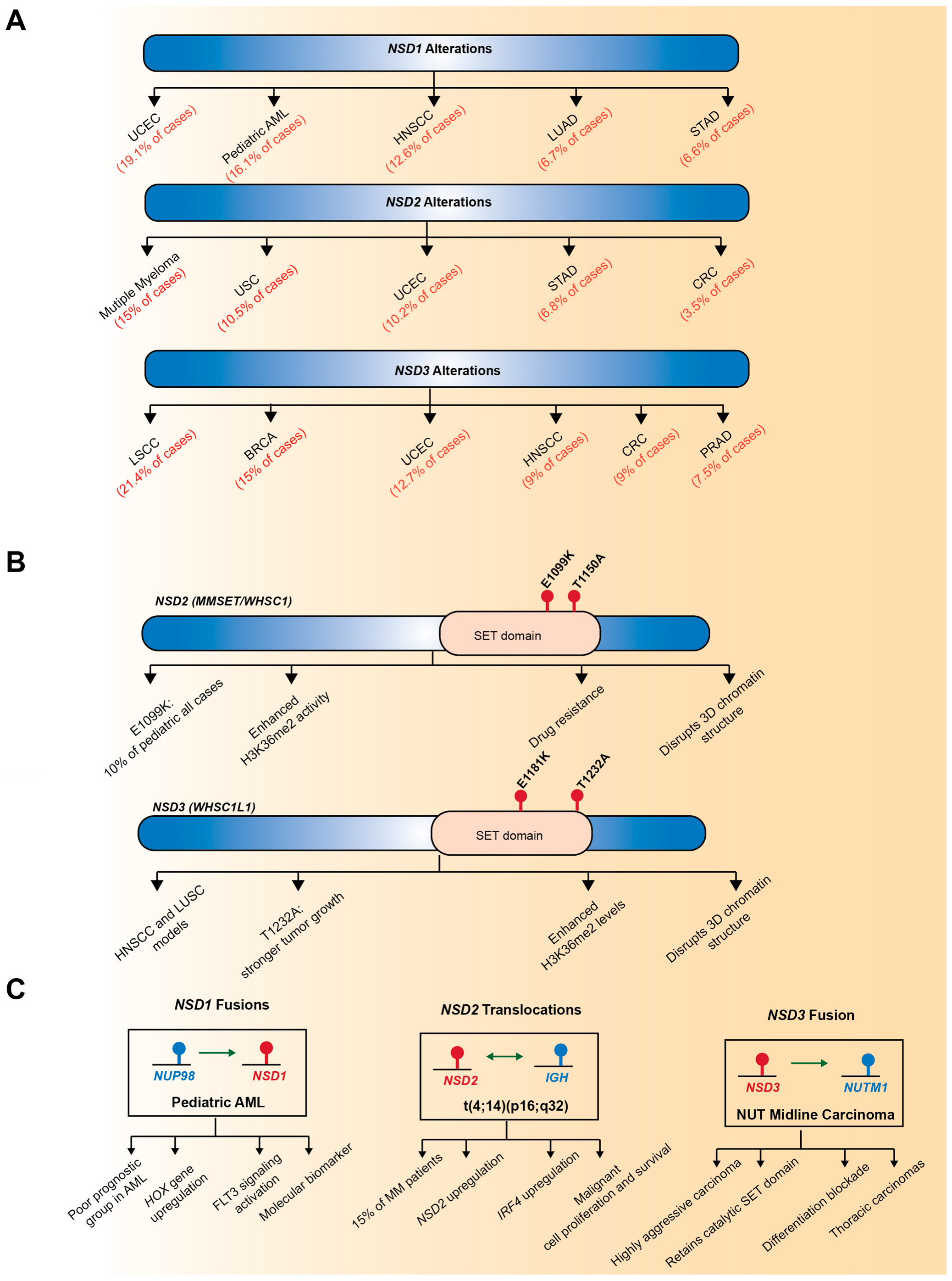
3. Genetic Alterations of NSD Family Genes in Human Cancers
3.1. Gene Amplification
3.2. Point Mutations
3.3. Chromosomal Translocation
4. Diverse Role of NSD Family Methyltransferases in Human Cancer
4.1. Tumorigenesis, Cancer Cell Proliferation, and Anti-Apoptosis
4.1.1. Roles of NSD1
4.1.2. Roles of NSD2
4.1.3. Roles of NSD3
4.2. Tumor Angiogenesis, Invasion and Metastasis
4.3. Metabolic Reprogramming
4.4. Tumor Immune Microenvironment
4.5. Regulation of Genomic Stability and Response to Chemotherapy
4.6. Therapeutic Resistance
| Malignant Phenotypes | NSD Family Member | Cancer Types | Functional Roles | Molecular Mechanisms | Refs. | |
|---|---|---|---|---|---|---|
| Type of Mechanism | Mode of Action (Target Genes, Proteins, and Signaling) | |||||
| Tumorigenesis, Proliferation, and Survival | NSD1 | acute myeloid leukemia (NUP98-NSD1 fusion) | tumor-promoting | histone modification, chromatin remodeling | NUP98-NSD1 fusion promotes leukemogenesis by H3K36me2-mediated HOXA gene upregulation; inhibits EZH2-mediated H3K27me3; interacts with chromatin remodeler complexes | [17,104,105] |
| hepatocellular carcinoma, breast cancer | tumor-promoting | histone modification | upregulates Wnt10b expression by inhibiting H3K27me3 and subsequently activates WNT/β-catenin signaling pathway | [106,107] | ||
| colorectal cancer | tumor-promoting | protein methylation | methylates p65 at K218/221 and activates NF-κB signaling to promote cell proliferation | [108] | ||
| erythroleukemia | tumor-suppressive | histone modification, transcriptional regulation | NSD1 inactivation impairs GATA1-mediated erythroid differentiation, leading to leukemogenesis | [109] | ||
| head and neck squamous cell carcinoma | tumor-suppressive | histone modification | Impaired NSD1 activity by loss-of-function mutation or H3K36M blocks cellular differentiation, thus promoting oncogenesis | [110] | ||
| NSD2 | multiple myeloma harboring t(4;14) | tumor-promoting | histone modification | NSD2-induced H3K36me2 drives oncogenic transcriptional program to promote tumorigenesis, proliferation, and anti-apoptosis; inhibits H3K27me3 | [41,52] | |
| acute lymphoblastic leukemia, osteosarcoma | tumor-promoting | histone modification | NSD2 hyperactivation by gain-of-function mutation (E1099K/Y1179A) drives H3K36me2-dependent oncogenic transcriptional program | [20,57,95] | ||
| bladder cancer, lung cancer | tumor-promoting | protein interaction, histone modification | promotes cell proliferation by interacting with WNT pathway components and activating WNT/β-catenin/TCF4 signaling (cyclin D1 upregulation) | [112] | ||
| prostate cancer | tumor-promoting | protein interaction, histone modification | upregulates cyclin D, BCL2, and survivin via NF-κB signaling; activates androgen receptor (AR)-dependent transcription to drive prostate tumorigenesis | [113,114] | ||
| colorectal cancer, cervical cancer | tumor-promoting | histone modification | upregulates cancer-promoting genes (e.g., ADAM9, EGFR, and MET) by H3K36me2; activates PI3K-AKT pathway | [115,116] | ||
| lung adenocarcinoma | tumor-promoting | histone modification | regulates KRAS-driven transcriptional program, activates KRAS-MEK pathway | [117,118] | ||
| pancreatic cancer | tumor-suppressive | histone modification | inhibits tumorigenesis by H3K36me2-mediated IκBα upregulation and subsequent inactivation of NF-κB signaling | [119] | ||
| NSD2-S | t(4;14) multiple myeloma | tumor-promoting | transcriptional regulation | NSD2S alone is sufficient to drive tumorigenesis; upregulates glyoxalase 1 (GLO1) to induce anti-apoptotic genes (MCL2 and BCL2) | [27] | |
| NSD3 | breast cancer | tumor-promoting | histone modification | NSD3-induced H3K36me2 promotes tumorigenesis by upregulating NOTCH pathway and EMT-related genes | [22] | |
| lung squamous cell carcinoma | tumor-promoting | histone modification | NSD3 amplification or GOF mutation (T1232A) induces H3K36me2-dependent oncogenic transcriptional program (MYC upregulation, mTOR activation) | [23] | ||
| head and neck squamous cell carcinoma | tumor-promoting | histone modification, protein methylation | NSD3 GOF mutation drives oncogenic transcriptional program; upregulates cell cycle regulators (CDC6, CDK2); methylates EGFR at K721 for its activation | [46,89,121] | ||
| NUT midline carcinoma (NSD3-NUT fusion) | tumor-promoting | protein interaction, transcriptional regulation | NSD3-NUT fusion blocks cell differentiation and maintains proliferation by interacting with BRD4 for BRD4-dependent transcription | [50] | ||
| pancreatic cancer | tumor-promoting | histone modification | upregulates MYC, ADAM12, and NOTCH3; activates mTOR, EGFR/ERK signaling | [100,122] | ||
| NSD3-S | acute myeloid leukemia | tumor-promoting | chromatin remodeling | BRD4-NSD3S-CHD8 complex binds to super-enhancer of MYC gene for its transcription that is required for sustaining AML | [26] | |
| breast cancer, lung cancer | tumor-promoting | transcriptional regulation, protein interaction | regulates ER expression and its estrogen-independent activity; interacts with MYC for its transcriptional activation | [160,161] | ||
| Tumor Angiogenesis | NSD1 | esophageal cancer | pro-angiogenic | histone modification | upregulates HIF-1α and VEGF-A expression by recruiting STAT3 to HIF-1α promoter and facilitating H3K36me2 | [123] |
| NSD2 | prostate cancer | pro-angiogenic | histone modification | induces H3K36me2-mediated VEGF-A expression | [113] | |
| colorectal cancer | pro-angiogenic | protein methylation | methylates STAT3 at K163 for activation of STAT3/VEGF-A axis | [44] | ||
| Tumor Invasion & Metastasis | NSD1 | breast cancer | pro-metastatic | protein methylation | promotes EMT, migration, and invasion by inhibiting FBXL11 to promote p65 methylation and subsequent NF-κB signaling activation | [124] |
| hepatocellular carcinoma | pro-metastatic | histone modification | promotes migration, invasion, and metastasis by epigenetic upregulation of Wnt10b for WNT/β-catenin signaling activation | [106] | ||
| head and neck squamous cell carcinoma | pro-metastatic | unknown | is associated with occult lymph node metastasis in HNSCC patients | [125] | ||
| NSD2 | t(4;14)+ multiple myeloma | pro-metastatic | transcriptional regulation | upregulates TWIST1 transcription to induce EMT and tumor dissemination | [127] | |
| prostate cancer | pro-metastatic | histone modification, posttranslational modification | drives EMT, invasion, and metastasis; upregulates H3K36me2-mediated TWIST1 expression; NSD2 protein stabilization by AKT upregulates RICTOR and Rac1 expression via H3K36me2 to drive metastasis | [126,128,129] | ||
| breast cancer | pro-metastatic | histone modification, transcriptional regulation | facilitates EMT, migration, and invasion by activating FOXM1-mediated WNT/β-catenin signaling; upregulates MMP9 and ULK1 expression via H3K36me2 | [130,131,132] | ||
| renal cell carcinoma, osteosarcoma | pro-metastatic | transcriptional regulation | promotes EMT, migration, and invasion by downregulating E-cadherin | [133,134] | ||
| NSD3 | breast cancer | pro-metastatic | histone modification | promotes EMT, invasion, and metastasis by H3K36me2-dependent activation of NOTCH signaling and EMT program | [22] | |
5. Role of NSD Short Isoforms in Regulating Cancer
5.1. NSD3 Short Isoform
5.2. NSD2 Short Isoform
6. Targeting NSD Family for Cancer Treatment
6.1. Catalytic Inhibitors for NSD Family
6.2. Other Types of NSD Family Inhibitors
| Types | Drugs | Targets | Inhibitory Efficacy | In Vitro/In Vivo Anti-Tumor Efficacy | Refs. |
|---|---|---|---|---|---|
