Dendritic Cell-Derived Exosomes: Next Generation of Cancer Immunotherapy
Abstract
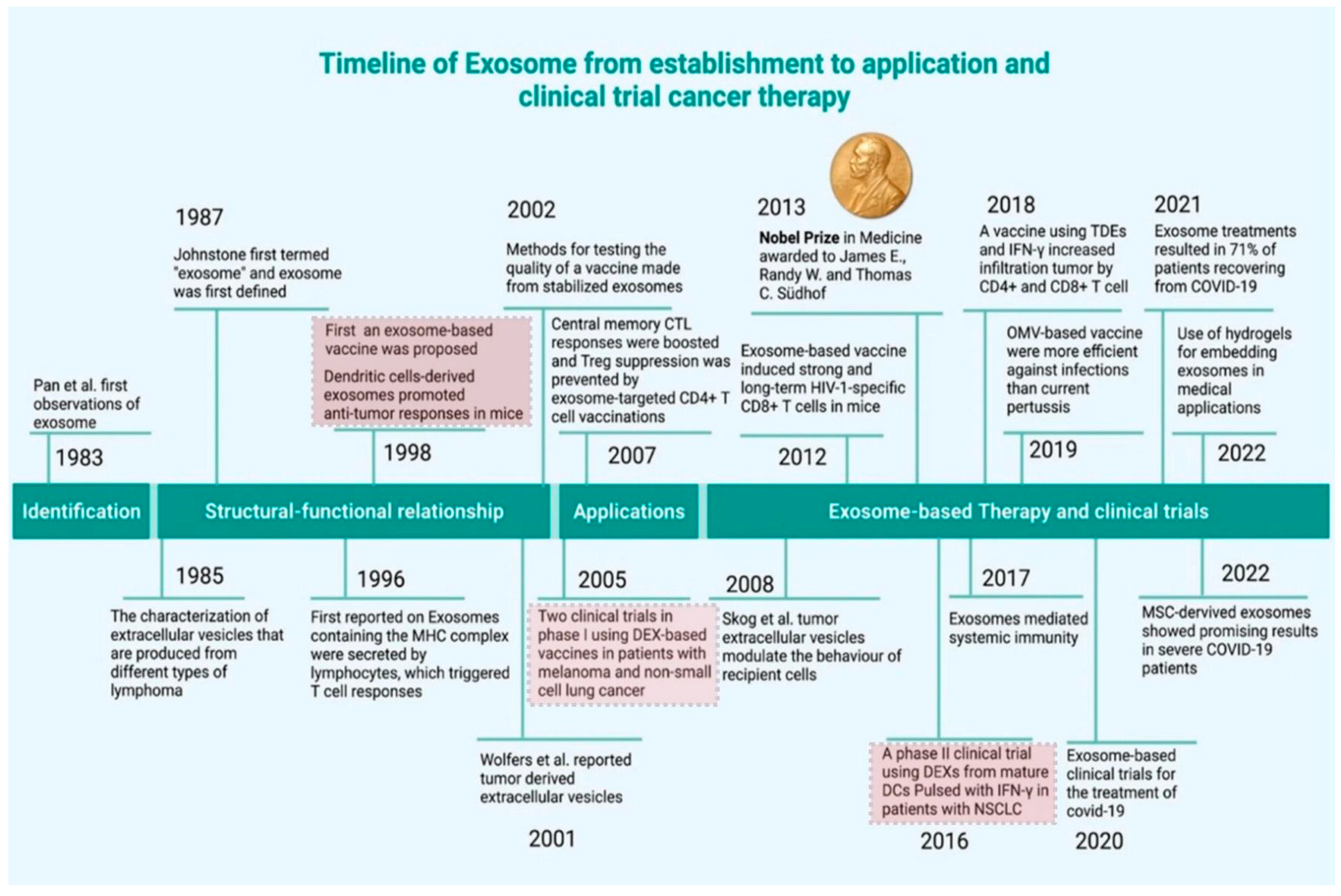
1. Introduction
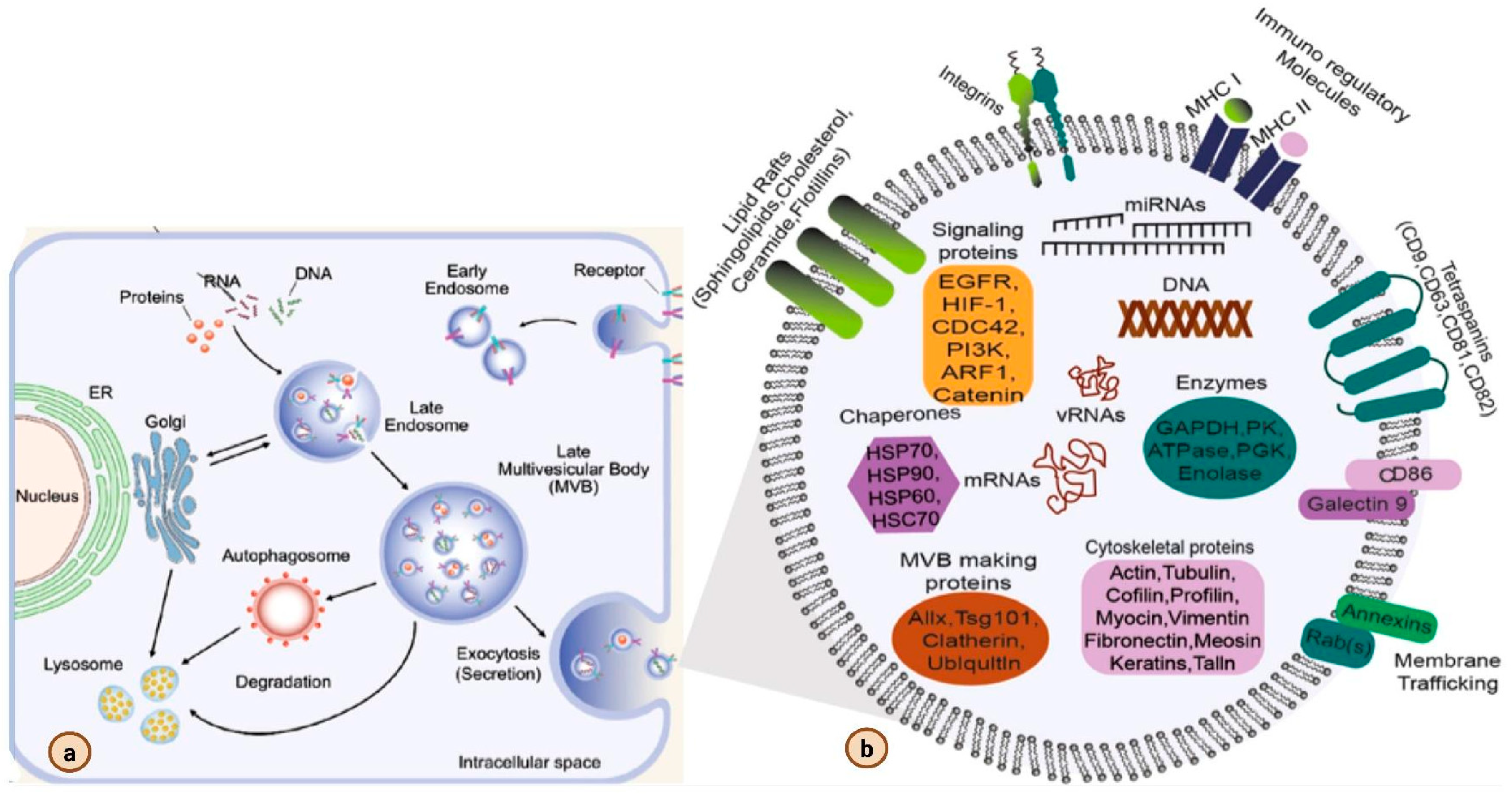
2. Biogenesis of Exosomes
3. Exosome Isolation and Characterization
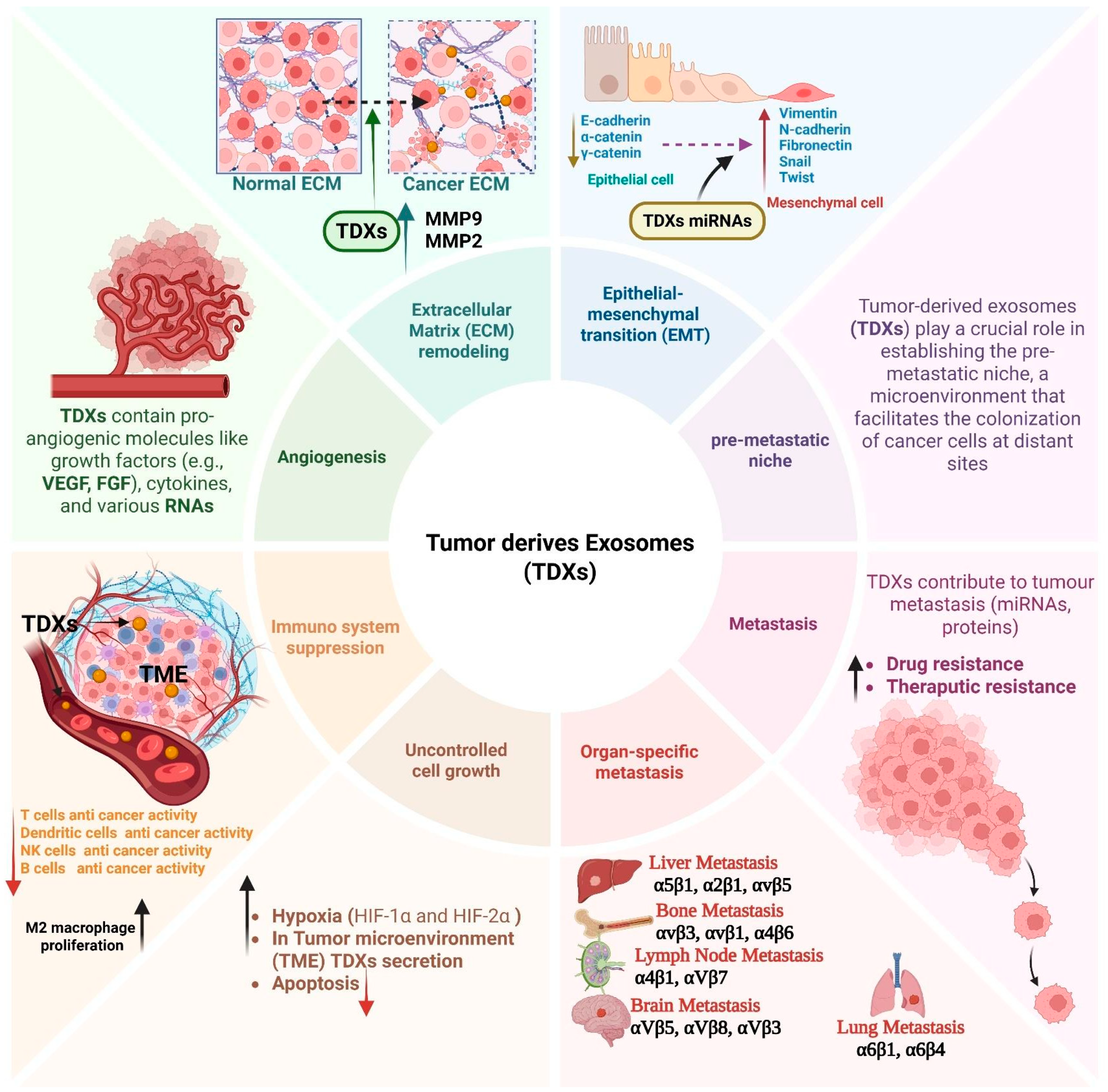
4. Role of Exosomes in Cancer
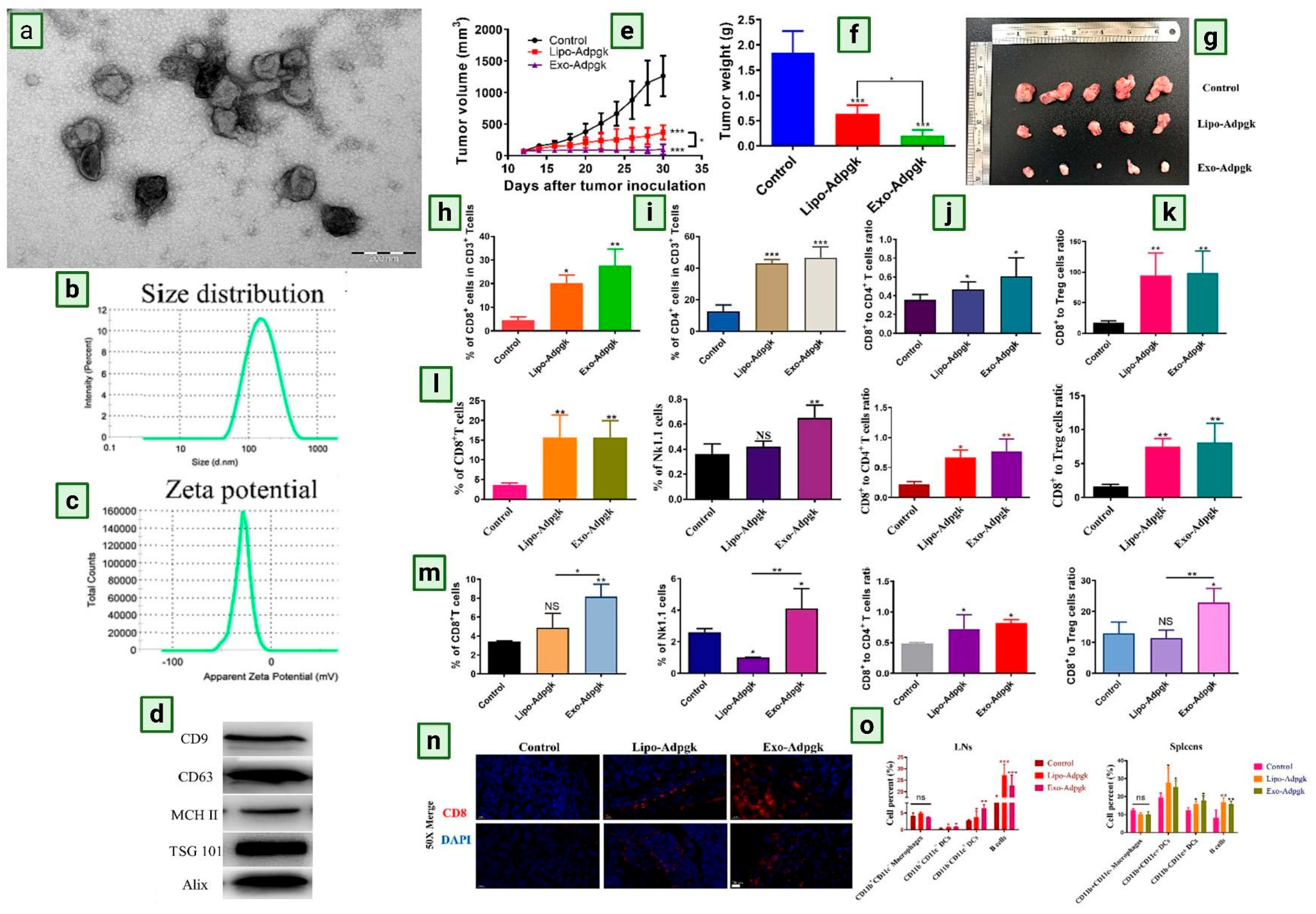
5. DC-Derived Exosomes in Therapeutic Application
6. Clinical Trial of Dendritic Cell-Derived Exosomes
7. Challenges and Future Prospects
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Guida, F.; Kidman, R.; Ferlay, J.; Schüz, J.; Soerjomataram, I.; Kithaka, B.; Ginsburg, O.; Vega, R.B.M.; Galukande, M.; Parham, G.; et al. Global and regional estimates of orphans attributed to maternal cancer mortality in 2020. Nat. Med. 2022, 28, 2563–2572. [Google Scholar] [CrossRef] [PubMed]
- Zafar, A.; Khatoon, S.; Khan, M.J.; Abu, J.; Naeem, A. Advancements and limitations in traditional anti-cancer therapies: A comprehensive review of surgery, chemotherapy, radiation therapy, and hormonal therapy. Discov. Oncol. 2025, 16, 607. [Google Scholar] [CrossRef] [PubMed]
- Ghemrawi, R.; Abuamer, L.; Kremesh, S.; Hussien, G.; Ahmed, R.; Mousa, W.; Khoder, G.; Khair, M. Revolutionizing Cancer Treatment: Recent Advances in Immunotherapy. Biomedicines 2024, 12, 2158. [Google Scholar] [CrossRef] [PubMed]
- Theivendren, P.; Kunjiappan, S.; Pavadai, P.; Ravi, K.; Murugavel, A.; Dayalan, A.; Kumar, A.S.K. Revolutionizing Cancer Immunotherapy: Emerging Nanotechnology-Driven Drug Delivery Systems for Enhanced Therapeutic Efficacy. ACS Meas. Sci. Au 2024, 5, 31–55. [Google Scholar] [CrossRef]
- Santos, P.; Almeida, F. Exosome-Based Vaccines: History, Current State, and Clinical Trials. Front. Immunol. 2021, 12, 711565. [Google Scholar] [CrossRef]
- Sonar, S.; Das, A.; Kalele, K.; Subramaniyan, V. Exosome-based cancer vaccine: A cell-free approach. Mol. Biol. Rep. 2025, 52, 421. [Google Scholar] [CrossRef]
- Mehta, N.K.; Moynihan, K.D.; Irvine, D.J. Engineering New Approaches to Cancer Vaccines. Cancer Immunol. Res. 2015, 3, 836–843. [Google Scholar] [CrossRef]
- Wang, H.; Najibi, A.J.; Sobral, M.C.; Seo, B.R.; Lee, J.Y.; Wu, D.; Li, A.W.; Verbeke, C.S.; Mooney, D.J. Biomaterial-based scaffold for in situ chemo-immunotherapy to treat poorly immunogenic tumors. Nat. Commun. 2020, 11, 5696. [Google Scholar] [CrossRef]
- Zhou, Y.; Wei, Y.; Tian, X.; Wei, X. Cancer vaccines: Current status and future directions. J. Hematol. Oncol. 2025, 18, 18. [Google Scholar] [CrossRef]
- Kaur, P.; Mehrotra, R.; Rengaswamy, S.; Kaur, T.; Hariprasad, R.; Mehendale, S.M.; Rajaraman, P.; Rath, G.K.; Bhatla, N.; Krishnan, S.; et al. Human papillomavirus vaccine for cancer cervix prevention: Rationale & recommendations for implementation in India. Indian J. Med. Res. 2017, 146, 153–157. [Google Scholar]
- Lee, K.-W.; Yam, J.W.P.; Mao, X. Dendritic Cell Vaccines: A Shift from Conventional Approach to New Generations. Cells 2023, 12, 2147. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Fu, C.; Zhou, L.; Mi, Q.-S.; Jiang, A. DC-Derived Exosomes for Cancer Immunotherapy. Cancers 2021, 13, 3667. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, J.; Peng, Y.; Du, Y.; Yang, Z.; Qi, X. Dendritic cell derived exosomes loaded neoantigens for personalized cancer immunotherapies. J. Control. Release 2022, 353, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Shin, K.J.; Chae, Y.C. Regulation of cargo selection in exosome biogenesis and its biomedical applications in cancer. Exp. Mol. Med. 2024, 56, 877–889. [Google Scholar] [CrossRef]
- Tenchov, R.; Sasso, J.M.; Wang, X.; Liaw, W.-S.; Chen, C.-A.; Zhou, Q.A. Exosomes─Nature’s Lipid Nanoparticles, a Rising Star in Drug Delivery and Diagnostics. ACS Nano 2022, 16, 17802–17846. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Z.-K.; Duan, X.; Zhang, H.-J.; Xiao, B.-L.; Wang, K.-M.; Chen, G. Targeted inhibition of tumor-derived exosomes as a novel therapeutic option for cancer. Exp. Mol. Med. 2022, 54, 1379–1389. [Google Scholar] [CrossRef]
- Sohrabi, B.; Dayeri, B.; Zahedi, E.; Khoshbakht, S.; Pour, N.N.; Ranjbar, H.; Nejad, A.D.; Noureddini, M.; Alani, B. Mesenchymal stem cell (MSC)-derived exosomes as novel vehicles for delivery of miRNAs in cancer therapy. Cancer Gene Ther. 2022, 29, 1105–1116. [Google Scholar] [CrossRef]
- Jung, I.; Shin, S.; Baek, M.-C.; Yea, K. Modification of immune cell-derived exosomes for enhanced cancer immunotherapy: Current advances and therapeutic applications. Exp. Mol. Med. 2024, 56, 19–31. [Google Scholar] [CrossRef]
- Sonar, S.; Anand, K. Plant-derived exosomes: A Green Nanomedicine for Cancer. Clin. Transl. Discov. 2024, 4, e333. [Google Scholar] [CrossRef]
- Das, A.; Sonar, S.; Kalele, K.; Subramaniyan, V. Milk exosomes: Harnessing nature’s duality for cancer therapy. Clin. Transl. Discov. 2024, 4, e349. [Google Scholar] [CrossRef]
- Pitt, J.M.; André, F.; Amigorena, S.; Soria, J.-C.; Eggermont, A.; Kroemer, G.; Zitvogel, L. Dendritic cell–derived exosomes for cancer therapy. J. Clin. Investig. 2016, 126, 1224–1232. [Google Scholar] [CrossRef] [PubMed]
- Nikfarjam, S.; Rezaie, J.; Kashanchi, F.; Jafari, R. Dexosomes as a cell-free vaccine for cancer immunotherapy. J. Exp. Clin. Cancer Res. 2020, 39, 258. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Shi, Y. Exosomes Derived from Immune Cells: The New Role of Tumor Immune Microenvironment and Tumor Therapy. Int. J. Nanomed. 2022, 17, 6527–6550. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Yao, Z.; Ke, X.; Hu, M.; Ren, H.; Gao, S.; Zhang, H. Extracellular vesicles-based vaccines: Emerging immunotherapies against cancer. J. Control. Release 2024, 378, 438–459. [Google Scholar] [CrossRef]
- Besse, B.; Charrier, M.; Lapierre, V.; Dansin, E.; Lantz, O.; Planchard, D.; Le Chevalier, T.; Livartoski, A.; Barlesi, F.; Laplanche, A.; et al. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. OncoImmunology 2016, 5, 1071008. [Google Scholar] [CrossRef]
- Blumenschein, G.R.; Devarakonda, S.; Johnson, M.; Moreno, V.; Gainor, J.; Edelman, M.J.; Heymach, J.V.; Govindan, R.; Bachier, C.; de Spéville, B.D.; et al. Phase I clinical trial evaluating the safety and efficacy of ADP-A2M10 SPEAR T cells in patients with MAGE-A10+ advanced non-small cell lung cancer. J. Immunother. Cancer 2022, 10, e003581. [Google Scholar] [CrossRef]
- Tian, H.; Li, W. Dendritic cell-derived exosomes for cancer immunotherapy: Hope and challenges. Ann. Transl. Med. 2017, 5, 221. [Google Scholar] [CrossRef]
