Molecular Mechanisms of the Ubiquitin-Specific Proteases (USPs) Family in Biliary Tract Cancer and Targeted Intervention Strategies
Abstract
1. Introduction
2. The Ubiquitin-Proteasome System (UPS) and USPs
2.1. UPS
2.2. USPs
3. USPs and CCA
3.1. USP1
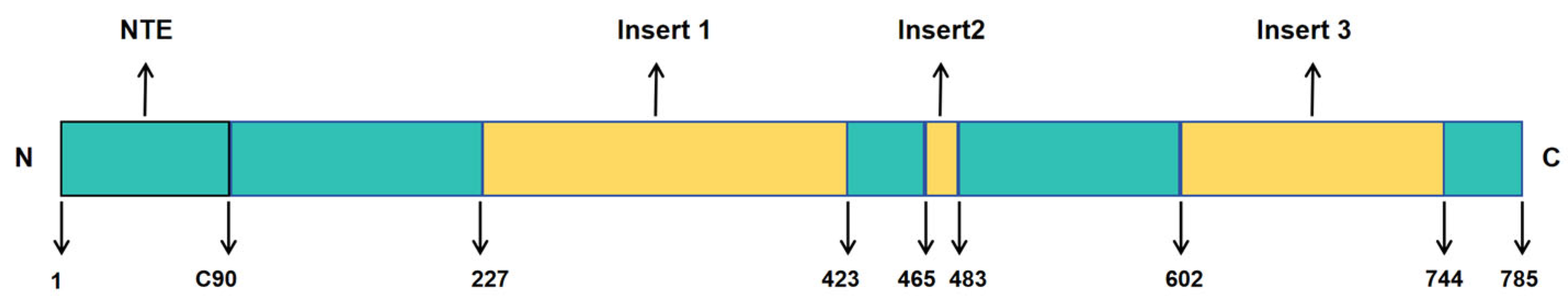
3.1.1. Molecular Characteristics and Functions of USP1
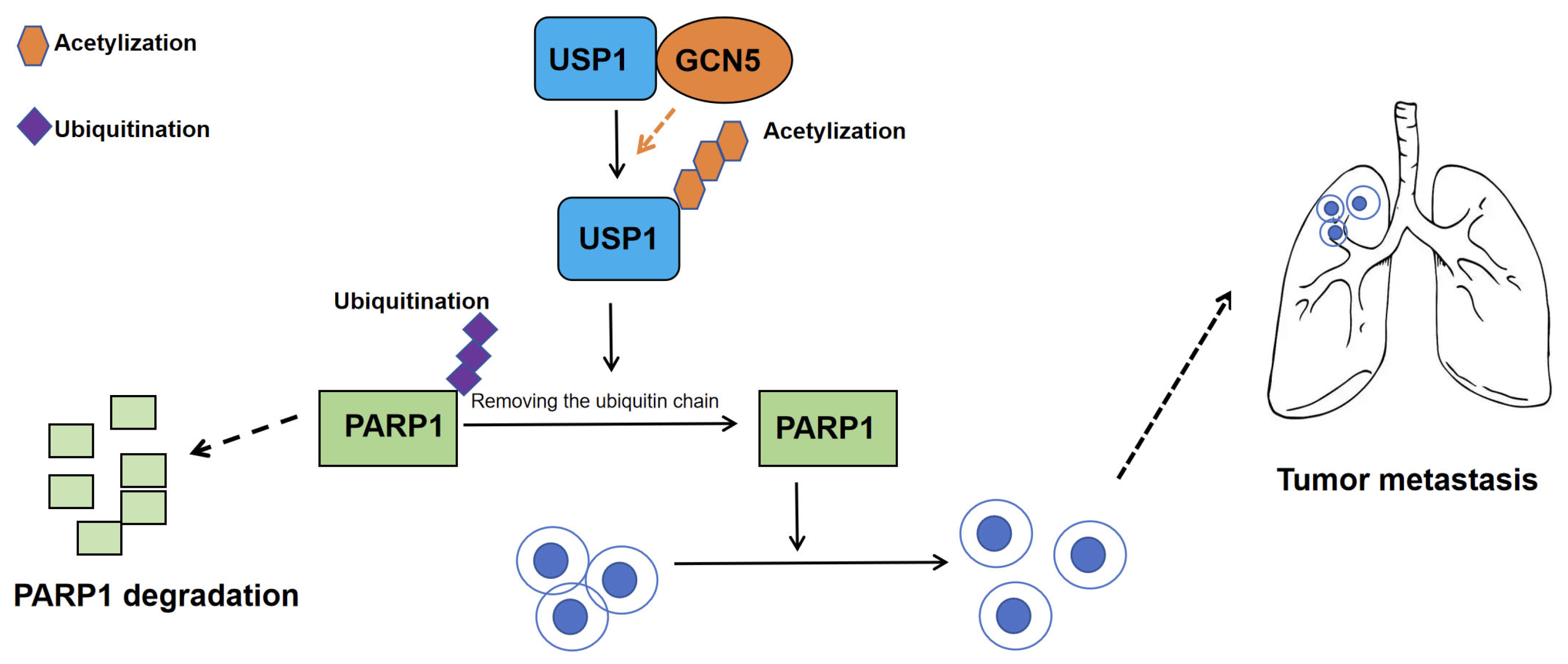
3.1.2. Role and Molecular Mechanisms of USP1 Family in CCA
3.2. USP8
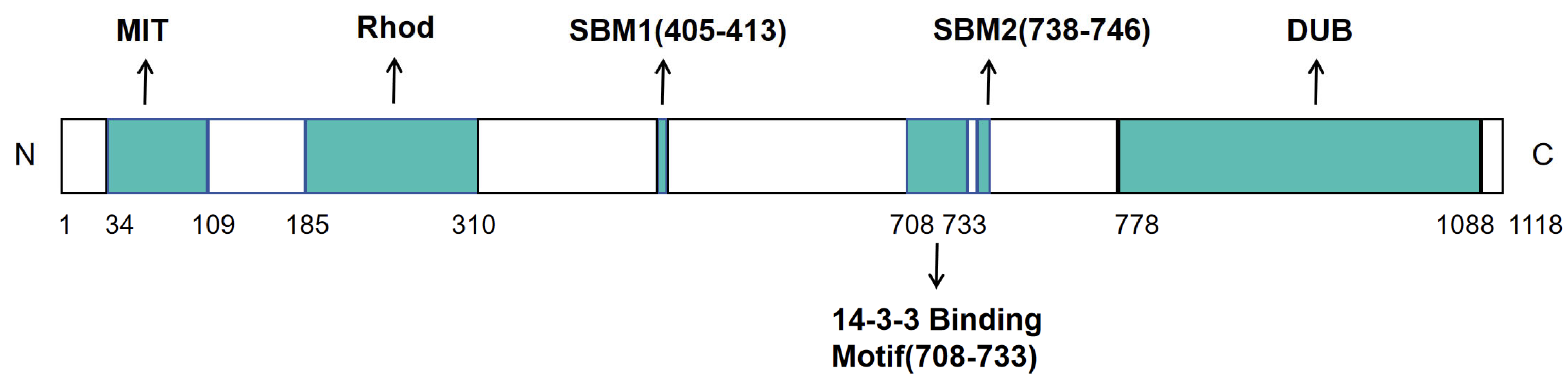
3.2.1. Molecular Characteristics and Molecular Functions of USP8
3.2.2. USP8 Stabilizes O-GlcNAc Transferase (OGT) via Deubiquitination to Drive iCCA Progression and Pemigatinib Resistance
3.3. USP9X
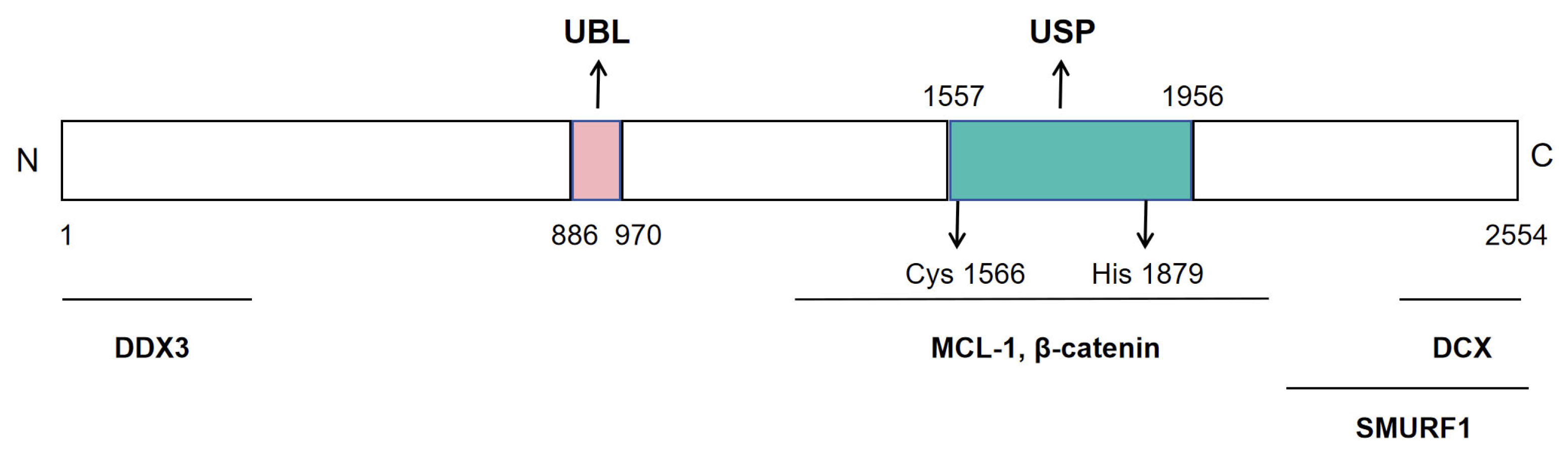
3.3.1. Molecular Characteristics and Functions of USP9X
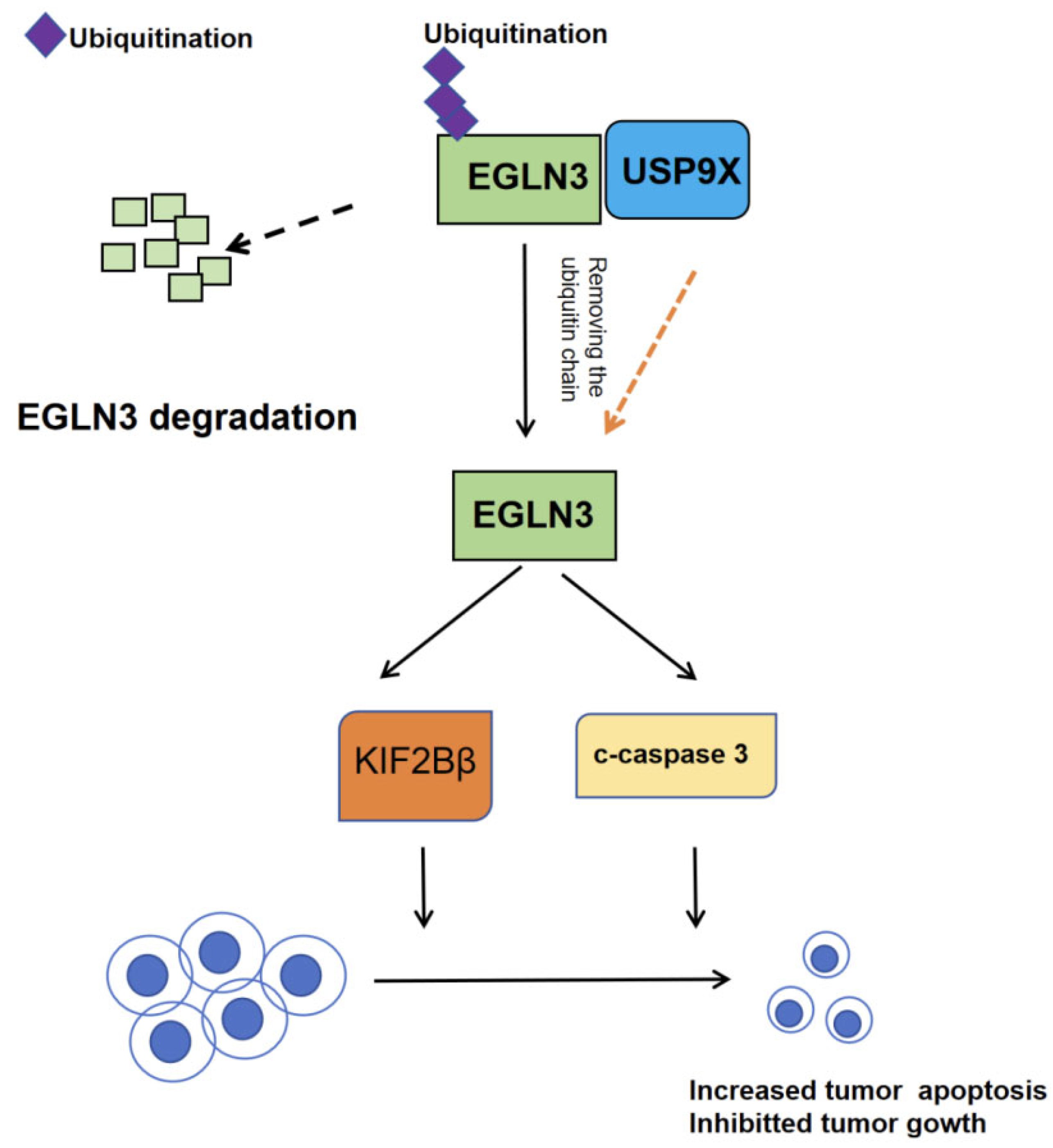
3.3.2. USP9X Regulates Apoptosis in Cholangiocarcinoma
3.4. USP21
3.4.1. Molecular Characteristics and Molecular Functions of USP21
3.4.2. Role and Molecular Mechanisms of USP21 in CCA
3.5. USP22
3.5.1. Molecular Characteristics and Molecular Functions of USP22
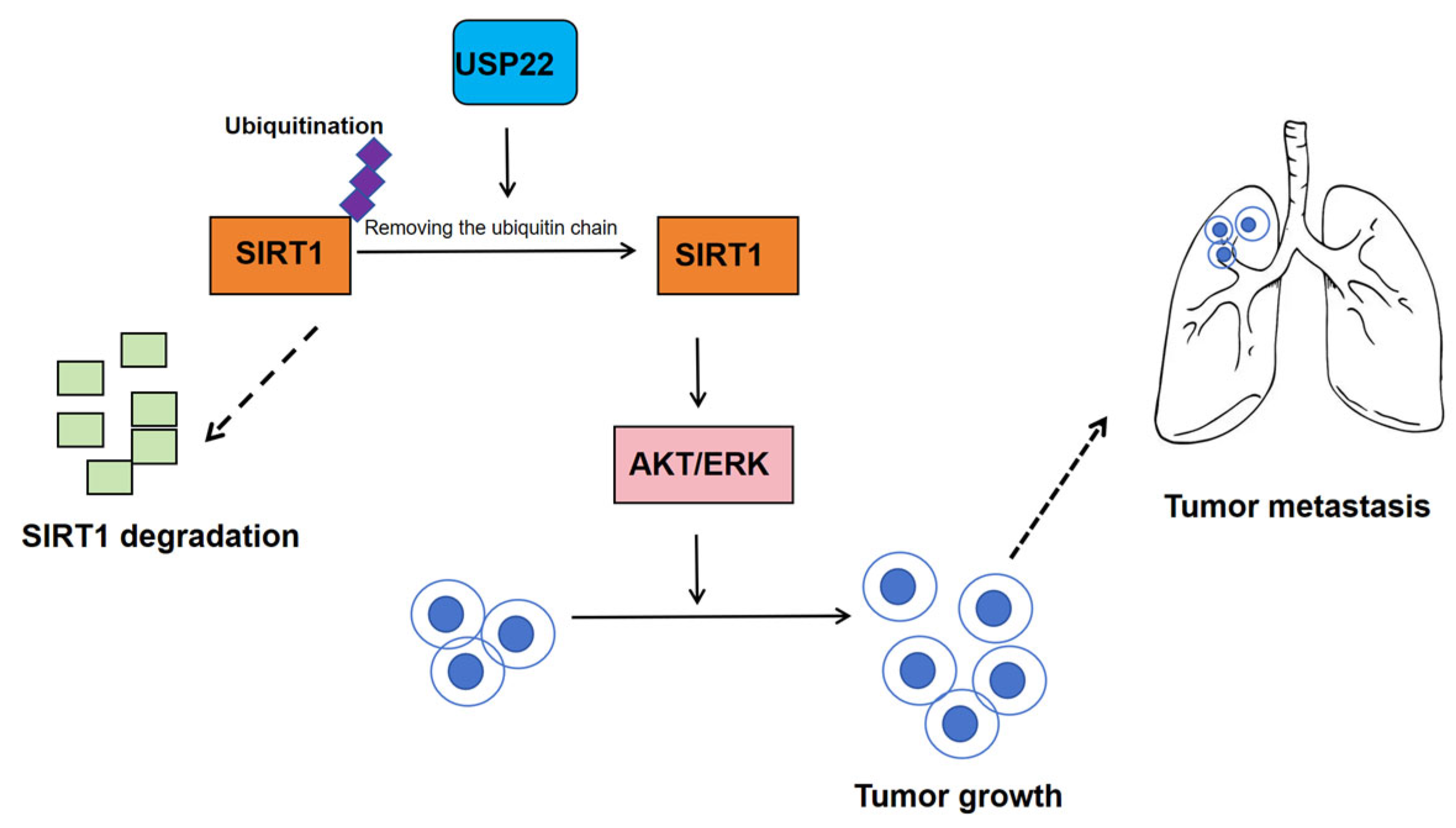
3.5.2. Role and Molecular Mechanisms of USP22 in CCA
4. USPs and GBC
4.1. USP3
4.1.1. Molecular Characteristics and Molecular Functions of USP3
4.1.2. Role and Molecular Mechanisms of USP3 in GBC
5. Strategies for Targeting USP1, USP3, USP8, USP9X, USP21 and USP22
5.1. Targeting the GCN5–USP1–PARP1 Axis in CCA
5.2. Targeting the USPs–DNM1L Axis in GBC
5.3. USP8 Inhibitors DUB-IN-3 Suppress the Malignant Progression of iCCA by Disrupting the USP8–OGT Axis
5.4. The Dual Role of USP9X in Tumorigenesis and Its Therapeutic Potential
5.5. Therapeutic Potential of Targeting the USP21–HSP90 Axis in CCA
5.6. Therapeutic Prospects of Targeting the USP22–SIRT1 Signaling Axis in CCA
6. Future Prospects
6.1. Elucidating Tissue-Specific USP Networks
6.2. The Relationship of USP Family and Tumor Environment
6.3. Mechanistic Exploration of USP-Driven Therapeutic Resistance
6.4. Translation and Intervention
6.5. Clinical Significance and Precision Medicine
7. Conclusions
7.1. The Relationship Between BTC and Tumor Progression
7.2. The Relationship Between USPs and BTC Metabolism
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACSS2 | Acetyl-CoA synthetase 2 |
| ALDOC | Aldolase c |
| ATP | Adenosine triphosphate |
| AXIN | Axis inhibition protein |
| BACE1 | Beta-secretase 1 |
| BTC | Biliary tract cancer |
| CAFs | Cancer-associated fibroblasts |
| CCA | Cholangiocarcinoma |
| CHMP1B | Charged multivesicular body protein 1B |
| dCCA | Distal cholangiocarcinoma |
| DNM1L | Dynamin 1 like |
| DUBs | Deubiquitinating enzymes |
| EGFR | Epidermal growth factor receptor |
| EGLN3 | Prolyl 4-hydroxylase domain protein 3 |
| ENO2 | Enolase 2 |
| ENO3 | Enolase 3 |
| ESCRT-II | Endosomal sorting complexes required for transport III |
| FDA | Food and drug administration |
| GBC | Gallbladder cancer |
| GCN5 | General control of amino-acid synthesis 5-like 2 |