| Catalytic SET inhibitor | BT5 | NSD1 | IC50 = 5.8 ± 1.4 μM (NSD1), IC50 = 26.7 ± 7.1 μM (NSD2), IC50 = 14.3 ± 6.1 μM (NSD3), (4 h incubation) | Anti-proliferative effect in NUP98-NSD1 leukemia cell line (GI50 = 1.3 μM at day 3) and primary samples from patients with NUP98-NSD1 fusion | [174] |
| LEM-06 | NSD2 | IC50 = 0.8 mM | [166] | ||
| DA-3003-1 PF-03882845 Chaetocin TC LPA54 ABT-199 | NSD2 (no selectivity to NSD2) | IC50 = 0.17 μM (DA3003-1) IC50 = 7.6 μM (PF-03882845) IC50 = 0.13 μM (Chaetocin) IC50 = 8.5 μM (TC LPA54) IC50 = 1.7 μM (ABT-199) | [168] | ||
| LEM-14 | NSD2 | IC50 = 132 μM | [167] | ||
| LEM-14-1189 | NSD1/2/3 | IC50 = 418 μM (NSD1), 111 μM (NSD2), and 60 μM (NSD3) | [167] | ||
| Compound 15a (structural modification of DA-3003-1) | NSD2 | IC50 = 0.23 μM | Apoptosis induction in cells; Anti-tumor effects in tumor xenograft (KMS-11 multiple myeloma) | [169] | |
| Compound 3 | NSD2, NSD3 | IC50 = 0.81 μM (NSD2) and 0.84 μM (NSD3) | Anti-proliferative effect in vitro (non-small cell lung cancer cells) | [175] | |
| Compound 42 | NSD2 | IC50 = 0.017 μM | Apoptosis induction in vitro; Anti-tumor effects in xenograft model (RS4;11 ALL cells) | [170] | |
| KTX-1001 | NSD2 | IC50 = 0.460 nM | Phase I clinical trial in multiple myeloma | [171,172] | |
| IACS-17596 IACS-17817 | NSD2 | IC50 = 8.8 nM IC50 = 19 nM | Clinical-grade; Anti-tumor effects in cells, xenograft, and PDX models (KRAS-driven lung and pancreatic cancers) | [173] | |
| 13i | NSD3 | IC50 = 287 μM | Anti-proliferative effect in JIMT-1 breast cancer cells | [176] | |
| PWWP inhibitor | 3f | NSD2 | IC50 = 17.3 μM | [184] | |
| UNC6934 | NSD2 | IC50 = 104 nM | Anti-proliferative effects in t(4;14) multiple myeloma cells | [185] | |
| Compound 38 | NSD2 | IC50 = 0.11 μM | Anti-proliferative effects in vitro (KMS11 multiple myeloma, ALL cells) | [186] | |
| Compound 34 | NSD2 | pIC50 = 8.2 (IC50 ≈ 6 nM) | [187] | ||
| BI-9321 | NSD3 | IC50 = 0.2 μM | Anti-proliferative effects in vitro (MOML-13, RN-2 AML cells) | [183] | |
| PROTAC degrader | MS159 (E3 ligase: CRBN) | NSD2 | DC50 = 5.2 ± 0.9 μM | Anti-proliferative effects in vitro (KMS11 and H929 multiple myeloma) | [180] |
| UNC8732 (E3 ligase: FBXO22) | NSD2 | DC50 = 60 nM | Anti-proliferative effects in vitro (NSD2 E1099K ALL cells) | [179] | |
| UNC8153 (E3 ligase: FBXO22) | NSD2 | IC50 = 0.35 μM | Anti-proliferative effects in multiple myeloma cells | [181] | |
| LLC0424 (E3 ligase: CRBN) | NSD2 | DC50 = 20 nM | Anti-proliferative effects in vitro (ALL cells); NSD2 degradation in vivo | [182] | |
| MS9715 (E3 ligase: VHL) | NSD3 | DC50 = 4.9 ± 0.4 μM | Anti-proliferative effects in vitro (MLL-rearranged AML and multiple myeloma cells) | [177] | |
| Compound 8 (E3 ligase: VHL) | NSD3 | DC50 = 0.94 ~ 1.43 μM (reduces H3K36me2) | Induces apoptosis and cell cycle arrest in lung cancer cells | [178] | |
| LLC0150 | NSD1/2 | IC50 = 0.274 ~ 69.68 nM | Anti-proliferative effects in vitro and in vivo (prostate cancer) | [114] |
6.3. Indirect Inhibition of NSD Family Function
6.4. Potential Toxicity and Safety Consideration of NSD Inhibition
7. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AK2 | Adenylate kinase 2 |
| AML | Acute myeloid leukemia |
| ALL | Acute lymphoblastic leukemia |
| AR | Androgen receptor |
| AROS | Active regulator of SIRT1 |
| ASH1L | ASH1-like |
| ATRA | All-trans retinoic acid |
| BET | Bromodomain and extraterminal |
| CN | Cytogenetically normal |
| DDR | DNA damage response |
| DSB | Double-strand break |
| EMT | Epithelial–mesenchymal transition |
| ERα | Estrogen receptor alpha |
| G6PD | Glucose-6-phosphate dehydrogenase |
| HCC | Hepatocellular carcinoma |
| HGSC | High-grade serous carcinoma |
| HK2 | Hexokinase 2 |
| H3K36 | Histone lysine 36 |
| H3K36me1 | Histone lysine 36 monomethylation |
| H3K36me2 | Histone lysine 36 dimethylation |
| H3K36me3 | Histone lysine 36 trimethylation |
| HMG | High mobility group |
| HNSCC | Head and neck squamous cell carcinoma |
| HR | Homologous recombination |
| IRF3 | Interferon regulatory factor 3 |
| JHDM | Jumonji C (JmjC) domain-containing histone demethylase |
| LCA | Laryngeal cancer |
| LUAD | Lung adenocarcinoma |
| LUSC | Lung squamous cell carcinoma |
| MLL1 | Mixed lineage leukemia 1 |
| NHEJ | Non-homologous end joining |
| NSL | Non-specific lethal |
| NUP98 | Nucleoporin 98 |
| NSD | Nuclear receptor-binding SET domain-containing protein |
| NSRP1 | Nuclear speckle splicing regulatory protein 1 |
| PHD | Plant homeodomain |
| PROTAC | PROteolysis-Targeting Chimera |
| PWWP | Proline-tryptophan-tryptophan-proline |
| SAM | S-adenosylmethionine |
| SET | Su(var)3-9, enhancer of zeste and trithorax |
| SETMAR | SET domain and mariner transposase fusion gene-containing |
| SMYD2 | SET and MYND domain-containing 2 |
| TCGA | The Cancer Genome Atlas |
| TMB | Tumor mutation burden |
| TNBC | Triple-negative breast cancer |
| VEGF-A | Vascular endothelial growth factor-A |
References
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33 (Suppl. 3), 245–254. [Google Scholar] [CrossRef]
- Millan-Zambrano, G.; Burton, A.; Bannister, A.J.; Schneider, R. Histone post-translational modifications—Cause and consequence of genome function. Nat. Rev. Genet. 2022, 23, 563–580. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Allis, C.D.; Wang, G.G. The language of chromatin modification in human cancers. Nat. Rev. Cancer 2021, 21, 413–430. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef]
- Wagner, E.J.; Carpenter, P.B. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 2012, 13, 115–126. [Google Scholar] [CrossRef]
- Topchu, I.; Pangeni, R.P.; Bychkov, I.; Miller, S.A.; Izumchenko, E.; Yu, J.; Golemis, E.; Karanicolas, J.; Boumber, Y. The role of NSD1, NSD2, and NSD3 histone methyltransferases in solid tumors. Cell. Mol. Life Sci. 2022, 79, 285. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.L.; Swaroop, A.; Troche, C.; Licht, J.D. The Role of Nuclear Receptor-Binding SET Domain Family Histone Lysine Methyltransferases in Cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a026708. [Google Scholar] [CrossRef]
- He, L.; Cao, Y.; Sun, L. NSD family proteins: Rising stars as therapeutic targets. Cell Insight 2024, 3, 100151. [Google Scholar] [CrossRef]
- Cavalli, G.; Heard, E. Advances in epigenetics link genetics to the environment and disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef]
- Mohammad, H.P.; Barbash, O.; Creasy, C.L. Targeting epigenetic modifications in cancer therapy: Erasing the roadmap to cancer. Nat. Med. 2019, 25, 403–418. [Google Scholar] [CrossRef]
- Lu, C.; Jain, S.U.; Hoelper, D.; Bechet, D.; Molden, R.C.; Ran, L.; Murphy, D.; Venneti, S.; Hameed, M.; Pawel, B.R.; et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 2016, 352, 844–849. [Google Scholar] [CrossRef]
- Li, J.; Ahn, J.H.; Wang, G.G. Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell. Mol. Life Sci. 2019, 76, 2899–2916. [Google Scholar] [CrossRef] [PubMed]
- Jaju, R.J.; Fidler, C.; Haas, O.A.; Strickson, A.J.; Watkins, F.; Clark, K.; Cross, N.C.; Cheng, J.F.; Aplan, P.D.; Kearney, L.; et al. A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood 2001, 98, 1264–1267. [Google Scholar] [CrossRef]
- Hollink, I.H.; van den Heuvel-Eibrink, M.M.; Arentsen-Peters, S.T.; Pratcorona, M.; Abbas, S.; Kuipers, J.E.; van Galen, J.F.; Beverloo, H.B.; Sonneveld, E.; Kaspers, G.J.; et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood 2011, 118, 3645–3656. [Google Scholar] [CrossRef] [PubMed]