- Xia, J.; Miao, Y.; Wang, X.; Huang, X.; Dai, J. Recent progress of dendritic cell-derived exosomes (Dex) as an anti-cancer nanovaccine. Biomed. Pharmacother. 2022, 152, 113250. [Google Scholar] [CrossRef]
- Hosseini, R.; Asef-Kabiri, L.; Yousefi, H.; Sarvnaz, H.; Salehi, M.; Akbari, M.E.; Eskandari, N. The roles of tumor-derived exosomes in altered differentiation, maturation and function of dendritic cells. Mol. Cancer 2021, 20, 83. [Google Scholar] [CrossRef]
- Zhang, F.; Guo, J.; Zhang, Z.; Qian, Y.; Wang, G.; Duan, M.; Zhao, H.; Yang, Z.; Jiang, X. Mesenchymal stem cell-derived exosome: A tumor regulator and carrier for targeted tumor therapy. Cancer Lett. 2022, 526, 29–40. [Google Scholar] [CrossRef]
- Jin, X.; Zhang, J.; Zhang, Y.; He, J.; Wang, M.; Hei, Y.; Guo, S.; Xu, X.; Liu, Y. Different origin-derived exosomes and their clinical advantages in cancer therapy. Front. Immunol. 2024, 15, 1401852. [Google Scholar] [CrossRef]
- Redkin, T.; Turubanova, V. Dendritic cell-derived exosomes as anti-cancer cell-free agents: New insights into enhancing immunogenic effects. Front. Immunol. 2025, 16, 1586892. [Google Scholar] [CrossRef] [PubMed]
- Hussen, B.M.; Faraj, G.S.H.; Rasul, M.F.; Hidayat, H.J.; Salihi, A.; Baniahmad, A.; Taheri, M.; Ghafouri-Frad, S. Strategies to overcome the main challenges of the use of exosomes as drug carrier for cancer therapy. Cancer Cell Int. 2022, 22, 323. [Google Scholar] [CrossRef] [PubMed]
- Krylova, S.V.; Feng, D. The Machinery of Exosomes: Biogenesis, Release, and Uptake. Int. J. Mol. Sci. 2023, 24, 1337. [Google Scholar] [CrossRef] [PubMed]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef]
- Han, Q.-F.; Li, W.-J.; Hu, K.-S.; Gao, J.; Zhai, W.-L.; Yang, J.-H.; Zhang, S.-J. Exosome biogenesis: Machinery, regulation, and therapeutic implications in cancer. Mol. Cancer 2022, 21, 207. [Google Scholar] [CrossRef]
- Woodman, P.G.; E Futter, C. Multivesicular bodies: Co-ordinated progression to maturity. Curr. Opin. Cell Biol. 2008, 20, 408–414. [Google Scholar] [CrossRef]
- Rädler, J.; Gupta, D.; Zickler, A.; EL Andaloussi, S. Exploiting the biogenesis of extracellular vesicles for bioengineering and therapeutic cargo loading. Mol. Ther. 2023, 31, 1231–1250. [Google Scholar] [CrossRef]
- Tschuschke, M.; Kocherova, I.; Bryja, A.; Mozdziak, P.; Volponi, A.A.; Janowicz, K.; Sibiak, R.; Piotrowska-Kempisty, H.; Iżycki, D.; Bukowska, D.; et al. Inclusion Biogenesis, Methods of Isolation and Clinical Application of Human Cellular Exosomes. J. Clin. Med. 2020, 9, 436. [Google Scholar] [CrossRef]
- Mageswaran, S.K.; Dixon, M.G.; Curtiss, M.; Keener, J.P.; Babst, M. Binding to Any ESCRT Can Mediate Ubiquitin-Independent Cargo Sorting. Traffic 2013, 15, 212–229. [Google Scholar] [CrossRef]
- Shim, S.; Merrill, S.A.; Hanson, P.I. Novel Interactions of ESCRT-III with LIP5 and VPS4 and their Implications for ESCRT-III Disassembly. Mol. Biol. Cell 2008, 19, 2661–2672. [Google Scholar] [CrossRef]
- Christ, L.; Raiborg, C.; Wenzel, E.M.; Campsteijn, C.; Stenmark, H. Cellular Functions and Molecular Mechanisms of the ESCRT Membrane-Scission Machinery. Trends Biochem. Sci. 2017, 42, 42–56. [Google Scholar] [CrossRef] [PubMed]
- McAndrews, K.M.; Kalluri, R. Mechanisms associated with biogenesis of exosomes in cancer. Mol. Cancer 2019, 18, 52. [Google Scholar] [CrossRef]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Catalano, M.; O’Driscoll, L. Inhibiting extracellular vesicles formation and release: A review of EV inhibitors. J. Extracell. Vesicles 2020, 9, 1703244. [Google Scholar] [CrossRef]
- Essola, J.M.; Zhang, M.; Yang, H.; Li, F.; Xia, B.; Mavoungou, J.F.; Hussain, A.; Huang, Y. Exosome regulation of immune response mechanism: Pros and cons in immunotherapy. Bioact. Mater. 2024, 32, 124–146. [Google Scholar] [CrossRef]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef]
- Chaput, N.; Angevin, E.; Zitvogel, L.; Taïeb, J.; Schartz, N.E.C.; André, F. Exosome-based immunotherapy. Cancer Immunol. Immunother. 2004, 53, 234–239. [Google Scholar] [CrossRef]
- Yan, W.; Jiang, S. Immune Cell-Derived Exosomes in the Cancer-Immunity Cycle. Trends Cancer 2020, 6, 506–517. [Google Scholar] [CrossRef]
- Reiners, K.S.; Dassler, J.; Coch, C.; von Strandmann, E.P. Role of Exosomes Released by Dendritic Cells and/or by Tumor Targets: Regulation of NK Cell Plasticity. Front. Immunol. 2014, 5, 91. [Google Scholar] [CrossRef]
- Das, A.; Saha, P.; Kalele, K.; Sonar, S. Clinical signature of exosomal tetraspanin proteins in cancer. Clin. Transl. Discov. 2024, 4, e341. [Google Scholar] [CrossRef]
- Horbay, R.; Hamraghani, A.; Ermini, L.; Holcik, S.; Beug, S.T.; Yeganeh, B. Role of Ceramides and Lysosomes in Extracellular Vesicle Biogenesis, Cargo Sorting and Release. Int. J. Mol. Sci. 2022, 23, 15317. [Google Scholar] [CrossRef]
- Arya, S.B.; Chen, S.; Jordan-Javed, F.; Parent, C.A. Ceramide-rich microdomains facilitate nuclear envelope budding for non-conventional exosome formation. Nat. Cell Biol. 2022, 24, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Jankovičová, J.; Sečová, P.; Michalková, K.; Antalíková, J. Tetraspanins, More than Markers of Extracellular Vesicles in Reproduction. Int. J. Mol. Sci. 2020, 21, 7568. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Li, J.; Liu, S.; Wang, T.; Ianni, A.; Bober, E.; Braun, T.; Xiang, R.; Yue, S. Exosomal tetraspanins mediate cancer metastasis by altering host microenvironment. Oncotarget. 2017, 8, 62803–62815. [Google Scholar] [CrossRef] [PubMed]
- Egea-Jimenez, A.L.; Zimmermann, P. Phospholipase D and phosphatidic acid in the biogenesis and cargo loading of extracellular vesicles. J. Lipid Res. 2018, 59, 1554–1560. [Google Scholar] [CrossRef]
- Wolf, A.; Tanguy, E.; Wang, Q.; Gasman, S.; Vitale, N. Phospholipase D and cancer metastasis: A focus on exosomes. Adv. Biol. Regul. 2022, 87, 100924. [Google Scholar] [CrossRef]
- Schorey, J.S.; Cheng, Y.; Singh, P.P.; Smith, V.L. Exosomes and other extracellular vesicles in host–pathogen interactions. Embo. Rep. 2014, 16, 24–43. [Google Scholar] [CrossRef]
- Martinez-Arroyo, O.; Selma-Soriano, E.; Ortega, A.; Cortes, R.; Redon, J. Small Rab GTPases in Intracellular Vesicle Trafficking: The Case of Rab3A/Raphillin-3A Complex in the Kidney. Int. J. Mol. Sci. 2021, 22, 7679. [Google Scholar] [CrossRef]
- Arya, S.B.; Collie, S.P.; Parent, C.A. The ins-and-outs of exosome biogenesis, secretion, and internalization. Trends Cell Biol. 2023, 34, 90–108. [Google Scholar] [CrossRef]
- Wei, H.; Chen, Q.; Lin, L.; Sha, C.; Li, T.; Liu, Y.; Yin, X.; Xu, Y.; Chen, L.; Gao, W.; et al. Regulation of exosome production and cargo sorting. Int. J. Biol. Sci. 2021, 17, 163–177. [Google Scholar] [CrossRef]
