Preclinical Diagnosis of Type 1 Diabetes: Reality or Utopia
Abstract
1. Introduction
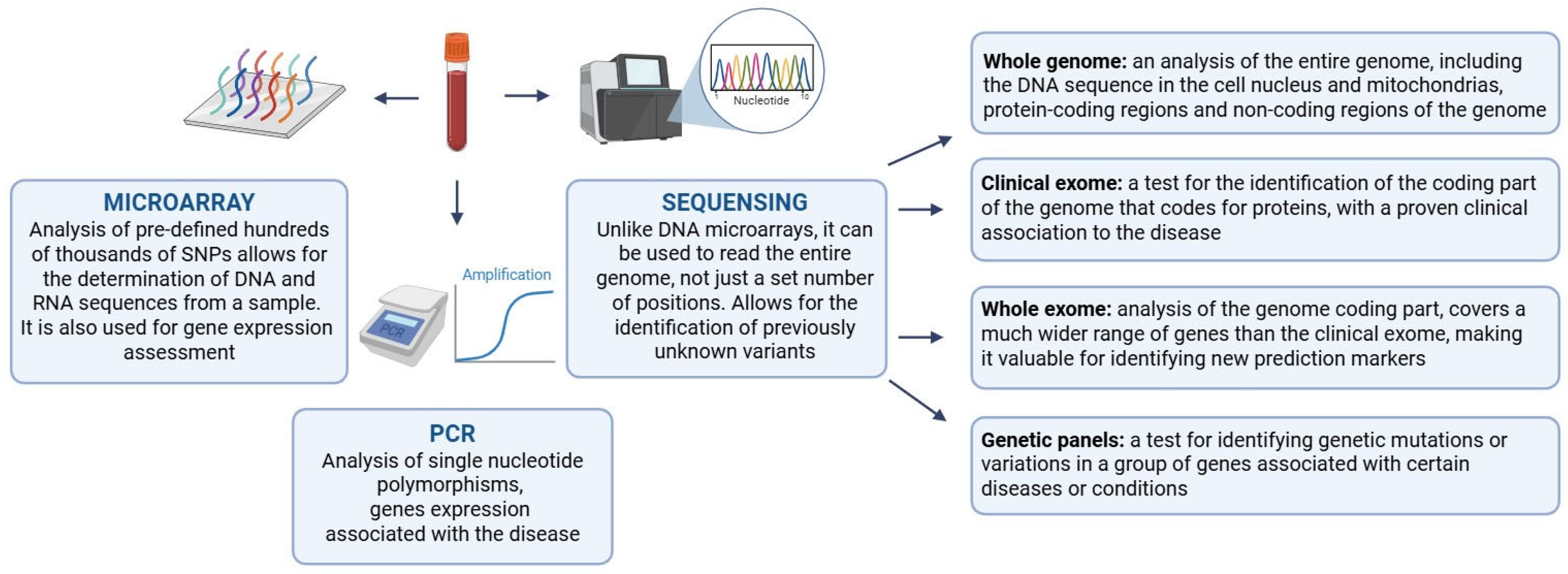
2. T1D Classic Biomarkers
2.1. Assessment of Genetic Predisposition to T1D
| PGS | Number of Variants | Source of Variant Associations (GWAS) | Development/ Training | PGS Evaluation | Original Genome | Reference Build |
|---|---|---|---|---|---|---|
| PGS000022 | 37 | — | — | African: 33.3%; Hispanic or Latin American: 33.3%; European: 33.3%; 3 Sample Sets | NR | [41] |
| PGS000869 | 48 | — | — | European, 29,652 individuals | hg19 | [42] |
| PGS001817 | 825 | — | European, 391,124 individuals | European: 37.5%; African: 25%; Eastern asian: 12.5%; Middle Eastern: 12.5%; South Asian: 12.5%; 8 Sample Sets | GRCh37 | [33] |
| PGS002025 | 1068 | — | European, 391,124 individuals | European: 37.5%; African: 25%; Eastern asian: 12.5%; Middle Eastern: 12.5%; South Asian: 12.5%; 8 Sample Sets | GRCh37 | [33] |
| PGS003993 | 63,162 | European: 89.3%; African: 7.1%; Additional Diverse Ancestries: 3.6%; 59,527 individuals (100%) | European: 100%; 804 individuals (100%) | European: 80%; South Asian: 20%; 5 Sample Sets | GRCh38 | [43] |
| PGS004009 | 4031 | European: 89.3%; African: 7.1%; Additional Diverse Ancestries: 3.6%; 59,527 individuals | European, 804 individuals | European: 80%; South Asian: 20%; 5 Sample Sets | GRCh38 | [43] |
| PGS004020 | 6682 | European: 89.3%; African: 7.1%; Additional Diverse Ancestries: 3.6%; 59,527 individuals | European, 804 individuals | European: 80%; South Asian: 20%; 5 Sample Sets | GRCh38 | [43] |
| PGS004035 | 56,562 | European: 89.3%; African: 7.1%; Additional Diverse Ancestries: 3.6%; 59,527 individuals | European, 804 individuals | European: 80%; South Asian: 20%; 5 Sample Sets | GRCh38 | [43] |
| PGS004063 | 56,288 | European: 89.3%; African: 7.1%; Additional Diverse Ancestries: 3.6%; 59,527 individuals | European, 804 individuals | European: 80%; South Asian: 20%; 5 Sample Sets | GRCh38 | [43] |
| PGS004078 | 56,288 | European: 89.3%; African: 7.1%; Additional Diverse Ancestries: 3.6%; 59,527 individuals | European, 804 individuals | European: 80%; South Asian: 20%; 5 Sample Sets | GRCh38 | [43] |
| PGS004093 | 61,651 | European: 89.3%; African: 7.1%; Additional Diverse Ancestries: 3.6%; 59,527 individuals | European, 804 individuals | European: 80%; South Asian: 20%; 5 Sample Sets | GRCh38 | [43] |
| PGS004102 | 61,651 | European: 89.3%; African: 7.1%; Additional Diverse Ancestries: 3.6%; 59,527 individuals | European, 804 individuals | European: 80%; South Asian: 20%; 5 Sample Sets | GRCh38 | [43] |
| PGS004117 | 131 | European: 89.3%; African: 7.1%; Additional Diverse Ancestries: 3.6%; 59,527 individuals | European, 804 individuals | European: 80%; South Asian: 20%; 5 Sample Sets | GRCh38 | [43] |
| PGS004132 | 354 | European: 89.3%; African: 7.1%; Additional Diverse Ancestries: 3.6%; 59,527 individuals | European, 804 individuals | European: 80%; South Asian: 20%; 5 Sample Sets | GRCh38 | [43] |
| PGS004162 | 62,645 | European: 89.3%; African: 7.1%; Additional Diverse Ancestries: 3.6%; 59,527 individuals | European, 804 individuals | European: 80%; South Asian: 20%; 5 Sample Sets | GRCh38 | [43] |
| PGS004171 | 520 | — | European, 200,000 individuals | European: 100%; 1 Sample Set | GRCh37 | [44] |
| PGS004172 | 70 | — | European, 200,000 individuals | European: 100%; 1 Sample Set | GRCh37 | [44] |
| PGS004173 | 295 | — | European, 200,000 individuals | European: 100%; 1 Sample Set | GRCh37 | [44] |
| PGS004174 | 49 | — | European, 200,000 individuals | European: 100%; 1 Sample Set | GRCh37 | [44] |
| PGS004175 | 315 | — | European, 200,000 individuals | European: 100%; 1 Sample Set | GRCh37 | [44] |
| PGS004874 | 56,916 | European: 89.3%; African: 7.1%; Additional Diverse Ancestries: 3.6%; 59,527 individuals | European, 404 individuals | European: 100%; 8 Sample Sets | GRCh37 | [45] |
2.2. Limitations and Prospects of T1D Genetic Screening
| SNP | Region | Gene | Autoimmune Disease | Source |
|---|---|---|---|---|
| rs6679677 | 1p13.2 | PTPN22 | THY, PSOR, T1D, JIA | [28] |
| rs62324212 | 4q27 | IL21 | THY, AS, CEL, CVID, UC, T1D, JIA, CD | [28] |
| rs706778 | 10p15.1 | IL2RA | THY, AS, PSOR, CEL, T1D, JIA | [28] |
| rs1689510 | 12q13.2 | SUOX | PSOR, T1D | [28] |
| rs2641348 | 1p12 | NOTCH2 | CD, T1D | [30] |
| rs78037977 | 1q24.3 | FASLG | Asthma, vitiligo, allergic sensitization, T1D | [30] |
| rs7582694 | 2q32.2-q32.3 | STAT4 | SLE, hypothyroidism, CEL, RA, T1D | [30] |
| rs10213692 | 5q11.2 | ANKRD55/IL6ST | Body fat percentage, T1D | [30] |
| rs212408 | 6q25.3 | TAGAP | RA, CD, MS, T1D | [30] |
| rs10245867 | 7p15.2-p15.1 | JAZF1 | MS, CD, eczema, T1D | [30] |
| rs11033048 | 11p13 | SLC1A2 | Eczema, hay fever, MS, SLE, T1D | [30] |
| rs968567 | 11q12.2 | FADS2 | Vitiligo, T1D | [30] |
| rs911263 | 14q24.1 | RAD51B | RA, T1D | [30] |
| rs1052553 | 17q21.31 | MAPT | PBC, SLE, RA, T1D | [30] |
| rs10795791 | 10p15.1 | IL2RA | RA, T1D | [49] |
| rs11203203 | 21q22.3 | UBASH3A | RA, T1D, Vitiligo | [49] |
| rs12720356 | 19p13.2 | TYK2 | CRD, PSOR, T1D, UC | [49] |
| rs12927355 | 16p13.13 | CLEC16A | MS, T1D | [49] |
| rs2076530 | 6p21.32 | BTNL2 | RA, AS, T1D | [53] |
| rs3129953 | 6p21.32 | BTNL2 | T1D, THY | [53] |
| rs887464 | 6p21.33 | PSORS1C3 | RA, AS, T1D, THY | [53] |
| rs12708716 | 16p13.13 | CLEC16A | MS, Primary Billiary Cirrosis, T1D | [54] |
| rs2292239 | 12q13.2 | ERBB3 | T1D, allergic sensitization | [54] |
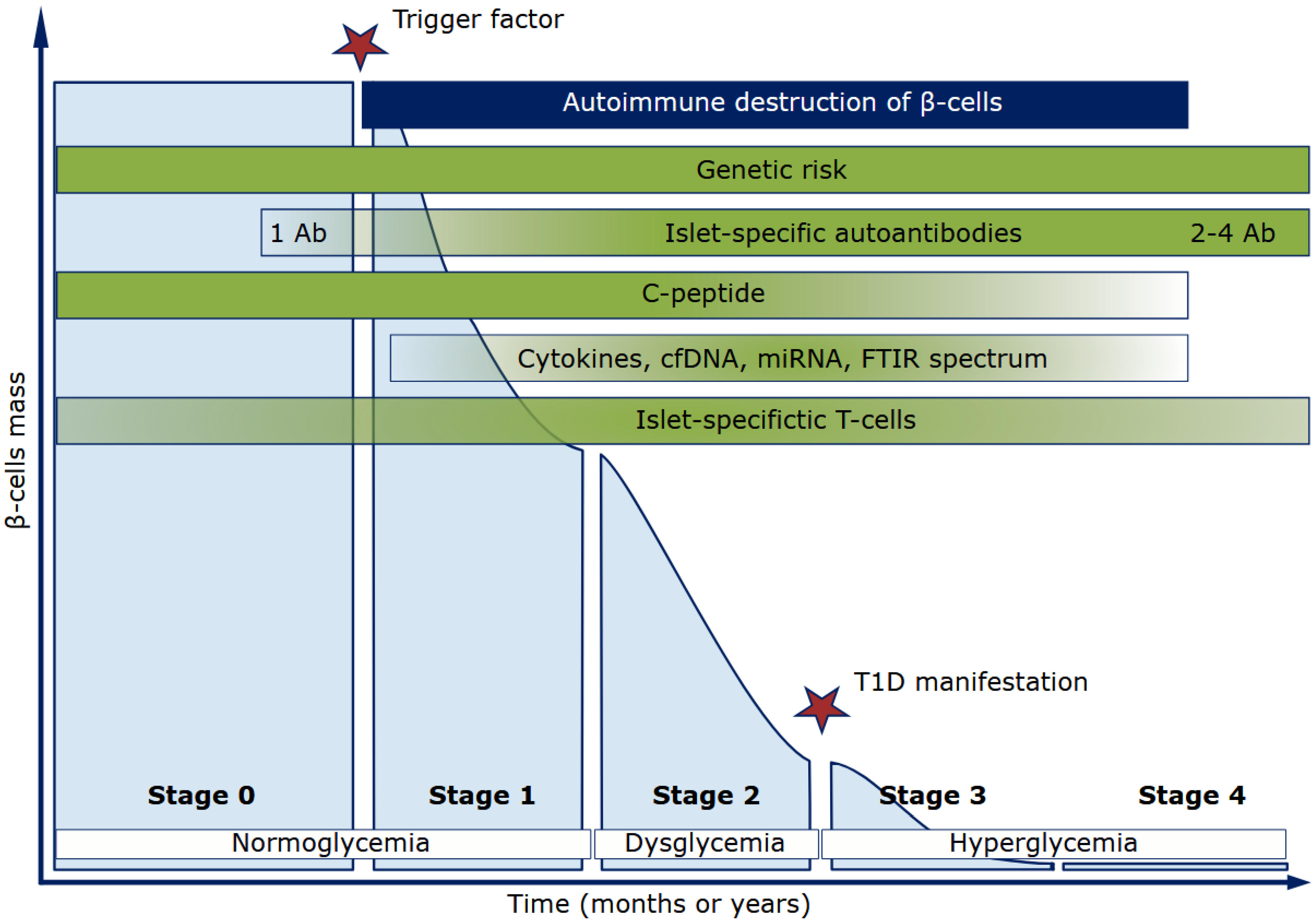
2.3. Markers of the T1D Autoimmune Process
2.3.1. Autoantibodies to Islet Antigens
2.3.2. C-Peptide
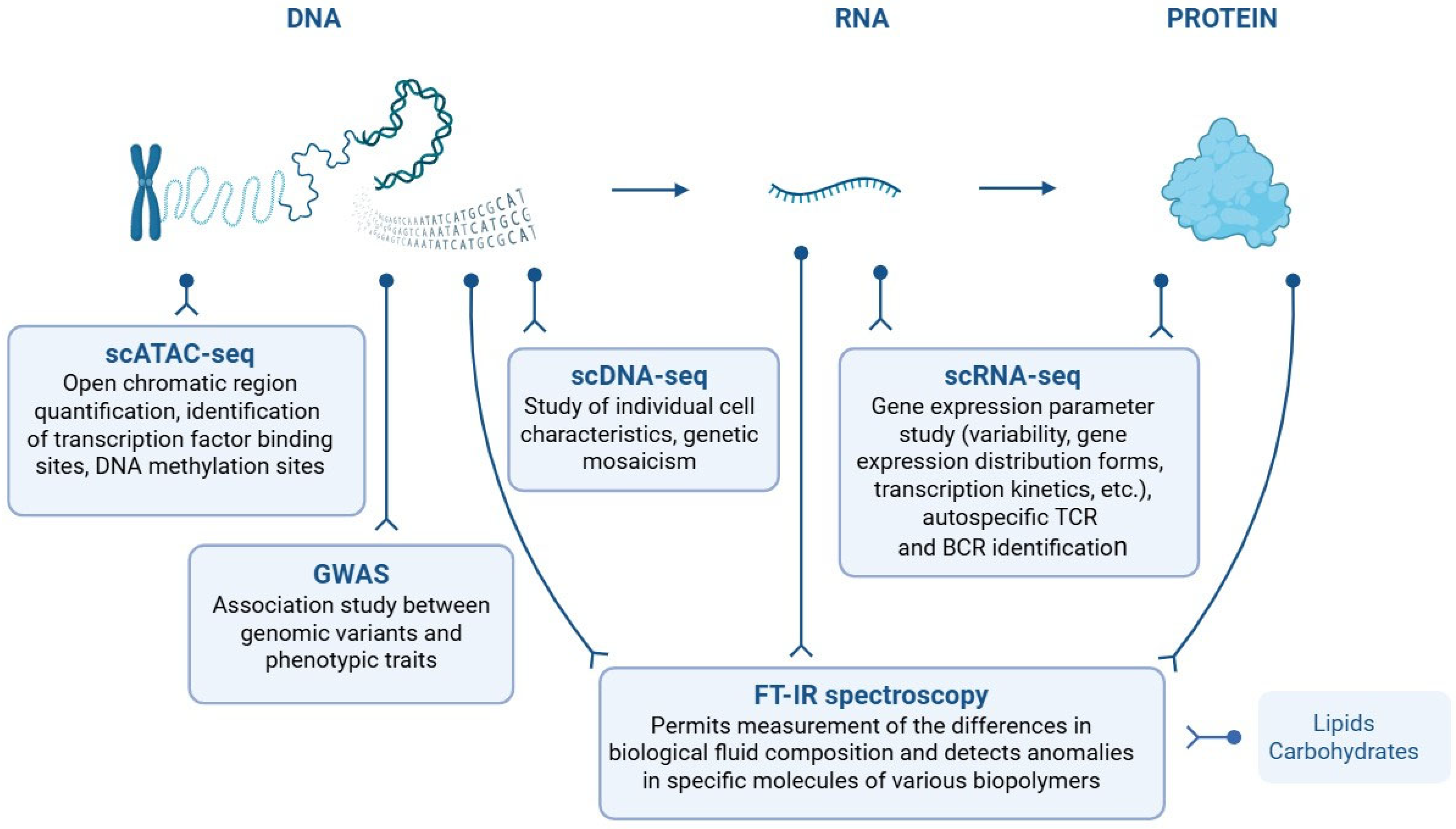
3. T1D Novel Biomarkers
3.1. Cytokines in T1D Pathogenesis
| Cytokine | Level Compared to Control Group * | T1D Duration | Biomaterial | Comparison Group, N, Age | Reference |
|---|---|---|---|---|---|
| IL-10 | ↓ | Preclinical stages | Small intestine cells | NOD mice, 4–6 weeks | [99] |
| IL-12 | ↑ | Preclinical stages | Cells of the small and large intestines | NOD mice, 4–6 weeks | [99] |
| IL-6 | ↑ | Preclinical stages | Colon cells | NOD mice, 4–6 weeks | [99] |
| IL-17 | ↑ | Preclinical stages | Pancreatic cells | NOD mice, 4–6 weeks | [99] |
| IL1-RA | ↓ | 2 weeks, Stage 3 | Blood serum | 100 patients up to 18 years old with newly diagnosed T1D | [103] |
| CXCL10 | ↑ | 4–9 weeks, Stage 3 | Pancreatic islet cells | 6 patients, 24–35 years old | [104] |
| CXCL10 | ↑ | 4–9 weeks, Stage 3 | Pancreatic islet cells | NOD mice, 8 weeks | [104] |
| EGF | ↑ | 4–16 years, Stages 3–4 | Blood serum | 52 patients with T1D, 8–18 years old | [105] |
| eotaxin/CCL11 | ↑ | 4–16 years, Stages 3–4 | Blood serum | 52 patients with T1D, 8–18 years old | [105] |
| MDC/CCL22 | ↑ | 4–16 years, Stages 3–4 | Blood serum | 52 patients with T1D, 8–18 years old | [105] |
| sCD40L | ↑ | 4–16 years, Stages 3–4 | Blood serum | 52 patients with T1D, 8–18 years old | [105] |
| TGF-α | ↑ | 4–16 years, Stages 3–4 | Blood serum | 52 patients with T1D, 8–18 years old | [105] |
| TNF-α | ↑ | 4–16 years, Stages 3–4 | Blood serum | 52 patients with T1D, 8–18 years old | [105] |
| M-CSF | ↓ | 1–23 years, Stages 3–4 | Blood serum | 25 patients (13 female and 12 male), 11–25 years old | [106] |
| IL-6 | ↓ | 1–23 years, Stages 3–4 | Blood serum | 25 patients (13 female and 12 male), 11–25 years old | [106] |
| CXCL1 | ↓ | 1–23 years, Stages 3–4 | Blood serum | 25 patients (13 female and 12 male), 11–25 years old | [106] |
| TGF-α | ↓ | 1–23 years, Stages 3–4 | Blood serum | Of these, 13 are female | [106] |
| IL-1α | ↓ | 1–23 years, Stages 3–4 | Blood serum | Of these, 13 are female | [106] |
| IL-4 | ↓ | 1–23 years, Stages 3–4 | Blood serum | Of these, 13 are female | [106] |
| IL-13 | ↓ | 1–23 years, Stages 3–4 | Blood serum | Of these, 13 are female | [106] |
| IL-22 | ↓ | 1–23 years, Stages 3–4 | Blood serum | Of these, 13 are female | [106] |
| MIP-1α | ↓ | 1–23 years, Stages 3–4 | Blood serum | Of these, 13 are female | [106] |
| CCL5 (RANTES) | ↓ | 1–23 years, Stages 3–4 | Blood serum | Of these, 13 are female | [106] |
| MIP-3 | ↓ | 1–23 years, Stages 3–4 | Blood serum | Of these, 13 are female | [106] |
| IL-22 | ↑ | 1–23 years, Stages 3–4 | Blood serum | Of these, 12 are male | [106] |
| EGF | ↑ | 1–23 years, Stages 3–4 | Blood serum | Of these, 12 are male | [106] |
| PDGF-AB/BB | ↑ | 1–23 years, Stages 3–4 | Blood serum | Of these, 12 are male | [106] |
| IL-12 | ↑ | 8.0 ± 6.0 years, Stage 4 | Blood serum | 29 patients with T1D without microvascular complications, 21.5 ± 11.0 years old, receiving insulin and antihypertensive drugs | [107] |
| IL-33 | ↑ | 8.0 ± 6.0 years, Stage 4 | Blood serum | 29 patients with T1D without microvascular complications, 21.5 ± 11.0 years old, receiving insulin and antihypertensive drugs | [107] |
| IL-4 | ↑ | 8.0 ± 6.0 years, Stage 4 | Blood serum | 29 patients with T1D without microvascular complications, 21.5 ± 11.0 years old, receiving insulin and antihypertensive drugs | [107] |
| IL-10 | ↑ | 8.0 ± 6.0 years, Stage 4 | Blood serum | 29 patients with T1D without microvascular complications, 21.5 ± 11.0 years old, receiving insulin and antihypertensive drugs | [107] |
| IL-17 | ↑ | 8.0 ± 6.0 years, Stage 4 | Blood serum | 29 patients with T1D without microvascular complications, 21.5 ± 11.0 years old, receiving insulin and antihypertensive drugs | [107] |