- Struski, S.; Lagarde, S.; Bories, P.; Puiseux, C.; Prade, N.; Cuccuini, W.; Pages, M.P.; Bidet, A.; Gervais, C.; Lafage-Pochitaloff, M.; et al. NUP98 is rearranged in 3.8% of pediatric AML forming a clinical and molecular homogenous group with a poor prognosis. Leukemia 2017, 31, 565–572. [Google Scholar] [CrossRef]
- Wang, G.G.; Cai, L.; Pasillas, M.P.; Kamps, M.P. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat. Cell Biol. 2007, 9, 804–812. [Google Scholar] [CrossRef]
- Xie, Z.; Bi, C.; Chooi, J.Y.; Chan, Z.L.; Mustafa, N.; Chng, W.J. MMSET regulates expression of IRF4 in t(4;14) myeloma and its silencing potentiates the effect of bortezomib. Leukemia 2015, 29, 2347–2354. [Google Scholar] [CrossRef] [PubMed]
- Oyer, J.A.; Huang, X.; Zheng, Y.; Shim, J.; Ezponda, T.; Carpenter, Z.; Allegretta, M.; Okot-Kotber, C.I.; Patel, J.P.; Melnick, A.; et al. Point mutation E1099K in MMSET/NSD2 enhances its methyltranferase activity and leads to altered global chromatin methylation in lymphoid malignancies. Leukemia 2014, 28, 198–201. [Google Scholar] [CrossRef]
- Jaffe, J.D.; Wang, Y.; Chan, H.M.; Zhang, J.; Huether, R.; Kryukov, G.V.; Bhang, H.E.; Taylor, J.E.; Hu, M.; Englund, N.P.; et al. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat. Genet. 2013, 45, 1386–1391. [Google Scholar] [CrossRef]
- Li, J.; Hlavka-Zhang, J.; Shrimp, J.H.; Piper, C.; Dupere-Richer, D.; Roth, J.S.; Jing, D.; Casellas Roman, H.L.; Troche, C.; Swaroop, A.; et al. PRC2 Inhibitors Overcome Glucocorticoid Resistance Driven by NSD2 Mutation in Pediatric Acute Lymphoblastic Leukemia. Cancer Discov. 2022, 12, 186–203. [Google Scholar] [CrossRef]
- Jeong, G.Y.; Park, M.K.; Choi, H.J.; An, H.W.; Park, Y.U.; Choi, H.J.; Park, J.; Kim, H.Y.; Son, T.; Lee, H.; et al. NSD3-Induced Methylation of H3K36 Activates NOTCH Signaling to Drive Breast Tumor Initiation and Metastatic Progression. Cancer Res. 2021, 81, 77–90. [Google Scholar] [CrossRef]
- Yuan, G.; Flores, N.M.; Hausmann, S.; Lofgren, S.M.; Kharchenko, V.; Angulo-Ibanez, M.; Sengupta, D.; Lu, X.; Czaban, I.; Azhibek, D.; et al. Elevated NSD3 histone methylation activity drives squamous cell lung cancer. Nature 2021, 590, 504–508. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; McGee, J.; Chen, X.; Doman, T.N.; Gong, X.; Zhang, Y.; Hamm, N.; Ma, X.; Higgs, R.E.; Bhagwat, S.V.; et al. Identification of druggable cancer driver genes amplified across TCGA datasets. PLoS ONE 2014, 9, e98293. [Google Scholar] [CrossRef]
- Angrand, P.O.; Apiou, F.; Stewart, A.F.; Dutrillaux, B.; Losson, R.; Chambon, P. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics 2001, 74, 79–88. [Google Scholar] [CrossRef]
- Shen, C.; Ipsaro, J.J.; Shi, J.; Milazzo, J.P.; Wang, E.; Roe, J.S.; Suzuki, Y.; Pappin, D.J.; Joshua-Tor, L.; Vakoc, C.R. NSD3-Short Is an Adaptor Protein that Couples BRD4 to the CHD8 Chromatin Remodeler. Mol. Cell 2015, 60, 847–859. [Google Scholar] [CrossRef]
- Xie, Z.; Chooi, J.Y.; Toh, S.H.M.; Yang, D.; Basri, N.B.; Ho, Y.S.; Chng, W.J. MMSET I acts as an oncoprotein and regulates GLO1 expression in t(4;14) multiple myeloma cells. Leukemia 2019, 33, 739–748. [Google Scholar] [CrossRef]
- Kooistra, S.M.; Helin, K. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 2012, 13, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Kiefer, C.M.; Dean, A. Distinctive signatures of histone methylation in transcribed coding and noncoding human beta-globin sequences. Mol. Cell. Biol. 2007, 27, 1271–1279. [Google Scholar] [CrossRef]
- Bannister, A.J.; Schneider, R.; Myers, F.A.; Thorne, A.W.; Crane-Robinson, C.; Kouzarides, T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 2005, 280, 17732–17736. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Gan, H.; Lee, J.H.; Han, J.; Wang, Z.; Riester, S.M.; Jin, L.; Chen, J.; Zhou, H.; Wang, J.; et al. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 2016, 352, 1344–1348. [Google Scholar] [CrossRef]
- Popovic, R.; Martinez-Garcia, E.; Giannopoulou, E.G.; Zhang, Q.; Zhang, Q.; Ezponda, T.; Shah, M.Y.; Zheng, Y.; Will, C.M.; Small, E.C.; et al. Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLoS Genet. 2014, 10, e1004566. [Google Scholar] [CrossRef] [PubMed]
- Zaghi, M.; Broccoli, V.; Sessa, A. H3K36 Methylation in Neural Development and Associated Diseases. Front. Genet. 2019, 10, 1291. [Google Scholar] [CrossRef] [PubMed]
- Edmunds, J.W.; Mahadevan, L.C.; Clayton, A.L. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008, 27, 406–420. [Google Scholar] [CrossRef]
- Xiao, C.; Fan, T.; Tian, H.; Zheng, Y.; Zhou, Z.; Li, S.; Li, C.; He, J. H3K36 trimethylation-mediated biological functions in cancer. Clin. Epigenet. 2021, 13, 199. [Google Scholar] [CrossRef]
- Tsukada, Y.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006, 439, 811–816. [Google Scholar] [CrossRef]
- Stec, I.; Wright, T.J.; van Ommen, G.J.; de Boer, P.A.; van Haeringen, A.; Moorman, A.F.; Altherr, M.R.; den Dunnen, J.T. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum. Mol. Genet. 1998, 7, 1071–1082. [Google Scholar] [CrossRef]
- Qiao, Q.; Li, Y.; Chen, Z.; Wang, M.; Reinberg, D.; Xu, R.M. The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation. J. Biol. Chem. 2011, 286, 8361–8368. [Google Scholar] [CrossRef]
- Graham, S.E.; Tweedy, S.E.; Carlson, H.A. Dynamic behavior of the post-SET loop region of NSD1: Implications for histone binding and drug development. Protein Sci. 2016, 25, 1021–1029. [Google Scholar] [CrossRef]
- Li, Y.; Trojer, P.; Xu, C.F.; Cheung, P.; Kuo, A.; Drury, W.J., III; Qiao, Q.; Neubert, T.A.; Xu, R.M.; Gozani, O.; et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J. Biol. Chem. 2009, 284, 34283–34295. [Google Scholar] [CrossRef] [PubMed]
- Kuo, A.J.; Cheung, P.; Chen, K.; Zee, B.M.; Kioi, M.; Lauring, J.; Xi, Y.; Park, B.H.; Shi, X.; Garcia, B.A.; et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol. Cell 2011, 44, 609–620. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Q.; Xu, X.; Xie, B.; Zhao, Y.; Li, N.; Cao, X. The methyltransferase NSD3 promotes antiviral innate immunity via direct lysine methylation of IRF3. J. Exp. Med. 2017, 214, 3597–3610. [Google Scholar] [CrossRef] [PubMed]
- Weirich, S.; Kusevic, D.; Schnee, P.; Reiter, J.; Pleiss, J.; Jeltsch, A. Discovery of NSD2 non-histone substrates and design of a super-substrate. Commun. Biol. 2024, 7, 707. [Google Scholar] [CrossRef]
- Song, D.; Lan, J.; Chen, Y.; Liu, A.; Wu, Q.; Zhao, C.; Feng, Y.; Wang, J.; Luo, X.; Cao, Z.; et al. NSD2 promotes tumor angiogenesis through methylating and activating STAT3 protein. Oncogene 2021, 40, 2952–2967. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Song, D.; Chen, Y.; Huang, C.; Liu, A.; Wu, Q.; She, X.; Li, K.; Wan, K.; Yu, C.; et al. NSD2 methylates AROS to promote SIRT1 activation and regulates fatty acid metabolism-mediated cancer radiotherapy. Cell Rep. 2023, 42, 113126. [Google Scholar] [CrossRef]
- Saloura, V.; Vougiouklakis, T.; Zewde, M.; Deng, X.; Kiyotani, K.; Park, J.H.; Matsuo, Y.; Lingen, M.; Suzuki, T.; Dohmae, N.; et al. WHSC1L1-mediated EGFR mono-methylation enhances the cytoplasmic and nuclear oncogenic activity of EGFR in head and neck cancer. Sci. Rep. 2017, 7, 40664. [Google Scholar] [CrossRef]
- Wu, H.; Zeng, H.; Lam, R.; Tempel, W.; Amaya, M.F.; Xu, C.; Dombrovski, L.; Qiu, W.; Wang, Y.; Min, J. Structural and histone binding ability characterizations of human PWWP domains. PLoS ONE 2011, 6, e18919. [Google Scholar] [CrossRef]
- Sankaran, S.M.; Wilkinson, A.W.; Elias, J.E.; Gozani, O. A PWWP Domain of Histone-Lysine N-Methyltransferase NSD2 Binds to Dimethylated Lys-36 of Histone H3 and Regulates NSD2 Function at Chromatin. J. Biol. Chem. 2016, 291, 8465–8474. [Google Scholar] [CrossRef]
