Promising Approaches Based on Bioimaging Reporters for Direct Rapid Detection of Mycobacterium tuberculosis
Abstract
1. Introduction
2. Fluorophore‒Quencher Pair Probes
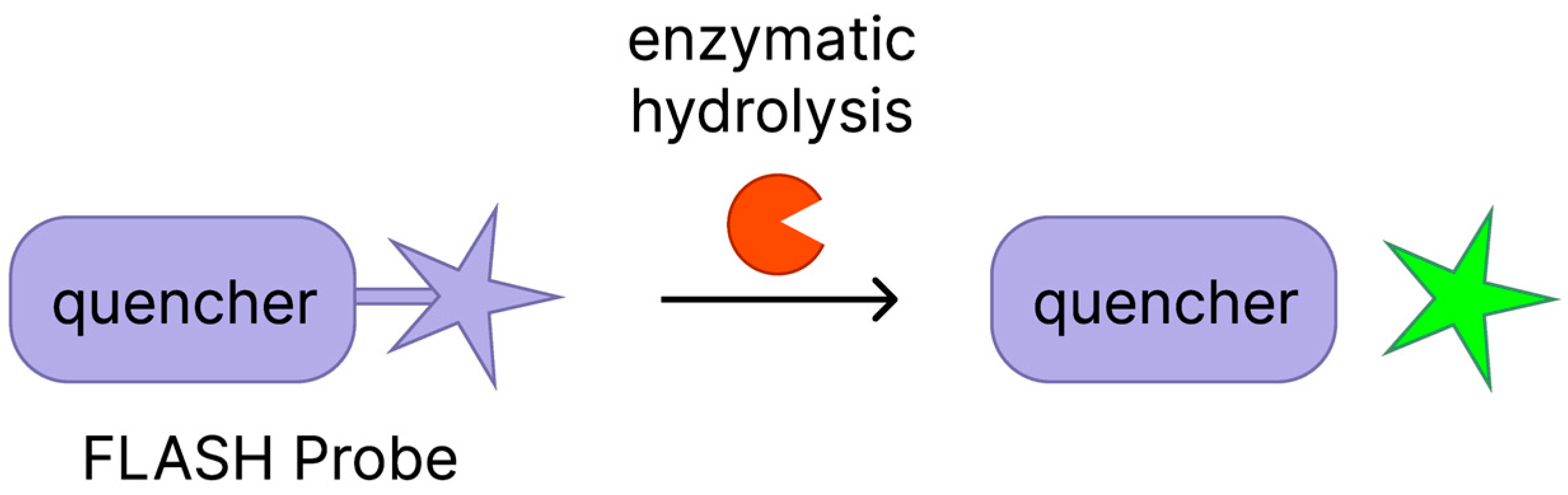
2.1. FLASH Hip1 Probe
2.2. CDG-DNB3 Probe
2.3. NFC-Probe
2.4. Cy3-NO2-Tre
2.5. N14G and N14G-Fe
3. Fluorogenic Probes
3.1. DMN-Tre
3.2. HC-3-Tre
3.3. RMR-Tre
4. Fluorescent Mycobacteriophages
4.1. Φ2GFP10 Reporter Phage
4.2. mCherrybomb Reporter Phage
4.3. Φ2DRM Reporter Phage
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Mtb | Mycobacterium tuberculosis |
| DST | Drug susceptibility testing |
| PCR | Polymerase chain reaction |
| NTM | Nontuberculous mycobacteria |
| CFU | Colony forming unit |
| TMM | Trehalose monomycolate |
| TDM | Trehalose dimycolate |
| IrtAB | Iron-regulated transporter AB |
| SID | Siderophore interaction domain |
| Hip1 | Hydrolase important for pathogenesis 1 |
| BlaC | β-lactamase |
| Dpre1 | Decaprenylphosphoryl-β-d-ribose-2′-epimerase |
| Ag85 | Antigen 85 complex |
References
- World Health Organization. Global Tuberculosis Report 2024; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Dong, B.; He, Z.; Li, Y.; Xu, X.; Wang, C.; Zeng, J. Improved Conventional and New Approaches in the Diagnosis of Tuberculosis. Front. Microbiol. 2022, 13, 924410. [Google Scholar] [CrossRef]
- Huang, Y.; Ai, L.; Wang, X.; Sun, Z.; Wang, F. Review and Updates on the Diagnosis of Tuberculosis. J. Clin. Med. 2022, 11, 5826. [Google Scholar] [CrossRef]
- Bartolomeu-Gonçalves, G.; de Souza, J.M.; Fernandes, B.T.; Spoladori, L.; Correia, G.; Castro, I.; Borges, P.; Silva-Rodrigues, G.; Tavares, E.; Yamauchi, L.; et al. Tuberculosis Diagnosis: Current, Ongoing, and Future Approaches. Diseases 2024, 12, 202. [Google Scholar] [CrossRef]
- Heemskerk, D.; Caws, M.; Marais, B.; Farrar, J. Diagnosis. In Tuberculosis in Adults and Children, 1st ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 28–30. ISBN 978-3-319-19132-4. [Google Scholar]
- Steingart, K.R.; Henry, M.; Ng, V.; Hopewell, P.; Ramsay, A.; Cunningham, J.; Urbanczik, R.; Perkins, M.; Aziz, M.; Pai, M. Fluorescence versus conventional sputum smear microscopy for tuberculosis: A systematic review. Lancet Infect. Dis. 2006, 6, 570–581. [Google Scholar] [CrossRef]
- Chen, W.C.; Chang, C.C.; Lin, Y.E. Pulmonary Tuberculosis Diagnosis Using an Intelligent Microscopy Scanner and Image Recognition Model for Improved Acid-Fast Bacilli Detection in Smears. Microorganisms 2024, 12, 1734. [Google Scholar] [CrossRef]
- Yang, Z.; Li, J.; Shen, J.; Cao, H.; Wang, Y.; Hu, S.; Du, Y.; Wang, Y.; Yan, Z.; Xie, L.; et al. Recent progress in tuberculosis diagnosis: Insights into blood-based biomarkers and emerging technologies. Front. Cell. Infect. Microbiol. 2025, 15, 1567592. [Google Scholar] [CrossRef]
- Wu, Q.; Zhu, Y.; Zhang, Y.; Liu, Z.; Zhang, M.; Chen, J.; Wu, B. Evaluation and Comparison of Laboratory Methods in Diagnosing Mycobacterium tuberculosis and Nontuberculous Mycobacteria in 3012 Sputum Samples. Clin. Respir. J. 2025, 19, e70071. [Google Scholar] [CrossRef]
- Wells, W.A.; Boehme, C.C.; Cobelens, F.G.; Daniels, C.; Dowdy, D.; Gardiner, E.; Gheuens, J.; Kim, P.; Kimerling, M.; Kreiswirth, B.; et al. Alignment of new tuberculosis drug regimens and DST: A framework for action. Lancet Infect. Dis. 2013, 13, 449–458. [Google Scholar] [CrossRef]
- Gutierrez, C. Benefits and challenges of molecular diagnostics for childhood tuberculosis. Int. J. Mycobacteriol. 2016, 5, S4–S5. [Google Scholar] [CrossRef]
- Nguyen, T.N.A.; Anton-Le Berre, V.; Bañuls, A.L.; Nguyen, T.V.A. Molecular Diagnosis of Drug-Resistant Tuberculosis; A Literature Review. Front. Microbiol. 2019, 10, 79. [Google Scholar] [CrossRef]
- Danchuk, S.N.; Solomon, O.E.; Kohl, T.A.; Dreyer, V.; Barilar, I.; Utpatel, C.; Niemann, S.; Soolingen, D.; Anthony, R.; van Ingen, J.; et al. Challenging the gold standard: The limitations of molecular assays for detection of Mycobacterium tuberculosis heteroresistance. Thorax 2024, 79, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Alebouyeh, S.; Weinrick, B.; Achkar, J.M.; García, M.J.; Prados-Rosales, R. Feasibility of novel approaches to detect viable Mycobacterium tuberculosis within the spectrum of the tuberculosis disease. Front. Med. 2022, 9, 965359. [Google Scholar] [CrossRef]
- Dicks, K.V.; Stout, J.E. Molecular Diagnostics for Mycobacterium tuberculosis Infection. Annu. Rev. Med. 2019, 70, 77–90. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Consolidated Guidelines on Tuberculosis. Module 3: Diagnosis—Rapid Diagnostics for Tuberculosis Detection, 3rd ed.; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Asmar, S.; Drancourt, M. Rapid culture-based diagnosis of pulmonary tuberculosis in developed and developing countries. Front. Microb. 2015, 6, 1184. [Google Scholar] [CrossRef]