| IL-9 | ↑ | 8.0 ± 6.0 years, Stage 4 | Blood serum | 29 patients with T1D without microvascular complications, 21.5 ± 11.0 years old, receiving insulin and antihypertensive drugs | [107] |
| IL-12 | ↑ | 8.0 ± 6.0 years, Stage 4 | Blood serum | 96 patients with T1D complicated by retinopathy and nephropathy, 29.0 ± 15.2 years old, receiving insulin and antihypertensive drugs | [107] |
| IL-33 | ↑ | 8.0 ± 6.0 years, Stage 4 | Blood serum | 96 patients with T1D complicated by retinopathy and nephropathy, 29.0 ± 15.2 years old, receiving insulin and antihypertensive drugs | [107] |
| IL-4 | ↑ | 8.0 ± 6.0 years, Stage 4 | Blood serum | 96 patients with T1D complicated by retinopathy and nephropathy, 29.0 ± 15.2 years old, receiving insulin and antihypertensive drugs | [107] |
| IL-10 | ↑ | 8.0 ± 6.0 years, Stage 4 | Blood serum | 96 patients with T1D complicated by retinopathy and nephropathy, 29.0 ± 15.2 years old, receiving insulin and antihypertensive drugs | [107] |
| IL-17 | ↑ | 8.0 ± 6.0 years, Stage 4 | Blood serum | 96 patients with T1D complicated by retinopathy and nephropathy, 29.0 ± 15.2 years old, receiving insulin and antihypertensive drugs | [107] |
| IL-9 | ↑ | 8.0 ± 6.0 years, Stage 4 | Blood serum | 96 patients with T1D complicated by retinopathy and nephropathy, 29.0 ± 15.2 years old, receiving insulin and antihypertensive drugs | [107] |
3.2. Circulating Cell-Free DNA as a Biomarker for T1D Diagnosis
3.3. T1D MicroRNA Expression Profile
4. New Approaches and Methods in T1D Diagnosis
4.1. Cellular Markers of T1D
4.1.1. Novel T1D-Specific Immune Cell Markers Accrued by Single-Cell Transcriptomics
4.1.2. Antigen-Specific T-Cells as T1D Biomarkers
4.2. Molecular Markers of T1D
T1D-Specific Key Molecular Markers, Accrued by FTIR Spectroscopy of Biological Fluids Combined with Deep Learning
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gregory, G.A.; Robinson, T.I.; Linklater, S.E.; Wang, F.; Colagiuri, S.; de Beaufort, C.; Donaghue, K.C.; Harding, J.L.; Wander, P.L.; Zhang, X.; et al. Global incidence, prevalence, and mortality of type 1 diabetes in 2021 with projection to 2040: A modelling study. Lancet Diabetes Endocrinol. 2022, 10, 741–760. [Google Scholar] [CrossRef]
- Carr, A.L.J.; Evans-Molina, C.; Oram, R.A. Precision medicine in type 1 diabetes. Diabetologia 2022, 67, 1854–1866. [Google Scholar] [CrossRef] [PubMed]
- Ma˘nescu, M.; Ma˘nescu, I.B.; Grama, A. A review of stage 0 biomarkers in type 1 diabetes: The holy grail of early detection and prevention? J. Pers. Med. 2024, 14, 878. [Google Scholar] [CrossRef] [PubMed]
- Besser, R.E.; Bell, K.J.; Couper, J.J.; Ziegler, A.G.; Wherrett, D.K.; Knip, M.; Speake, C.; Casteels, K.; Driscoll, K.A.; Jacobsen, L.; et al. ISPAD Clinical Practice Consensus Guidelines 2022: Stages of type 1 diabetes in children and adolescents. Pediatr. Diabetes 2022, 23, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Parviainen, A.; Härkönen, T.; Ilonen, J.; But, A.; Knip, M.; Register, F.P.D. Heterogeneity of type 1 diabetes at diagnosis supports existence of age-related endotypes. Diabetes Care 2022, 45, 871–879. [Google Scholar] [CrossRef]
- Evans-Molina, C.; Sims, E.K.; DiMeglio, L.A.; Ismail, H.M.; Steck, A.K.; Palmer, J.P.; Krischer, J.P.; Geyer, S.; Xu, P.; Sosenko, J.M.; et al. β Cell dysfunction exists more than 5 years before type 1 diabetes diagnosis. JCI Insight 2018, 3, e120877. [Google Scholar] [CrossRef]
- Phillip, M.; Achenbach, P.; Addala, A.; Albanese-O’Neill, A.; Battelino, T.; Bell, K.J.; Besser, R.E.; Bonifacio, E.; Colhoun, H.M.; Couper, J.J.; et al. Consensus guidance for monitoring individuals with islet autoantibody–positive pre-stage 3 type 1 diabetes. Diabetes Care 2024, 47, 1276–1298. [Google Scholar]
- Abdellaoui, A.; Yengo, L.; Verweij, K.J.; Visscher, P.M. 15 years of GWAS discovery: Realizing the promise. Am. J. Hum. Genet. 2023, 110, 179–194. [Google Scholar] [CrossRef]
- Leete, P.; Oram, R.A.; McDonald, T.J.; Shields, B.M.; Ziller, C.; TIGI study team; Hattersley, A.T.; Richardson, S.J.; Morgan, N.G. Studies of insulin and proinsulin in pancreas and serum support the existence of aetiopathological endotypes of type 1 diabetes associated with age at diagnosis. Diabetologia 2020, 63, 1258–1267. [Google Scholar]
- Weston, C.S.; Boehm, B.O.; Pozzilli, P. Type 1 diabetes: A new vision of the disease based on endotypes. Diabetes/Metab. Res. Rev. 2024, 40, e3770. [Google Scholar] [CrossRef]
- Torabi, F.; Vadakekolathu, J.; Wyatt, R.; Leete, P.; Tombs, M.A.; Richardson, C.C.; Boocock, D.J.; Turner, M.D.; Morgan, N.G.; Richardson, S.J.; et al. Differential expression of genes controlling lymphocyte differentiation and migration in two distinct endotypes of type 1 diabetes. Diabet. Med. 2023, 40, e15155. [Google Scholar] [CrossRef]
- Redondo, M.; Yu, L.; Hawa, M.; Mackenzie, T.; Pyke, D.; Eisenbarth, G.; Leslie, R. Heterogeneity of type I diabetes: Analysis of monozygotic twins in Great Britain and the United States. Diabetologia 2001, 44, 354–362. [Google Scholar] [CrossRef]
- Noble, J.A.; Valdes, A.M. Genetics of the HLA region in the prediction of type 1 diabetes. Curr. Diabetes Rep. 2011, 11, 533–542. [Google Scholar] [CrossRef]
- Alekseev, L.; Dedov, I.; Khaitov, R.; Boldyreva, M.; Trofimov, D.Y.; Peterkova, V.; Kuraeva, T.; Abramov, D. Immunogenetics of type 1 diabetes mellitus—From fundamental ideas to medical practice. Ann. Russ. Acad. Med. Sci. 2012, 67, 75–80. [Google Scholar] [CrossRef]
- Sharp, S.A.; Rich, S.S.; Wood, A.R.; Jones, S.E.; Beaumont, R.N.; Harrison, J.W.; Schneider, D.A.; Locke, J.M.; Tyrrell, J.; Weedon, M.N.; et al. Development and standardization of an improved type 1 diabetes genetic risk score for use in newborn screening and incident diagnosis. Diabetes Care 2019, 42, 200–207. [Google Scholar] [CrossRef]
- Herold, K.C.; Usmani-Brown, S.; Ghazi, T.; Lebastchi, J.; Beam, C.A.; Bellin, M.D.; Ledizet, M.; Sosenko, J.M.; Krischer, J.P.; Palmer, J.P.; et al. β cell death and dysfunction during type 1 diabetes development in at-risk individuals. J. Clin. Investig. 2015, 125, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Redondo, M.J.; Steck, A.K.; Pugliese, A. Genetics of type 1 diabetes. Pediatr. Diabetes 2018, 19, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Aly, T.A.; Ide, A.; Humphrey, K.; Barker, J.M.; Steck, A.; Erlich, H.A.; Yu, L.; Miao, D.; Redondo, M.J.; McFann, K.; et al. Genetic prediction of autoimmunity: Initial oligogenic prediction of anti-islet autoimmunity amongst DR3/DR4–DQ8 relatives of patients with type 1A diabetes. J. Autoimmun. 2005, 25, 40–45. [Google Scholar] [CrossRef]
- Feil, R.; Fraga, M.F. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef]
- Jones, E.Y.; Fugger, L.; Strominger, J.L.; Siebold, C. MHC class II proteins and disease: A structural perspective. Nat. Rev. Immunol. 2006, 6, 271–282. [Google Scholar] [CrossRef]
- Kiani, J.; Hajilooi, M.; Furst, D.; Rezaei, H.; Shahryari-Hesami, S.; Kowsarifard, S.; Zamani, A.; Solgi, G. HLA class II susceptibility pattern for type 1 diabetes (T1D) in an Iranian population. Int. J. Immunogenet. 2015, 42, 279–286. [Google Scholar] [CrossRef]
- Robino, A.; Bevilacqua, E.; Aldegheri, L.; Conti, A.; Bazzo, V.; Tornese, G.; Catamo, E. Next-generation sequencing reveals additional HLA class I and class II alleles associated with type 1 diabetes and age at onset. Front. Immunol. 2024, 15, 1427349. [Google Scholar] [CrossRef]
- Varney, M.D.; Valdes, A.M.; Carlson, J.A.; Noble, J.A.; Tait, B.D.; Bonella, P.; Lavant, E.; Fear, A.L.; Louey, A.; Moonsamy, P.; et al. HLA DPA1, DPB1 alleles and haplotypes contribute to the risk associated with type 1 diabetes: Analysis of the type 1 diabetes genetics consortium families. Diabetes 2010, 59, 2055–2062. [Google Scholar] [CrossRef] [PubMed]
- Michalek, D.A.; Tern, C.; Zhou, W.; Robertson, C.C.; Farber, E.; Campolieto, P.; Chen, W.M.; Onengut-Gumuscu, S.; Rich, S.S. A multi-ancestry genome-wide association study in type 1 diabetes. Hum. Mol. Genet. 2024, 33, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Ilonen, J.; Kiviniemi, M.; El-Amir, M.I.; Nygård, L.; Härkönen, T.; Lempainen, J.; Knip, M. Increased Frequency of the HLA-DRB1* 04: 04-DQA1* 03-DQB1* 03: 02 Haplotype Among HLA-DQB1* 06: 02–Positive Children With Type 1 Diabetes. Diabetes 2024, 73, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Sugihara, S.; Ogata, T.; Kawamura, T.; Urakami, T.; Takemoto, K.; Kikuchi, N.; Takubo, N.; Tsubouchi, K.; Horikawa, R.; Kobayashi, K.; et al. HLA-class II and class I genotypes among Japanese children with Type 1A diabetes and their families. Pediatr. Diabetes 2012, 13, 33–44. [Google Scholar] [CrossRef]
- Howson, J.; Roy, M.; Zeitels, L.; Stevens, H.; Todd, J. HLA class II gene associations in African American Type 1 diabetes reveal a protective HLA-DRB1* 03 haplotype. Diabet. Med. 2013, 30, 710–716. [Google Scholar] [CrossRef]
- Li, Y.R.; Li, J.; Zhao, S.D.; Bradfield, J.P.; Mentch, F.D.; Maggadottir, S.M.; Hou, C.; Abrams, D.J.; Chang, D.; Gao, F.; et al. Meta-analysis of shared genetic architecture across ten pediatric autoimmune diseases. Nat. Med. 2015, 21, 1018–1027. [Google Scholar] [CrossRef]
- Chiou, J.; Geusz, R.J.; Okino, M.L.; Han, J.Y.; Miller, M.; Melton, R.; Beebe, E.; Benaglio, P.; Huang, S.; Korgaonkar, K.; et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature 2021, 594, 398–402. [Google Scholar] [CrossRef]