- Pasillas, M.P.; Shah, M.; Kamps, M.P. NSD1 PHD domains bind methylated H3K4 and H3K9 using interactions disrupted by point mutations in human sotos syndrome. Hum. Mutat. 2011, 32, 292–298. [Google Scholar] [CrossRef]
- French, C.A.; Rahman, S.; Walsh, E.M.; Kuhnle, S.; Grayson, A.R.; Lemieux, M.E.; Grunfeld, N.; Rubin, B.P.; Antonescu, C.R.; Zhang, S.; et al. NSD3-NUT fusion oncoprotein in NUT midline carcinoma: Implications for a novel oncogenic mechanism. Cancer Discov. 2014, 4, 928–941. [Google Scholar] [CrossRef] [PubMed]
- Morishita, M.; Mevius, D.; di Luccio, E. In vitro histone lysine methylation by NSD1, NSD2/MMSET/WHSC1 and NSD3/WHSC1L. BMC Struct. Biol. 2014, 14, 25. [Google Scholar] [CrossRef]
- Martinez-Garcia, E.; Popovic, R.; Min, D.J.; Sweet, S.M.; Thomas, P.M.; Zamdborg, L.; Heffner, A.; Will, C.; Lamy, L.; Staudt, L.M.; et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood 2011, 117, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.S.; Lee, S.H.; Kan, P.Y.; Voigt, P.; Ma, L.; Shi, X.; Reinberg, D.; Lee, M.G. Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev. 2012, 26, 2749–2762. [Google Scholar] [CrossRef]
- Murphy, F.V.t.; Sweet, R.M.; Churchill, M.E. The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J. 1999, 18, 6610–6618. [Google Scholar] [CrossRef] [PubMed]
- Stros, M.; Launholt, D.; Grasser, K.D. The HMG-box: A versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell. Mol. Life Sci. 2007, 64, 2590–2606. [Google Scholar] [CrossRef]
- Kang, H.B.; Choi, Y.; Lee, J.M.; Choi, K.C.; Kim, H.C.; Yoo, J.Y.; Lee, Y.H.; Yoon, H.G. The histone methyltransferase, NSD2, enhances androgen receptor-mediated transcription. FEBS Lett. 2009, 583, 1880–1886. [Google Scholar] [CrossRef]
- Li, W.; Tian, W.; Yuan, G.; Deng, P.; Sengupta, D.; Cheng, Z.; Cao, Y.; Ren, J.; Qin, Y.; Zhou, Y.; et al. Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases. Nature 2021, 590, 498–503. [Google Scholar] [CrossRef]
- Lucio-Eterovic, A.K.; Singh, M.M.; Gardner, J.E.; Veerappan, C.S.; Rice, J.C.; Carpenter, P.B. Role for the nuclear receptor-binding SET domain protein 1 (NSD1) methyltransferase in coordinating lysine 36 methylation at histone 3 with RNA polymerase II function. Proc. Natl. Acad. Sci. USA 2010, 107, 16952–16957. [Google Scholar] [CrossRef] [PubMed]
- Kurotaki, N.; Harada, N.; Yoshiura, K.; Sugano, S.; Niikawa, N.; Matsumoto, N. Molecular characterization of NSD1, a human homologue of the mouse Nsd1 gene. Gene 2001, 279, 197–204. [Google Scholar] [CrossRef]
- Conteduca, G.; Testa, B.; Baldo, C.; Arado, A.; Malacarne, M.; Candiano, G.; Garbarino, A.; Coviello, D.A.; Cantoni, C. Identification of alternative transcripts of NSD1 gene in Sotos Syndrome patients and healthy subjects. Gene 2023, 851, 146970. [Google Scholar] [CrossRef]
- Chesi, M.; Nardini, E.; Lim, R.S.; Smith, K.D.; Kuehl, W.M.; Bergsagel, P.L. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 1998, 92, 3025–3034. [Google Scholar] [CrossRef]
- Garlisi, C.G.; Uss, A.S.; Xiao, H.; Tian, F.; Sheridan, K.E.; Wang, L.; Motasim Billah, M.; Egan, R.W.; Stranick, K.S.; Umland, S.P. A unique mRNA initiated within a middle intron of WHSC1/MMSET encodes a DNA binding protein that suppresses human IL-5 transcription. Am. J. Respir. Cell Mol. Biol. 2001, 24, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Woo Park, J.; Kim, K.B.; Kim, J.Y.; Chae, Y.C.; Jeong, O.S.; Seo, S.B. RE-IIBP Methylates H3K79 and Induces MEIS1-mediated Apoptosis via H2BK120 Ubiquitination by RNF20. Sci. Rep. 2015, 5, 12485. [Google Scholar] [CrossRef] [PubMed]
- Mirabella, F.; Murison, A.; Aronson, L.I.; Wardell, C.P.; Thompson, A.J.; Hanrahan, S.J.; Fok, J.H.; Pawlyn, C.; Kaiser, M.F.; Walker, B.A.; et al. A novel functional role for MMSET in RNA processing based on the link between the REIIBP isoform and its interaction with the SMN complex. PLoS ONE 2014, 9, e99493. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yu, X. REIIBP methylates nucleolar proteins and regulates pre-rRNA processing. J. Biol. Chem. 2025, 301, 110609. [Google Scholar] [CrossRef]
- Stec, I.; van Ommen, G.J.; den Dunnen, J.T. WHSC1L1, on human chromosome 8p11.2, closely resembles WHSC1 and maps to a duplicated region shared with 4p16.3. Genomics 2001, 76, 5–8. [Google Scholar] [CrossRef]
- Kim, S.M.; Kee, H.J.; Eom, G.H.; Choe, N.W.; Kim, J.Y.; Kim, Y.S.; Kim, S.K.; Kook, H.; Kook, H.; Seo, S.B. Characterization of a novel WHSC1-associated SET domain protein with H3K4 and H3K27 methyltransferase activity. Biochem. Biophys. Res. Commun. 2006, 345, 318–323. [Google Scholar] [CrossRef]
- Rayasam, G.V.; Wendling, O.; Angrand, P.O.; Mark, M.; Niederreither, K.; Song, L.; Lerouge, T.; Hager, G.L.; Chambon, P.; Losson, R. NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J. 2003, 22, 3153–3163. [Google Scholar] [CrossRef]
- Hajdu, I.; Ciccia, A.; Lewis, S.M.; Elledge, S.J. Wolf-Hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage. Proc. Natl. Acad. Sci. USA 2011, 108, 13130–13134. [Google Scholar] [CrossRef]
- Yuan, W.; Xu, M.; Huang, C.; Liu, N.; Chen, S.; Zhu, B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 2011, 286, 7983–7989. [Google Scholar] [CrossRef]
- Schmitges, F.W.; Prusty, A.B.; Faty, M.; Stutzer, A.; Lingaraju, G.M.; Aiwazian, J.; Sack, R.; Hess, D.; Li, L.; Zhou, S.; et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 2011, 42, 330–341. [Google Scholar] [CrossRef]
- Marango, J.; Shimoyama, M.; Nishio, H.; Meyer, J.A.; Min, D.J.; Sirulnik, A.; Martinez-Martinez, Y.; Chesi, M.; Bergsagel, P.L.; Zhou, M.M.; et al. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood 2008, 111, 3145–3154. [Google Scholar] [CrossRef]
- Huang, N.; vom Baur, E.; Garnier, J.M.; Lerouge, T.; Vonesch, J.L.; Lutz, Y.; Chambon, P.; Losson, R. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J. 1998, 17, 3398–3412. [Google Scholar] [CrossRef]
- Fang, Y.; Tang, Y.; Zhang, Y.; Pan, Y.; Jia, J.; Sun, Z.; Zeng, W.; Chen, J.; Yuan, Y.; Fang, D. The H3K36me2 methyltransferase NSD1 modulates H3K27ac at active enhancers to safeguard gene expression. Nucleic Acids Res. 2021, 49, 6281–6295. [Google Scholar] [CrossRef]
- Weinberg, D.N.; Papillon-Cavanagh, S.; Chen, H.; Yue, Y.; Chen, X.; Rajagopalan, K.N.; Horth, C.; McGuire, J.T.; Xu, X.; Nikbakht, H.; et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 2019, 573, 281–286. [Google Scholar] [CrossRef]
- Xu, W.; Li, J.; Rong, B.; Zhao, B.; Wang, M.; Dai, R.; Chen, Q.; Liu, H.; Gu, Z.; Liu, S.; et al. DNMT3A reads and connects histone H3K36me2 to DNA methylation. Protein Cell 2020, 11, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Nimura, K.; Ura, K.; Shiratori, H.; Ikawa, M.; Okabe, M.; Schwartz, R.J.; Kaneda, Y. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature 2009, 460, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Shirane, K.; Miura, F.; Ito, T.; Lorincz, M.C. NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing. Nat. Genet. 2020, 52, 1088–1098. [Google Scholar] [CrossRef]
- Campos-Sanchez, E.; Deleyto-Seldas, N.; Dominguez, V.; Carrillo-de-Santa-Pau, E.; Ura, K.; Rocha, P.P.; Kim, J.; Aljoufi, A.; Esteve-Codina, A.; Dabad, M.; et al. Wolf-Hirschhorn Syndrome Candidate 1 Is Necessary for Correct Hematopoietic and B Cell Development. Cell Rep. 2017, 19, 1586–1601. [Google Scholar] [CrossRef] [PubMed]
- Kurotaki, N.; Imaizumi, K.; Harada, N.; Masuno, M.; Kondoh, T.; Nagai, T.; Ohashi, H.; Naritomi, K.; Tsukahara, M.; Makita, Y.; et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet. 2002, 30, 365–366. [Google Scholar] [CrossRef]