- Singh, S. End Tuberculosis: Challenges and Opportunities. Tuberc. Res. Treat. 2024, 2024, 2307742. [Google Scholar] [CrossRef]
- Wang, W.H.; Takeuchi, R.; Jain, S.H.; Jiang, Y.H.; Watanuki, S.; Ohtaki, Y.; Nakaishi, K.; Watabe, S.; Lu, P.L.; Ito, E. A novel, rapid (within hours) culture-free diagnostic method for detecting live Mycobacterium tuberculosis with high sensitivity. EBioMedicine 2020, 60, 103007. [Google Scholar] [CrossRef] [PubMed]
- Cruciani, M.; Scarparo, C.; Malena, M.; Bosco, O.; Serpelloni, G.; Mengoli, C. Meta-Analysis of BACTEC MGIT 960 and BACTEC 460 TB, with or without Solid Media, for Detection of Mycobacteria. J. Clin. Microbiol. 2004, 42, 2321–2325. [Google Scholar] [CrossRef]
- Dinnes, J.; Deeks, J.; Kunst, H.; Gibson, A.; Cummins, E.; Waugh, N.; Drobniewski, F.; Lalvani, A. A systematic review of rapid diagnostic tests for the detection of tuberculosis infection. Health Technol. Assess. 2007, 11, 1–196. [Google Scholar] [CrossRef]
- Tortoli, E.; Cichero, P.; Piersimoni, C.; Simonetti, M.T.; Gesu, G.; Nista, D. Use of BACTEC MGIT 960 for Recovery of Mycobacteria from Clinical Specimens: Multicenter Study. J. Clin. Microbiol. 1999, 37, 3578–3582. [Google Scholar] [CrossRef]
- Lavrova, A.I.; Dogonadze, M.Z.; Sychev, A.V.; Manicheva, O.A.; Postnikov, E.B. Ensemble density-dependent synchronization of mycobacterial growth: BACTEC MGIT 960 fluorescence-based analysis and mathematical modeling of coupled biophysical and chemical processes. AIMS Microbiol. 2022, 8, 208–225. [Google Scholar] [CrossRef]
- Trousil, J.; Ulmann, V.; Hrubý, M. Fluorescence & Bioluminescence in the Quest for Imaging, Probing & Analysis of Mycobacterial Infections. Future Microbiol. 2018, 13, 933–951. [Google Scholar] [CrossRef]
- Lesur, E.; Rollando, P.; Guianvarc’h, D.; Bourdreux, Y. Synthesis of trehalose-based chemical tools for the study of the mycobacterial membrane. Comptes Rendus Chim. 2025, 26, 17–38. [Google Scholar] [CrossRef]
- Stavropoulou, K.; Papanastasiou, I.P. Overview of Small Molecules as Fluorescent Probes of Mycobacterium tuberculosis. ACS Omega 2024, 9, 31220–31227. [Google Scholar] [CrossRef]
- Banahene, N.; Swarts, B.M. Metabolic Labeling of Live Mycobacteria with Trehalose-Based Probes. In Mycobacteria Protocols. Methods in Molecular Biology; Parish, T., Kumar, A., Eds.; Humana Press: New York, NY, USA, 2021; Volume 2314, pp. 385–398. ISBN 978-1-0716-1460-0. [Google Scholar]
- Biegas, K.J.; Swarts, B.M. Chemical probes for tagging mycobacterial lipids. Curr. Opin. Chem. Biol. 2021, 65, 57–65. [Google Scholar] [CrossRef]
- Kumar, G.; Narayan, R.; Kapoor, S. Chemical Tools for Illumination of Tuberculosis Biology, Virulence Mechanisms, and Diagnosis. J. Med. Chem. 2020, 63, 15308–15332. [Google Scholar] [CrossRef] [PubMed]
- Meile, S.; Kilcher, S.; Loessner, M.J.; Dunne, M. Reporter Phage-Based Detection of Bacterial Pathogens: Design Guidelines and Recent Developments. Viruses 2020, 12, 944. [Google Scholar] [CrossRef] [PubMed]
- Banahene, N.; Kavunja, H.W.; Swarts, B.M. Chemical Reporters for Bacterial Glycans: Development and Applications. Chem. Rev. 2022, 122, 3336–3413. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kocaoglu, O.; Carlson, E.E. Progress and prospects for small-molecule probes of bacterial imaging. Nat. Chem. Biol. 2016, 12, 472–478. [Google Scholar] [CrossRef]
- Parker, M.F.L.; Flavell, R.R.; Luu, J.M.; Rosenberg, O.S.; Ohliger, M.A.; Wilson, D.M. Small Molecule Sensors Targeting the Bacterial Cell Wall. ACS Infect. Dis. 2020, 6, 1587–1598. [Google Scholar] [CrossRef]
- Kong, Y.; Jiang, Q.; Zhang, F.; Yang, Y. Small Molecular Fluorescent Probes: Application Progress of Specific Bacteria Detection and Antibacterial Phototherapy. Chem. Asian J. 2023, 18, e202300178. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, Z.; Guo, X.; Zhang, G.; Jin, Y.; Wang, Z. Recent advances in organic molecule fluorescent probes for microbial imaging. J. Mater. Chem. B 2025, 13, 1070–1071. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.A.; Park, S.Y.; Cha, Y.; Gopala, L.; Lee, M.H. Strategies of Detecting Bacteria Using Fluorescence-Based Dyes. Front. Chem. 2021, 9, 743923. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Li, G.; Wang, J.; Piao, C.; Zhou, X. Recent Progress in Identifying Bacteria with Fluorescent Probes. Molecules 2022, 27, 6440. [Google Scholar] [CrossRef]
- Bajar, B.; Wang, E.; Zhang, S.; Lin, M.; Chu, J. A Guide to Fluorescent Protein FRET Pairs. Sensors 2016, 16, 1488. [Google Scholar] [CrossRef]
- Wu, X.; Deng, Y.; Xu, Y.; Kang, H.; Hu, J.-J.; Yoon, J.; Liang, G. Activatable Fluorescence and Bio/Chemiluminescence Probes for Aminopeptidases: From Design to Biomedical Applications. Adv. Mater. 2024, 36, 2409893. [Google Scholar] [CrossRef]