- Dellar, E.R.; Hill, C.; Melling, G.E.; Carter, D.R.; Baena-Lopez, L.A. Unpacking extracellular vesicles: RNA cargo loading and function. J. Extracell. Biol. 2022, 1, e40. [Google Scholar] [CrossRef] [PubMed]
- Huda, N.; Nafiujjaman; Deaguero, I.G.; Okonkwo, J.; Hill, M.L.; Kim, T. Nurunnabi Potential Use of Exosomes as Diagnostic Biomarkers and in Targeted Drug Delivery: Progress in Clinical and Preclinical Applications. ACS Biomater. Sci. Eng. 2021, 7, 2106–2149. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.; Bajracharya, S.D.; Yuen, P.S.T.; Zhou, H.; Star, R.A.; Illei, G.G.; Alevizos, I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010, 16, 34–38. [Google Scholar] [CrossRef]
- Yakubovich, E.I.; Polischouk, A.G.; Evtushenko, V.I. Principles and Problems of Exosome Isolation from Biological Fluids. Biochem. (Moscow) Suppl. Ser. A Membr. Cell Biol. 2022, 16, 115–126. [Google Scholar] [CrossRef]
- Mirgh, D.; Krishnan, A.; Gorai, S. Sweat exosomes: A new horizon of liquid biopsy in cancer. J. Liq. Biopsy 2023, 2, 100122. [Google Scholar] [CrossRef]
- Daily, A.; Ravishankar, P.; Harms, S.; Klimberg, V.S. Using tears as a non-invasive source for early detection of breast cancer. PLoS ONE 2022, 17, e0267676. [Google Scholar] [CrossRef]
- Zhao, B.; Lin, H.; Jiang, X.; Li, W.; Gao, Y.; Li, M.; Yu, Y.; Chen, N.; Gao, J. Exosome-like nanoparticles derived from fruits, vegetables, and herbs: Innovative strategies of therapeutic and drug delivery. Theranostics 2024, 14, 4598–4621. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, T.; Zhou, M. Immune-Cell-Derived Exosomes for Cancer Therapy. Mol. Pharm. 2022, 19, 3042–3056. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Wu, Y.; Xu, Y.; Li, G.; Li, Z.; Liu, T. Mesenchymal stem cell-derived exosomes in cancer therapy resistance: Recent advances and therapeutic potential. Mol. Cancer 2022, 21, 179. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Luo, J.-D.; Jiang, H.; Duan, D.D. Tumor exosomes: A double-edged sword in cancer therapy. Acta Pharmacol. Sin. 2018, 39, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Li, J.; Huang, B.; Liu, J.; Chen, X.; Chen, X.-M.; Xu, Y.-M.; Huang, L.-F.; Wang, X.-Z. Exosomes: Novel Biomarkers for Clinical Diagnosis. Sci. World J. 2015, 2015, 657086. [Google Scholar] [CrossRef]
- Thomas, S.C.; Kim, J.-W.; Pauletti, G.M.; Hassett, D.J.; Kotagiri, N. Exosomes: Biological Pharmaceutical Nanovectors for Theranostics. Front. Bioeng. Biotechnol. 2022, 9, 808614. [Google Scholar] [CrossRef]
- Wang, J.; Huang, X.; Xie, J.; Han, Y.; Huang, Y.; Zhang, H. Exosomal analysis: Advances in biosensor technology. Clin. Chim. Acta 2021, 518, 142–150. [Google Scholar] [CrossRef]
- Janouskova, O.; Herma, R.; Semeradtova, A.; Poustka, D.; Liegertova, M.; Malinska, H.A.; Maly, J. Conventional and Nonconventional Sources of Exosomes–Isolation Methods and Influence on Their Downstream Biomedical Application. Front. Mol. Biosci. 2022, 9, 846650. [Google Scholar] [CrossRef]
- Gao, J.; Li, A.; Hu, J.; Feng, L.; Liu, L.; Shen, Z. Recent developments in isolating methods for exosomes. Front. Bioeng. Biotechnol. 2023, 10, 1100892. [Google Scholar] [CrossRef]
- Bari, S.M.I.; Hossain, F.B.; Nestorova, G.G. Advances in Biosensors Technology for Detection and Characterization of Extracellular Vesicles. Sensors 2021, 21, 7645. [Google Scholar] [CrossRef]
- Cui, L.; Song, Y.; Hou, Z.; Yang, L.; Guo, S.; Wang, C. From bench to bedside: The research status and application opportunity of extracellular vesicles and their engineering strategies in the treatment of skin defects. J. Nanobiotechnol. 2025, 23, 375. [Google Scholar] [CrossRef]
- Chou, C.-Y.; Chiang, P.-C.; Li, C.-C.; Chang, J.-W.; Lu, P.-H.; Hsu, W.-F.; Chang, L.-C.; Hsu, J.-L.; Wu, M.-S.; Wo, A.M. Improving the Purity of Extracellular Vesicles by Removal of Lipoproteins from Size Exclusion Chromatography- and Ultracentrifugation-Processed Samples Using Glycosaminoglycan-Functionalized Magnetic Beads. ACS Appl. Mater. Interfaces 2024, 16, 44386–44398. [Google Scholar] [CrossRef]
- Purushothaman, A. Exosomes from Cell Culture-Conditioned Medium: Isolation by Ultracentrifugation and Characterization. In The Extracellular Matrix; Humana Press: New York, NY, USA, 2019; Volume 1952, pp. 233–244. [Google Scholar] [CrossRef]
- Faur, C.I.; Rotaru, H.; Osan, C.; Jurj, A.; Roman, R.C.; Moldovan, M.; Chirila, M.; Hedesiu, M. Salivary exosomal microRNAs as biomarkers for head and neck cancer detection—A literature review. Maxillofac. Plast. Reconstr. Surg. 2021, 43, 19. [Google Scholar] [CrossRef]
- Yu, L.-L.; Zhu, J.; Liu, J.-X.; Jiang, F.; Ni, W.-K.; Qu, L.-S.; Ni, R.-Z.; Lu, C.-H.; Xiao, M.-B. A Comparison of Traditional and Novel Methods for the Separation of Exosomes from Human Samples. BioMed Res. Int. 2018, 2018, 3634563. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Yin, Z.; Chen, F.; Lei, P. Mesenchymal stem cell-derived exosome: A promising alternative in the therapy of Alzheimer’s disease. Alzheimer’s Res. Ther. 2020, 12, 109. [Google Scholar] [CrossRef] [PubMed]
- Contreras, H.; Alarcón-Zapata, P.; Nova-Lamperti, E.; Ormazabal, V.; Varas-Godoy, M.; Salomon, C.; Zuniga, F.A. Comparative study of size exclusion chromatography for isolation of small extracellular vesicle from cell-conditioned media, plasma, urine, and saliva. Front. Nanotechnol. 2023, 5, 1146772. [Google Scholar] [CrossRef]
- Gámez-Valero, A.; Monguió-Tortajada, M.; Carreras-Planella, L.; La Franquesa, M.; Beyer, K.; Borràs, F.E. Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles’ characteristics compared to precipitating agents. Sci. Rep. 2016, 6, 33641. [Google Scholar] [CrossRef]
- Guru, K.T.P.; Sreeja, J.S.; Dharmapal, D.; Sengupta, S.; Basu, P.K. Novel Gold Nanoparticle-Based Quick Small-Exosome Isolation Technique from Serum Sample at a Low Centrifugal Force. Nanomaterials 2022, 12, 1660. [Google Scholar] [CrossRef]
- Kim, J.; Lee, H.; Park, K.; Shin, S. Rapid and Efficient Isolation of Exosomes by Clustering and Scattering. J. Clin. Med. 2020, 9, 650. [Google Scholar] [CrossRef]
- Van Deun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; De Wever, O.; Hendrix, A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3, 24858. [Google Scholar] [CrossRef]
- Meng, Y.; Zhang, Y.; Bühler, M.; Wang, S.; Asghari, M.; Stürchler, A.; Mateescu, B.; Weiss, T.; Stavrakis, S.; Demello, A.J. Direct isolation of small extracellular vesicles from human blood using viscoelastic microfluidics. Sci. Adv. 2023, 9, eadi5296. [Google Scholar] [CrossRef]