- Robertson, C.C.; Inshaw, J.R.; Onengut-Gumuscu, S.; Chen, W.M.; Santa Cruz, D.F.; Yang, H.; Cutler, A.J.; Crouch, D.J.; Farber, E.; Bridges Jr, S.L.; et al. Fine-mapping, trans-ancestral and genomic analyses identify causal variants, cells, genes and drug targets for type 1 diabetes. Nat. Genet. 2021, 53, 962–971. [Google Scholar] [CrossRef]
- Lambert, S.A.; Gil, L.; Jupp, S.; Ritchie, S.C.; Xu, Y.; Buniello, A.; McMahon, A.; Abraham, G.; Chapman, M.; Parkinson, H.; et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat. Genet. 2021, 53, 420–425. [Google Scholar] [CrossRef]
- Cerezo, M.; Sollis, E.; Ji, Y.; Lewis, E.; Abid, A.; Bircan, K.O.; Hall, P.; Hayhurst, J.; John, S.; Mosaku, A.; et al. The NHGRI-EBI GWAS Catalog: Standards for reusability, sustainability and diversity. Nucleic Acids Res. 2025, 53, D998–D1005. [Google Scholar] [CrossRef]
- Privé, F.; Aschard, H.; Carmi, S.; Folkersen, L.; Hoggart, C.; O’Reilly, P.F.; Vilhjálmsson, B.J. Portability of 245 polygenic scores when derived from the UK Biobank and applied to 9 ancestry groups from the same cohort. Am. J. Hum. Genet. 2022, 109, 12–23. [Google Scholar] [CrossRef]
- Bonifacio, E.; Beyerlein, A.; Hippich, M.; Winkler, C.; Vehik, K.; Weedon, M.N.; Laimighofer, M.; Hattersley, A.T.; Krumsiek, J.; Frohnert, B.I.; et al. Genetic scores to stratify risk of developing multiple islet autoantibodies and type 1 diabetes: A prospective study in children. PLoS Med. 2018, 15, e1002548. [Google Scholar] [CrossRef]
- Oram, R.A.; Patel, K.; Hill, A.; Shields, B.; McDonald, T.J.; Jones, A.; Hattersley, A.T.; Weedon, M.N. A type 1 diabetes genetic risk score can aid discrimination between type 1 and type 2 diabetes in young adults. Diabetes Care 2016, 39, 337–344. [Google Scholar] [CrossRef]
- Winkler, C.; Krumsiek, J.; Buettner, F.; Angermüller, C.; Giannopoulou, E.Z.; Theis, F.J.; Ziegler, A.G.; Bonifacio, E. Feature ranking of type 1 diabetes susceptibility genes improves prediction of type 1 diabetes. Diabetologia 2014, 57, 2521–2529. [Google Scholar] [CrossRef] [PubMed]
- Laine, A.P.; Valta, M.; Toppari, J.; Knip, M.; Veijola, R.; Ilonen, J.; Lempainen, J. Non-HLA gene polymorphisms in the pathogenesis of type 1 diabetes: Phase and endotype specific effects. Front. Immunol. 2022, 13, 909020. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.B.; Lynch, K.F.; Roth, R.; Lundgren, M.; Parikh, H.M.; Akolkar, B.; Hagopian, W.; Krischer, J.; Rewers, M.; She, J.X.; et al. First-appearing islet autoantibodies for type 1 diabetes in young children: Maternal life events during pregnancy and the child’s genetic risk. Diabetologia 2021, 64, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.R.; Kanai, M.; Kamatani, Y.; Okada, Y.; Neale, B.M.; Daly, M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019, 51, 584–591. [Google Scholar] [CrossRef]
- Thompson, D.J.; Wells, D.; Selzam, S.; Peneva, I.; Moore, R.; Sharp, K.; Tarran, W.A.; Beard, E.J.; Riveros-Mckay, F.; Giner-Delgado, C.; et al. A systematic evaluation of the performance and properties of the UK Biobank Polygenic Risk Score (PRS) Release. PLoS ONE 2024, 19, e0307270. [Google Scholar] [CrossRef]
- Perry, D.J.; Wasserfall, C.H.; Oram, R.A.; Williams, M.D.; Posgai, A.; Muir, A.B.; Haller, M.J.; Schatz, D.A.; Wallet, M.A.; Mathews, C.E.; et al. Application of a genetic risk score to racially diverse type 1 diabetes populations demonstrates the need for diversity in risk-modeling. Sci. Rep. 2018, 8, 4529. [Google Scholar] [CrossRef] [PubMed]
- Aksit, M.A.; Pace, R.G.; Vecchio-Pagán, B.; Ling, H.; Rommens, J.M.; Boelle, P.Y.; Guillot, L.; Raraigh, K.S.; Pugh, E.; Zhang, P.; et al. Genetic modifiers of cystic fibrosis-related diabetes have extensive overlap with type 2 diabetes and related traits. J. Clin. Endocrinol. Metab. 2020, 105, 1401–1415. [Google Scholar] [CrossRef] [PubMed]
- Monti, R.; Eick, L.; Hudjashov, G.; Läll, K.; Kanoni, S.; Wolford, B.N.; Wingfield, B.; Pain, O.; Wharrie, S.; Jermy, B.; et al. Evaluation of polygenic scoring methods in five biobanks shows larger variation between biobanks than methods and finds benefits of ensemble learning. Am. J. Hum. Genet. 2024, 111, 1431–1447. [Google Scholar] [CrossRef] [PubMed]
- Raben, T.G.; Lello, L.; Widen, E.; Hsu, S.D. Biobank-scale methods and projections for sparse polygenic prediction from machine learning. Sci. Rep. 2023, 13, 11662. [Google Scholar] [CrossRef] [PubMed]
- Jermy, B.; Läll, K.; Wolford, B.N.; Wang, Y.; Zguro, K.; Cheng, Y.; Kanai, M.; Kanoni, S.; Yang, Z.; Hartonen, T.; et al. A unified framework for estimating country-specific cumulative incidence for 18 diseases stratified by polygenic risk. Nat. Commun. 2024, 15, 5007. [Google Scholar] [CrossRef]
- Dorman, J.; Steenkiste, A.; O’leary, L.; McCarthy, B.; Lorenzen, T.; Foley, T. Type 1 diabetes in offspring of parents with type 1 diabetes: The tip of an autoimmune iceberg? Pediatr. Diabetes 2000, 1, 17–22. [Google Scholar] [CrossRef]
- Turtinen, M.; Härkönen, T.; Parkkola, A.; Ilonen, J.; Knip, M.; Register, F.P.D. Characteristics of familial type 1 diabetes: Effects of the relationship to the affected family member on phenotype and genotype at diagnosis. Diabetologia 2019, 62, 2025–2039. [Google Scholar] [CrossRef]
- Gillespie, K.M.; Aitken, R.J.; Wilson, I.; Williams, A.J.; Bingley, P.J. Early onset of diabetes in the proband is the major determinant of risk in HLA DR3-DQ2/DR4-DQ8 siblings. Diabetes 2014, 63, 1041–1047. [Google Scholar] [CrossRef]
- Gokuladhas, S.; Schierding, W.; Golovina, E.; Fadason, T.; O’Sullivan, J. Unravelling the shared genetic mechanisms underlying 18 autoimmune diseases using a systems approach. Front. Immunol. 2021, 12, 693142. [Google Scholar] [CrossRef]
- Demela, P.; Pirastu, N.; Soskic, B. Cross-disorder genetic analysis of immune diseases reveals distinct gene associations that converge on common pathways. Nat. Commun. 2023, 14, 2743. [Google Scholar] [CrossRef]
- Alshiekh, S.; Maziarz, M.; Geraghty, D.E.; Larsson, H.E.; Agardh, D. High-resolution genotyping indicates that children with type 1 diabetes and celiac disease share three HLA class II loci in DRB3, DRB4 and DRB5 genes. Hla 2021, 97, 44–51. [Google Scholar] [CrossRef]
- Magistrelli, G.; Jeannin, P.; Herbault, N.; Benoit de Coignac, A.; Gauchat, J.F.; Bonnefoy, J.Y.; Delneste, Y. A soluble form of CTLA-4 generated by alternative splicing is expressed by nonstimulated human T cells. Eur. J. Immunol. 1999, 29, 3596–3602. [Google Scholar] [CrossRef]
- Sirota, M.; Schaub, M.A.; Batzoglou, S.; Robinson, W.H.; Butte, A.J. Autoimmune disease classification by inverse association with SNP alleles. PLoS Genet. 2009, 5, e1000792. [Google Scholar] [CrossRef] [PubMed]
- Kreiner, E.; Waage, J.; Standl, M.; Brix, S.; Pers, T.H.; Alves, A.C.; Warrington, N.M.; Tiesler, C.M.; Fuertes, E.; Franke, L.; et al. Shared genetic variants suggest common pathways in allergy and autoimmune diseases. J. Allergy Clin. Immunol. 2017, 140, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Al Yafei, Z.; Mack, S.J.; Alvares, M.; Ali, B.R.; Afandi, B.; Beshyah, S.A.; Sharma, C.; Osman, W.; Mirghani, R.; Nasr, A.; et al. HLA-DRB1 and–DQB1 alleles, haplotypes and genotypes in Emirati patients with type 1 diabetes underscores the benefits of evaluating understudied populations. Front. Genet. 2022, 13, 841879. [Google Scholar] [CrossRef] [PubMed]
- Stayoussef, M.; Benmansour, J.; Al-Jenaidi, F.A.; Nemr, R.; Ali, M.E.; Mahjoub, T.; Almawi, W.Y. Influence of common and specific HLA-DRB1/DQB1 haplotypes on genetic susceptibilities of three distinct Arab populations to type 1 diabetes. Clin. Vaccine Immunol. 2009, 16, 136–138. [Google Scholar] [CrossRef]
- Azulay, R.S.d.S.; Porto, L.C.; Silva, D.A.; Tavares, M.d.G.; Reis, R.M.D.F.; Nascimento, G.C.; Damianse, S.d.S.P.; Rocha, V.C.d.C.; Magalhães, M.; Rodrigues, V.; et al. Genetic ancestry inferred from autosomal and Y chromosome markers and HLA genotypes in Type 1 Diabetes from an admixed Brazilian population. Sci. Rep. 2021, 11, 14157. [Google Scholar] [CrossRef]
- Boldyreva, M.N.; Khaitov, R.M.; Dedov, I.I.; Bogatova, O.V.; Guskova, I.A.; Yankevich, T.E.; Zilov, A.V.; Osokina, I.V.; Evseeva, I.V.; Ganicheva, L.L.; et al. A New Look at the Mechanism of HLA-Associated Predisposition to Type 1 Diabetes Mellitus. Theoretical and Applied Aspects. Immunologiya 2005, 26, 324–328. (In Russian) [Google Scholar]
- Khdair, S.I.; Jarrar, W.; Jarrar, Y.B.; Bataineh, S.; Al-Khaldi, O. Association of HLA-DRB1 and-DQ alleles and haplotypes with type 1 diabetes in Jordanians. Endocr. Metab. Immune Disord.-Drug Targets 2020, 20, 895–902. [Google Scholar] [CrossRef]
- Sayad, A.; Akbari, M.; Pajouhi, M.; Mostafavi, F.; Zamani, M. The influence of the HLA-DRB, HLA-DQB and polymorphic positions of the HLA-DRβ1 and HLA-DQβ1 molecules on risk of Iranian type 1 diabetes mellitus patients. Int. J. Immunogenet. 2012, 39, 429–436. [Google Scholar] [CrossRef]
- Zhao, L.P.; Papadopoulos, G.K.; Lybrand, T.P.; Moustakas, A.K.; Bondinas, G.P.; Carlsson, A.; Larsson, H.E.; Ludvigsson, J.; Marcus, C.; Persson, M.; et al. The KAG motif of HLA-DRB1 (β71, β74, β86) predicts seroconversion and development of type 1 diabetes. EBioMedicine 2021, 69, 103431. [Google Scholar] [CrossRef]
- Naito, T.; Suzuki, K.; Hirata, J.; Kamatani, Y.; Matsuda, K.; Toda, T.; Okada, Y. A deep learning method for HLA imputation and trans-ethnic MHC fine-mapping of type 1 diabetes. Nat. Commun. 2021, 12, 1639. [Google Scholar] [CrossRef]
- Lee, Y.H.; Kim, J.H.; Song, G.G. Interleukin-18 promoter-607 C/A and-137 G/C polymorphisms and susceptibility to type 1 diabetes: A meta-analysis. Hum. Immunol. 2015, 76, 537–545. [Google Scholar] [CrossRef]