- Baujat, G.; Rio, M.; Rossignol, S.; Sanlaville, D.; Lyonnet, S.; Le Merrer, M.; Munnich, A.; Gicquel, C.; Cormier-Daire, V.; Colleaux, L. Paradoxical NSD1 mutations in Beckwith-Wiedemann syndrome and 11p15 anomalies in Sotos syndrome. Am. J. Hum. Genet. 2004, 74, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Jacques-Fricke, B.T.; Gammill, L.S. Neural crest specification and migration independently require NSD3-related lysine methyltransferase activity. Mol. Biol. Cell 2014, 25, 4174–4186. [Google Scholar] [CrossRef]
- Jacques-Fricke, B.T.; Roffers-Agarwal, J.; Hussein, A.O.; Yoder, K.J.; Gearhart, M.D.; Gammill, L.S. Profiling NSD3-dependent neural crest gene expression reveals known and novel candidate regulatory factors. Dev. Biol. 2021, 475, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.S.; Gammill, L.S.; Bronner-Fraser, M. Discovery of transcription factors and other candidate regulators of neural crest development. Dev. Dyn. 2008, 237, 1021–1033. [Google Scholar] [CrossRef]
- Fnu, S.; Williamson, E.A.; De Haro, L.P.; Brenneman, M.; Wray, J.; Shaheen, M.; Radhakrishnan, K.; Lee, S.H.; Nickoloff, J.A.; Hromas, R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 2011, 108, 540–545. [Google Scholar] [CrossRef]
- de Krijger, I.; van der Torre, J.; Peuscher, M.H.; Eder, M.; Jacobs, J.J.L. H3K36 dimethylation by MMSET promotes classical non-homologous end-joining at unprotected telomeres. Oncogene 2020, 39, 4814–4827. [Google Scholar] [CrossRef]
- Zhang, J.; Lee, Y.R.; Dang, F.; Gan, W.; Menon, A.V.; Katon, J.M.; Hsu, C.H.; Asara, J.M.; Tibarewal, P.; Leslie, N.R.; et al. PTEN Methylation by NSD2 Controls Cellular Sensitivity to DNA Damage. Cancer Discov. 2019, 9, 1306–1323. [Google Scholar] [CrossRef]
- Pei, H.; Zhang, L.; Luo, K.; Qin, Y.; Chesi, M.; Fei, F.; Bergsagel, P.L.; Wang, L.; You, Z.; Lou, Z. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 2011, 470, 124–128. [Google Scholar] [CrossRef]
- Caeiro, L.D.; Nakata, Y.; Borges, R.L.; Zha, M.; Garcia-Martinez, L.; Banuelos, C.P.; Stransky, S.; Liu, T.; Chan, H.L.; Brabson, J.; et al. Methylation of histone H3 lysine 36 is a barrier for therapeutic interventions of head and neck squamous cell carcinoma. Genes Dev. 2024, 38, 46–69. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Gao, H.; Jiang, D.; Guo, G.; Hou, J.; Han, Y.; Zhang, C.; Hu, X.; Indulkar, S.; Kloeber, J.A.; et al. Isoform-specific function of NSD3 in DNA replication stress confers resistance to PARP inhibitors in prostate cancer. Mol. Cell 2025, 85, 2673–2687.E8. [Google Scholar] [CrossRef]
- Voutsadakis, I.A. Characteristics and Prognosis of 8p11.23-Amplified Squamous Lung Carcinomas. J. Clin. Med. 2023, 12, 1711. [Google Scholar] [CrossRef]
- Voutsadakis, I.A. Amplification of 8p11.23 in cancers and the role of amplicon genes. Life Sci. 2021, 264, 118729. [Google Scholar] [CrossRef]
- Voutsadakis, I.A. 8p11.23 Amplification in Breast Cancer: Molecular Characteristics, Prognosis and Targeted Therapy. J. Clin. Med. 2020, 9, 3079. [Google Scholar] [CrossRef]
- Yang, Z.Q.; Liu, G.; Bollig-Fischer, A.; Giroux, C.N.; Ethier, S.P. Transforming properties of 8p11-12 amplified genes in human breast cancer. Cancer Res. 2010, 70, 8487–8497. [Google Scholar] [CrossRef]
- Swaroop, A.; Oyer, J.A.; Will, C.M.; Huang, X.; Yu, W.; Troche, C.; Bulic, M.; Durham, B.H.; Wen, Q.J.; Crispino, J.D.; et al. An activating mutation of the NSD2 histone methyltransferase drives oncogenic reprogramming in acute lymphocytic leukemia. Oncogene 2019, 38, 671–686. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Kumar, A.; Hamada, K.; Okada, C.; Oguni, A.; Machiyama, A.; Sakuraba, S.; Nishizawa, T.; Nureki, O.; Kono, H.; et al. Structural basis of the regulation of the normal and oncogenic methylation of nucleosomal histone H3 Lys36 by NSD2. Nat. Commun. 2021, 12, 6605. [Google Scholar] [CrossRef] [PubMed]
- Bea, S.; Valdes-Mas, R.; Navarro, A.; Salaverria, I.; Martin-Garcia, D.; Jares, P.; Gine, E.; Pinyol, M.; Royo, C.; Nadeu, F.; et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 18250–18255. [Google Scholar] [CrossRef] [PubMed]
- Khella, M.S.; Schnee, P.; Weirich, S.; Bui, T.; Brohm, A.; Bashtrykov, P.; Pleiss, J.; Jeltsch, A. The T1150A cancer mutant of the protein lysine dimethyltransferase NSD2 can introduce H3K36 trimethylation. J. Biol. Chem. 2023, 299, 104796. [Google Scholar] [CrossRef]
- Moon, S.W.; Son, H.J.; Mo, H.Y.; Choi, E.J.; Yoo, N.J.; Lee, S.H. Mutation and expression alterations of histone methylation-related NSD2, KDM2B and SETMAR genes in colon cancers. Pathol. Res. Pract. 2021, 219, 153354. [Google Scholar] [CrossRef]
- Sun, Y.; Xie, J.; Cai, S.; Wang, Q.; Feng, Z.; Li, Y.; Lu, J.J.; Chen, W.; Ye, Z. Elevated expression of nuclear receptor-binding SET domain 3 promotes pancreatic cancer cell growth. Cell Death Dis. 2021, 12, 913. [Google Scholar] [CrossRef]
- Sibley, K.; Fenton, J.A.; Dring, A.M.; Ashcroft, A.J.; Rawstron, A.C.; Morgan, G.J. A molecular study of the t(4;14) in multiple myeloma. Br. J. Haematol. 2002, 118, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Rosati, R.; La Starza, R.; Veronese, A.; Aventin, A.; Schwienbacher, C.; Vallespi, T.; Negrini, M.; Martelli, M.F.; Mecucci, C. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15). Blood 2002, 99, 3857–3860. [Google Scholar] [CrossRef]
- Suzuki, S.; Kurabe, N.; Ohnishi, I.; Yasuda, K.; Aoshima, Y.; Naito, M.; Tanioka, F.; Sugimura, H. NSD3-NUT-expressing midline carcinoma of the lung: First characterization of primary cancer tissue. Pathol. Res. Pract. 2015, 211, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Jevtic, Z.; Matafora, V.; Casagrande, F.; Santoro, F.; Minucci, S.; Garre, M.; Rasouli, M.; Heidenreich, O.; Musco, G.; Schwaller, J.; et al. SMARCA5 interacts with NUP98-NSD1 oncofusion protein and sustains hematopoietic cells transformation. J. Exp. Clin. Cancer Res. 2022, 41, 34. [Google Scholar] [CrossRef]
- Xu, H.; Valerio, D.G.; Eisold, M.E.; Sinha, A.; Koche, R.P.; Hu, W.; Chen, C.W.; Chu, S.H.; Brien, G.L.; Park, C.Y.; et al. NUP98 Fusion Proteins Interact with the NSL and MLL1 Complexes to Drive Leukemogenesis. Cancer Cell 2016, 30, 863–878. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, F.; Chen, Q.; Wan, C.; Xiong, J.; Xu, J. CRISPR/Cas9-mediated knockout of NSD1 suppresses the hepatocellular carcinoma development via the NSD1/H3/Wnt10b signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 467. [Google Scholar] [CrossRef]
- Chen, Y.; Li, X.; Xu, J.; Xiao, H.; Tang, C.; Liang, W.; Zhu, X.; Fang, Y.; Wang, H.; Shi, J. Knockdown of nuclear receptor binding SET domain-containing protein 1 (NSD1) inhibits proliferation and facilitates apoptosis in paclitaxel-resistant breast cancer cells via inactivating the Wnt/beta-catenin signaling pathway. Bioengineered 2022, 13, 3526–3536. [Google Scholar] [CrossRef]
- Lu, T.; Jackson, M.W.; Wang, B.; Yang, M.; Chance, M.R.; Miyagi, M.; Gudkov, A.V.; Stark, G.R. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc. Natl. Acad. Sci. USA 2010, 107, 46–51. [Google Scholar] [CrossRef]
- Leonards, K.; Almosailleakh, M.; Tauchmann, S.; Bagger, F.O.; Thirant, C.; Juge, S.; Bock, T.; Mereau, H.; Bezerra, M.F.; Tzankov, A.; et al. Nuclear interacting SET domain protein 1 inactivation impairs GATA1-regulated erythroid differentiation and causes erythroleukemia. Nat. Commun. 2020, 11, 2807. [Google Scholar] [CrossRef]
- Papillon-Cavanagh, S.; Lu, C.; Gayden, T.; Mikael, L.G.; Bechet, D.; Karamboulas, C.; Ailles, L.; Karamchandani, J.; Marchione, D.M.; Garcia, B.A.; et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat. Genet. 2017, 49, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Hudlebusch, H.R.; Santoni-Rugiu, E.; Simon, R.; Ralfkiaer, E.; Rossing, H.H.; Johansen, J.V.; Jorgensen, M.; Sauter, G.; Helin, K. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin. Cancer Res. 2011, 17, 2919–2933. [Google Scholar] [CrossRef]