- MacGilvary, N.J.; Tan, S. Fluorescent Mycobacterium tuberculosis reporters: Illuminating host–pathogen interactions. Pathog. Dis. 2018, 76, fty017. [Google Scholar] [CrossRef]
- Zeynali kelishomi, F.; Khanjani, S.; Fardsanei, F.; Saghi Sarabi, H.; Nikkhahi, F.; Dehghani, B. Bacteriophages of Mycobacterium tuberculosis, their diversity, and potential therapeutic uses: A review. BMC Infect. Dis. 2022, 22, 957. [Google Scholar] [CrossRef]
- Cheng, Y.; Xie, H.; Sule, P.; Hassounah, H.; Graviss, E.; Kong, Y.; Cirillo, J.; Rao, J. Fluorogenic Probes with Substitutions at the 2 and 7 Positions of Cephalosporin are Highly BlaC-Specific for Rapid Mycobacterium tuberculosis Detection. Angew. Chem. Int. Ed. 2014, 53, 9360–9364. [Google Scholar] [CrossRef]
- Jiang, T.; Bai, X.; Li, M. Advances in the Development of Bacterial Bioluminescence Imaging. Annu. Rev. Anal. Chem. 2024, 17, 265–288. [Google Scholar] [CrossRef]
- Tang, X.; Qi, Q.; Li, B.; Zhu, Z.; Lu, J.; Liu, L. Recent Advances on Fluorescent Sensors for Detection of Pathogenic Bacteria. Chemosensors 2025, 13, 182. [Google Scholar] [CrossRef]
- Chen, N.; Wu, Q.; Yuan, X.; Xie, H. Sensitive detection and labelling of proteins with a molecular rotor-based fluorogenic probe. Org. Biomol. Chem. 2025, 23, 5773–5777. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, X.; Wu, L.; Wu, M.; James, T.D.; Zhang, R. Bioorthogonally activated probes for precise fluorescence imaging. Chem. Soc. Rev. 2025, 54, 201–265. [Google Scholar] [CrossRef]
- Loving, G.S.; Sainlos, M.; Imperiali, B. Monitoring protein interactions and dynamics with solvatochromic fluorophores. Trends Biotechnol. 2010, 28, 73–83. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Klymchenko, A.S.; Mely, Y. Fluorescent Environment-Sensitive Dyes as Reporters of Biomolecular Interactions. In Progress in Molecular Biology and Translational Science, 1st ed.; Morris, M.C., Ed.; Academic Press: London, UK, 2013; Volume 113, pp. 35–58. ISBN 978-0-12-386932-6. [Google Scholar]
- Haidekker, M.A.; Brady, T.P.; Lichlyter, D.; Theodorakis, E.A. Effects of solvent polarity and solvent viscosity on the fluorescent properties of molecular rotors and related probes. Bioorg. Chem. 2005, 33, 415–425. [Google Scholar] [CrossRef]
- He, J.; Han, X.; Yue, Y. Recent progress of dual-responsive fluorescent probes for polarity and analytes. Analyst 2025, 150, 3045–3070. [Google Scholar] [CrossRef]
- Kozma, E.; Kele, P. Bioorthogonal Reactions in Bioimaging. Top. Curr. Chem. 2024, 382, 7. [Google Scholar] [CrossRef]
- Martynov, V.I.; Pakhomov, A.A. BODIPY derivatives as fluorescent reporters of molecular activities in living cells. Russ. Chem. Rev. 2021, 90, 1213–1262. [Google Scholar] [CrossRef]
- Shieh, P.; Bertozzi, C.R. Design strategies for bioorthogonal smart probes. Org. Biomol. Chem. 2014, 12, 9307–9320. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, H.; Hao, T.; Zhang, N.; Li, M.; Wang, X.; Wang, X.; Wei, W.; Zhao, J. Fluorogenic Reactions in Chemical Biology: Seeing Chemistry in Cells. Chem. Biomed. Imaging 2023, 1, 590–619. [Google Scholar] [CrossRef] [PubMed]
- Nadler, A.; Schultz, C. The Power of Fluorogenic Probes. Angew. Chem. Int. Ed. 2013, 52, 2408–2410. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; He, Y.; Lo, Y.; Song, H.; Lu, J. Fluorescent probes based on bioorthogonal reactions: Construction strategies and applications. TrAC Trends Anal. Chem. 2023, 169, 117388. [Google Scholar] [CrossRef]
- Kobayashi, H.; Ogawa, M.; Alford, R.; Choyke, P.L.; Urano, Y. New Strategies for Fluorescent Probe Design in Medical Diagnostic Imaging. Chem. Rev. 2010, 110, 2620–2640. [Google Scholar] [CrossRef]
- Yuan, L.; Lin, W.; Zheng, K.; Zhu, S. FRET-Based Small-Molecule Fluorescent Probes: Rational Design and Bioimaging Applications. Acc. Chem. Res. 2013, 46, 1462–1473. [Google Scholar] [CrossRef]
- Li, Y.X.; Xie, D.T.; Yang, Y.X.; Chen, Z.; Guo, W.Y.; Yang, W.C. Development of Small-Molecule Fluorescent Probes Targeting Enzymes. Molecules 2022, 27, 4501. [Google Scholar] [CrossRef]
- Chyan, W.; Raines, R.T. Enzyme-Activated Fluorogenic Probes for Live-Cell and in Vivo Imaging. ACS Chem. Biol. 2018, 13, 1810–1823. [Google Scholar] [CrossRef]
- Ashton, T.D.; Jolliffe, K.A.; Pfeffer, F.M. Luminescent probes for the bioimaging of small anionic species in vitro and in vivo. Chem. Soc. Rev. 2015, 44, 4547–4595. [Google Scholar] [CrossRef] [PubMed]
- Gui, Q.; Lawson, T.; Shan, S.; Yan, L.; Liu, Y. The Application of Whole Cell-Based Biosensors for Use in Environmental Analysis and in Medical Diagnostics. Sensors 2017, 17, 1623. [Google Scholar] [CrossRef] [PubMed]
- Meile, S.; Du, J.; Staubli, S.; Grossmann, S.; Koliwer-Brandl, H.; Piffaretti, P.; Leitner, L.; Matter, C.; Baggenstos, J.; Hunold, L.; et al. Engineered reporter phages for detection of Escherichia coli, Enterococcus, and Klebsiella in urine. Nat. Commun. 2023, 14, 4336. [Google Scholar] [CrossRef] [PubMed]