- Zhai, N.; Bidares, R.; Makoui, M.H.; Aslani, S.; Mohammadi, P.; Razi, B.; Imani, D.; Yazdchi, M.; Mikaeili, H. Vitamin D receptor gene polymorphisms and the risk of the type 1 diabetes: A meta-regression and updated meta-analysis. BMC Endocr. Disord. 2020, 20, 121. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Wenzlau, J.M.; Zhang, C.; Dong, F.; Waugh, K.; Leslie, R.D.; Rewers, M.J.; Michels, A.W.; Yu, L.; Diabetes Autoimmunity Study in the Young (DAISY); et al. Strong Association of Autoantibodies Targeting Deamidated Extracellular Epitopes of Insulinoma Antigen-2 with Clinical Onset of Type 1 Diabetes. Diabetes 2025, 74, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Haller, M.J.; Bell, K.J.; Besser, R.E.; Casteels, K.; Couper, J.J.; Craig, M.E.; Elding Larsson, H.; Jacobsen, L.; Lange, K.; Oron, T.; et al. ISPAD clinical practice consensus guidelines 2024: Screening, staging, and strategies to preserve beta-cell function in children and adolescents with type 1 diabetes. Horm. Res. Paediatr. 2025, 97, 529–545. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, L.M.; Bocchino, L.; Evans-Molina, C.; DiMeglio, L.; Goland, R.; Wilson, D.M.; Atkinson, M.A.; Aye, T.; Russell, W.E.; Wentworth, J.M.; et al. The risk of progression to type 1 diabetes is highly variable in individuals with multiple autoantibodies following screening. Diabetologia 2020, 63, 588–596. [Google Scholar] [CrossRef]
- Kwon, B.C.; Anand, V.; Achenbach, P.; Dunne, J.L.; Hagopian, W.; Hu, J.; Koski, E.; Lernmark, Å.; Lundgren, M.; Ng, K.; et al. Progression of type 1 diabetes from latency to symptomatic disease is predicted by distinct autoimmune trajectories. Nat. Commun. 2022, 13, 1514. [Google Scholar] [CrossRef]
- Krischer, J.P.; Liu, X.; Vehik, K.; Akolkar, B.; Hagopian, W.A.; Rewers, M.J.; She, J.X.; Toppari, J.; Ziegler, A.G.; Lernmark, Å.; et al. Predicting islet cell autoimmunity and type 1 diabetes: An 8-year TEDDY study progress report. Diabetes Care 2019, 42, 1051–1060. [Google Scholar] [CrossRef]
- Mansachs, S.J.; Villumsen, S.O.; Johannesen, J.; Lind, A.; Kaur, S.; Pociot, F. Genetic variants associated with neuropeptide Y autoantibody levels in newly diagnosed individuals with type 1 diabetes. Genes 2022, 13, 869. [Google Scholar] [CrossRef]
- Bonser, A.M.; Garcia-Webb, P.; Harrison, L.C. C-peptide measurement: Methods and clinical utility. CRC Crit. Rev. Clin. Lab. Sci. 1984, 19, 297–352. [Google Scholar] [CrossRef]
- Martinez, M.M.; Salami, F.; Larsson, H.E.; Toppari, J.; Lernmark, Å.; Kero, J.; Veijola, R.; Koskenniemi, J.J.; Tossavainen, P.; Lundgren, M.; et al. Beta cell function in participants with single or multiple islet autoantibodies at baseline in the TEDDY Family Prevention Study: TEFA. Endocrinol. Diabetes Metab. 2021, 4, e00198. [Google Scholar] [CrossRef]
- Januszewski, A.S.; Cho, Y.H.; Joglekar, M.V.; Farr, R.J.; Scott, E.S.; Wong, W.K.; Carroll, L.M.; Loh, Y.W.; Benitez-Aguirre, P.Z.; Keech, A.C.; et al. Insulin micro-secretion in Type 1 diabetes and related microRNA profiles. Sci. Rep. 2021, 11, 11727. [Google Scholar] [CrossRef]
- Krivova, Y.; Proshchina, A.; Otlyga, D.; Kharlamova, A.; Saveliev, S. Detection of Insulin in Insulin-Deficient Islets of Patients with Type 1 Diabetes. Life 2025, 15, 125. [Google Scholar] [CrossRef]
- Thunander, M.; Törn, C.; Petersson, C.; Ossiansson, B.; Fornander, J.; Landin-Olsson, M. Levels of C-peptide, body mass index and age, and their usefulness in classification of diabetes in relation to autoimmunity, in adults with newly diagnosed diabetes in Kronoberg, Sweden. Eur. J. Endocrinol. 2012, 166, 1021–1029. [Google Scholar] [CrossRef]
- Ludvigsson, J.; Carlsson, A.; Forsander, G.; Ivarsson, S.; Kockum, I.; Lernmark, Å.; Lindblad, B.; Marcus, C.; Samuelsson, U. C-peptide in the classification of diabetes in children and adolescents. Pediatr. Diabetes 2012, 13, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Besser, R.E.; Shepherd, M.H.; McDonald, T.J.; Shields, B.M.; Knight, B.A.; Ellard, S.; Hattersley, A.T. Urinary C-peptide creatinine ratio is a practical outpatient tool for identifying hepatocyte nuclear factor 1-α/hepatocyte nuclear factor 4-α maturity-onset diabetes of the young from long-duration type 1 diabetes. Diabetes Care 2011, 34, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Hattersley, A.; Bruining, J.; Shield, J.; Njolstad, P.; Donaghue, K. ISPAD Clinical Practice Consensus Guidelines 2006–2007. The diagnosis and management of monogenic diabetes in children. Pediatr. Diabetes 2006, 7, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Barthson, J.; Germano, C.M.; Moore, F.; Maida, A.; Drucker, D.J.; Marchetti, P.; Gysemans, C.; Mathieu, C.; Nuñez, G.; Jurisicova, A.; et al. Cytokines tumor necrosis factor-α and interferon-γ induce pancreatic β-cell apoptosis through STAT1-mediated Bim protein activation. J. Biol. Chem. 2011, 286, 39632–39643. [Google Scholar] [CrossRef]
- Lee, L.F.; Xu, B.; Michie, S.A.; Beilhack, G.F.; Warganich, T.; Turley, S.; McDevitt, H.O. The role of TNF-α in the pathogenesis of type 1 diabetes in the nonobese diabetic mouse: Analysis of dendritic cell maturation. Proc. Natl. Acad. Sci. USA 2005, 102, 15995–16000. [Google Scholar] [CrossRef]
- Lombardi, A.; Tsomos, E.; Hammerstad, S.S.; Tomer, Y. Interferon alpha: The key trigger of type 1 diabetes. J. Autoimmun. 2018, 94, 7–15. [Google Scholar] [CrossRef]
- Lu, J.; Liu, J.; Li, L.; Lan, Y.; Liang, Y. Cytokines in type 1 diabetes: Mechanisms of action and immunotherapeutic targets. Clin. Transl. Immunol. 2020, 9, e1122. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Kaminga, A.C.; Kinra, S.; Wen, S.W.; Liu, H.; Tan, X.; Liu, A. Chemokines in type 1 diabetes mellitus. Front. Immunol. 2022, 12, 690082. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.A.; Jenkins, B.J. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat. Rev. Immunol. 2018, 18, 773–789. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Qiao, Y.C.; Pan, Y.H.; Xu, Y.; Huang, Y.C.; Wang, Y.H.; Geng, L.J.; Zhao, H.L.; Zhang, X.X. Correlation between serum interleukin-6 level and type 1 diabetes mellitus: A systematic review and meta-analysis. Cytokine 2017, 94, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Apaolaza, P.S.; Balcacean, D.; Zapardiel-Gonzalo, J.; Nelson, G.; Lenchik, N.; Akhbari, P.; Gerling, I.; Richardson, S.J.; Rodriguez-Calvo, T.; nPOD Virus Group. Islet expression of type I interferon response sensors is associated with immune infiltration and viral infection in type 1 diabetes. Sci. Adv. 2021, 7, eabd6527. [Google Scholar] [CrossRef]
- Rodrigues, K.B.; Dufort, M.J.; Llibre, A.; Speake, C.; Rahman, M.J.; Bondet, V.; Quiel, J.; Linsley, P.S.; Greenbaum, C.J.; Duffy, D.; et al. Innate immune stimulation of whole blood reveals IFN-1 hyper-responsiveness in type 1 diabetes. Diabetologia 2020, 63, 1576–1587. [Google Scholar] [CrossRef]
- Sarvetnick, N.; Liggitt, D.; Pitts, S.L.; Hansen, S.E.; Stewart, T.A. Insulin-dependent diabetes mellitus induced in transgenic mice by ectopic expression of class II MHC and interferon-gamma. Cell 1988, 52, 773–782. [Google Scholar] [CrossRef]
- Hu, F.; Guo, F.; Zhu, Y.; Zhou, Q.; Li, T.; Xiang, H.; Shang, D. IL-17 in pancreatic disease: Pathogenesis and pharmacotherapy. Am. J. Cancer Res. 2020, 10, 3551. [Google Scholar]
- Klatzmann, D.; Abbas, A.K. The promise of low-dose interleukin-2 therapy for autoimmune and inflammatory diseases. Nat. Rev. Immunol. 2015, 15, 283–294. [Google Scholar] [CrossRef]
- Russell, M.A.; Morgan, N. The impact of anti-inflammatory cytokines on the pancreatic β-cell. Islets 2014, 6, e950547. [Google Scholar] [CrossRef]
- Leslie, K.A.; Russell, M.A.; Taniguchi, K.; Richardson, S.J.; Morgan, N.G. The transcription factor STAT6 plays a critical role in promoting beta cell viability and is depleted in islets of individuals with type 1 diabetes. Diabetologia 2019, 62, 87–98. [Google Scholar] [CrossRef]
- Hillhouse, E.E.; Beauchamp, C.; Chabot-Roy, G.; Dugas, V.; Lesage, S. Interleukin-10 limits the expansion of immunoregulatory CD4- CD8- T cells in autoimmune-prone non-obese diabetic mice. Immunol. Cell Biol. 2010, 88, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Torres-Aguilar, H.; Sánchez-Torres, C.; Jara, L.J.; Blank, M.; Shoenfeld, Y. IL-10/TGF-β-treated dendritic cells, pulsed with insulin, specifically reduce the response to insulin of CD4+ effector/memory T cells from type 1 diabetic individuals. J. Clin. Immunol. 2010, 30, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 2009, 27, 519–550. [Google Scholar] [CrossRef] [PubMed]
- Ishigame, H.; Zenewicz, L.A.; Sanjabi, S.; Licona-Limón, P.; Nakayama, M.; Leonard, W.J.; Flavell, R.A. Excessive Th1 responses due to the absence of TGF-β signaling cause autoimmune diabetes and dysregulated Treg cell homeostasis. Proc. Natl. Acad. Sci. USA 2013, 110, 6961–6966. [Google Scholar] [CrossRef]
- Matsuda, E.; Obama, Y.; Kosai, K.I. Safe and low-dose but therapeutically effective adenovirus-mediated hepatocyte growth factor gene therapy for type 1 diabetes in mice. Life Sci. 2021, 268, 119014. [Google Scholar] [CrossRef]
- Schroderus, A.M.; Pitkänen, V.; Ekman, I.; Stevens, D.; Rytkönen-Nissinen, M.; Rintamäki, R.; Pihlajamäki, J.; Knip, M.; Veijola, R.; Toppari, J.; et al. Temporal alterations in CD8+ T cells during the progression from stage 1 to stage 3 type 1 diabetes. Diabetes 2024, 73, 1705–1715. [Google Scholar] [CrossRef]