- Chernyshev, V.S.; Chuprov-Netochin, R.N.; Tsydenzhapova, E.; Svirshchevskaya, E.V.; Poltavtseva, R.A.; Merdalimova, A.; Yashchenok, A.; Keshelava, A.; Sorokin, K.; Keshelava, V.; et al. Asymmetric depth-filtration: A versatile and scalable method for high-yield isolation of extracellular vesicles with low contamination. J. Extracell. Vesicles 2022, 11, e12256. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Yang, X.; Gao, Z.; Effah, C.Y.; Zhang, X.; Wu, Y.; Qu, L. A Holistic Review of the State-of-the-Art Microfluidics for Exosome Separation: An Overview of the Current Status, Existing Obstacles, and Future Outlook. Small 2021, 17, e2007174. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Li, H.; Chen, W.; Qin, J. Microfluidic strategies for label-free exosomes isolation and analysis. TrAC Trends Anal. Chem. 2019, 118, 686–698. [Google Scholar] [CrossRef]
- Yang, F.; Liao, X.; Tian, Y.; Li, G. Exosome separation using microfluidic systems: Size-based, immunoaffinity-based and dynamic methodologies. Biotechnol. J. 2017, 12, 1600699. [Google Scholar] [CrossRef]
- Soares Martins, T.; Catita, J.; Martins Rosa, I.; A B da Cruz e Silva, O.; Henriques, A.G. Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS ONE 2018, 13, e0198820. [Google Scholar] [CrossRef]
- Sun, Z.; Yang, J.; Li, H.; Wang, C.; Fletcher, C.; Li, J.; Zhan, Y.; Du, L.; Wang, F.; Jiang, Y. Progress in the research of nanomaterial-based exosome bioanalysis and exosome-based nanomaterials tumor therapy. Biomaterials 2021, 274, 120873. [Google Scholar] [CrossRef]
- Ma, X.; Hao, Y.; Liu, L. Progress in Nanomaterials-Based Optical and Electrochemical Methods for the Assays of Exosomes. Int. J. Nanomed. 2021, 16, 7575–7608. [Google Scholar] [CrossRef]
- Li, Y.; Meng, L.; Li, B.; Li, Y.; Shen, T.; Zhao, B. The Exosome Journey: From Biogenesis to Regulation and Function in Cancers. J. Oncol. 2022, 2022, 9356807. [Google Scholar] [CrossRef]
- Welsh, J.A.; Arkesteijn, G.J.A.; Bremer, M.; Cimorelli, M.; Dignat-George, F.; Giebel, B.; Görgens, A.; Hendrix, A.; Kuiper, M.; Lacroix, R.; et al. A compendium of single extracellular vesicle flow cytometry. J. Extracell. Vesicles 2023, 12, e12299. [Google Scholar] [CrossRef]
- Abhange, K.; King, S.; Peterson, N.; Sahai, V.; Cuneo, K.C.; Lubman, D.M. The Use of dSTORM-Based Single Exosome Analysis To Study Tetraspanin Abundance in Extracellular Vesicles. ACS Omega 2025, 10, 34659–34665. [Google Scholar] [CrossRef]
- McNamara, R.P.; Zhou, Y.; Eason, A.B.; Landis, J.T.; Chambers, M.G.; Willcox, S.; Peterson, T.A.; Schouest, B.; Maness, N.J.; MacLean, A.G.; et al. Imaging of surface microdomains on individual extracellular vesicles in 3-D. J. Extracell. Vesicles 2022, 11, e12191. [Google Scholar] [CrossRef]
- Breitwieser, K.; Koch, L.F.; Tertel, T.; Proestler, E.; Burgers, L.D.; Lipps, C.; Adjaye, J.; Fürst, R.; Giebel, B.; Saul, M.J. Detailed Characterization of Small Extracellular Vesicles from Different Cell Types Based on Tetraspanin Composition by ExoView R100 Platform. Int. J. Mol. Sci. 2022, 23, 8544. [Google Scholar] [CrossRef] [PubMed]
- Breyne, K.; Ughetto, S.; Rufino-Ramos, D.; Mahjoum, S.; Grandell, E.A.; de Almeida, L.P.; Breakefield, X.O. Exogenous loading of extracellular vesicles, virus-like particles, and lentiviral vectors with supercharged proteins. Commun. Biol. 2022, 5, 485. [Google Scholar] [CrossRef] [PubMed]
- Bandu, R.; Oh, J.W.; Kim, K.P. Mass spectrometry-based proteome profiling of extracellular vesicles and their roles in cancer biology. Exp. Mol. Med. 2019, 51, 2516. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, M.; Kar, R.; Ghosh, S.; Sonar, S.; Mirgh, D.; Sivakumar, I.; Nayak, A.; Muthusamy, R. Clinical Impact of Multi-omics profiling of extracellular vesicles in cancer Liquid Biopsy. J. Liq. Biopsy 2024, 3, 100138. [Google Scholar] [CrossRef]
- Hartjes, T.A.; Mytnyk, S.; Jenster, G.W.; van Steijn, V.; van Royen, M.E. Extracellular Vesicle Quantification and Characterization: Common Methods and Emerging Approaches. Bioengineering 2019, 6, 7. [Google Scholar] [CrossRef]
- Horibe, S.; Tanahashi, T.; Kawauchi, S.; Murakami, Y.; Rikitake, Y. Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer 2018, 18, 47. [Google Scholar] [CrossRef]
- Arneth, B. Tumor Microenvironment. Medicina 2019, 56, 15. [Google Scholar] [CrossRef]
- Maheshwari, S.; Singh, A.K.; Arya, R.K.; Pandey, D.; Singh, A.; Datta, D. Exosomes: Emerging Players of Intercellular Communication in Tumor Microenvironment. Discoveries 2014, 2, e26. [Google Scholar] [CrossRef]
- Maia, J.; Caja, S.; Moraes, M.C.S.; Couto, N.; Costa-Silva, B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Petrova, V.; Annicchiarico-Petruzzelli, M.; Melino, G.; Amelio, I. The hypoxic tumour microenvironment. Oncogenesis 2018, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Han, F.; Du, Y.; Shi, H.; Zhou, W. Hypoxic microenvironment in cancer: Molecular mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2023, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Hypoxia-inducible factors: Mediators of cancer progression and targets for cancer therapy. Trends Pharmacol. Sci. 2012, 33, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P. The Role of Hypoxia-Induced Factors in Tumor Progression. Oncologist 2004, 9, 10–17. [Google Scholar] [CrossRef]
- Meng, W.; Hao, Y.; He, C.; Li, L.; Zhu, G. Exosome-orchestrated hypoxic tumor microenvironment. Mol. Cancer 2019, 18, 57. [Google Scholar] [CrossRef]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef]
- Zhou, J.; Schmid, T.; Schnitzer, S.; Brüne, B. Tumor hypoxia and cancer progression. Cancer Lett. 2006, 237, 10–21. [Google Scholar] [CrossRef]
- Chiang, A.C.; Massagué, J. Molecular Basis of Metastasis. N. Engl. J. Med. 2008, 359, 2814–2823. [Google Scholar] [CrossRef]
- Shao, C.; Yang, F.; Miao, S.; Liu, W.; Wang, C.; Shu, Y.; Shen, H. Role of hypoxia-induced exosomes in tumor biology. Mol. Cancer 2018, 17, 120. [Google Scholar] [CrossRef]
- Park, J.E.; Tan, H.S.; Datta, A.; Lai, R.C.; Zhang, H.; Meng, W.; Lim, S.K.; Sze, S.K. Hypoxic Tumor Cell Modulates Its Microenvironment to Enhance Angiogenic and Metastatic Potential by Secretion of Proteins and Exosomes. Mol. Cell. Proteom. 2010, 9, 1085–1099. [Google Scholar] [CrossRef]
- Das, A.; Sonar, S.; Dhar, R.; Subramaniyan, V. Exosomes in melanoma: Future potential for clinical theranostics. Pathol. Res. Pr. 2025, 269, 155950. [Google Scholar] [CrossRef]