- Schofield, D.; Bull, C.T.; Rubio, I.; Wechter, W.P.; Westwater, C.; Molineux, I.J. “Light-tagged” bacteriophage as a diagnostic tool for the detection of phytopathogens. Bioengineered 2013, 4, 50–54. [Google Scholar] [CrossRef]
- Yuan, M.; Wu, Y.; Zhao, C.; Chen, Z.; Su, L.; Yang, H.; Song, J. Activated molecular probes for enzyme recognition and detection. Theranostics 2022, 12, 1459–1485. [Google Scholar] [CrossRef]
- Marshall, A.P.; Shirley, J.D.; Carlson, E.E. Enzyme-targeted fluorescent small-molecule probes for bacterial imaging. Curr. Opin. Chem. Biol. 2020, 57, 155–165. [Google Scholar] [CrossRef]
- Ji, Y.; Wang, Y.; Zhang, N.; Xu, S.; Zhang, L.; Wang, Q.; Zhang, Q.; Hu, H. Cell-Permeable Fluorogenic Probes for Identification and Imaging Nitroreductases in Live Bacterial Cells. J. Org. Chem. 2019, 84, 1299–1309. [Google Scholar] [CrossRef]
- Pala, L.; Sirec, T.; Spitz, U. Modified Enzyme Substrates for the Detection of Bacteria: A Review. Molecules 2020, 25, 3690. [Google Scholar] [CrossRef]
- Lian, J.; Wang, Y.; Sun, X.; Shi, Q.; Meng, F. Progress on Multifunction Enzyme-Activated Organic Fluorescent Probes for Bioimaging. Front. Chem. 2022, 10, 935586. [Google Scholar] [CrossRef] [PubMed]
- Bronstein, I.; Olesen, C.E.M. Detection Methods Using Chemiluminescence. In Molecular Methods for Virus Detection, 1st ed.; Wiedbrauk, D.L., Farkas, D.H., Eds.; Academic Press: San Diego, CA, USA, 1995; pp. 147–174. ISBN 978-0-12-748920-9. [Google Scholar]
- Dunuweera, A.N.; Dunuweera, S.P.; Ranganathan, K. A Comprehensive Exploration of Bioluminescence Systems, Mechanisms, and Advanced Assays for Versatile Applications. Biochem. Res. Int. 2024, 2024, 273237. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Shi, P.; Song, W.; Bi, S. Chemiluminescence and Bioluminescence Imaging for Biosensing and Therapy: In Vitro and In Vivo Perspectives. Theranostics 2019, 9, 4047–4065. [Google Scholar] [CrossRef]
- Roda, A.; Mirasoli, M.; Michelini, E.; Di Fusco, M.; Zangheri, M.; Cevenini, L.; Roda, B.; Simoni, P. Progress in chemical luminescence-based biosensors: A critical review. Biosens. Bioelectron. 2016, 76, 164–179. [Google Scholar] [CrossRef]
- Prosser, J.I.; Killham, K.; Glover, L.A.; Rattray, E.A. Luminescence-Based Systems for Detection of Bacteria in the Environment. Crit. Rev. Biotechnol. 1996, 16, 157–183. [Google Scholar] [CrossRef]
- Madan-Lala, R.; Peixoto, K.V.; Re, F.; Rengarajan, J. Mycobacterium tuberculosis Hip1 Dampens Macrophage Proinflammatory Responses by Limiting Toll-Like Receptor 2 Activation. Infect. Immun. 2011, 79, 4828–4838. [Google Scholar] [CrossRef] [PubMed]
- Naffin-Olivos, J.L.; Georgieva, M.; Goldfarb, N.; Madan-Lala, R.; Dong, L.; Bizzell, E.; Valinetz, E.; Brandt, G.S.; Yu, S.; Shabashvili, D.E.; et al. Mycobacterium tuberculosis Hip1 Modulates Macrophage Responses through Proteolysis of GroEL2. PLoS Pathog. 2014, 10, e1004132. [Google Scholar] [CrossRef]
- Rastogi, S.; Briken, V. Interaction of Mycobacteria with Host Cell Inflammasomes. Front. Immunol. 2022, 13, 791136. [Google Scholar] [CrossRef]
- Lentz, C.S.; Ordonez, A.A.; Kasperkiewicz, P.; La Greca, F.; O’Donoghue, A.J.; Schulze, C.J.; Powers, J.C.; Craik, C.S.; Drag, M.; Jain, S.K.; et al. Design of Selective Substrates and Activity-Based Probes for Hydrolase Important for Pathogenesis 1 (HIP1) from Mycobacterium tuberculosis. ACS Infect. Dis. 2016, 2, 807–815. [Google Scholar] [CrossRef]
- Lin, H.; Xing, J.; Wang, H.; Wang, S.; Fang, R.; Li, X.; Li, Z.; Song, N. Roles of Lipolytic enzymes in Mycobacterium tuberculosis pathogenesis. Front. Microbiol. 2024, 15, 1329715. [Google Scholar] [CrossRef] [PubMed]
- Babin, B.M.; Fernandez-Cuervo, G.; Sheng, J.; Green, O.; Ordonez, A.A.; Turner, M.L.; Keller, L.J.; Jain, S.K.; Shabat, D.; Bogyo, M. Chemiluminescent Protease Probe for Rapid, Sensitive, and Inexpensive Detection of Live Mycobacterium tuberculosis. ACS Cent. Sci. 2021, 7, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Xie, J.; Lee, K.H.; Gaur, R.L.; Song, A.; Dai, T.; Ren, H.; Wu, J.; Sun, Z.; Banaei, N.; et al. Rapid and specific labeling of single live Mycobacterium tuberculosis with a dual-targeting fluorogenic probe. Sci. Transl. Med. 2018, 10, eaar4470. [Google Scholar] [CrossRef]
- Zhuang, Q.; Guo, H.; Peng, T.; Ding, E.; Zhao, H.; Liu, Q.; He, S.; Zhao, G. Advances in the detection of β-lactamase: A review. Int. J. Biol. Macromol. 2023, 251, 126159. [Google Scholar] [CrossRef]
- Wang, F.; Cassidy, C.; Sacchettini, J.C. Crystal Structure and Activity Studies of the Mycobacterium tuberculosis β-Lactamase Reveal Its Critical Role in Resistance to β-Lactam Antibiotics. Antimicrob. Agents Chemother. 2006, 50, 2762–2771. [Google Scholar] [CrossRef]