- Miranda, M.C.G.; Oliveira, R.P.; Torres, L.; Aguiar, S.L.F.; Pinheiro-Rosa, N.; Lemos, L.; Guimarães, M.A.; Reis, D.; Silveira, T.; Ferreira, Ê.; et al. Frontline Science: Abnormalities in the gut mucosa of non-obese diabetic mice precede the onset of type 1 diabetes. J. Leukoc. Biol. 2019, 106, 513–529. [Google Scholar] [CrossRef]
- Ferreira, R.C.; Guo, H.; Coulson, R.M.; Smyth, D.J.; Pekalski, M.L.; Burren, O.S.; Cutler, A.J.; Doecke, J.D.; Flint, S.; McKinney, E.F.; et al. A type I interferon transcriptional signature precedes autoimmunity in children genetically at risk for type 1 diabetes. Diabetes 2014, 63, 2538–2550. [Google Scholar] [CrossRef]
- Rigby, M.R.; Hayes, B.; Li, Y.; Vercruysse, F.; Hedrick, J.A.; Quattrin, T. Two-year follow-up from the T1GER study: Continued off- therapy metabolic improvements in children and young adults with new-onset T1D treated with golimumab and characterization of responders. Diabetes Care 2023, 46, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Al-Dubayee, M.; Babiker, A.; Alkewaibeen, A.; Alkhalifah, A.; Alanazi, T.; Nogoud, M.; Alotaibi, A.; Alotaibi, F.; Almetairi, F.; Alrowaily, M.A.; et al. Correlation analysis between cytokines’ profile, autoimmune antibodies and the duration of type 1 diabetes: A case control study in a specialized children’s centre in Riyadh. Int. J. Immunopathol. Pharmacol. 2023, 37, 03946320231209821. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Toda, K.; He, L.; Miao, D.; Yamada, S.; Yu, L.; Kodama, K. Expression-based genome-wide association study links OPN and IL1-RA with newly diagnosed type 1 diabetes in children. J. Clin. Endocrinol. Metab. 2022, 107, 1825–1832. [Google Scholar] [CrossRef] [PubMed]
- Nigi, L.; Brusco, N.; Grieco, G.E.; Licata, G.; Krogvold, L.; Marselli, L.; Gysemans, C.; Overbergh, L.; Marchetti, P.; Mathieu, C.; et al. Pancreatic alpha-cells contribute together with beta-cells to CXCL10 expression in type 1 diabetes. Front. Endocrinol. 2020, 11, 630. [Google Scholar] [CrossRef]
- Wołoszyn-Durkiewicz, A.; Iwaszkiewicz-Grześ, D.; Świętoń, D.; Kujawa, M.J.; Jankowska, A.; Durawa, A.; Glasner, P.; Tr-zonkowski, P.; Glasner, L.; Szurowska, E.; et al. The complex network of cytokines and chemokines in pediatric patients with long-standing type 1 diabetes. Int. J. Mol. Sci. 2024, 25, 1565. [Google Scholar] [CrossRef]
- Girdhar, K.; Mine, K.; DaCosta, J.M.; Atkinson, M.A.; Ludvigsson, J.; Altindis, E. Sex-specific cytokine, chemokine, and growth factor signatures in T1D patients and progressors. FASEB J. 2024, 38, e70270. [Google Scholar] [CrossRef]
- Shruthi, S.; Mohan, V.; Amutha, A.; Aravindhan, V. Increased serum levels of novel T cell cytokines IL-33, IL-9 and IL-17 in subjects with type-1 diabetes. Cytokine 2016, 86, 6–9. [Google Scholar] [CrossRef]
- Melanitou, E.; Devendra, D.; Liu, E.; Miao, D.; Eisenbarth, G.S. Early and quantal (by litter) expression of insulin autoantibodies in the nonobese diabetic mice predict early diabetes onset. J. Immunol. 2004, 173, 6603–6610. [Google Scholar] [CrossRef]
- Melanitou, E. Investigation of type 1 diabetes in NOD mice knockout for the osteopontin gene. Gene 2020, 753, 144785. [Google Scholar] [CrossRef]
- Talaat, I.; Nasr, A.; Alsulaimani, A.; Alghamdi, H.; Alswat, K.; Almalki, D.; Abushouk, A.; Saleh, A.; Allam, G. Association between type 1, type 2 cytokines, diabetic autoantibodies and 25-hydroxyvitamin D in children with type 1 diabetes. J. Endocrinol. Investig. 2016, 39, 1425–1434. [Google Scholar] [CrossRef]
- Simeunovic, A.; Brunborg, C.; Heier, M.; Seljeflot, I.; Dahl-Jørgensen, K.; Margeirsdottir, H.D. Sustained low-grade inflammation in young participants with childhood onset type 1 diabetes: The Norwegian atherosclerosis and childhood diabetes (ACD) study. Atherosclerosis 2023, 379, 117151. [Google Scholar] [CrossRef]
- Schloot, N.; Hanifi-Moghaddam, P.; Aabenhus-Andersen, N.; Alizadeh, B.; Saha, M.; Knip, M.; Devendra, D.; Wilkin, T.; Bonifacio, E.; Roep, B.; et al. Association of immune mediators at diagnosis of Type 1 diabetes with later clinical remission. Diabet. Med. 2007, 24, 512–520. [Google Scholar] [CrossRef]
- Gomez-Muñoz, L.; Perna-Barrull, D.; Caroz-Armayones, J.M.; Murillo, M.; Rodriguez-Fernandez, S.; Valls, A.; Vazquez, F.; Perez, J.; Corripio, R.; Castaño, L.; et al. Candidate biomarkers for the prediction and monitoring of partial remission in pediatric type 1 diabetes. Front. Immunol. 2022, 13, 825426. [Google Scholar] [CrossRef]
- Krishnamurthy, B.; Lacorcia, M.; Kay, T.W.; Thomas, H.E.; Mannering, S.I. Monitoring immunomodulation strategies in type 1 diabetes. Front. Immunol. 2023, 14, 1206874. [Google Scholar] [CrossRef] [PubMed]
- Marro, B.S.; Ware, B.C.; Zak, J.; De la Torre, J.C.; Rosen, H.; Oldstone, M.B. Progression of type 1 diabetes from the prediabetic stage is controlled by interferon-α signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 3708–3713. [Google Scholar] [CrossRef] [PubMed]
- Peters, D.L.; Pretorius, P.J. Origin, translocation and destination of extracellular occurring DNA—A new paradigm in genetic behaviour. Clin. Chim. Acta 2011, 412, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.; Schur, P.; Carr, R.; Kunkel, H. Deoxybonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J. Clin. Investig. 1966, 45, 1732–1740. [Google Scholar] [CrossRef]
- Stawski, R.; Walczak, K.; Kosielski, P.; Meissner, P.; Budlewski, T.; Padula, G.; Nowak, D. Repeated bouts of exhaustive exercise increase circulating cell free nuclear and mitochondrial DNA without development of tolerance in healthy men. PLoS ONE 2017, 12, e0178216. [Google Scholar] [CrossRef]
- Nielsen, H.G.; Øktedalen, O.; Opstad, P.K.; Lyberg, T. Plasma cytokine profiles in long-term strenuous exercise. J. Sports Med. 2016, 2016, 7186137. [Google Scholar] [CrossRef]
- Suzuki, K.; Nakaji, S.; Yamada, M.; Totsuka, M.; Sato, K.; Sugawara, K. Systemic inflammatory response to exhaustive exercise. Cytokine kinetics. Exerc. Immunol. Rev. 2002, 8, 6–48. [Google Scholar]
- Jestkova, E.M.; Ershova, E.S.; Martynov, A.V.; Zakharova, N.V.; Kostyuk, G.P.; Veiko, N.N.; Kostyuk, S.V. Concentration of circulating cell-free DNA in the peripheral blood plasma of patients with acute endogenous and exogenous etiology psychoses. Psikhiatriya 2021, 19, 6–14. (In Russian) [Google Scholar] [CrossRef]
- Akirav, E.M.; Lebastchi, J.; Galvan, E.M.; Henegariu, O.; Akirav, M.; Ablamunits, V.; Lizardi, P.M.; Herold, K.C. Detection of β cell death in diabetes using differentially methylated circulating DNA. Proc. Natl. Acad. Sci. USA 2011, 108, 19018–19023. [Google Scholar] [CrossRef] [PubMed]
- Lebastchi, J.; Deng, S.; Lebastchi, A.H.; Beshar, I.; Gitelman, S.; Willi, S.; Gottlieb, P.; Akirav, E.M.; Bluestone, J.A.; Herold, K.C. Immune therapy and β-cell death in type 1 diabetes. Diabetes 2013, 62, 1676–1680. [Google Scholar] [CrossRef] [PubMed]
- Usmani-Brown, S.; Perdigoto, A.L.; Lavoie, N.; Clark, P.; Korah, M.; Rui, J.; Betancur, G.; Herold, K.C. β cell responses to inflammation. Mol. Metab. 2019, 27, S104–S113. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.M.; Perez Chumbiauca, C.N.; Mather, K.J.; Mirmira, R.G.; Tersey, S.A. Detection of islet β-cell death in vivo by multiplex PCR analysis of differentially methylated DNA. Endocrinology 2013, 154, 3476–3481. [Google Scholar] [CrossRef]
- Husseiny, M.I.; Kuroda, A.; Kaye, A.N.; Nair, I.; Kandeel, F.; Ferreri, K. Development of a quantitative methylation-specific polymerase chain reaction method for monitoring beta cell death in type 1 diabetes. PLoS ONE 2012, 7, e47942. [Google Scholar] [CrossRef]
- Walczak, K.; Stawski, R.; Perdas, E.; Brzezinska, O.; Kosielski, P.; Galczynski, S.; Budlewski, T.; Padula, G.; Nowak, D. Circulating cell free DNA response to exhaustive exercise in average trained men with type I diabetes mellitus. Sci. Rep. 2021, 11, 4639. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, Y.; Zhong, L.; Ye, D.; Zhang, J.; Tu, Y.; Bornstein, S.R.; Zhou, Z.; Lam, K.S.; Xu, A. Increased neutrophil elastase and proteinase 3 and augmented NETosis are closely associated with β-cell autoimmunity in patients with type 1 diabetes. Diabetes 2014, 63, 4239–4248. [Google Scholar] [CrossRef]
- Gala-Lopez, B.L.; Neiman, D.; Kin, T.; O’Gorman, D.; Pepper, A.R.; Malcolm, A.J.; Pianzin, S.; Senior, P.A.; Campbell, P.; Glaser, B.; et al. Beta cell death by cell-free DNA and outcome after clinical islet transplantation. Transplantation 2018, 102, 978–985. [Google Scholar] [CrossRef]
- Lehmann-Werman, R.; Neiman, D.; Zemmour, H.; Moss, J.; Magenheim, J.; Vaknin-Dembinsky, A.; Rubertsson, S.; Nellgård, B.; Blennow, K.; Zetterberg, H.; et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl. Acad. Sci. USA 2016, 113, E1826–E1834. [Google Scholar] [CrossRef]
- Syed, F.; Tersey, S.A.; Turatsinze, J.V.; Felton, J.L.; Kang, N.J.; Nelson, J.B.; Sims, E.K.; Defrance, M.; Bizet, M.; Fuks, F.; et al. Circulating unmethylated CHTOP and INS DNA fragments provide evidence of possible islet cell death in youth with obesity and diabetes. Clin. Epigenet. 2020, 12, 116. [Google Scholar] [CrossRef] [PubMed]
- Husseiny, M.I.; Kaye, A.; Zebadua, E.; Kandeel, F.; Ferreri, K. Tissue-specific methylation of human insulin gene and PCR assay for monitoring beta cell death. PLoS ONE 2014, 9, e94591. [Google Scholar] [CrossRef] [PubMed]