- Gowda, R.; Robertson, B.M.; Iyer, S.; Barry, J.; Dinavahi, S.S.; Robertson, G.P. The role of exosomes in metastasis and progression of melanoma. Cancer Treat. Rev. 2020, 85, 101975. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-Y.; Chandramoorthy, H.C.; Mohammed, J.S.; Al-Hasnaawei, S.; Yaqob, M.; Kundlas, M.; Samikan, K.; Sahoo, S.; Sunori, S.K.; Abbas, Z.A. Exosomes as key mediators in immune and cancer cell interactions: Insights in melanoma progression and therapy. Arch. Dermatol. Res. 2025, 317, 729. [Google Scholar] [CrossRef] [PubMed]
- Roma-Rodrigues, C.; Fernandes, A.R.; Baptista, P.V. Exosome in Tumour Microenvironment: Overview of the Crosstalk between Normal and Cancer Cells. BioMed Res. Int. 2014, 2014, 179486. [Google Scholar] [CrossRef]
- Aslan, C.; Maralbashi, S.; Salari, F.; Kahroba, H.; Sigaroodi, F.; Kazemi, T.; Kharaziha, P. Tumor-derived exosomes: Implication in angiogenesis and antiangiogenesis cancer therapy. J. Cell Physiol. 2019, 234, 16885–16903. [Google Scholar] [CrossRef]
- Fasanaro, P.; D’ALessandra, Y.; Di Stefano, V.; Melchionna, R.; Romani, S.; Pompilio, G.; Capogrossi, M.C.; Martelli, F. MicroRNA-210 Modulates Endothelial Cell Response to Hypoxia and Inhibits the Receptor Tyrosine Kinase Ligand Ephrin-A3 *. J. Biol. Chem. 2008, 283, 15878–15883. [Google Scholar] [CrossRef]
- Landskroner-Eiger, S.; Moneke, I.; Sessa, W.C. miRNAs as Modulators of Angiogenesis. Cold Spring Harb. Perspect. Med. 2012, 3, a006643. [Google Scholar] [CrossRef]
- Shao, X.; Hua, S.; Feng, T.; Ocansey, D.K.W.; Yin, L. Hypoxia-Regulated TEXs and Tumor Progression: A Focus on Immune Evasion. Int. J. Mol. Sci. 2022, 23, 11789. [Google Scholar] [CrossRef]
- Guo, W.; Qiao, T.; Dong, B.; Li, T.; Liu, Q.; Xu, X. The Effect of Hypoxia-Induced Exosomes on Anti-Tumor Immunity and Its Implication for Immunotherapy. Front. Immunol. 2022, 13, 915985. [Google Scholar] [CrossRef]
- Whiteside, T.L. Tumor-Derived Exosomes and Their Role in Tumor-Induced Immune Suppression. Vaccines 2016, 4, 35. [Google Scholar] [CrossRef]
- Morrissey, S.M.; Zhang, F.; Ding, C.; Montoya-Durango, D.E.; Hu, X.; Yang, C.; Wang, Z.; Yuan, F.; Fox, M.; Zhang, H.-G.; et al. Tumor-derived exosomes drive immunosuppressive macrophages in a pre-metastatic niche through glycolytic dominant metabolic reprogramming. Cell Metab. 2021, 33, 2040–2058.e10. [Google Scholar] [CrossRef]
- Li, C.; Zhou, T.; Chen, J.; Li, R.; Chen, H.; Luo, S.; Chen, D.; Cai, C.; Li, W. The role of Exosomal miRNAs in cancer. J. Transl. Med. 2022, 20, 6. [Google Scholar] [CrossRef]
- Tian, X.; Shen, H.; Li, Z.; Wang, T.; Wang, S. Tumor-derived exosomes, myeloid-derived suppressor cells, and tumor microenvironment. J. Hematol. Oncol. 2019, 12, 84. [Google Scholar] [CrossRef]
- Bhardwaj, V.; Ansell, S.M. Modulation of T-cell function by myeloid-derived suppressor cells in hematological malignancies. Front. Cell Dev. Biol. 2023, 11, 1129343. [Google Scholar] [CrossRef]
- Olejarz, W.; Dominiak, A.; Żołnierzak, A.; Kubiak-Tomaszewska, G.; Lorenc, T. Tumor-Derived Exosomes in Immunosuppression and Immunotherapy. J. Immunol. Res. 2020, 2020, 6272498. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. The effect of tumor-derived exosomes on immune regulation and cancer immunotherapy. Future Oncol. 2017, 13, 2583–2592. [Google Scholar] [CrossRef] [PubMed]
- Gulley, J.L.; Schlom, J.; Barcellos-Hoff, M.H.; Wang, X.J.; Seoane, J.; Audhuy, F.; Lan, Y.; Dussault, I.; Moustakas, A. Dual inhibition of TGF-β and PD-L1: A novel approach to cancer treatment. Mol. Oncol. 2022, 16, 2117–2134. [Google Scholar] [CrossRef] [PubMed]
- Stefańska, K.; Józkowiak, M.; Volponi, A.A.; Shibli, J.A.; Golkar-Narenji, A.; Antosik, P.; Bukowska, D.; Piotrowska-Kempisty, H.; Mozdziak, P.; Dzięgiel, P.; et al. The Role of Exosomes in Human Carcinogenesis and Cancer Therapy—Recent Findings from Molecular and Clinical Research. Cells 2023, 12, 356. [Google Scholar] [CrossRef]
- Jiang, J.; Li, J.; Zhou, X.; Zhao, X.; Huang, B.; Qin, Y. Exosomes Regulate the Epithelial–Mesenchymal Transition in Cancer. Front. Oncol. 2022, 12, 864980. [Google Scholar] [CrossRef]
- Yang, C.; Dou, R.; Wei, C.; Liu, K.; Shi, D.; Zhang, C.; Liu, Q.; Wang, S.; Xiong, B. Tumor-derived exosomal microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis. Mol. Ther. 2021, 29, 2088–2107. [Google Scholar] [CrossRef]
- Shen, Y.-Q.; Sun, L.; Wang, S.-M.; Zheng, X.-Y.; Xu, R. Exosomal integrins in tumor progression, treatment and clinical prediction (Review). Int. J. Oncol. 2024, 65, 118. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Pan, W.; Miao, Q.; Yin, W.; Li, X.; Ye, W.; Zhang, D.; Deng, L.; Zhang, J.; Chen, M. The role and clinical applications of exosomes in cancer drug resistance. Cancer Drug Resist. 2024, 7, 43. [Google Scholar] [CrossRef]
- Steinbichler, T.B.; Dudás, J.; Skvortsov, S.; Ganswindt, U.; Riechelmann, H.; Skvortsova, I.I. Therapy resistance mediated by exosomes. Mol. Cancer 2019, 18, 58. [Google Scholar] [CrossRef]
- Das, A.; Sonar, S.; Kalele, K.; Subramaniyan, V. Fruit exosomes: A sustainable green cancer therapeutic. Sustain. Food Technol. 2024, 3, 145–160. [Google Scholar] [CrossRef]
- Zhang, Z.; Lin, F.; Wu, W.; Jiang, J.; Zhang, C.; Qin, D.; Xu, Z. Exosomal microRNAs in lung cancer: A narrative review. Transl. Cancer Res. 2024, 13, 3090–3105. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Yu, S.; Zhou, L.; Shi, M.; Hu, Y.; Xu, X.; Shen, B.; Liu, S.; Yan, D.; Feng, J. Cisplatin-resistant lung cancer cell–derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100–5p-dependent manner. Int. J. Nanomed. 2017, 12, 3721–3733. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Bai, X.; Ni, J.; Zhang, H.; Duan, W.; Graham, P.; Li, Y. Exosomes and breast cancer drug resistance. Cell Death Dis. 2020, 11, 987. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Jiang, D.; Bai, S.; Zhang, X.; Kang, Y. Research progress of exosomes in drug resistance of breast cancer. Front. Bioeng. Biotechnol. 2024, 11, 1214648. [Google Scholar] [CrossRef]
- Mishra, A.; Bharti, P.S.; Rani, N.; Nikolajeff, F.; Kumar, S. A tale of exosomes and their implication in cancer. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188908. [Google Scholar] [CrossRef]
- Wu, Y.; Cao, Y.; Chen, L.; Lai, X.; Zhang, S.; Wang, S. Role of Exosomes in Cancer and Aptamer-Modified Exosomes as a Promising Platform for Cancer Targeted Therapy. Biol. Proced. Online 2024, 26, 15. [Google Scholar] [CrossRef]
- Dhar, R.; Kumarasamy, V.; Subramaniyan, V. Signature of exosomes in cancer translational medicine. Int. J. Surg. 2025, 111, 4138–4139. [Google Scholar] [CrossRef]