| GPX4 | Glutathione peroxidase 4 |
| GST | Glutathione S-transferase |
| HCC | Hepatocellularcarcinoma |
| HIF1α | Hypoxia-inducible factor 1-alpha |
| HSP90 | Heat shock protein 90 |
| iCCA | Intrahepatic cholangiocarcinoma |
| IFN-β | Interferon-beta |
| IHC | Immunohistochemistry |
| IRF3 | Interferon regulatory factor 3 signaling |
| ITCH | Itchy E3 ubiquitin protein ligase |
| IP | Immunoprecipitation |
| JAMMs | JAMM/MPN+metalloproteases |
| KAT2A | Lysine acetyltransferase 2A |
| KDM4C | lysine-specific demethylase 4C |
| KIF1Bβ | Kinesin family member 1B Beta isoform |
| KPNA2 | Karyopherin subunit alpha 2 |
| LATS2 | Large tumor suppressor kinase 2 |
| MAPK | Mitogen-activated protein kinase |
| MCL-1 | Myeloid cell leukemia-1 |
| MCPIPs | Monocyte chemotactic protein-induced proteins |
| MDA5 | Melanoma differentiation-associated protein 5 |
| MINDYs | MIU-containing novel DUB family |
| MIT | Microtubule-interacting and trafficking |
| mtDNA | Mitochondrial DNA |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NLSs | Nuclear localization sequence |
| OGT | O-GlcNAc transferase |
| OTUs | Ovarian tumor proteases |
| UCHs | Ubiquitin C-terminal hydrolases |
| PARP1 | Poly (ADP-ribose) polymerase 1 |
| pCCA | Perihilar cholangiocarcinoma |
| PD-L1 | Programmed death-ligand 1 |
| PDX | Patient-derived xenograft |
| PI3K/AKT | Phosphatidylinositol 3-kinase/protein kinase B |
| PKLR | Pyruvate kinase L/R |
| PPPDEs | Papain-like peptidases of dsRNA viruses and eukaryotes |
| PROTAC | Proteolysis targeting chimera |
| RIP1 | Receptor-interacting protein 1 |
| ROS | Reactive oxygen species |
| RPS16 | Ribosomal protein S16 |
| SBM | SH3-binding motifs |
| STING | Stimulator of interferon genes |
| SIRT1 | Sirtuin 1 |
| TBK1 | TANK-binding kinase 1 |
| TGF-β | Transforming growth factor-beta |
| TME | Tumor microenvironment |
| TNM | Tumor-node-metastasis |
| USPs | Ubiquitin-specific proteases |
| USP1 | Ubiquitin specific peptidase 1 |
| USP3 | Ubiquitin specific peptidase 3 |
| USP8 | Ubiquitin specific peptidase 8 |
| USP9X | Ubiquitin specific peptidase 9, X-linked |
| USP21 | Ubiquitin specific peptidase 21 |
| USP22 | Ubiquitin specific peptidase 22 |
| Wnt | Wingless-type |
| ZNF | Zinc finger protein |
| ZUP1 | zinc finger-containing ubiquitin peptidase 1 |
References
- Shroff, R.T.; Bachini, M. Treatment options for biliary tract cancer: Unmet needs, new targets and opportunities from both physicians’ and patients’ perspectives. Future Oncol. 2024, 20, 1435–1450. [Google Scholar] [CrossRef]
- Valle, J.W.; Kelley, R.K.; Nervi, B.; Oh, D.Y.; Zhu, A.X. Biliary tract cancer. Lancet 2021, 397, 428–444. [Google Scholar] [CrossRef]
- Banales, J.M.; Marin, J.J.G.; Lamarca, A.; Rodrigues, P.M.; Khan, S.A.; Roberts, L.R.; Cardinale, V.; Carpino, G.; Andersen, J.B.; Braconi, C.; et al. Cholangiocarcinoma 2020: The next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 557–588. [Google Scholar] [CrossRef] [PubMed]