- Gezer, U.; Özgür, E.; Yörüker, E.E.; Polatoglou, E.; Holdenrieder, S.; Bronkhorst, A. LINE-1 cfDNA methylation as an emerging biomarker in solid cancers. Cancers 2024, 16, 3725. [Google Scholar] [CrossRef]
- Kurdyukov, S.; Bullock, M. DNA methylation analysis: Choosing the right method. Biology 2016, 5, 3. [Google Scholar] [CrossRef]
- Tabakov, D.V.; Maznina, A.A.; Astakhova, E.A.; Egorova, A.E.; Zakharova, E.N.; Glushkova, O.V.; Petriaikina, E.S.; Svetlichnyy, D.; Krupinova, J.A.; Bogdanov, V.P.; et al. Diagnosis of cancer, autoimmune and infectious diseases and prediction of the therapy effectiveness based on the individual’s immunotype. Front. Immunol. 2025, 16, 1658970. [Google Scholar] [CrossRef]
- Linares-Pineda, T.M.; Gutiérrez-Repiso, C.; Peña-Montero, N.; Molina-Vega, M.; Rubio, F.L.; Arana, M.S.; Tinahones, F.J.; Picón- César, M.J.; Morcillo, S. Higher β cell death in pregnant women, measured by DNA methylation patterns of cell-free DNA, compared to new-onset type 1 and type 2 diabetes subjects: A cross-sectional study. Diabetol. Metab. Syndr. 2023, 15, 115. [Google Scholar] [CrossRef]
- Neiman, D.; Gillis, D.; Piyanzin, S.; Cohen, D.; Fridlich, O.; Moss, J.; Zick, A.; Oron, T.; Sundberg, F.; Forsander, G.; et al. Multiplexing DNA methylation markers to detect circulating cell-free DNA derived from human pancreatic β cells. JCI Insight 2020, 5, e136579. [Google Scholar] [CrossRef]
- Sims, E.K.; Evans-Molina, C.; Tersey, S.A.; Eizirik, D.L.; Mirmira, R.G. Biomarkers of islet beta cell stress and death in type 1 diabetes. Diabetologia 2018, 61, 2259–2265. [Google Scholar] [CrossRef]
- Makarova, J.; Turchinovich, A.; Shkurnikov, M.; Tonevitsky, A. Extracellular miRNAs and cell-cell communication: Problems and prospects. Trends Biochem. Sci. 2021, 46, 640–651. [Google Scholar] [CrossRef]
- Angelescu, M.A.; Andronic, O.; Dima, S.O.; Popescu, I.; Meivar-Levy, I.; Ferber, S.; Lixandru, D. miRNAs as biomarkers in diabetes: Moving towards precision medicine. Int. J. Mol. Sci. 2022, 23, 12843. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, P.; Branco, R.C.S.; Weaver, S.A.; Chang, G.; Lee, C.C.; Syed, F.; Evans-Molina, C. miR-146a-5p mediates inflammation- induced β cell mitochondrial dysfunction and apoptosis. J. Biol. Chem. 2024, 300, 107827. [Google Scholar] [CrossRef]
- Sims, E.K.; Lakhter, A.J.; Anderson-Baucum, E.; Kono, T.; Tong, X.; Evans-Molina, C. MicroRNA 21 targets BCL2 mRNA to increase apoptosis in rat and human beta cells. Diabetologia 2017, 60, 1057–1065. [Google Scholar] [CrossRef]
- Filios, S.R.; Xu, G.; Chen, J.; Hong, K.; Jing, G.; Shalev, A. MicroRNA-200 is induced by thioredoxin-interacting protein and regulates Zeb1 protein signaling and beta cell apoptosis. J. Biol. Chem. 2014, 289, 36275–36283. [Google Scholar] [CrossRef]
- Yoshihara, E. TXNIP/TBP-2: A master regulator for glucose homeostasis. Antioxidants 2020, 9, 765. [Google Scholar] [CrossRef] [PubMed]
- Ventriglia, G.; Mancarella, F.; Sebastiani, G.; Cook, D.P.; Mallone, R.; Mathieu, C.; Gysemans, C.; Dotta, F. miR-409-3p is reduced in plasma and islet immune infiltrates of NOD diabetic mice and is differentially expressed in people with type 1 diabetes. Diabetologia 2020, 63, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Cao, Y.; Chen, S.; Ruiz, M.; Chakrabarti, S. Reprint of: MiRNA-1 regulates endothelin-1 in diabetes. Life Sci. 2014, 118, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.S.; Ferreira, L.R.P.; da Silva, A.C.; Alves, L.I.; Damasceno, J.G.; Kulikowski, L.; Cunha-Neto, E.; da Silva, M.E.R. Progression of type 1 diabetes: Circulating microRNA expression profiles changes from preclinical to overt disease. J. Immunol. Res. 2022, 2022, 2734490. [Google Scholar] [CrossRef]
- Taheri, M.; Eghtedarian, R.; Dinger, M.E.; Ghafouri-Fard, S. Emerging roles of non-coding RNAs in the pathogenesis of type 1 diabetes mellitus. Biomed. Pharmacother. 2020, 129, 110509. [Google Scholar] [CrossRef]
- Garavelli, S.; Bruzzaniti, S.; Tagliabue, E.; Di Silvestre, D.; Prattichizzo, F.; Mozzillo, E.; Fattorusso, V.; La Sala, L.; Ceriello, A.; Puca, A.A.; et al. Plasma circulating miR-23˜ 27˜ 24 clusters correlate with the immunometabolic derangement and predict C-peptide loss in children with type 1 diabetes. Diabetologia 2020, 63, 2699–2712. [Google Scholar] [CrossRef]
- Frørup, C.; Mirza, A.H.; Yarani, R.; Nielsen, L.B.; Mathiesen, E.R.; Damm, P.; Svare, J.; Engelbrekt, C.; Størling, J.; Johannesen, J.; et al. Plasma exosome-enriched extracellular vesicles from lactating mothers with type 1 diabetes contain aberrant levels of miRNAs during the postpartum period. Front. Immunol. 2021, 12, 744509. [Google Scholar] [CrossRef]
- Mirza, A.H.; Kaur, S.; Nielsen, L.B.; Størling, J.; Yarani, R.; Roursgaard, M.; Mathiesen, E.R.; Damm, P.; Svare, J.; Mortensen, H.B.; et al. Breast milk-derived extracellular vesicles enriched in exosomes from mothers with type 1 diabetes contain aberrant levels of microRNAs. Front. Immunol. 2019, 10, 2543. [Google Scholar] [CrossRef]
- Aljani, B.; Lindner, A.; Weigelt, M.; Zhao, M.; Sharma, V.; Bonifacio, E.; Jones, P.; Eugster, A. Small RNA-Seq and real time rt-qPCR reveal islet miRNA released under stress conditions. Islets 2024, 16, 2392343. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; How Huang, K.; Jen Lee, M.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Mustafin, R.N. Relationship of MicroRNAs with Transposable Elements in the Type 1 Diabetes Development. Russ. Arch. Intern. Med. 2024, 13, 413–421. (In Russian) [Google Scholar] [CrossRef]
- Osipova, J.; Fischer, D.C.; Dangwal, S.; Volkmann, I.; Widera, C.; Schwarz, K.; Lorenzen, J.M.; Schreiver, C.; Jacoby, U.; Heimhalt, M.; et al. Diabetes-associated microRNAs in pediatric patients with type 1 diabetes mellitus: A cross-sectional cohort study. J. Clin. Endocrinol. Metab. 2014, 99, E1661–E1665. [Google Scholar] [CrossRef] [PubMed]
- Blood collection tube and RNA purification method recommendations for extracellular RNA transcriptome profiling. Nat. Commun. 2025, 16, 4513. [CrossRef]
- Swolin-Eide, D.; Forsander, G.; Pundziute Lyckå, A.; Novak, D.; Grillari, J.; Diendorfer, A.B.; Hackl, M.; Magnusson, P. Circulating microRNAs in young individuals with long-duration type 1 diabetes in comparison with healthy controls. Sci. Rep. 2023, 13, 11634. [Google Scholar] [CrossRef]
- Nizam, R.; Malik, M.Z.; Jacob, S.; Alsmadi, O.; Koistinen, H.A.; Tuomilehto, J.; Alkandari, H.; Al-Mulla, F.; Thanaraj, T.A. Circulating hsa-miR-320a and its regulatory network in type 1 diabetes mellitus. Front. Immunol. 2024, 15, 1376416. [Google Scholar] [CrossRef]
- Larsson, A.J.; Johnsson, P.; Hagemann-Jensen, M.; Hartmanis, L.; Faridani, O.R.; Reinius, B.; Segerstolpe, Å.; Rivera, C.M.; Ren, B.; Sandberg, R. Genomic encoding of transcriptional burst kinetics. Nature 2019, 565, 251–254. [Google Scholar] [CrossRef]
- Ngara, M.; Wierup, N. Lessons from single-cell RNA sequencing of human islets. Diabetologia 2022, 65, 1241–1250. [Google Scholar] [CrossRef]
- Van de Sande, B.; Lee, J.S.; Mutasa-Gottgens, E.; Naughton, B.; Bacon, W.; Manning, J.; Wang, Y.; Pollard, J.; Mendez, M.; Hill, J.; et al. Applications of single-cell RNA sequencing in drug discovery and development. Nat. Rev. Drug Discov. 2023, 22, 496–520. [Google Scholar] [CrossRef] [PubMed]
- Pu, Z.; Chen, S.; Lu, Y.; Wu, Z.; Cai, Z.; Mou, L. Exploring the molecular mechanisms of macrophages in islet transplantation using single-cell analysis. Front. Immunol. 2024, 15, 1407118. [Google Scholar] [CrossRef] [PubMed]
- Ashton, M.P.; Eugster, A.; Dietz, S.; Loebel, D.; Lindner, A.; Kuehn, D.; Taranko, A.E.; Heschel, B.; Gavrisan, A.; Ziegler, A.G.; et al. Association of Dendritic cell signatures with autoimmune inflammation revealed by Single-Cell profiling. Arthritis Rheumatol. 2019, 71, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Fasolino, M.; Schwartz, G.W.; Patil, A.R.; Mongia, A.; Golson, M.L.; Wang, Y.J.; Morgan, A.; Liu, C.; Schug, J.; Liu, J.; et al. Single-cell multi-omics analysis of human pancreatic islets reveals novel cellular states in type 1 diabetes. Nat. Metab. 2022, 4, 284–299. [Google Scholar] [CrossRef]
- Ji, L.; Guo, W. Single-cell RNA sequencing highlights the roles of C1QB and NKG7 in the pancreatic islet immune microenviron- ment in type 1 diabetes mellitus. Pharmacol. Res. 2023, 187, 106588. [Google Scholar] [CrossRef]
- Nakayama, M.; Michels, A.W. Using the T cell receptor as a biomarker in type 1 diabetes. Front. Immunol. 2021, 12, 777788. [Google Scholar] [CrossRef]
- Mitchell, A.M.; Baschal, E.E.; McDaniel, K.A.; Simmons, K.M.; Pyle, L.; Waugh, K.; Steck, A.K.; Yu, L.; Gottlieb, P.A.; Rewers, M.J.; et al. Temporal development of T cell receptor repertoires during childhood in health and disease. JCI Insight 2022, 7, e161885. [Google Scholar] [CrossRef]
- Pogorelyy, M.V.; Kirk, A.M.; Adhikari, S.; Minervina, A.A.; Sundararaman, B.; Vegesana, K.; Brice, D.C.; Scott, Z.B.; Thomas, P.G.; Team, S.S.; et al. TIRTL-seq: Deep, quantitative, and affordable paired TCR repertoire sequencing. bioRxiv 2024. [Google Scholar] [CrossRef]
- Hanna, S.J.; Bonami, R.H.; Corrie, B.; Westley, M.; Posgai, A.L.; Luning Prak, E.T.; Breden, F.; Michels, A.W.; Brusko, T.M.; Type 1 Diabetes AIRR Consortium. The Type 1 Diabetes T Cell Receptor and B Cell Receptor Repository in the AIRR Data Commons: A practical guide for access, use and contributions through the Type 1 Diabetes AIRR Consortium. Diabetologia 2025, 68, 186–202. [Google Scholar] [CrossRef]