- Kong, Y.; Yao, H.; Ren, H.; Subbian, S.; Cirillo, S.L.; Sacchettini, J.C.; Rao, J.; Cirillo, J.D. Imaging tuberculosis with endogenous beta-lactamase reporter enzyme fluorescence in live mice. Proc. Natl. Acad. Sci. USA 2010, 107, 12239–12244. [Google Scholar] [CrossRef]
- Yang, H.J.; Kong, Y.; Cheng, Y.; Janagama, H.; Hassounah, H.; Xie, H.; Rao, J.; Cirillo, J. Real-time Imaging of Mycobacterium tuberculosis, Using a Novel Near-Infrared Fluorescent Substrate. J. Infect. Dis. 2017, 215, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Thai, H.B.; Yu, J.K.; Park, B.S.; Park, Y.J.; Min, S.J.; Ahn, D.R. A fluorogenic substrate of beta-lactamases and its potential as a probe to detect the bacteria resistant to the third-generation oxyimino-cephalosporins. Biosens. Bioelectron. 2016, 77, 1026–1031. [Google Scholar] [CrossRef]
- Mao, W.; Wang, Y.; Qian, X.; Xia, L.; Xie, H. A Carbapenem-Based Off–On Fluorescent Probe for Specific Detection of Metallo-β-Lactamase Activities. ChemBioChem 2019, 20, 511–515. [Google Scholar] [CrossRef]
- Mao, W.; Xia, L.; Wang, Y.; Xie, H. A Self-Immobilizing and Fluorogenic Probe for β-Lactamase Detection. Chem. Asian J. 2016, 11, 3493–3497. [Google Scholar] [CrossRef]
- Shao, Q.; Zheng, Y.; Dong, X.; Tang, K.; Yan, X.; Xing, B. A Covalent Reporter of β-Lactamase Activity for Fluorescent Imaging and Rapid Screening of Antibiotic-Resistant Bacteria. Chem. Eur. J. 2013, 19, 10903–10910. [Google Scholar] [CrossRef] [PubMed]
- Gholap, S.P.; Yao, C.; Green, O.; Babjak, M.; Jakubec, P.; Malatinský, T.; Ihssen, J.; Wick, L.; Spitz, U.; Shabat, D. Chemiluminescence Detection of Hydrogen Sulfide Release by β-Lactamase-Catalyzed β-Lactam Biodegradation: Unprecedented Pathway for Monitoring β-Lactam Antibiotic Bacterial Resistance. Bioconjug. Chem. 2021, 32, 991–1000. [Google Scholar] [CrossRef]
- Hu, L.; Liu, R.; Ma, Z.; Yu, T.; Li, Z.; Zou, Y.; Yuan, C.; Chen, F.; Xie, H. Specific detection of IMP-1 β-lactamase activity using a trans cephalosporin-based fluorogenic probe. Chem. Commun. 2021, 57, 13586–13589. [Google Scholar] [CrossRef]
- Chen, F.; Li, Y.; Peng, Y.; Zhu, Y.; He, G.; Zhang, Z.; Xie, H. Highly Sensitive In Vivo Imaging of Bacterial Infections with a Hydrophilicity-Switching, Self-Immobilizing, Near-Infrared Fluorogenic β-Lactamase Probe Enriched within Bacteria. Adv. Sci. 2025, 12, e2408559. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Mu, R.; Fang, M.; Cheng, Y.; Senchyna, F.; Moreno, A.; Banaei, N.; Rao, J. A dual-caged resorufin probe for rapid screening of infections resistant to lactam antibiotics. Chem. Sci. 2021, 12, 9153–9161. [Google Scholar] [CrossRef] [PubMed]
- Negri, A.; Javidnia, P.; Mu, R.; Zhang, X.; Vendome, J.; Gold, B.; Roberts, J.; Barman, D.; Ioerger, T.; Sacchettini, J.C.; et al. Identification of a Mycothiol-Dependent Nitroreductase from Mycobacterium tuberculosis. ACS Infect. Dis. 2018, 4, 771–787. [Google Scholar] [CrossRef]
- Eke, I.E.; Abramovitch, R.B. Functions of nitroreductases in mycobacterial physiology and drug susceptibility. J. Bacteriol. 2025, 207, e0032624. [Google Scholar] [CrossRef]
- Mu, R.; Kong, C.; Yu, W.; Wang, H.; Ma, Y.; Li, X.; Wu, J.; Somersan-Karakaya, S.; Li, H.; Sun, Z.; et al. A Nitrooxidoreductase Rv2466c-dependent Fluorescent Probe for Rapid Mycobacterium tuberculosis Diagnosis and Drug Susceptibility Testing. ACS Infect. Dis. 2019, 5, 1210–1219. [Google Scholar] [CrossRef]
- Liu, G.; Li, X.; Hong, X.; Geng, P.; Sun, Z. Detecting Mycobacterium tuberculosis using a nitrofuranyl calanolide-trehalose probe based on nitroreductase Rv2466c. Chem. Commun. 2021, 57, 12688–12691. [Google Scholar] [CrossRef] [PubMed]
- Geng, P.; Hong, X.; Li, X.; Ni, D.; Liu, G. Optimization of nitrofuranyl calanolides for the fluorescent detection of Mycobacterium tuberculosis. Eur. J. Med. Chem. 2022, 244, 114835. [Google Scholar] [CrossRef]
- Hong, X.; Geng, P.; Tian, N.; Li, X.; Gao, M.; Nie, L.; Sun, Z.; Liu, G. From Bench to Clinic: A Nitroreductase Rv3368c-Responsive Cyanine-Based Probe for the Specific Detection of Live Mycobacterium tuberculosis. Anal. Chem. 2024, 96, 1576–1586. [Google Scholar] [CrossRef]
- Murugasu-Oei, B.; Tay, A.; Dick, T. Upregulation of stress response genes and ABC transporters in anaerobic stationary-phase Mycobacterium smegmatis. Mol. Gen. Genet. 1999, 262, 677–682. [Google Scholar] [CrossRef]
- Ratledge, C. Iron, mycobacteria and tuberculosis. Tuberculosis 2004, 84, 110–130. [Google Scholar] [CrossRef] [PubMed]
- Gobin, J.; Horwitz, M.A. Exochelins of Mycobacterium tuberculosis remove iron from human iron-binding proteins and donate iron to mycobactins in the M. tuberculosis cell wall. J. Exp. Med. 1996, 183, 1527–1532. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Fang, R.; Wang, H.; Li, X.; Xing, J.; Li, Z.; Song, N. The role of transcriptional regulators in metal ion homeostasis of Mycobacterium tuberculosis. Front. Cell. Infect. Microbiol. 2024, 14, 1360880. [Google Scholar] [CrossRef]