- Toyokawa, G.; Cho, H.S.; Masuda, K.; Yamane, Y.; Yoshimatsu, M.; Hayami, S.; Takawa, M.; Iwai, Y.; Daigo, Y.; Tsuchiya, E.; et al. Histone lysine methyltransferase Wolf-Hirschhorn syndrome candidate 1 is involved in human carcinogenesis through regulation of the Wnt pathway. Neoplasia 2011, 13, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Guo, L.; Duan, Z.J.; Tepper, C.G.; Xue, L.; Chen, X.; Kung, H.J.; Gao, A.C.; Zou, J.X.; Chen, H.W. Histone methyltransferase NSD2/MMSET mediates constitutive NF-kappaB signaling for cancer cell proliferation, survival, and tumor growth via a feed-forward loop. Mol. Cell. Biol. 2012, 32, 3121–3131. [Google Scholar] [CrossRef] [PubMed]
- Parolia, A.; Eyunni, S.; Verma, B.K.; Young, E.; Liu, Y.; Liu, L.; George, J.; Aras, S.; Das, C.K.; Mannan, R.; et al. NSD2 is a requisite subunit of the AR/FOXA1 neo-enhanceosome in promoting prostate tumorigenesis. Nat. Genet. 2024, 56, 2132–2143. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.H.; Li, Q.; Huang, Z.J.; Sun, M.X.; Lu, J.J.; Zhang, X.H.; Li, G.; Wu, F. Identification of histone methyltransferase NSD2 as an important oncogenic gene in colorectal cancer. Cell Death Dis. 2021, 12, 974. [Google Scholar] [CrossRef]
- Yin, Z.; Sun, Y.; Ge, S.; Sun, J. Epigenetic activation of WHSC1 functions as an oncogene and is associated with poor prognosis in cervical cancer. Oncol. Rep. 2017, 37, 2286–2294. [Google Scholar] [CrossRef]
- Garcia-Carpizo, V.; Sarmentero, J.; Han, B.; Grana, O.; Ruiz-Llorente, S.; Pisano, D.G.; Serrano, M.; Brooks, H.B.; Campbell, R.M.; Barrero, M.J. NSD2 contributes to oncogenic RAS-driven transcription in lung cancer cells through long-range epigenetic activation. Sci. Rep. 2016, 6, 32952. [Google Scholar] [CrossRef]
- Sengupta, D.; Zeng, L.; Li, Y.; Hausmann, S.; Ghosh, D.; Yuan, G.; Nguyen, T.N.; Lyu, R.; Caporicci, M.; Morales Benitez, A.; et al. NSD2 dimethylation at H3K36 promotes lung adenocarcinoma pathogenesis. Mol. Cell 2021, 81, 4481–4492.E9. [Google Scholar] [CrossRef]
- Feng, W.; Niu, N.; Lu, P.; Chen, Z.; Rao, H.; Zhang, W.; Ma, C.; Liu, C.; Xu, Y.; Gao, W.Q.; et al. Multilevel Regulation of NF-kappaB Signaling by NSD2 Suppresses Kras-Driven Pancreatic Tumorigenesis. Adv. Sci. 2024, 11, e2309387. [Google Scholar] [CrossRef]
- Turner-Ivey, B.; Smith, E.L.; Rutkovsky, A.C.; Spruill, L.S.; Mills, J.N.; Ethier, S.P. Development of mammary hyperplasia, dysplasia, and invasive ductal carcinoma in transgenic mice expressing the 8p11 amplicon oncogene NSD3. Breast Cancer Res. Treat. 2017, 164, 349–358. [Google Scholar] [CrossRef]
- Saloura, V.; Vougiouklakis, T.; Zewde, M.; Kiyotani, K.; Park, J.H.; Gao, G.; Karrison, T.; Lingen, M.; Nakamura, Y.; Hamamoto, R. WHSC1L1 drives cell cycle progression through transcriptional regulation of CDC6 and CDK2 in squamous cell carcinoma of the head and neck. Oncotarget 2016, 7, 42527–42538. [Google Scholar] [CrossRef]
- Xiong, Q.; Zhou, Y.; Zhang, S.; Zhang, Y.; Xu, Y.; Yang, Y.; Zhou, C.; Zeng, Z.; Han, J.; Zhu, Q. NSD3, a member of nuclear receptor-binding SET domain family, is a potential prognostic biomarker for pancreatic cancer. Cancer Med. 2023, 12, 10961–10978. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Xiao, H.; Cai, Y.; Zhang, N. NSD1 promotes esophageal cancer tumorigenesis via HIF1alpha signaling. Cell Biol. Toxicol. 2023, 39, 1835–1850. [Google Scholar] [CrossRef]
- Chen, Y.; Tang, W.; Zhu, X.; Zhang, L.; Zhu, Y.; Xiao, H.; Xu, J.; Fang, Y.; Li, X.; Tang, C.; et al. Nuclear receptor binding SET domain protein 1 promotes epithelial-mesenchymal transition in paclitaxel-resistant breast cancer cells via regulating nuclear factor kappa B and F-box and leucine-rich repeat protein 11. Bioengineered 2021, 12, 11506–11519. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Y.; Li, X.; Lin, Y.; Chen, Y.; Yang, H.; Shen, Y. Clinical and molecular characterizations of HNSCC patients with occult lymph node metastasis. Sci. Rep. 2025, 15, 25263. [Google Scholar] [CrossRef] [PubMed]
- Ezponda, T.; Popovic, R.; Shah, M.Y.; Martinez-Garcia, E.; Zheng, Y.; Min, D.J.; Will, C.; Neri, A.; Kelleher, N.L.; Yu, J.; et al. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer. Oncogene 2013, 32, 2882–2890. [Google Scholar] [CrossRef]
- Cheong, C.M.; Mrozik, K.M.; Hewett, D.R.; Bell, E.; Panagopoulos, V.; Noll, J.E.; Licht, J.D.; Gronthos, S.; Zannettino, A.C.W.; Vandyke, K. Twist-1 is upregulated by NSD2 and contributes to tumour dissemination and an epithelial-mesenchymal transition-like gene expression signature in t(4;14)-positive multiple myeloma. Cancer Lett. 2020, 475, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xue, W.; Yuan, H.; Dong, B.; Ding, Y.; Liu, Y.; Jiang, M.; Kan, S.; Sun, T.; Ren, J. AKT-mediated stabilization of histone methyltransferase WHSC1 promotes prostate cancer metastasis. J. Clin. Investig. 2017, 127, 1284–1302. [Google Scholar] [CrossRef]
- Aytes, A.; Giacobbe, A.; Mitrofanova, A.; Ruggero, K.; Cyrta, J.; Arriaga, J.; Palomero, L.; Farran-Matas, S.; Rubin, M.A.; Shen, M.M.; et al. NSD2 is a conserved driver of metastatic prostate cancer progression. Nat. Commun. 2018, 9, 5201. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, J.; Chen, Y.; Li, H.; Lin, L. WHSC1 promotes wnt/beta-catenin signaling in a FoxM1-dependent manner facilitating proliferation, invasion and epithelial-mesenchymal transition in breast cancer. J. Recept. Signal Transduct. Res. 2020, 40, 410–418. [Google Scholar] [CrossRef]
- Kochumon, S.; Al-Sayyar, A.; Jacob, T.; Bahman, F.; Akhter, N.; Wilson, A.; Sindhu, S.; Hannun, Y.A.; Ahmad, R.; Al-Mulla, F. TGF-beta and TNF-alpha interaction promotes the expression of MMP-9 through H3K36 dimethylation: Implications in breast cancer metastasis. Front. Immunol. 2024, 15, 1430187. [Google Scholar] [CrossRef]
- Chen, D.; Chen, X.; Yang, M.; Li, Q.; Weng, S.; Kou, S.; Liu, X.; Jiang, G.; Liu, H. H3K36me2 methyltransferase NSD2/WHSC1 promotes triple-negative breast cancer metastasis via activation of ULK1-dependent autophagy. Autophagy 2025, 21, 1824–1842. [Google Scholar] [CrossRef]
- Han, X.; Piao, L.; Yuan, X.; Wang, L.; Liu, Z.; He, X. Knockdown of NSD2 Suppresses Renal Cell Carcinoma Metastasis by Inhibiting Epithelial-Mesenchymal Transition. Int. J. Med. Sci. 2019, 16, 1404–1411. [Google Scholar] [CrossRef]
- Lu, M.H.; Fan, M.F.; Yu, X.D. NSD2 promotes osteosarcoma cell proliferation and metastasis by inhibiting E-cadherin expression. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 928–936. [Google Scholar] [PubMed]
- Zheng, D.; Pu, X.; Deng, X.; Liu, C.; Li, S. Metabolic profiles in laryngeal cancer defined two distinct molecular subtypes with divergent prognoses. Front. Immunol. 2025, 16, 1512502. [Google Scholar] [CrossRef]
- Tang, S.; Wang, Q.; Wang, Z.; Cai, L.; Pan, D.; Li, J.; Chen, Q.; Zhou, Y.; Shen, Y.Q. NSD1 mutation status determines metabolic inhibitor sensitivity in head and neck squamous cell carcinomas by regulating mitochondrial respiration. J. Pathol. 2025, 266, 306–321. [Google Scholar] [CrossRef]
- Wang, J.; Duan, Z.; Nugent, Z.; Zou, J.X.; Borowsky, A.D.; Zhang, Y.; Tepper, C.G.; Li, J.J.; Fiehn, O.; Xu, J.; et al. Reprogramming metabolism by histone methyltransferase NSD2 drives endocrine resistance via coordinated activation of pentose phosphate pathway enzymes. Cancer Lett. 2016, 378, 69–79. [Google Scholar] [CrossRef]