- Nogueira, M.S.; Barreto, A.L.; Furukawa, M.; Rovai, E.S.; Bastos, A.; Bertoncello, G.; e Silva, L.F.d.C. FTIR spectroscopy as a point of care diagnostic tool for diabetes and periodontitis: A saliva analysis approach. Photodiagn. Photodyn. Ther. 2022, 40, 103036. [Google Scholar] [CrossRef]
- Kondrakhova, D.; Unger, M.; Stadler, H.; Zakut’anská, K.; Tomašovicˇová, N.; Tomecˇková, V.; Horák, J.; Kimákova, T.; Komanicky, V. Determination diabetes mellitus disease markers in tear fluid by photothermal AFM-IR analysis. Nanomed. Nanotechnol. Biol. Med. 2025, 64, 102803. [Google Scholar] [CrossRef] [PubMed]
- Pang, W.; Xing, Y.; Morais, C.L.; Lao, Q.; Li, S.; Qiao, Z.; Li, Y.; Singh, M.N.; Barauna, V.G.; Martin, F.L.; et al. Serum-based ATR-FTIR spectroscopy combined with multivariate analysis for the diagnosis of pre-diabetes and diabetes. Analyst 2024, 149, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Rostoka, E.; Shvirksts, K.; Salna, E.; Trapina, I.; Fedulovs, A.; Grube, M.; Sokolovska, J. Prediction of type 1 diabetes with machine learning algorithms based on FTIR spectral data in peripheral blood mononuclear cells. Anal. Methods 2023, 15, 4926–4937. [Google Scholar] [CrossRef] [PubMed]
- Synytsya, A.; Janstova, D.; Šmidová, M.; Synytsya, A.; Petrtýl, J. Evaluation of IR and Raman spectroscopic markers of human collagens: Insides for indicating colorectal carcinogenesis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 296, 122664. [Google Scholar] [CrossRef]
- Woldaregay, A.Z.; Årsand, E.; Walderhaug, S.; Albers, D.; Mamykina, L.; Botsis, T.; Hartvigsen, G. Data-driven modeling and prediction of blood glucose dynamics: Machine learning applications in type 1 diabetes. Artif. Intell. Med. 2019, 98, 109–134. [Google Scholar] [CrossRef]
- Yang, X.; Fang, T.; Li, Y.; Guo, L.; Li, F.; Huang, F.; Li, L. Pre-diabetes diagnosis based on ATR-FTIR spectroscopy combined with CART and XGBoots. Optik 2019, 180, 189–198. [Google Scholar] [CrossRef]
- Wu, X.; Shuai, W.; Chen, C.; Chen, X.; Luo, C.; Chen, Y.; Shi, Y.; Li, Z.; Lv, X.; Chen, C.; et al. Rapid screening for autoimmune diseases using Fourier transform infrared spectroscopy and deep learning algorithms. Front. Immunol. 2023, 14, 1328228. [Google Scholar] [CrossRef]
- American Diabetes Association Professional Practice Committee. 2. Diagnosis and classification of diabetes: Standards of care in diabetes—2025. Diabetes Care 2025, 48, S27–S49. [Google Scholar] [CrossRef]
- Sims, E.K.; Besser, R.E.; Dayan, C.; Geno Rasmussen, C.; Greenbaum, C.; Griffin, K.J.; Hagopian, W.; Knip, M.; Long, A.E.; Martin, F.; et al. Screening for type 1 diabetes in the general population: A status report and perspective. Diabetes 2022, 71, 610–623. [Google Scholar] [CrossRef]
- INNODIA. Available online: https://www.innodia.eu/ (accessed on 12 May 2025).
- Balmas, E.; Chen, J.; Hu, A.K.; DeBerg, H.A.; Rosasco, M.G.; Gersuk, V.H.; Serti, E.; Speake, C.; Greenbaum, C.J.; Nepom, G.T.; et al. Islet-autoreactive CD4+ T cells are linked with response to alefacept in type 1 diabetes. JCI Insight 2023, 8, e167881. [Google Scholar] [CrossRef]
- Edwards, T.H.; Chen, J.; Dufort, M.; Hartley, R.; Speake, C.; Greenbaum, C.; Linsley, P.; Cerosaletti, K. 93-OR: Clonal Expansion of Islet Reactive CD4 T-Cells with Early Disease Progression in Type 1 Diabetes. Diabetes 2024, 73, 93–OR. [Google Scholar] [CrossRef]
- Speake, C.; Bahnson, H.T.; Wesley, J.D.; Perdue, N.; Friedrich, D.; Pham, M.N.; Lanxon-Cookson, E.; Kwok, W.W.; Sehested Hansen, B.; von Herrath, M.; et al. Systematic assessment of immune marker variation in type 1 diabetes: A prospective longitudinal study. Front. Immunol. 2019, 10, 2023. [Google Scholar] [CrossRef]
- Abdullatypov, A.V.; Glushkova, O.V.; Petriaikina, E.S.; Bogdanov, V.P.; Antysheva, Z.G.; Tabakov, D.V.; Akimov, V.E.; Yudin, V.S.; Keskinov, A.A.; Yudin, S.M.; et al. Single-cell RNA sequencing (scRNA-seq) in studies of type 1 diabetes mellitus (T1D): Modern state-of-the-art and technical peculiarities. Front. Endocrinol. 2025, 16, 1663728. [Google Scholar] [CrossRef]
| Haplotype | Effect | Hetero/Homo | Source |
|---|---|---|---|
| HLA-DQA1*05:01/DQB1*02:01 | Susceptibility | - | [20] |
| HLA-DRA1*01:01/DRB1*04:01 | Susceptibility | - | [20] |
| HLA-DRA1*01:01/DRB1*04:05 | Susceptibility | - | [20] |
| HLA-DQA1*03:01/DQB1*03:02 | Susceptibility | both | [21] |
| HLA-DRB1*03:04 | Susceptibility | both | [21] |
| HLA-DRB1*03:01 | Susceptibility | both | [21] |
| HLA-DRB1*04:04 | Susceptibility | both | [21] |
| HLA-DRB1*04:01 | Susceptibility | both | [21] |
| HLA-DRB1*08:01 | Susceptibility | - | [22] |
| HLA-DPA1*01:03/DPB1*03:01 | Susceptibility | - | [23] |
| HLA-DRB1*08:01/DQA1*04:01/DQB1*04:02 | Susceptibility | - | [24] |
| HLA-DRB1*04:01/DQA1*03/DQB1*03:02 | Susceptibility | - | [25] |
| HLA-DRB1*03:01/DQA1*05:01/DQB1*02:01 | Susceptibility | - | [13] |
| HLA-DRB1*04:05 | Susceptibility | - | [13] |
| HLA-DRB1*09:01/DQB1*03:03 | Susceptibility | homo | [26] |
| HLA-DRB1*04:05/DQB1*04:01 | Susceptibility | hetero | [26] |
| HLA-DRB1*08:02/DQB1*03:02 | Susceptibility | hetero | [26] |
| HLA-DRB1*04:04/DQA1*03:01/DQB1*03:02 | Susceptibility | - | [25] |
| HLA-DRB1*04:05/DQA1*03:01/DQB1*03:02 | Susceptibility | - | [13] |
| HLA-DRB1*04:01/DQA1*03:01/DQB1*03:02 | Susceptibility | - | [13] |
| HLA-DRB1*04:02/DQA1*03:01/DQB1*03:02 | Susceptibility | - | [13] |
| HLA-DRB1*03:01/DQA1*05:01/DQB1*02:01 | Susceptibility | - | [13] |
| HLA-DRA1*01:01/DRB1*04:03 | Protection | - | [20] |
| HLA-DQA1*01:02/DQB1*06:02 | Protection | - | [20] |
| HLA-DPA1*01:03/DPB1*04:02 | Protection | - | [23] |
| HLA-DPA1*01:03/DPB1*01:01 | Protection | - | [23] |
| HLA-DRB1*15:01 | Protection | - | [27] |
| HLA-DRB1*13:03/DQA1*05:01/DQB1*03:01 | Protection | - | [13] |
| HLA-DRB1*11:04/DQA1*05:01/DQB1*03:01 | Protection | - | [13] |
| HLA-DRB1*15:01/DQA1*01:02/DQB1*06:02 | Protection | - | [13] |
| HLA-DRB1*07:01/DQA1*02:01/DQB1*03:03 | Protection | - | [13] |
| HLA-DRB1*14:01/DQA1*01:01/DQB1*05:03 | Protection | - | [13] |
| HLA-DQB1*06:02 | Protection | - | [25] |
| SNP | Gene | Gene Product Function | T1D Patients/ Control, N | Population According to Author Data | Sex, Age | Source |
|---|---|---|---|---|---|---|
| rs17885785 | INS | Insulin is involved in carbohydrate metabolism regulation | 1590/10,718 | Canadian, Caucasians | Males and females, 3–17 years, with a mean age of onset of 7.9 years | [28] |
| rs7795896 | CFTR | Cystic fibrosis transmembrane regulator involved in the transport of chloride ions across the cell membrane | 18,942/501,638 | European ancestry, Caucasians | Males and females, exact ages not specified | [29] |
| rs2269241 | PGM1 | Phosphoglucomutase 1 catalyzes the transfer of phosphate between positions 1 and 6 of glucose | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs34090353 | RPAP2 | RNA polymerase II-associated protein 2 is involved in dephosphorylation of the RNA polymerase II C-terminal domain and snRNA transcription | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs2229238 | IL6R | Subunit of the IL6 receptor complex, regulator of IL6 signaling pathways | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs2816313 | RGS1 | Regulator of G protein signaling 1, a critical mediator of T-cell regulatory function | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs11120029 | TATDN3 | TATDN3 protein provides metal ion binding activity and nuclease activity | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs10169963 | AC096559.1 | Non-coding RNA | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs12712067 | AFF3 | Protein AFF3—nuclear transcriptional activator of lymphoid tissue | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs10933559 | FARP2 | Provides activity of guanine nucleotide exchange factor | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [28] |
| rs1876142 | PTGER4 | Prostaglandin E2 (PGE2) receptor, participates in T-cell activation | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [29] |
| rs9405661 | IRF4 | Interferon regulatory factor 4, plays an important role in antiviral responses and in the regulation of interferon-induced genes | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs12665429 | TNFAIP3 | Possesses both ubiquitin ligase and deubiquitinase activity, inhibits the activation of transcription factors NF-kB and AP-1 and cytokine-induced apoptosis | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs17143056 | ABCB5 | Involved in ATP-dependent transmembrane transport of structurally diverse molecules | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs2250903 | CTSB | Cathepsin B, lysosomal cysteine protease with both endopeptidase and exopeptidase activity | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs1405209 | NR4A3 | Transcriptional activator | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs722988 | NRP1 | Neuropilin 1, mediates insulin signaling pathways, involved in signaling pathways controlling cell migration | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs79538630 | CD5/CD6 | CD5 and CD6 are receptors at the interface of the innate and adaptive immune responses of T-lymphocytes, involved in cell adhesion and important for the continued activation of T-cells | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs645078 | CCDC88B | Protein CCDC88B, involved in the binding of organelles to microtubules | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs605093 | FLI1 | Transcription factor containing an ETS DNA-binding domain | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs7313065 | ITGB7 | Integrin beta-7, involved in signaling from the extracellular matrix to the cell, migration of lymphocytes to the intestine | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs74537115 | AKAP11 | Anchor proteins of A-kinase are involved in binding to the regulatory subunit of protein kinase A and localization of holoenzyme within the cell; participant in the cell cycle control system | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs4238595 | UMOD | Uromodulin, inhibitor of calcium crystallization in renal fluids | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs2597169 | PRR15L | Proline-rich 15-like protein: associated with pancreatic cancer subtypes | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs56178904 | ICOSLG | Involved in the T-cell receptor signaling pathway and positive regulation of interleukin-4 production | 25,193/35,476 | European, African American, East Asian, Finnish, mixed ancestry | Males and females, exact ages not specified | [30] |