- Rodriguez, G.M. Control of iron metabolism in Mycobacterium tuberculosis. Trends Microbiol. 2006, 14, 320–327. [Google Scholar] [CrossRef]
- Zhang, L.; Hendrickson, R.C.; Meikle, V.; Lefkowitz, E.J.; Ioerger, T.R.; Niederweis, M. Comprehensive analysis of iron utilization by Mycobacterium tuberculosis. PLoS Pathog. 2020, 16, e1008337. [Google Scholar] [CrossRef]
- Theriault, M.E.; Pisu, D.; Wilburn, K.M.; Lê-Bury, G.; MacNamara, C.; Petrassi, H.M.; Love, M.; Rock, J.; VanderVen, B.; Russell, D. Iron limitation in M. tuberculosis has broad impact on central carbon metabolism. Commun. Biol. 2022, 5, 685. [Google Scholar] [CrossRef]
- Gonda, I.; Sorrentino, S.; Galazzo, L.; Lichti, N.; Arnold, F.; Mehdipour, A.; Bordignon, E.; Seeger, M. The mycobacterial ABC transporter IrtAB employs a membrane-facing crevice for siderophore-mediated iron uptake. Nat. Commun. 2025, 16, 1133. [Google Scholar] [CrossRef]
- Arnold, F.M.; Weber, M.S.; Gonda, I.; Gallenito, M.J.; Adenau, S.; Egloff, P.; Zimmermann, I.; Hutter, C.A.J.; Hürlimann, L.M.; Peters, E.E.; et al. The ABC exporter IrtAB imports and reduces mycobacterial siderophores. Nature 2020, 580, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Shyam, M.; Shilkar, D.; Verma, H.; Dev, A.; Sinha, B.N.; Brucoli, F.; Bhakta, S.; Jayaprakash, V. The Mycobactin Biosynthesis Pathway: A Prospective Therapeutic Target in the Battle against Tuberculosis. J. Med. Chem. 2021, 64, 71–100. [Google Scholar] [CrossRef] [PubMed]
- Chao, A.; Sieminski, P.J.; Owens, C.P.; Goulding, C.W. Iron Acquisition in Mycobacterium tuberculosis. Chem. Rev. 2019, 119, 1193–1220. [Google Scholar] [CrossRef] [PubMed]
- Sritharan, M. Iron Homeostasis in Mycobacterium tuberculosis: Mechanistic Insights into Siderophore-Mediated Iron Uptake. J. Bacteriol. 2016, 198, 2399–2409. [Google Scholar] [CrossRef]
- Ni, D.; Hong, X.; Liu, D.; Li, X.; Li, L.; Liu, W.; Sun, Z.; Liu, G. Insights into IrtAB: Iron Transport Facilitates Ultrasensitive Detection of Mycobacteria in Both Cellular and Clinical Environments. ACS Cent. Sci. 2025, 11, 261–271. [Google Scholar] [CrossRef]
- Zhang, Z.J.; Wang, Y.C.; Yang, X.; Hang, H.C. Chemical Reporters for Exploring Microbiology and Microbiota Mechanisms. ChemBioChem 2020, 21, 19–32. [Google Scholar] [CrossRef]
- Garcia-Vilanova, A.; Chan, J.; Torrelles, J.B. Underestimated Manipulative Roles of Mycobacterium tuberculosis Cell Envelope Glycolipids During Infection. Front. Immunol. 2019, 10, 2909. [Google Scholar] [CrossRef]
- Verschoor, J.A.; Baird, M.S.; Grooten, J. Toward understanding the functional diversity of cell wall mycolic acids of Mycobacterium tuberculosis. Prog. Lipid Res. 2012, 51, 325–339. [Google Scholar] [CrossRef]
- Matsunaga, I.; Naka, T.; Talekar, R.S.; McConnell, M.J.; Katoh, K.; Nakao, H.; Otsuka, A.; Behar, S.M.; Yano, I.; Moody, D.B.; et al. Mycolyltransferase-mediated Glycolipid Exchange in Mycobacteria. J. Biol. Chem. 2008, 283, 28835–28841. [Google Scholar] [CrossRef]
- Angala, S.K.; Belardinelli, J.M.; Huc-Claustre, E.; Wheat, W.H.; Jackson, M. The cell envelope glycoconjugates of Mycobacterium tuberculosis. Crit. Rev. Biochem. 2014, 49, 361–399. [Google Scholar] [CrossRef]
- Brown, T.; Chavent, M.; Im, W. Molecular Modeling and Simulation of the Mycobacterial Cell Envelope: From Individual Components to Cell Envelope Assemblies. J. Phys. Chem. B 2023, 127, 10941–10949. [Google Scholar] [CrossRef]
- Babu Sait, M.R.; Koliwer-Brandl, H.; Stewart, J.A.; Swarts, B.; Jacobsen, M.; Ioerger, T.; Kalscheuer, R. PPE51 mediates uptake of trehalose across the mycomembrane of Mycobacterium tuberculosis. Sci. Rep. 2022, 12, 2097. [Google Scholar] [CrossRef] [PubMed]
- Kalscheuer, R.; Koliwer-Brandl, H. Genetics of Mycobacterial Trehalose Metabolism. Microbiol. Spectr. 2014, 2, MGM2-0002-2013. [Google Scholar] [CrossRef]
- Liang, J.; Liu, F.; Xu, P.; Shangguan, W.; Hu, T.; Wang, S.; Yang, X.; Xiong, Z.; Yang, X.; Guddat, L.W.; et al. Molecular recognition of trehalose and trehalose analogs by Mycobacterium tuberculosis LpqY-SugABC. Proc. Natl. Acad. Sci. USA 2023, 120, e2307625120. [Google Scholar] [CrossRef] [PubMed]
- Furze, C.M.; Delso, I.; Casal, E.; Guy, C.S.; Seddon, C.; Brown, C.M.; Parker, H.L.; Radhakrishnan, A.; Pacheco-Gomez, R.; Stansfeld, P.J.; et al. Structural basis of trehalose recognition by the mycobacterial LpqY-SugABC transporter. J. Biol. Chem. 2021, 296, 100307. [Google Scholar] [CrossRef] [PubMed]
- Kalscheuer, R.; Weinrick, B.; Veeraraghavan, U.; Besra, G.S.; Jacobs, W.R., Jr. Trehalose-recycling ABC transporter LpqY-SugA-SugB-SugC is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2010, 107, 21761–21766. [Google Scholar] [CrossRef]