- Moris, D.; Palta, M.; Kim, C.; Allen, P.J.; Morse, M.A.; Lidsky, M.E. Advances in the treatment of intrahepatic cholangiocarcinoma: An overview of the current and future therapeutic landscape for clinicians. CA Cancer J. Clin. 2023, 73, 198–222. [Google Scholar] [CrossRef]
- Huang, F.; Liu, Z.; Song, Y.; Wang, G.; Shi, A.; Chen, T.; Huang, S.; Lian, S.; Li, K.; Tang, Y.; et al. Bile acids activate cancer-associated fibroblasts and induce an immunosuppressive microenvironment in cholangiocarcinoma. Cancer Cell 2025, 43, 1460–1475.e10. [Google Scholar] [CrossRef] [PubMed]
- Roa, J.C.; García, P.; Kapoor, V.K.; Maithel, S.K.; Javle, M.; Koshiol, J. Gallbladder cancer. Nat. Rev. Dis. Primers 2022, 8, 69. [Google Scholar] [CrossRef]
- Pellegrino, N.E.; Guven, A.; Gray, K.; Shah, P.; Kasture, G.; Nastke, M.D.; Thakurta, A.; Gesta, S.; Vishnudas, V.K.; Narain, N.R.; et al. The next frontier: Translational development of Ubiquitination, SUMOylation, and NEDDylation in cancer. Int. J. Mol. Sci. 2022, 23, 3480. [Google Scholar] [CrossRef]
- Dagar, G.; Kumar, R.; Yadav, K.K.; Singh, M.; Pandita, T.K. Ubiquitination and deubiquitination: Implications on cancer therapy. Biochim. Biophys. Acta Gene Regul. Mech. 2023, 1866, 194979. [Google Scholar] [CrossRef]
- Li, K.Q.; Bai, X.; Ke, A.T.; Ding, S.Q.; Zhang, C.D.; Dai, D.Q. Ubiquitin-specific proteases: From biological functions to potential therapeutic applications in gastric cancer. Biomed. Pharmacother. 2024, 173, 116323. [Google Scholar] [CrossRef]
- Han, S.; Wang, R.; Zhang, Y.; Li, X.; Gan, Y.; Gao, F.; Rong, P.; Wang, W.; Li, W. The role of ubiquitination and deubiquitination in tumor invasion and metastasis. Int. J. Biol. Sci. 2022, 18, 2292–2303. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Wu, H. Ubiquitination-Proteasome System (UPS) and Autophagy Two Main Protein Degradation Machineries in Response to Cell Stress. Cells 2022, 11, 851. [Google Scholar] [CrossRef]
- French, M.E.; Koehler, C.F.; Hunter, T. Emerging functions of branched ubiquitin chains. Cell Discov. 2021, 7, 6. [Google Scholar] [CrossRef]
- Kim, Y.; Kim, N.; Lee, W.; Kim, Y.; Kim, D.; Park, J.; Jung, Y.K. More 26S and 30S proteasomes are beneficial in proteinopathy. Proc. Natl. Acad. Sci. USA 2025, 122, e2422570122. [Google Scholar] [CrossRef]
- Li, S.; Song, Y.; Wang, K.; Liu, G.; Dong, X.; Yang, F.; Chen, G.; Cao, C.; Zhang, H.; Wang, M.; et al. USP32 deubiquitinase: Cellular functions, regulatory mechanisms, and potential as a cancer therapy target. Cell Death Discov. 2023, 9, 338. [Google Scholar] [CrossRef]
- He, S.; Wen, S.; Wang, Z.; Qu, Y.; Xu, C.; Li, D.; Hu, J. The Ubiquitin-Proteasome System in Asthma: Mechanisms and Therapeutic Possibilities. Clin. Rev. Allergy Immunol. 2025, 68, 86. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Yu, P.; Liu, S.; Li, R.; Niu, X.; Chen, Y.; Zhang, Z.; Zhou, F.; Zhang, L. Deubiquitylating Enzymes in Cancer and Immunity. Adv. Sci. 2023, 10, e2303807. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Einig, E.; Xu, W.; Kollampally, R.B.; Schlosser, A.; Flentje, M.; Popov, N. The dimeric deubiquitinase USP28 integrates 53BP1 and MYC functions to limit DNA damage. Nucleic Acids Res. 2024, 52, 3011–3030. [Google Scholar] [CrossRef]