- Luo, S.; Chen, J.; Xu, F.; Chen, H.; Li, Y.; Li, W. Dendritic Cell-Derived Exosomes in Cancer Immunotherapy. Pharmaceutics 2023, 15, 2070. [Google Scholar] [CrossRef] [PubMed]
- André, F.; Chaput, N.; Schartz, N.E.; Flament, C.; Aubert, N.; Bernard, J.; Lemonnier, F.; Raposo, G.; Escudier, B.; Hsu, D.H.; et al. Exosomes as potent cell-free peptide-based vaccine. I. Dendritic cell-derived exosomes transfer functional MHC class I/peptide complexes to dendritic cells. J. Immunol. 2004, 172, 2126–2136. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, J.; Huang, Y.; Zhang, X.; Zhang, G.; Kong, S.; Gao, J.; Zhang, X.; Ding, B. Drug Delivery Systems Based on Dendritic-Cell-Derived Exosomes. Pharmaceutics 2025, 17, 326. [Google Scholar] [CrossRef] [PubMed]
- Shahir, M.; Hashemi, S.M.; Asadirad, A.; Varahram, M.; Kazempour-Dizaji, M.; Folkerts, G.; Garssen, J.; Adcock, I.; Mortaz, E. Effect of mesenchymal stem cell-derived exosomes on the induction of mouse tolerogenic dendritic cells. J. Cell. Physiol. 2020, 235, 7043–7055. [Google Scholar] [CrossRef]
- Aldahlawi, A.M.; Abdullah, S.T. Dendritic Cell-Based Immunotherapies and their Potential use in Colorectal Cancer Immunotherapy. J. Microsc. Ultrastruct. 2021, 10, 107–113. [Google Scholar] [CrossRef]
- Ghorbaninezhad, F.; Alemohammad, H.; Najafzadeh, B.; Masoumi, J.; Shadbad, M.A.; Shahpouri, M.; Saeedi, H.; Rahbarfarzam, O.; Baradaran, B. Dendritic cell-derived exosomes: A new horizon in personalized cancer immunotherapy? Cancer Lett. 2023, 562, 216168. [Google Scholar] [CrossRef]
- Dutta, A. Exosomes-based cell-free cancer therapy: A novel strategy for targeted therapy. Immunol. Med. 2020, 44, 116–123. [Google Scholar] [CrossRef]
- Wei, B.; Huang, H.; Cao, Q.; Song, X.; Zhang, Z. Bibliometric and visualized analysis of the applications of exosomes based drug delivery. Biomed. Pharmacother. 2024, 176, 116803. [Google Scholar] [CrossRef] [PubMed]
- Piao, Y.J.; Kim, H.S.; Moon, W.K. Noninvasive Photoacoustic Imaging of Dendritic Cell Stimulated with Tumor Cell-Derived Exosome. Mol. Imaging Biol. 2019, 22, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Rehman, F.U.; Liu, Y.; Zheng, M.; Shi, B. Exosomes based strategies for brain drug delivery. Biomaterials 2022, 293, 121949. [Google Scholar] [CrossRef] [PubMed]
- Xi, X.-M.; Chen, M.; Xia, S.-J.; Lu, R. Drug loading techniques for exosome-based drug delivery systems. Pharmazie 2021, 76, 61–67. [Google Scholar] [CrossRef]
- Kumar, D.N.; Chaudhuri, A.; Kumar, D.; Singh, S.; Agrawal, A.K. Impact of the Drug Loading Method on the Drug Distribution and Biological Efficacy of Exosomes. AAPS Pharmscitech 2023, 24, 166. [Google Scholar] [CrossRef]
- Morse, M.A.; Garst, J.; Osada, T.; Khan, S.; Hobeika, A.; Clay, T.M.; Valente, N.; Shreeniwas, R.; Sutton, M.A.; Delcayre, A.; et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J. Transl. Med. 2005, 3, 9. [Google Scholar] [CrossRef]
- Yu, S.; Sha, H.; Qin, X.; Chen, Y.; Li, X.; Shi, M.; Feng, J. EGFR E746-A750 deletion in lung cancer represses antitumor immunity through the exosome-mediated inhibition of dendritic cells. Oncogene 2020, 39, 2643–2657. [Google Scholar] [CrossRef]
- Lu, Z.; Zuo, B.; Jing, R.; Gao, X.; Rao, Q.; Liu, Z.; Qi, H.; Guo, H.; Yin, H. Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J. Hepatol. 2017, 67, 739–748. [Google Scholar] [CrossRef]
- Schioppa, T.; Gaudenzi, C.; Zucchi, G.; Piserà, A.; Vahidi, Y.; Tiberio, L.; Sozzani, S.; Del Prete, A.; Bosisio, D.; Salvi, V. Extracellular vesicles at the crossroad between cancer progression and immunotherapy: Focus on dendritic cells. J. Transl. Med. 2024, 22, 691. [Google Scholar] [CrossRef]
- Elsayed, R.; Elashiry, M.; Tran, C.; Yang, T.; Carroll, A.; Liu, Y.; Hamrick, M.; Cutler, C.W. Engineered Human Dendritic Cell Exosomes as Effective Delivery System for Immune Modulation. Int. J. Mol. Sci. 2023, 24, 11306. [Google Scholar] [CrossRef]
- Barnwal, A.; Gaur, V.; Sengupta, A.; Tyagi, W.; Das, S.; Bhattacharyya, J. Tumor Antigen-Primed Dendritic Cell-Derived Exosome Synergizes with Colony Stimulating Factor-1 Receptor Inhibitor by Modulating the Tumor Microenvironment and Systemic Immunity. ACS Biomater. Sci. Eng. 2023, 9, 6409–6424. [Google Scholar] [CrossRef]
- Tuluwengjiang, G.; Rasulova, I.; Ahmed, S.; Kiasari, B.A.; Sârbu, I.; Ciongradi, C.I.; Omar, T.M.; Hussain, F.; Jawad, M.J.; Castillo-Acobo, R.Y.; et al. Dendritic cell-derived exosomes (Dex): Underlying the role of exosomes derived from diverse DC subtypes in cancer pathogenesis. Pathol. Res. Pr. 2024, 254, 155097. [Google Scholar] [CrossRef] [PubMed]
- Al-Hawary, S.I.S.; Almajidi, Y.Q.; Bansal, P.; Ahmad, I.; Kaur, H.; Hjazi, A.; Deorari, M.; Zwamel, A.H.; Hamzah, H.F.; Mohammed, B.A. Dendritic cell-derived exosome (DEX) therapy for digestive system cancers: Recent advances and future prospect. Pathol. Res. Pr. 2024, 257, 155288. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, X.; Qin, J.; Xue, Y.; Zhang, P.; Liu, Y.; Chen, M.; Zhu, G.; Song, X.; Cheng, L.; et al. Locoregional Immune Checkpoint Blockade and Remodeling of Lymph Nodes by Engineered Dendritic Cell-Derived Exosomes for Suppressing Tumor Progression and Metastasis. Adv. Sci. 2025, 12, e2500139. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Pu, Y.; Bao, Y.; He, S. Investigation of Potential Molecular Biomarkers for Diagnosis and Prognosis of AFP-Negative HCC. Int. J. Gen. Med. 2021, 14, 4369–4380. [Google Scholar] [CrossRef]
- Chen, T.; Dai, X.; Dai, J.; Ding, C.; Zhang, Z.; Lin, Z.; Hu, J.; Lu, M.; Wang, Z.; Qi, Y.; et al. AFP promotes HCC progression by suppressing the HuR-mediated Fas/FADD apoptotic pathway. Cell Death Dis. 2020, 11, 822. [Google Scholar] [CrossRef]
- Shi, S.; Wang, L.; Wang, C.; Xu, J.; Niu, Z. Serum-derived exosomes function as tumor antigens in patients with advanced hepatocellular carcinoma. Mol. Immunol. 2021, 134, 210–217. [Google Scholar] [CrossRef]
- Chang, C.; Pei, Y.; Zhang, C.; Zhang, W.; Qin, Y.; Shi, S. Combination therapy with dendritic cell loaded-exosomes supplemented with PD-1 inhibition at different time points have superior antitumor effect in hepatocellular carcinoma. Cancer Immunol. Immunother. 2023, 72, 3727–3738. [Google Scholar] [CrossRef]
- Escudier, B.; Dorval, T.; Chaput, N.; André, F.; Caby, M.-P.; Novault, S.; Flament, C.; Leboulaire, C.; Borg, C.; Amigorena, S.; et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: Results of thefirst phase I clinical trial. J. Transl. Med. 2005, 3, 10. [Google Scholar] [CrossRef]
- Morales, R.-T.T.; Ko, J. Future of Digital Assays to Resolve Clinical Heterogeneity of Single Extracellular Vesicles. ACS Nano 2022, 16, 11619–11645. [Google Scholar] [CrossRef]