- Chong, P.S.Y.; Chooi, J.Y.; Lim, J.S.L.; Leow, A.C.Y.; Toh, S.H.M.; Azaman, I.; Koh, M.Y.; Teoh, P.J.; Tan, T.Z.; Chung, T.H.; et al. Histone Methyltransferase NSD2 Activates PKCalpha to Drive Metabolic Reprogramming and Lenalidomide Resistance in Multiple Myeloma. Cancer Res. 2023, 83, 3414–3427. [Google Scholar] [CrossRef] [PubMed]
- Sobh, A.; Encinas, E.; Patel, A.; Surapaneni, G.; Bonilla, E.; Kaestner, C.; Poullard, J.; Clerio, M.; Vasan, K.; Freeman, T.; et al. NSD2 drives t(4;14) myeloma cell dependence on adenylate kinase 2 by diverting one-carbon metabolism to the epigenome. Blood 2024, 144, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Peng, X.; Fang, C.; Peng, X.; Tang, J.; Wang, Z.; Long, Y.; Chen, J.; Peng, Y.; Zhang, Z.; et al. Histones Methyltransferase NSD3 Inhibits Lung Adenocarcinoma Glycolysis Through Interacting with PPP1CB to Decrease STAT3 Signaling Pathway. Adv Sci. 2024, 11, e2400381. [Google Scholar] [CrossRef] [PubMed]
- Rona, G.B.; Almeida, D.S.G.; Pinheiro, A.S.; Eleutherio, E.C.A. The PWWP domain of the human oncogene WHSC1L1/NSD3 induces a metabolic shift toward fermentation. Oncotarget 2017, 8, 54068–54081. [Google Scholar] [CrossRef] [PubMed]
- Chava, S.; Wajapeyee, N. NSD proteins in anti-tumor immunity and their therapeutic targeting by protein degraders. Cell. Mol. Life Sci. 2025, 82, 268. [Google Scholar] [CrossRef]
- Li, Y.; Goldberg, E.M.; Chen, X.; Xu, X.; McGuire, J.T.; Leuzzi, G.; Karagiannis, D.; Tate, T.; Farhangdoost, N.; Horth, C.; et al. Histone methylation antagonism drives tumor immune evasion in squamous cell carcinomas. Mol. Cell 2022, 82, 3901–3918 e3907. [Google Scholar] [CrossRef]
- Chen, C.; Shin, J.H.; Fang, Z.; Brennan, K.; Horowitz, N.B.; Pfaff, K.L.; Welsh, E.L.; Rodig, S.J.; Gevaert, O.; Gozani, O.; et al. Targeting KDM2A Enhances T-cell Infiltration in NSD1-Deficient Head and Neck Squamous Cell Carcinoma. Cancer Res. 2023, 83, 2645–2655. [Google Scholar] [CrossRef]
- Li, M.; Gao, X.; Wang, X. Identification of tumor mutation burden-associated molecular and clinical features in cancer by analyzing multi-omics data. Front. Immunol. 2023, 14, 1090838. [Google Scholar] [CrossRef] [PubMed]
- Want, M.Y.; Tsuji, T.; Singh, P.K.; Thorne, J.L.; Matsuzaki, J.; Karasik, E.; Gillard, B.; Cortes Gomez, E.; Koya, R.C.; Lugade, A.; et al. WHSC1/NSD2 regulates immune infiltration in prostate cancer. J. Immunother. Cancer 2021, 9, e001374. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, J.; Zhang, Y.; Pan, Y.; Li, Z.; Wang, M.; Gao, Y.; Feng, D.; He, X.; Zhang, C. Association of WHSC1/NSD2 and T-cell infiltration with prostate cancer metastasis and prognosis. Sci. Rep. 2023, 13, 21629. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Li, N.; Pei, S.; Lian, Y.; Li, L.; Peng, Y.; Liu, Q.; Guo, J.; Wang, X.; Han, Y.; et al. Histone methyltransferase WHSC1 loss dampens MHC-I antigen presentation pathway to impair IFN-gamma-stimulated antitumor immunity. J. Clin. Investig. 2022, 132, e153167. [Google Scholar] [CrossRef]
- Yan, J.; Zhang, M.Y.; Lin, J.; Li, K.X.; Zhao, Z.M.; Gao, Y.M.; Deng, X.L.; Wang, C.S.; Wang, H.S. WHSC1 is involved in DNA damage, cellular senescence and immune response in hepatocellular carcinoma progression. J. Cell. Mol. Med. 2023, 27, 1436–1441. [Google Scholar] [CrossRef]
- Gladstein, A.C.; Poltorack, C.D.; Solomon, A.M.C.; Venkatesh, S.; Adler, K.M.; Robertson, M.R.; Stransky, S.; Irizarry-Negron, V.M.; Ruiz, D.A.; Freeburg, N.F.; et al. The H3 (K36M) oncohistone inhibits NSD2 to activate a SETD2-dependent antiviral-like immune response in KRAS-driven lung cancer. bioRxiv 2025. [Google Scholar] [CrossRef]
- Kim, H.S.; Min, K.W.; Kim, D.H.; Son, B.K.; Kwon, M.J.; Hong, S.M. High WHSC1L1 Expression Reduces Survival Rates in Operated Breast Cancer Patients with Decreased CD8+ T Cells: Machine Learning Approach. J. Pers. Med. 2021, 11, 636. [Google Scholar] [CrossRef]
- Xu, D.; Liu, S.; Wu, X.; Marti, T.M.; Dorn, P.; Schmid, R.A.; Peng, R.W.; Shu, Y. Dissecting the Immunological Profiles in NSD3-Amplified LUSC through Integrative Multi-Scale Analyses. Cancers 2022, 14, 4997. [Google Scholar] [CrossRef] [PubMed]
- Bui, N.; Huang, J.K.; Bojorquez-Gomez, A.; Licon, K.; Sanchez, K.S.; Tang, S.N.; Beckett, A.N.; Wang, T.; Zhang, W.; Shen, J.P.; et al. Disruption of NSD1 in Head and Neck Cancer Promotes Favorable Chemotherapeutic Responses Linked to Hypomethylation. Mol. Cancer Ther. 2018, 17, 1585–1594. [Google Scholar] [CrossRef]
- Shah, M.Y.; Martinez-Garcia, E.; Phillip, J.M.; Chambliss, A.B.; Popovic, R.; Ezponda, T.; Small, E.C.; Will, C.; Phillip, M.P.; Neri, P.; et al. MMSET/WHSC1 enhances DNA damage repair leading to an increase in resistance to chemotherapeutic agents. Oncogene 2016, 35, 5905–5915. [Google Scholar] [CrossRef]
- Wang, Q.; Zheng, J.; Zou, J.X.; Xu, J.; Han, F.; Xiang, S.; Liu, P.; Chen, H.W.; Wang, J. S-adenosylhomocysteine (AdoHcy)-dependent methyltransferase inhibitor DZNep overcomes breast cancer tamoxifen resistance via induction of NSD2 degradation and suppression of NSD2-driven redox homeostasis. Chem. Biol. Interact. 2020, 317, 108965. [Google Scholar] [CrossRef]
- Yu, S.; Si, Y.; Xu, M.; Wang, Y.; Liu, C.; Bi, C.; Sun, M.; Sun, H. Downregulation of the splicing regulator NSRP1 confers resistance to CDK4/6 inhibitors via activation of interferon signaling in breast cancer. J. Biol. Chem. 2025, 301, 108070. [Google Scholar] [CrossRef]
- Liu, J.; Xie, Y.; Guo, J.; Li, X.; Wang, J.; Jiang, H.; Peng, Z.; Wang, J.; Wang, S.; Li, Q.; et al. Targeting NSD2-mediated SRC-3 liquid-liquid phase separation sensitizes bortezomib treatment in multiple myeloma. Nat. Commun. 2021, 12, 1022. [Google Scholar] [CrossRef]
- Peng, Z.; Wang, J.; Guo, J.; Li, X.; Wang, S.; Xie, Y.; Jiang, H.; Wang, Y.; Wang, M.; Hu, M.; et al. All-trans retinoic acid improves NSD2-mediated RARalpha phase separation and efficacy of anti-CD38 CAR T-cell therapy in multiple myeloma. J. Immunother. Cancer 2023, 11, e006325. [Google Scholar] [CrossRef] [PubMed]
- De Santis, A.; De Santis, L.; Rossi, F.; Gasparini, S.; Licursi, V.; Amico, V.A.; Capone, I.; Fragale, A.; D’Atri, S.; Gabriele, L.; et al. NSD2 and miRNAs as Key Regulators of Melanoma Response to Romidepsin and Interferon-alpha2b Treatment. Cancer Med. 2025, 14, e70917. [Google Scholar] [CrossRef]
- Irish, J.C.; Mills, J.N.; Turner-Ivey, B.; Wilson, R.C.; Guest, S.T.; Rutkovsky, A.; Dombkowski, A.; Kappler, C.S.; Hardiman, G.; Ethier, S.P. Amplification of WHSC1L1 regulates expression and estrogen-independent activation of ERalpha in SUM-44 breast cancer cells and is associated with ERalpha over-expression in breast cancer. Mol. Oncol. 2016, 10, 850–865. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ivanov, A.A.; Su, R.; Gonzalez-Pecchi, V.; Qi, Q.; Liu, S.; Webber, P.; McMillan, E.; Rusnak, L.; Pham, C.; et al. The OncoPPi network of cancer-focused protein-protein interactions to inform biological insights and therapeutic strategies. Nat. Commun. 2017, 8, 14356. [Google Scholar] [CrossRef]
- Jones, D.H.; Lin, D.I. Amplification of the NSD3-BRD4-CHD8 pathway in pelvic high-grade serous carcinomas of tubo-ovarian and endometrial origin. Mol. Clin. Oncol. 2017, 7, 301–307. [Google Scholar] [CrossRef]
- Gonzalez-Pecchi, V.; Kwan, A.K.; Doyle, S.; Ivanov, A.A.; Du, Y.; Fu, H. NSD3S stabilizes MYC through hindering its interaction with FBXW7. J. Mol. Cell. Biol. 2020, 12, 438–447. [Google Scholar] [CrossRef]
- Todoerti, K.; Ronchetti, D.; Agnelli, L.; Castellani, S.; Marelli, S.; Deliliers, G.L.; Zanella, A.; Lombardi, L.; Neri, A. Transcription repression activity is associated with the type I isoform of the MMSET gene involved in t(4;14) in multiple myeloma. Br. J. Haematol. 2005, 131, 214–218. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kee, H.J.; Choe, N.W.; Kim, S.M.; Eom, G.H.; Baek, H.J.; Kook, H.; Kook, H.; Seo, S.B. Multiple-myeloma-related WHSC1/MMSET isoform RE-IIBP is a histone methyltransferase with transcriptional repression activity. Mol. Cell. Biol. 2008, 28, 2023–2034. [Google Scholar] [CrossRef]