- Li, J.; Peng, J.; Wu, L.; Shen, X.; Zhen, X.; Zhang, Y.; Ma, H.; Xu, Y.; Xiong, Q.; Zhu, Q.; et al. The deubiquitinase USP28 maintains the expression of the transcription factor MYCN and is essential in neuroblastoma cells. J. Biol. Chem. 2023, 299, 104856. [Google Scholar] [CrossRef]
- Mi, Z.; Graham, S.H. Role of UCHL1 in the pathogenesis of neurodegenerative diseases and brain injury. Ageing Res. Rev. 2023, 86, 101856. [Google Scholar] [CrossRef]
- Parihar, N.; Bhatt, L.K. Deubiquitylating enzymes: Potential target in autoimmune diseases. Inflammopharmacology 2021, 29, 1683–1699. [Google Scholar] [CrossRef]
- Dong, L.; Chang, Q.; Ma, J.; Liu, C.; Guo, D.; Li, X.; Yang, D.; Fan, Y.; Liang, K.; Li, D.; et al. Associations of blood UCH-L1 and NfL levels with cognitive dysfunction in Parkinson’s disease patients. Neurosci. Lett. 2023, 804, 137219. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.L.; Xi, Z.; Dai, J.W.; Xue, J.Q.; Guan, X.; Zhao, L.; Chen, Z.G.; Xing, F. Drug resistance mechanisms and treatment strategies mediated by Ubiquitin-Specific Proteases (USPs) in cancers: New directions and therapeutic options. Mol. Cancer 2024, 23, 88. [Google Scholar] [CrossRef]
- Snyder, N.A.; Silva, G.M. Deubiquitinating Enzymes (DUBs): Regulation, Homeostasis, and Oxidative Stress Response. J. Biol. Chem. 2021, 297, 101077. [Google Scholar] [CrossRef] [PubMed]
- Mennerich, D.; Kubaichuk, K.; Kietzmann, T. DUBs, hypoxia, and cancer. Trends Cancer 2019, 5, 632–653. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, H. Ubiquitin-Specific Proteases (USPs) and Metabolic Disorders. Int. J. Mol. Sci. 2023, 24, 3219. [Google Scholar] [CrossRef]
- Cruz, L.; Soares, P.; Correia, M. Ubiquitin-Specific Proteases: Players in Cancer Cellular Processes. Pharmaceuticals 2021, 14, 848. [Google Scholar] [CrossRef]
- Fujiwara, T.; Saito, A.; Suzuki, M.; Suzuki, T.; Takahashi, E.; Tanigami, A.; Ichiyama, A.; Chung, C.H.; Nakamura, Y.; Tanaka, K. Identification and chromosomal assignment of USP1, a novel gene encoding a human ubiquitin-specific protease. Genomics 1998, 54, 155–158. [Google Scholar] [CrossRef]
- Joo, H.Y.; Jones, A.; Yang, C.; Zhai, L.; Smith, A.D., IV; Zhang, Z.; Chandrasekharan, M.B.; Sun, Z.W.; Renfrow, M.B.; Wang, Y.M.; et al. Regulation of histone H2A and H2B deubiquitination and Xenopus development by USP12 and USP46. J. Biol. Chem. 2011, 286, 7190–7201. [Google Scholar] [CrossRef]
- Dharadhar, S.; Dijk, W.J.; Scheffers, S.; Fish, A.; Sixma, T.K. Insert L1 is a central hub for allosteric regulation of USP1 activity. EMBO Rep. 2021, 22, e51749. [Google Scholar] [CrossRef]