- Koh, H.B.; Kim, H.J.; Kang, S.-W.; Yoo, T.-H. Exosome-Based Drug Delivery: Translation from Bench to Clinic. Pharmaceutics 2023, 15, 2042. [Google Scholar] [CrossRef]
- Dhar, R.; Devi, A.; Patil, S.; Tovani-Palone, M.R. Exosomes in cancer therapy: Advances and current challenges. Electron J Gen Med. 2023, 20, em524. [Google Scholar] [CrossRef]
- Kashkoulinejad Kouhi, T. Exosome-mediated communication between T cells and dendritic cells: Implications for therapeutic strategies. Cytokine 2025, 189, 156914. [Google Scholar] [CrossRef] [PubMed]
- Hazrati, A.; Soudi, S.; Malekpour, K.; Mahmoudi, M.; Rahimi, A.; Hashemi, S.M.; Varma, R.S. Immune cells-derived exosomes function as a double-edged sword: Role in disease progression and their therapeutic applications. Biomark Res. 2022, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Wang, Y.; Wang, S.; Liu, B. Nanomaterials assisted exosomes isolation and analysis towards liquid biopsy. Mater. Today Bio 2022, 16, 100371. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef]
- Yang, X.; Chen, J.; Wang, N.; Liu, Z.; Li, Y. Clinical use of dendritic cell-derived exosomes for hepatocellular carcinoma immunotherapy: How far we are? J. Hepatol. 2018, 69, 984–986. [Google Scholar] [CrossRef]
- Chen, J.; Tan, Q.; Yang, Z.; Chen, W.; Zhou, E.; Li, M.; Deng, J.; Wu, Y.; Liu, J.; Xu, J.; et al. Dendritic Cell Derived-Extracellular Vesicles Engineered to Express Interleukin-12 and Anti-CTLA-4 on Their Surface for Combinational Cancer Immunotherapy. J. Extracell. Vesicles 2025, 14, e70068. [Google Scholar] [CrossRef]
| Exosome for Liquid Biopsy for Cancer | ||
|---|---|---|
| Exosome Source | Clinical Significance | References |
| Blood (Plasma/Serum) | Blood is a complex biofluid containing exosomes from various cell types. Plasma and serum are commonly used for exosome isolation due to their accessibility. However, they also contain abundant proteins like albumin and lipoproteins that can contaminate exosome preparations. | [65,66,67] |
| Urine | Exosomes are present in urine and can be valuable for studying kidney function and urological diseases. | [65] |
| Saliva | Saliva offers a non-invasive source of exosomes, useful for oral and systemic disease diagnostics. | [66] |
| Breast Milk | Breast milk is rich in exosomes, which play a role in infant immunity and development. | [65] |
| Amniotic Fluid | Exosomes in amniotic fluid can provide insights into fetal development and pregnancy-related complications. | [65] |
| Cerebrospinal Fluid | CSF-derived exosomes are valuable for studying neurological disorders. | [65] |
| Ascites Fluid | This fluid, found in the abdominal cavity of patients with certain cancers, contains exosomes that can provide information about the tumor microenvironment. | [65] |
| Sweat | Biomarker source | [68] |
| Tear | Biomarker source | [69] |
| Exosomes sources for therapeutic development | ||
| Exosome source | Clinical significance | References |
| Plant cell | Use as a therapeutic tool | [70] |
| Immune cell | Use as a therapeutic tool | [71] |
| Stem cell | Use as a therapeutic tool | [72] |
| Tumor cell | Use as a therapeutic tool (not recommended due to its enrichment of oncogenic cargo) | [73] |
| Methods | Principle | Procedure | Advantages | Disadvantages | References |
|---|---|---|---|---|---|
| Differential Ultracentrifugation | This method involves multiple rounds of centrifugation at progressively higher speeds. Each step pellets particles of a certain size and density, allowing for the enrichment of exosomes. |
| Can be used for large volumes, relatively low cost, and with no need for additional chemical reagents | Can be time-consuming and may result in low purity due to the co-purification of other molecules. Exosomes might be damaged during the process | [67,82,83] |
| Density Gradient Ultracentrifugation | This method separates particles based on their buoyant density. The sample is layered on top of a density gradient medium (e.g., sucrose or iodixanol) and centrifuged until particles reach their equilibrium density. |
| Higher purity compared to differential ultracentrifugation | More complex and time-consuming than differential ultracentrifugation, as well as low yields | [77,84,85] |
| Size Exclusion Chromatography (SEC) | SEC separates molecules based on their size as they pass through a porous matrix. A column is packed with a stationary phase consisting of porous beads. Smaller molecules enter the pores and take a longer, more tortuous path, eluting later. Larger particles, like exosomes, cannot enter the pores and elute earlier. |
| SEC can separate exosomes based on their size Gentle method that preserves exosome integrity | A limited quantity of EVs recovered. | [78,86,87] |
| Filtration | Filtration methods use filters with defined pore sizes to separate particles based on size. Ultrafiltration membranes with specific molecular weight cut-offs are commonly used to enrich exosomes |
| Relatively simple and rapid. Can be used to concentrate exosome samples | Membrane clogging can be an issue. Exosomes may be damaged by shear forces during filtration. | [77,80,88] |
| Precipitation with Polymers | This method involves using polymers to reduce the solubility of exosomes in solution. The polymers bind to water molecules, effectively forcing exosomes to aggregate and precipitate out of the solution |
| Simple and relatively inexpensive. Does not require specialized equipment like ultracentrifuges | Co-precipitation of other non-exosomal contaminants, such as proteins and polymeric materials, is unavoidable. May be less pure compared to other methods like ultracentrifugation or size exclusion chromatography. The choice of polymer and precipitation conditions can affect exosome yield and purity. | [89,90,91] |
| Immunoaffinity Capture | This method uses the specific binding between an antibody and an exosomal surface protein to selectively capture exosomes. Antibodies against specific exosomal markers (e.g., CD9, CD63, CD81) are immobilized on a solid support (e.g., beads, columns, or microplates). When a sample containing exosomes is incubated with the antibody-conjugated support, exosomes expressing the target protein are captured. |
| High specificity for exosomes expressing the target protein. Can be used to isolate specific subpopulations of exosomes. | Requires knowledge of exosomal surface markers. Antibody availability and cost can be limiting factors Elution steps can result in sample loss, making the method less suitable for downstream analysis. | [78,91,92]. |