- di Luccio, E. Inhibition of Nuclear Receptor Binding SET Domain 2/Multiple Myeloma SET Domain by LEM-06 Implication for Epigenetic Cancer Therapies. J. Cancer Prev. 2015, 20, 113–120. [Google Scholar] [CrossRef]
- Shen, Y.; Morishita, M.; Lee, D.; Kim, S.; Lee, T.; Mevius, D.; Roh, Y.; di Luccio, E. Identification of LEM-14 inhibitor of the oncoprotein NSD2. Biochem. Biophys. Res. Commun. 2019, 508, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Coussens, N.P.; Kales, S.C.; Henderson, M.J.; Lee, O.W.; Horiuchi, K.Y.; Wang, Y.; Chen, Q.; Kuznetsova, E.; Wu, J.; Chakka, S.; et al. High-throughput screening with nucleosome substrate identifies small-molecule inhibitors of the human histone lysine methyltransferase NSD2. J. Biol. Chem. 2018, 293, 13750–13765. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Yu, A.; Xing, L.; Chen, X.; Ding, H.; Yang, H.; Song, Z.; Shi, Q.; Geng, M.; Huang, X.; et al. Structural Modification and Pharmacological Evaluation of Substituted Quinoline-5,8-diones as Potent NSD2 Inhibitors. J. Med. Chem. 2023, 66, 1634–1651. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Shi, Q.; Li, B.; Yang, H.; Liu, L.; Zhou, R.; Feng, Z.; Yang, Z.; Zhan, J.; Xiong, X.F.; et al. Discovery of a Highly Potent and Selective Inhibitor Targeting Protein Lysine Methyltransferase NSD2. J. Med. Chem. 2024, 67, 16056–16071. [Google Scholar] [CrossRef]
- Lewis, C.A.; Schmidt, C.; Beebe, L.; Connolly, T.J. Characterization of the activity of KTX-1001, a small molecule inhibitor of multiple myeloma SET domain using surface plasmon resonance. J. Biol. Chem. 2025, 301, 110382. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Bolinger, A.A.; Chen, H.; Zhou, J. Drug Discovery Targeting Nuclear Receptor Binding SET Domain Protein 2 (NSD2). J. Med. Chem. 2023, 66, 10991–11026. [Google Scholar] [CrossRef]
- Jeong, J.; Hausmann, S.; Dong, H.; Szczepski, K.; Flores, N.M.; Garcia Gonzalez, A.; Shi, L.; Lu, X.; Lempiainen, J.; Jakab, M.; et al. NSD2 inhibitors rewire chromatin to treat lung and pancreatic cancers. Nature 2025. [Google Scholar] [CrossRef]
- Huang, H.; Howard, C.A.; Zari, S.; Cho, H.J.; Shukla, S.; Li, H.; Ndoj, J.; Gonzalez-Alonso, P.; Nikolaidis, C.; Abbott, J.; et al. Covalent inhibition of NSD1 histone methyltransferase. Nat. Chem. Biol. 2020, 16, 1403–1410. [Google Scholar] [CrossRef]
- Piao, L.; Gao, Y.; Xu, X.; Su, Y.; Wang, Y.D.; Zhou, J.; Gao, Y.; Fang, J.; Li, Q.; Chang, S.; et al. Discovery of potent small molecule inhibitors of histone lysine methyltransferase NSDs. Eur. J. Med. Chem. 2024, 268, 116264. [Google Scholar] [CrossRef]
- Kim, S.; Hwang, I.; Kim, S.H.; Chung, H.W.; Ji, M.J.; Moon, S.; Park, H.M.; Kong, G.; Hur, W. Identification of novel class inhibitors of NSD3 methyltransferase showing a unique, bivalent binding mode in the SET domain. Chem. Biol. Drug Des. 2023, 102, 500–513. [Google Scholar] [CrossRef]
- Xu, C.; Meng, F.; Park, K.S.; Storey, A.J.; Gong, W.; Tsai, Y.H.; Gibson, E.; Byrum, S.D.; Li, D.; Edmondson, R.D.; et al. A NSD3-targeted PROTAC suppresses NSD3 and cMyc oncogenic nodes in cancer cells. Cell Chem. Biol. 2022, 29, 386–397.E9. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, Y.; Chen, X.; Yu, A.; Du, W.; Huang, Y.; Wu, F.; Yu, L.; Li, J.; Wen, C.; et al. Discovery of a potent and selective proteolysis targeting chimera (PROTAC) degrader of NSD3 histone methyltransferase. Eur. J. Med. Chem. 2022, 239, 114528. [Google Scholar] [CrossRef]
- Nie, D.Y.; Tabor, J.R.; Li, J.; Kutera, M.; St-Germain, J.; Hanley, R.P.; Wolf, E.; Paulakonis, E.; Kenney, T.M.G.; Duan, S.; et al. Recruitment of FBXO22 for targeted degradation of NSD2. Nat. Chem. Biol. 2024, 20, 1597–1607. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Xu, C.; Park, K.S.; Kaniskan, H.U.; Wang, G.G.; Jin, J. Discovery of a First-in-Class Degrader for Nuclear Receptor Binding SET Domain Protein 2 (NSD2) and Ikaros/Aiolos. J. Med. Chem. 2022, 65, 10611–10625. [Google Scholar] [CrossRef] [PubMed]
- Hanley, R.P.; Nie, D.Y.; Tabor, J.R.; Li, F.; Sobh, A.; Xu, C.; Barker, N.K.; Dilworth, D.; Hajian, T.; Gibson, E.; et al. Discovery of a Potent and Selective Targeted NSD2 Degrader for the Reduction of H3K36me2. J. Am. Chem. Soc. 2023, 145, 8176–8188. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Parolia, A.; Liu, Y.; Hou, C.; He, T.; Qiao, Y.; Eyunni, S.; Luo, J.; Li, C.; Wang, Y.; et al. Discovery of LLC0424 as a Potent and Selective in Vivo NSD2 PROTAC Degrader. J. Med. Chem. 2024, 67, 6938–6951. [Google Scholar] [CrossRef]
- Bottcher, J.; Dilworth, D.; Reiser, U.; Neumuller, R.A.; Schleicher, M.; Petronczki, M.; Zeeb, M.; Mischerikow, N.; Allali-Hassani, A.; Szewczyk, M.M.; et al. Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3. Nat. Chem. Biol. 2019, 15, 822–829. [Google Scholar] [CrossRef]
- Ferreira de Freitas, R.; Liu, Y.; Szewczyk, M.M.; Mehta, N.; Li, F.; McLeod, D.; Zepeda-Velazquez, C.; Dilworth, D.; Hanley, R.P.; Gibson, E.; et al. Discovery of Small-Molecule Antagonists of the PWWP Domain of NSD2. J. Med. Chem. 2021, 64, 1584–1592. [Google Scholar] [CrossRef]
- Dilworth, D.; Hanley, R.P.; Ferreira de Freitas, R.; Allali-Hassani, A.; Zhou, M.; Mehta, N.; Marunde, M.R.; Ackloo, S.; Carvalho Machado, R.A.; Khalili Yazdi, A.; et al. A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization. Nat. Chem. Biol. 2022, 18, 56–63. [Google Scholar] [CrossRef]
- Li, N.; Yang, H.; Liu, K.; Zhou, L.; Huang, Y.; Cao, D.; Li, Y.; Sun, Y.; Yu, A.; Du, Z.; et al. Structure-Based Discovery of a Series of NSD2-PWWP1 Inhibitors. J. Med. Chem. 2022, 65, 9459–9477. [Google Scholar] [CrossRef] [PubMed]
- Carlino, L.; Astles, P.C.; Ackroyd, B.; Ahmed, A.; Chan, C.; Collie, G.W.; Dale, I.L.; O’Donovan, D.H.; Fawcett, C.; di Fruscia, P.; et al. Identification of Novel Potent NSD2-PWWP1 Ligands Using Structure-Based Design and Computational Approaches. J. Med. Chem. 2024, 67, 8962–8987. [Google Scholar] [CrossRef]
- Wang, J.J.; Zou, J.X.; Wang, H.; Duan, Z.J.; Wang, H.B.; Chen, P.; Liu, P.Q.; Xu, J.Z.; Chen, H.W. Histone methyltransferase NSD2 mediates the survival and invasion of triple-negative breast cancer cells via stimulating ADAM9-EGFR-AKT signaling. Acta Pharmacol. Sin. 2019, 40, 1067–1075. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.E.; Nguyen, M.T.; Kim, J.; Lee, C.H.; Nam, J.-W.; Chung, H.; Park, M.K.; Lee, J.-Y. NSD Family-Mediated H3K36 Methylation in Human Cancer: Mechanisms and Therapeutic Opportunities. Biomedicines 2025, 13, 2749. https://doi.org/10.3390/biomedicines13112749
Park JE, Nguyen MT, Kim J, Lee CH, Nam J-W, Chung H, Park MK, Lee J-Y. NSD Family-Mediated H3K36 Methylation in Human Cancer: Mechanisms and Therapeutic Opportunities. Biomedicines. 2025; 13(11):2749. https://doi.org/10.3390/biomedicines13112749
Chicago/Turabian StylePark, Jae Eun, Minh Tuan Nguyen, Jaehee Kim, Chang Hoon Lee, Jin-Wu Nam, Heekyoung Chung, Mi Kyung Park, and Jeong-Yeon Lee. 2025. "NSD Family-Mediated H3K36 Methylation in Human Cancer: Mechanisms and Therapeutic Opportunities" Biomedicines 13, no. 11: 2749. https://doi.org/10.3390/biomedicines13112749
APA StylePark, J. E., Nguyen, M. T., Kim, J., Lee, C. H., Nam, J.-W., Chung, H., Park, M. K., & Lee, J.-Y. (2025). NSD Family-Mediated H3K36 Methylation in Human Cancer: Mechanisms and Therapeutic Opportunities. Biomedicines, 13(11), 2749. https://doi.org/10.3390/biomedicines13112749