- Antonenko, S.; Zavelevich, M.; Telegeev, G. The role of USP1 deubiquitinase in the pathogenesis and therapy of cancer. Acta Biochim. Pol. 2023, 70, 219–231. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Zhu, Y.; Wu, Q.; Ma, S.; Ma, Y.; Shen, Z.C.; Wang, Z.; Sun, W.; Zhou, Y.C.; Wang, D.; et al. USP1 promotes cholangiocarcinoma progression by deubiquitinating PARP1 to prevent its proteasomal degradation. Cell Death Dis. 2023, 14, 669. [Google Scholar] [CrossRef]
- Ma, A.; Tang, M.; Zhang, L.; Wang, B.; Yang, Z.; Liu, Y.; Xu, G.; Wu, L.; Jing, T.; Xu, X.; et al. USP1 inhibition destabilizes KPNA2 and suppresses breast cancer metastasis. Oncogene 2019, 38, 2405–2419. [Google Scholar] [CrossRef]
- Liao, Y.; Shao, Z.; Liu, Y.; Xia, X.; Deng, Y.; Yu, C.; Sun, W.; Kong, W.; He, X.; Liu, F.; et al. USP1-dependent RPS16 protein stability drives growth and metastasis of human hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. 2021, 40, 201. [Google Scholar] [CrossRef]
- D’Angelo, D.; De Martino, M.; Arra, C.; Fusco, A. Emerging Role of USP8, HMGA, and Non-Coding RNAs in Pituitary Tumorigenesis. Cancers 2019, 11, 1302. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, X.; Lao, M.; Sun, K.; He, L.; Xu, J.; Duan, Y.; Chen, Y.; Ying, H.; Li, M.; et al. Targeting ubiquitin-specific protease 8 sensitizes anti-programmed death-ligand 1 immunotherapy of pancreatic cancer. Cell Death Differ. 2023, 30, 560–575. [Google Scholar] [CrossRef] [PubMed]
- Hashemi-Madani, N.; Cheraghi, S.; Emami, Z.; Mehrjardi, A.Z.; Kaynama, M.R.; Khamseh, M.E. Targeted analysis of Ubiquitin-Specific Peptidase (USP8) in a population of Iranian people with Cushing’s disease and a systematic review of the literature. BMC Endocr. Disord. 2024, 24, 86. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Huang, S.; He, Y.; Wang, W.; Tong, C.; Ma, M.; Zhao, M.; Yi, L.; Knobeloch, K.P.; Zhang, P. USP8-Governed MDA5 Homeostasis Promotes Innate Immunity and Autoimmunity. Adv. Sci. 2025, 12, e03865. [Google Scholar] [CrossRef] [PubMed]
- Journet, A.; Barette, C.; Aubry, L.; Soleilhac, E.; Fauvarque, M.O. Identification of chemicals breaking the USP8 interaction with its endocytic substrate CHMP1B. SLAS Discov. 2022, 27, 395–404. [Google Scholar] [CrossRef]
- Mauri, S.; Bernardo, G.; Martinez, A.; Favaro, M.; Trevisan, M.; Cobraiville, G.; Fillet, M.; Caicci, F.; Whitworth, A.J.; Ziviani, E. USP8 Down-Regulation Promotes Parkin-Independent Mitophagy in the Drosophila Brain and in Human Neurons. Cells 2023, 12, 1143. [Google Scholar] [CrossRef]
- Aduke Yeates, E.F.; Tesco, G. The Endosome-associated Deubiquitinating Enzyme USP8 Regulates BACE1 Enzyme Ubiquitination and Degradation. J. Biol. Chem. 2016, 291, 15753–15766. [Google Scholar] [CrossRef]
- Ma, T.; Ge, X.; Zhu, J.; Song, C.; Wang, P.; Cai, J. Dioscin Impedes Proliferation, Metastasis and Enhances Autophagy of Gastric Cancer Cells via Regulating the USP8/TGM2 Pathway. Mol. Biotechnol. 2024, 66, 3700–3711. [Google Scholar] [CrossRef]
- Tang, J.; Long, G.; Xiao, L.; Zhou, L. USP8 positively regulates hepatocellular carcinoma tumorigenesis and confers ferroptosis resistance through beta-catenin stabilization. Cell Death Dis. 2023, 14, 360. [Google Scholar] [CrossRef]