| Microfluidic | Microfluidic exosome separation leverages the unique physical and chemical properties of exosomes in a controlled microenvironment. These properties include size, surface markers, deformability, and electrical characteristics. | Size-based separation: This method separates exosomes based on their size using microchannels with precisely controlled dimensions. Dynamic methodologies: Separation based on other properties, such as electrical characteristics. | Enhanced purity: Microfluidic systems can achieve higher purity compared to ultracentrifugation. Cost-effectiveness: Microfluidic technologies offer a cost-effective solution for exosome isolation. | Technological immaturity: Exosome research using microfluidics is still in its early stages. Lack of standardization: The absence of standardized methods for exosome separation can lead to suboptimal inter-laboratory correlation and difficulty in comparing studies. Challenges in isolating exosomes: The inherent heterogeneity of exosomes and the complexity of biofluids pose significant challenges for their isolation. | [93,94,95] |
| Characterization Types | Importance | Reference |
|---|---|---|
| Physical Characterization | ||
| Nanoparticle Tracking Analysis | NTA is utilized for concentration measurements and size distribution curves of exosomes. NTA operates by tracking the Brownian motion of individual particles in a sample using light scattering. By measuring the rate of movement, the software calculates the hydrodynamic diameter of each particle using the Stokes-Einstein equation. | [96,97,98] |
| Dynamic Light Scattering | DLS, as well as NTA, fluorescence signals, and flow cytometry, are optical methods used to characterize vesicles. | [99] |
| Electron Microscopy (TEM/SEM): | Traditional detection techniques utilized to quantify the isolated exosomes include scanning electron microscopes and transmission electron microscopes. | [98] |
| NanoFCM | Nano-flow cytometry is a quantitative and qualitative measurement of single EVs, like exosomes (it is applicable for cell culture suspension and body fluid). | [100] |
| Super-Resolution Microscopy (SRM) | SRM works with Oxford Nano Imaging (ONI) and supports the decoding of exosomes’ morphology, tracking of EV uptake, cargo composition, and heterogeneity. | [101,102] |
| Characterization of Molecules | ||
| Western Blotting | Western blotting can be performed using exosomal markers such as CD9, CD63, and CD81. | [98] |
| Flow Cytometry | It is one of the optical methods for characterizing vesicles. It can identify and characterize the cytoplasmic or surface proteins of EVs. | [99] |
| Exoview Chip | This is a microarray-based method where affinity-based antibodies are captured for exosomes’ surface markers, such as CD9, CD63, and CD81. This capture visualization by the supporter of ExoViewR100 | [103,104] |
| Analysis of Proteomes and Genomes | Mass Spectrometry, RNA-seq: These techniques are used for in-depth analysis of exosome contents. | [105,106] |
| Quantification and Evaluation of Purity | Quantifying protein (BCA/Bradford test): These are standard methods for determining the overall protein concentration in the exosome sample.
| [107] |
| Functional Analysis | Studies on uptake (cell interaction tests and fluorescent labeling): These assays help to understand how exosomes interact with and are taken up by target cells.
| [108] |
| Clinical Trials ID | Status | Cancer Types | Clinical Significant | Sponsor |
|---|---|---|---|---|
| NCT01159288 | Completed | advanced non-small cell lung cancer (NSCLC) | Trial of a Vaccination With Tumor Antigen-loaded Dendritic Cell-derived Exosomes (CSET 1437) | Gustave Roussy, Cancer Campus, Grand Paris |
| Key insights from the clinical trial: Phase: Phase II Objective: To assess the efficacy of IFN-γ-DEXs as maintenance immunotherapy after platinum-based chemotherapy in advanced NSCLC. Patient Groups Number Enrolled/Treated: 26 enrolled/22 treated Diagnosis: Advanced (stage IIIB/IV) unresectable NSCLC Key Criteria: HLA-A2 positive, no progression after 4 cycles of platinum-based chemotherapy. Baseline: 64% had adenocarcinoma, 82% had stage IV disease. Interventions Treatment: IFN-γ-matured dendritic cell-derived exosomes (IFN-γ-DEXs) loaded with MAGE tumor antigens. Dosing: 0.13 μg MHC class II molecules per injection, administered intravenously. Schedule: Weekly for the first 4 vaccinations, followed by boosters administered every 2 weeks for 3 boosters, then monthly boosters. Adjunct Therapy: Oral metronomic cyclophosphamide (50 mg/day) to inhibit T-regs. Endpoint Primary: Progression-free survival (PFS) rate at 4 months. Secondary: Overall survival (OS), time to progression (TTP), safety, and immunological response. Key Findings Efficacy (Primary Endpoint): Not met. Only 32% (7/22) of patients were progression-free at 4 months (target was >50%). Median PFS was 2.2 months. Overall Survival (OS): Median OS was 15 months. Safety: Treatment was well-tolerated. One patient experienced a Grade 3 dose-limiting hepatotoxicity. Immunological Response: No MAGE-specific CD8+ T-cell responses were detected. An increase in NKp30-dependent Natural Killer (NK) cell function was observed and was correlated with longer PFS. Clinical benefit was associated with higher levels of the NK ligand BAG6 on the exosomes. | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhar, R.; Sonar, S.; Das, A.; Tajul Akmal, N.A.S.; Hawa Jasni, A.; RMT Balasubramaniam, V.; Narayanan, K.; Subramaniyan, V. Dendritic Cell-Derived Exosomes: Next Generation of Cancer Immunotherapy. Biomedicines 2025, 13, 2497. https://doi.org/10.3390/biomedicines13102497
Dhar R, Sonar S, Das A, Tajul Akmal NAS, Hawa Jasni A, RMT Balasubramaniam V, Narayanan K, Subramaniyan V. Dendritic Cell-Derived Exosomes: Next Generation of Cancer Immunotherapy. Biomedicines. 2025; 13(10):2497. https://doi.org/10.3390/biomedicines13102497
Chicago/Turabian StyleDhar, Rajib, Swarup Sonar, Asmit Das, Nur Aliaa Sorfina Tajul Akmal, Ainil Hawa Jasni, Vinod RMT Balasubramaniam, Kumaran Narayanan, and Vetriselvan Subramaniyan. 2025. "Dendritic Cell-Derived Exosomes: Next Generation of Cancer Immunotherapy" Biomedicines 13, no. 10: 2497. https://doi.org/10.3390/biomedicines13102497
APA StyleDhar, R., Sonar, S., Das, A., Tajul Akmal, N. A. S., Hawa Jasni, A., RMT Balasubramaniam, V., Narayanan, K., & Subramaniyan, V. (2025). Dendritic Cell-Derived Exosomes: Next Generation of Cancer Immunotherapy. Biomedicines, 13(10), 2497. https://doi.org/10.3390/biomedicines13102497