- Backus, K.M.; Boshoff, H.I.; Barry, C.S.; Boutureira, O.; Patel, M.K.; D’Hooge, F.; Lee, S.S.; Via, L.E.; Tahlan, K.; Barry, C.E., III; et al. Uptake of unnatural trehalose analogs as a reporter for Mycobacterium tuberculosis. Nat. Chem. Biol. 2011, 7, 228–235. [Google Scholar] [CrossRef]
- Swarts, B.M.; Holsclaw, C.M.; Jewett, J.C.; Alber, M.; Fox, D.M.; Siegrist, M.S.; Leary, J.A.; Kalscheuer, R.; Bertozzi, C.R. Probing the Mycobacterial Trehalome with Bioorthogonal Chemistry. J. Am. Chem. Soc. 2012, 134, 16123–16126. [Google Scholar] [CrossRef]
- Belisle, J.T.; Vissa, V.D.; Sievert, T.; Takayama, K.; Brennan, P.J.; Besra, G.S. Role of the Major Antigen of Mycobacterium tuberculosis in Cell Wall Biogenesis. Science 1997, 276, 1420–1422. [Google Scholar] [CrossRef]
- Vanaporn, M.; Titball, R.W. Trehalose and bacterial virulence. Virulence 2020, 11, 1192–1202. [Google Scholar] [CrossRef]
- Kamariza, M.; Keyser, S.G.L.; Utz, A.; Knapp, B.D.; Ealand, C.; Ahn, G.; Cambier, C.J.; Chen, T.; Kana, B.; Huang, K.C.; et al. Toward Point-of-Care Detection of Mycobacterium tuberculosis: A Brighter Solvatochromic Probe Detects Mycobacteria within Minutes. JACS Au 2021, 1, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Banahene, N.; Gepford, D.M.; Biegas, K.J.; Swanson, D.H.; Hsu, Y.P.; Murphy, B.A.; Taylor, Z.E.; Lepori, I.; Siegrist, M.S.; Obregón-Henao, A.; et al. A Far-Red Molecular Rotor Fluorogenic Trehalose Probe for Live Mycobacteria Detection and DrugSusceptibility Testing. Angew. Chem. Int. Ed. 2023, 62, e202213563. [Google Scholar] [CrossRef] [PubMed]
- Kamariza, M.; Shieh, P.; Bertozzi, C.R. Imaging Mycobacterial Trehalose Glycolipids. In Methods in Enzymology, 1st ed.; Imperiali, B., Ed.; Academic Press: New York, NY, USA, 2018; Volume 598, pp. 355–369. ISBN 978-0-12-814419-0. [Google Scholar]
- Kamariza, M.; Shieh, P.; Ealand, C.S.; Peters, J.S.; Chu, B.; Rodriguez-Rivera, F.P.; Babu Sait, M.R.; Treuren, W.V.; Martinson, N.; Kalscheuer, R.; et al. Rapid detection of Mycobacterium tuberculosis in sputum with a solvatochromic trehalose probe. Sci. Transl. Med. 2018, 10, eaam6310. [Google Scholar] [CrossRef]
- Gil, J.; Paulson, J.; Zahn, H.; Brown, M.; Nguyen, M.M.; Erickson, S. Development of a Replication-Deficient Bacteriophage Reporter Lacking an Essential Baseplate Wedge Subunit. Viruses 2023, 16, 8. [Google Scholar] [CrossRef]
- Brown, M.; Hall, A.; Zahn, H.; Eisenberg, M.; Erickson, S. Bacteriophage-Based Detection of Staphylococcus aureus in Human Serum. Viruses 2022, 14, 1748. [Google Scholar] [CrossRef] [PubMed]
- Alonzo, L.F.; Jain, P.; Hinkley, T.; Clute-Reinig, N.; Garing, S.; Spencer, E.; Dinh, V.T.T.; Bell, D.; Nugen, S.; Nichols, K.P.; et al. Rapid, sensitive, and low-cost detection of Escherichia coli bacteria in contaminated water samples using a phage-based assay. Sci. Rep. 2022, 12, 7741. [Google Scholar] [CrossRef]
- Hussain, W.; Ullah, M.W.; Farooq, U.; Aziz, A.; Wang, S. Bacteriophage-based advanced bacterial detection: Concept, mechanisms, and applications. Biosens. Bioelectron. 2021, 177, 112973. [Google Scholar] [CrossRef]
- Vinay, M.; Franche, N.; Grégori, G.; Fantino, J.R.; Pouillot, F.; Ansaldi, M. Phage-Based Fluorescent Biosensor Prototypes to Specifically Detect Enteric Bacteria Such as E. coli and Salmonella enterica Typhimurium. PLoS ONE 2015, 10, e0131466. [Google Scholar] [CrossRef]
- Klumpp, J.; Loessner, M.J. Detection of Bacteria with Bioluminescent Reporter Bacteriophage. In Bioluminescence: Fundamentals and Applications in Biotechnology. Advances in Biochemical Engineering/Biotechnology, Volume 144; Thouand, G., Marks, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 1, pp. 155–171. ISBN 978-3-662-43385-0. [Google Scholar]
- Meile, S.; Sarbach, A.; Du, J.; Schuppler, M.; Saez, C.; Loessner, M.J.; Kilcher, S. Engineered reporter phages for rapid bioluminescence-based detection and differentiation of viable Listeria cells. Appl. Environ. Microbiol. 2020, 86, e00442-20. [Google Scholar] [CrossRef]
- Leprince, A.; Mahillon, J. Phage Adsorption to Gram-Positive Bacteria. Viruses 2023, 15, 196. [Google Scholar] [CrossRef]
- Jain, P.; Hartman, T.; Eisenberg, N.; O’Donnell, M.R.; Kriakov, J.; Govender, K.; Makume, M.; Thaler, D.S.; Hatfull, G.F.; Sturm, A.W.; et al. 2GFP10, a High-Intensity Fluorophage, Enables Detection and Rapid Drug Susceptibility Testing of Mycobacterium tuberculosis Directly from Sputum Samples. J. Clin. Microbiol. 2012, 50, 1362–1369. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, M.R.; Pym, A.; Jain, P.; Munsamy, V.; Wolf, A.; Karim, F.; Jacobs, W.; Larsen, M. A Novel Reporter Phage To Detect Tuberculosis and Rifampin Resistance in a High-HIV-Burden Population. J. Clin. Microbiol. 2015, 53, 2188–2194. [Google Scholar] [CrossRef] [PubMed]