- Tang, J.; Long, G.; Hu, K.; Xiao, D.; Liu, S.; Xiao, L.; Zhou, L.; Tao, Y. Targeting USP8 inhibits O-GlcNAcylation of SLC7A11 to promote ferroptosis of hepatocellular carcinoma via stabilization of OGT. Adv. Sci. 2023, 10, e2302953. [Google Scholar] [CrossRef]
- Long, G.; Wang, D.; Tang, J.; Hu, K.; Zhou, L. USP8 promotes the tumorigenesis of intrahepatic cholangiocarcinoma via stabilizing OGT. Cancer Cell Int. 2024, 24, 238. [Google Scholar] [CrossRef]
- Jing, X.; Chen, Y.; Chen, Y.; Shi, G.; Lv, S.; Cheng, N.; Feng, C.; Xin, Z.; Zhang, L.; Wu, J. Down-regulation of USP8 Inhibits cholangiocarcinoma cell proliferation and invasion. Cancer Manag. Res. 2020, 12, 2185–2194. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Hong, C.; Yang, S.; Qin, Z.; Yang, L.; Huang, Y. Roles of USP9X in cellular functions and tumorigenesis (Review). Oncol. Lett. 2023, 26, 506. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Song, J.; Liu, S.; Tang, B.; Shen, L.; Zhu, J.; Fang, S.; Wu, F.; Zheng, L.; Qiu, R.; et al. USP9X promotes apoptosis in cholangiocarcinoma by modulation expression of KIF1Bbeta via deubiquitinating EGLN3. J. Biomed. Sci. 2021, 28, 44. [Google Scholar] [CrossRef]
- Shi, Z.Y.; Li, C.Y.; Chen, R.Y.; Shi, J.J.; Liu, Y.J.; Lu, J.F.; Yang, G.J.; Chen, J. The emerging role of deubiquitylating enzyme USP21 as a potential therapeutic target in cancer. Bioorganic Chem. 2024, 147, 107400. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, Z.; Tang, Q.; Wang, Z.; Lu, J.; You, Y.; Wang, H. USP21 promotes self-renewal and tumorigenicity of mesenchymal glioblastoma stem cells by deubiquitinating and stabilizing FOXD1. Cell Death Dis. 2022, 13, 712. [Google Scholar] [CrossRef]
- Xu, G.; Tan, X.; Wang, H.; Sun, W.; Shi, Y.; Burlingame, S.; Gu, X.; Cao, G.; Zhang, T.; Qin, J.; et al. Ubiquitin-specific peptidase 21 inhibits tumor necrosis factor alpha-induced nuclear factor kappaB activation via binding to and deubiquitinating receptor-interacting protein 1. J. Biol. Chem. 2010, 285, 969–978. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Z.; Qiao, D.; Yuan, Y.; Han, C.; Yang, N.; Li, R.; Du, Q.; Tong, D.; Huang, Y. Porcine circovirus type 2 infection attenuates the K63-linked ubiquitination of STING to inhibit IFN-β induction via p38-MAPK pathway. Veter Microbiol. 2021, 258, 109098. [Google Scholar] [CrossRef]
- Xu, X.; Chen, Y.; Shao, S.; Wang, J.; Shan, J.; Wang, Y.; Wang, Y.; Chang, J.; Zhou, T.; Chen, R.; et al. USP21 deubiquitinates and stabilizes HSP90 and ENO1 to promote aerobic glycolysis and proliferation in cholangiocarcinoma. Int. J. Biol. Sci. 2024, 20, 1492–1508. [Google Scholar] [CrossRef]
- Jeusset, L.M.; McManus, K.J. Ubiquitin Specific Peptidase 22 Regulates Histone H2B Mono-Ubiquitination and Exhibits Both Oncogenic and Tumor Suppressor Roles in Cancer. Cancers 2017, 9, 167. [Google Scholar] [CrossRef]
- Devi, U.; Shukla, P.K. The structural, functional, and regulatory insight of deubiquitinating enzyme—USP22. Int. J. Biol. Macromol. 2025, 318 Pt 3, 145164. [Google Scholar] [CrossRef]
- Tian, Y.; Tang, B.; Wang, C.; Wang, Y.; Mao, J.; Yao, Y.; Gao, Z.; Liang, R.; Ye, M.; Cai, S.; et al. Operative ubiquitin-specific protease 22 deubiquitination confers a more invasive phenotype to cholangiocarcinoma. Cell Death Dis. 2021, 12, 678. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, W.; Wu, Y.; Chen, C. USP3: Key deubiquitylation enzyme in human diseases. Cancer Sci. 2024, 115, 2094–2106. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, Y.; Niu, K.; Yang, R.; Lv, Q.; Zhang, W.; Feng, K.; Zhang, Y. Ubiquitin specific peptidase 3: An emerging deubiquitinase that regulates physiology and diseases. Cell Death Discov. 2024, 10, 243. [Google Scholar] [CrossRef]
- Zhou, Q.; Wang, W.; Wu, J.; Qiu, S.; Yuan, S.; Fu, P.L.; Qian, Q.R.; Xu, Y.Z. Ubiquitin-specific protease 3 attenuates interleukin-1β-mediated chondrocyte senescence by deacetylating forkhead box O-3 via sirtuin-3. Bioengineered 2022, 13, 2017–2027. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zheng, B.; Wang, L.; Cui, W.; Jiang, C.; Li, Z.; Gao, W.; Zhang, W. Deubiquitinase ubiquitin-specific protease 3 (USP3) inhibits HIV-1 replication via promoting APOBEC3G (A3G) expression in both enzyme activity-dependent and -independent manners. Chin. Med. J. 2022, 135, 2706–2717. [Google Scholar] [CrossRef]
- Gao, S.J.; Li, J.D.; Song, L.P.; Wu, J.X.; Huang, W. Influenza A virus-induced downregulation of miR-26a contributes to reduced IFNα/β production. Virol. Sin. 2017, 32, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.P.; Zhang, X.X.; Zhao, J.; Zhu, R.T.; Wang, W.J.; Lu, Q.W.; Sun, Y.L. Ubiquitin-specific protease 3 facilitates cell proliferation by deubiquitinating pyruvate kinase L/R in gallbladder cancer. Lab. Investig. 2022, 102, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Zhang, X.; Wu, S.; Liu, J.; Zhai, Y.; Lin, C.; Wang, Z.; Zhang, Y.; Chen, H.; Zhu, R. Deubiquitination of DNM1L by USP3 triggers the development and metastasis of gallbladder carcinoma. Biol. Direct 2025, 20, 47. [Google Scholar] [CrossRef] [PubMed]
- Jie, X.; Fong, W.P.; Zhou, R.; Zhao, Y.; Zhao, Y.; Meng, R.; Zhang, S.; Dong, X.; Zhang, T.; Yang, K.; et al. USP9X-mediated KDM4C deubiquitination promotes lung cancer radioresistance by epigenetically inducing TGF-β2 transcription. Cell Death Differ. 2021, 28, 2095–2111. [Google Scholar] [CrossRef]
- Zhu, C.; Ji, X.; Zhang, H.; Zhou, Q.; Cao, X.; Tang, M.; Si, Y.; Yan, H.; Li, L.; Liang, T.; et al. Deubiquitylase USP9X suppresses tumorigenesis by stabilizing large tumor suppressor kinase 2 (LATS2) in the Hippo pathway. J. Biol. Chem. 2018, 293, 1178–1191. [Google Scholar] [CrossRef] [PubMed]






| USP Family Gene | BTC Type | Function | Substrates | Signal Pathway | Reference |
|---|---|---|---|---|---|
| USP1 | CCA | USP1 deubiquitinates and stabilizes PARP1, thereby promoting CCA growth and lung metastasis. | PARP1 | GCN5-USP1-PARP1 | [31] |