- Rondón, L.; Urdániz, E.; Latini, C.; Payaslian, F.; Matteo, M.; Sosa, E.; Do Porto, D.; Turjanski, A.; Nemirovsky, S.; Hatfull, G.; et al. Fluoromycobacteriophages Can Detect Viable Mycobacterium tuberculosis and Determine Phenotypic Rifampicin Resistance in 3–5 Days from Sputum Collection. Front. Microbiol. 2018, 9, 1471. [Google Scholar] [CrossRef] [PubMed]
- Urdániz, E.; Rondón, L.; Martí, M.A.; Hatfull, G.F.; Piuri, M. Rapid Whole-Cell Assay of Antitubercular Drugs Using Second-Generation Fluoromycobacteriophages. Antimicrob. Agents Chemother. 2016, 60, 3253–3256. [Google Scholar] [CrossRef]
- Jain, P.; Weinrick, B.C.; Kalivoda, E.J.; Yang, H.; Munsamy, V.; Vilcheze, C.; Weisbrod, T.; Larsen, M.; O’Donnell, M.; Pym, A.; et al. Dual-Reporter Mycobacteriophages (Φ2DRMs) Reveal Preexisting Mycobacterium tuberculosis Persistent Cells in Human Sputum. mBio 2016, 7, e01023-16. [Google Scholar] [CrossRef]



| Probe/Reporter | Probe Type | Target/Mechanism | Detection Mode | Sample | Time to Result | Detected Pathogen Level | References |
|---|---|---|---|---|---|---|---|
| FLASH Hip1 | Chemiluminescent enzymatic peptide probe | The fluorescence activated by cleavage of the peptide substrate by Hip1 serine protease of M. tuberculosis | Chemiluminescence | Processed sputum | 1 h | 15 × 103 CFU | [80] |
| CDG-DNB3 | Small molecule dual-enzyme-activated fluorogenic probe | Activated by β-lactamase BlaC cleavage; fluorescent product covalently binds DprE1 enzyme | Fluorescence | Processed sputum | 1 h | N/D | [81] |
| NFC Probe (Rv2466c-dependent) | Nitrofuranyl coumarin-based fluorescent probe | Fluorescence activation through elimination of nitro group quenching via enzymatic reduction | Fluorescence | Pure culture | 24 h (including incubation) | 1.5 × 104 CFU | [96] |
| Cy3-NO2-Tre | Cyanine-based fluorophore (Cy3) linked to trehalose with a nitro group (NO2) | Incorporation into mycobacterial cell wall via trehalose uptake pathway; Rv3368c nitroreductase-activated probe | Fluorescence | Processed sputum | 15 min | 4.3 × 102 CFU | [112] |
| N14G-Fe | Mycobactin-fluorophore conjugate | Active IrtAB-mediated uptake of iron-chelated probe, intracellular iron reduction releases fluorophore, leading to fluorescence activation | Fluorescence | Processed sputum | 10 min | 34 CFU | [99] |
| DMN-Tre | Fluorogenic trehalose analog (solvatochromic) | Incorporation into mycobacterial cell wall via trehalose uptake pathway; fluorescence activation in hydrophobic environment | Fluorescence | Processed sputum | 30 min | 104 CFU * | [131] |
| 3HC-3-Tre | Fluorogenic trehalose probe (solvatochromic) | Incorporation into mycobacterial cell wall via trehalose uptake pathway; fluorescence activation in hydrophobic environment | Fluorescence | Pure culture | 10 min | N/D | [128] |
| RMR-Tre | Fluorogenic trehalose probe (molecular rotor) | Incorporation into mycobacterial cell wall via trehalose uptake pathway; fluorescence activation in sterically constrained environment | Fluorescence | Pure culture | 10 min * | N/D | [129] |
| Φ2GFP10 | Genetically engineered fluorescent reporter phage | Infects of viable M. tuberculosis and expresses GFP for visualization | Fluorescence | Processed sputum | 12 h # | 104 CFU | [140,141] |
| mCherryBomb | Genetically engineered fluorescent reporter phage | Infects of viable M. tuberculosis and expresses red fluorescent protein mCherry | Fluorescence | Processed sputum | 16–18 h # | 1–19 CFU | [142,143] |
| Φ2DRM | Genetically engineered fluorescent reporter phage | Infects of viable M. tuberculosis and expresses mVenus constitutive, tdTomato for persister cells detection | Fluorescence | Processed sputum | 4 h | N/D | [144] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ambartsumyan, O.A.; Skuredina, O.A.; Eliseev, P.I.; Tiulkova, T.E.; Samoilova, A.G.; Vasilieva, I.A. Promising Approaches Based on Bioimaging Reporters for Direct Rapid Detection of Mycobacterium tuberculosis. Biomedicines 2025, 13, 2609. https://doi.org/10.3390/biomedicines13112609
Ambartsumyan OA, Skuredina OA, Eliseev PI, Tiulkova TE, Samoilova AG, Vasilieva IA. Promising Approaches Based on Bioimaging Reporters for Direct Rapid Detection of Mycobacterium tuberculosis. Biomedicines. 2025; 13(11):2609. https://doi.org/10.3390/biomedicines13112609
Chicago/Turabian StyleAmbartsumyan, Oganes A., Olesya A. Skuredina, Platon I. Eliseev, Tatiana E. Tiulkova, Anastasia G. Samoilova, and Irina A. Vasilieva. 2025. "Promising Approaches Based on Bioimaging Reporters for Direct Rapid Detection of Mycobacterium tuberculosis" Biomedicines 13, no. 11: 2609. https://doi.org/10.3390/biomedicines13112609
APA StyleAmbartsumyan, O. A., Skuredina, O. A., Eliseev, P. I., Tiulkova, T. E., Samoilova, A. G., & Vasilieva, I. A. (2025). Promising Approaches Based on Bioimaging Reporters for Direct Rapid Detection of Mycobacterium tuberculosis. Biomedicines, 13(11), 2609. https://doi.org/10.3390/biomedicines13112609