| USP3 | GBC | USP3 deubiquitinates PKLR, and DNM1L, promotes glycolytic flux, mitochondrial fission, GBC cell proliferation and hepatic metastasis. | PLKR, DNM1L | USP1-PLKR, USP1-DNM1L | [62] |
| USP8 | iCCA | USP8 drives tumor progression in iCCA through OGT stabilization | OGT | USP8-OGT | [44] |
| USP9X | CCA | USP9X promotes apoptosis in cholangiocarcinoma by deubiquitinating EGLN3, which upregulates KIF1Bβ expression | EGLN3 | USP9X-EGLN3-KIF1Bβ | [47] |
| USP21 | CCA | USP21 promotes aerobic glycolysis and proliferation in cholangiocarcinoma by deubiquitinating and stabilizing both HSP90 and ENO1 | HSP90, ENO1 | USP21-HSP90-HIF1α-ENO2, ENO3, ALDOC, ACSS2 USP21-ENO1 | [52] |
| USP22 | CCA | USP22 promotes CCA progression by inducing EMT and stabilizing SIRT1, which cooperates with USP22 to epigenetically modulate malignancy while exacerbating tumor growth via TAK1/Akt deacetylation. | SIRT1 | USP22-SRT1-TAK1/AKT–ERK | [55] |
| Feature | USP3 (GC) | USP21 (CCA) |
|---|---|---|
| Target protein(s) | DNM1L (mitochondrial fission protein) | HSP90, ENO1 (glycolysis-related) |
| Ubiquitin chain type | K48-linked ubiquitin chains | K48-linked ubiquitin chains |
| Metabolic Pathways Affected | Mitochondrial dysfunction, amino acid metabolism (e.g., glutamic acid) | Enhanced aerobic glycolysis (Warburg effect) |
| Main metabolic effects | Decreased mtDNA, disrupted nucleotide and amino acid metabolism | Increased glycolytic flux and energy supply |
| Functional consequences | Promotes proliferation, migration, and liver metastasis | Promotes proliferation, chemoresistance, and poor prognosis |
| Clinical significance | Potential therapeutic target involved in metabolic reprogramming and metastasis | Predictive of prognosis and involved in chemoresistance mechanisms |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Q.; Ma, D.; Zheng, S.; Hao, J.; Wang, G.; Ni, Y.; Zhu, J. Molecular Mechanisms of the Ubiquitin-Specific Proteases (USPs) Family in Biliary Tract Cancer and Targeted Intervention Strategies. Biomedicines 2025, 13, 2586. https://doi.org/10.3390/biomedicines13112586
Cheng Q, Ma D, Zheng S, Hao J, Wang G, Ni Y, Zhu J. Molecular Mechanisms of the Ubiquitin-Specific Proteases (USPs) Family in Biliary Tract Cancer and Targeted Intervention Strategies. Biomedicines. 2025; 13(11):2586. https://doi.org/10.3390/biomedicines13112586
Chicago/Turabian StyleCheng, Qian, Delin Ma, Shengmin Zheng, Jialing Hao, Gang Wang, Yanbin Ni, and Jiye Zhu. 2025. "Molecular Mechanisms of the Ubiquitin-Specific Proteases (USPs) Family in Biliary Tract Cancer and Targeted Intervention Strategies" Biomedicines 13, no. 11: 2586. https://doi.org/10.3390/biomedicines13112586
APA StyleCheng, Q., Ma, D., Zheng, S., Hao, J., Wang, G., Ni, Y., & Zhu, J. (2025). Molecular Mechanisms of the Ubiquitin-Specific Proteases (USPs) Family in Biliary Tract Cancer and Targeted Intervention Strategies. Biomedicines, 13(11), 2586. https://doi.org/10.3390/biomedicines13112586