| rs689 | INS | Insulin involved in carbohydrate metabolism regulation | 3532/3607 | African, European, mixed descent | Males and females, exact ages not specified | [24] |
| rs6679677 | PTPN22 | Non-receptor protein tyrosine phosphatase 22 involved in regulating CBL function in T-cell receptor signaling pathway, T-cell inhibitor | 3532/3607 | African, European, mixed descent | Males and females, exact ages not specified | [24] |
| rs61839660 | IL2RA | IL2 receptor alpha subunit, regulator of immune functions of regulatory T-cells | 3532/3607 | African, European, mixed descent | Males and females, exact ages not specified | [24] |
| rs737391 | RNLS | Renalase provides binding activities for NADH, adrenaline, and monoamine oxidase | 3532/3607 | African, European, mixed descent | Males and females, exact ages not specified | [24] |
| rs7302200 | IKZF4- RPS26- ERBB3 | Intergenic variant | 3532/3607 | African, European, mixed descent | Males and females, exact ages not specified | [24] |
| rs597808 | SH2B3 | Adapter protein encoded by the human gene SH2B3, a key negative regulator of cytokine signaling | 3532/3607 | African, European, mixed descent | Males and females, exact ages not specified | [24] |
| Population | Allele | Haplotype | Reference | ||
|---|---|---|---|---|---|
| Increased Risk | Decreased Risk | Increased Risk | Decreased Risk | ||
| Arabs | DRB1*03:01, DRB1*04:02, DQB1*02:01, DQB1*03:02 | DRB1*11:01, DRB1*16:02, DQB1*03:01, DQB1*06:01 | DRB1*03:01DQB1*02:01, DRB1*04:02DQB1*03:02, DRB1*04:05- DQB1*03:02 | DRB1*16:02DQB1*05:02 | [55] |
| Africans | DQA1*03:01 | — | HLA-DRB1*03:01- DQA1*05:01- DQB1*02:01 | — | [24] |
| Bahrainis | DRB1*03:01:01, DRB1*04:01:01, DQB1*02:01, DQB1*03:02 | DRB1*11:01:01, DQB1*03:01, DQB1*05:01:01 | DRB1*03:01:01- DQB1*02:01, DRB1*04:01:01- DQB1*03:02 | DRB1*10:01:01DQB1*05:01:01 | [56] |
| Brazilians | DRB1*03, DRB1*04, DQA1*03:01, DQA1*03:02, DQA1*05:03, DQA1*05:05 | DRB1*08, DRB1*11, DRB1*13, DRB1*14, DRB1*15, DQA1*01:01, DQA1*01:02, DQA1*01:03, DQA1*04:01 | DRB1*03:01 DQA1*05:01- DQB1*02:01 | — | [57] |
| Buryats | DRB1*04 | DRB1*01, DRB1*11, DRB1*13, DRB1*15 | — | — | [58] |
| Europeans | DQB1*03:02 | — | HLA-DRB1*04:01- DQA1*03:01DQB1*03:02, HLA-DRB1*08:01- DQA1*04:01- DQB1*04:02 | — | [24] |
| Jordanians | DRB1*04, DRB1*03:01, DQA1*03:01, DQA1*05:01, DQB1*02:01, DQB1*03:02 | DRB1*07:01, DRB1*11:01, DQA1*05:05, DQA1*01:03, DQA1*02:01, DQB1*03:01, DQB1*05:01 | DRB1*04DQA1*03:01- DQB1*03:02, DRB1*03:01DQA1*05:01- DQB1*02:01 | DRB1*11:01DQA1*05:05- DQB1*03:01 | [59] |
| Iranians | DRB1*04:01, DRB1*03:01, DQB1*03:02, DQB1*02:01 | DRB1*15:01, DRB1*:01, DQB1*03:01, DQB1*06:01 | DRB1*04:01DQB1*03:02, DRB1*03:01DQB1*02:01, DRB1*07:01- DQB1*03:03 | DRB1*15:01DQB1*06:01, DRB1*11:01- DQB1*03:01 | [60] |
| Kalmyks | DRB1*09 | DRB1*07, DRB1*11, DRB1*15 | — | — | [58] |
| Lebanese | DRB1*03:01:01, DRB1*13:07:01, DQB1*02:01 | DRB1*11:01:01, DQB1*03:01, DQB1*05:01:01 | DRB1*03:01:01 DQB1*02:01 | DRB1*15:01:01 DQB1*06:01:01 | [56] |
| Mari | DRB1*03, DRB1*04 | DRB1*07, DRB1*11, DRB1*13, DRB1*15 | — | — | [58] |
| Russians | DRB1*03, DRB1*04 | DRB1*07, DRB1*11, DRB1*13, DRB1*15, DRB1*16 | — | — | [58] |
| Tatars | DRB1*01, DRB1*03, DRB1*04 | DRB1*07, DRB1*13, DRB1*15 | — | — | [58] |
| Tunisians | DRB1*04:01:01 | DRB1*11:01:01, DQB1*03:01:01, DQB1*06:01:01 | DRB1*03:01:01- DQB1*02:01, DRB1*04:01:01 DQB1*03:02 | — | [56] |
| Tuvans | DRB1*03 | DRB1*13, DRB1*15 | — | — | [58] |
| Udmurts | DRB1*01, DRB1*03, DRB1*04 | DRB1*11, DRB1*13, DRB1*15 | — | — | [58] |
| Uzbeks | DRB1*03, DRB1*04 | DRB1*07, DRB1*13, DRB1*15 | — | — | [58] |
| Swedes | DRB1*04:01, DRB1*04:02, DRB1*04:04, DRB1*04:05 | DRB1*04:03, DRB1*04:07 | — | — | [61] |
| Japanese | HLA-B*54:01, HLA-A amino acid position 62 | — | — | — | [62] |
| Cytokine | Function | Reference |
|---|---|---|
| IL-1 | Causes dysfunction and death of β-cells. | [84] |
| IL-6 | Serves as a key regulator of the migration and inflammatory responses of effector T and B cells. | [85] |
| TNF-α | Enhances the expression of MHC-I molecules, thereby accelerating antigen presentation and apoptosis of β-cells, exhibiting direct cytotoxic effects. | [80] |
| IFN-1 | Induces increased presentation of autoantigens by islet cells, thereby enhancing the activation of effector T-cells. | [81] |
| IFN-α | Facilitates the presentation of self-antigens by islet cells, leading to recognition of these cells by cytotoxic T-lymphocytes. Induces the secretion of various chemokines involved in the recruitment of immune cells, such as T and NK lymphocytes and provokes oxidative stress. | [86,87] |
| IFN-γ | Mediates the destruction of β-cells in local islets and induces aberrant expression of MHC-I and MHC-II in local pancreatic cells, resulting in autoimmune β-cell death. | [88] |
| IL-17 | Through the IL-17RA and RC receptor complex, which is widely present on the surface of islet cells, IL-17A exacerbates islet inflammation by directly inducing apoptosis of β-cells and locally increasing levels of pro-inflammatory cytokines and chemokines. In interaction with IFN-γ and IL-1β, it synergistically induces inflammation and apoptosis in human pancreatic islet cells. | [89] |
| IL-2 | Exerts pleiotropic effects on various immune cell populations, including NK cells, effector T-cells, and Tregs. | [90] |
| IL-4 | Participates in the activation of the PI3K and JAK/STAT pathways, contributing to the viability of insulin-producing cells, as well as stimulating IL-2 synthesis and the activation and expansion of iNKT and Treg cells. | [91] |
| IL-13 | Activates the STAT signaling pathway, suppressing the ongoing destruction of β-cells and preventing the development of T1D. | [92] |
| IL-10 | Induces an increase in the number of Tregs, elevates levels of Th2-type cytokines (IL-4 and IL-10), and reduces the Th1 response (IL-2 and IFN-γ). Moreover, IL-10 is associated with a tolerant state of immature dendritic cells (DCs) and Bregs in humans and mice with T1D, promoting insulin-specific tolerance in effector and memory T-cells generated in T1D patients. | [93,94] |
| TGF-β | Induces the expression of Foxp3 and the differentiation of peripheral Tregs. | [95,96] |
| HGF | The signaling of HGF/c-Met in β-cells is essential for normal growth and function of β-cells under basal conditions and is critically important for the survival of β-cells in diabetes. | [97] |
| Biomarker | Research Only | Available Screening Assay | Guideline- Premorbid Supported Diagnostics | Early β-Cell Destruction Diagnostics | Preventive Therapy Choice | Cost- Effectiveness of Implementing Biomarker Panels * | |
|---|---|---|---|---|---|---|---|
| Clinical biomarkers | |||||||
| Genetic (HLA-haplotype, Non-HLA SNPs, PGS) | - | + [5,31,38] | + [38,178] | Yes, overdiagnosis [38,178] | No | Potential [38,178] | + |
| Islet Antigen Autoantibodies | - | + [7,178,179,180] | + [7,178,179,180] | No | Yes, overdiagnosis [7] | Potential [7] | +++ |
| C-Peptide | - | + [7,178] | + [7,178] | No | No | No | +++ |
| Exploratory biomarkers | |||||||
| Cytokines | + [101] | + [178] | - | No | Potential [81] | Potential [114,115] | +++ |
| cfDNA | + [130,135,136,137,138] | - | - | No | Potential [129,130] | Potential [129,130] | ++ [133,134,137] |
| MicroRNA | + [156] | - | - | No | Potential [149] | Potential [148] | + [147,157,158] |
| T1D specific immune cells | + [166,167] | - | - | Potential [163] | Potential [164,165] | Potential [181,182] | + [29] |
| Islet-TCR | + [167] | - | - | Potential [166] | No | Potential [168,169] | + |
| T1D specific vibrational bands | + [174,175,176] | - | - | Potential [175,176] | Potential [175,176] | Potential [177] | +++ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marakhovskaya, T.A.; Tabakov, D.V.; Glushkova, O.V.; Antysheva, Z.G.; Kiseleva, Y.S.; Petriaikina, E.S.; Bugaev-Makarovskiy, N.A.; Tashchilova, A.S.; Akimov, V.E.; Krupinova, J.A.; et al. Preclinical Diagnosis of Type 1 Diabetes: Reality or Utopia. Biomedicines 2025, 13, 2444. https://doi.org/10.3390/biomedicines13102444
Marakhovskaya TA, Tabakov DV, Glushkova OV, Antysheva ZG, Kiseleva YS, Petriaikina ES, Bugaev-Makarovskiy NA, Tashchilova AS, Akimov VE, Krupinova JA, et al. Preclinical Diagnosis of Type 1 Diabetes: Reality or Utopia. Biomedicines. 2025; 13(10):2444. https://doi.org/10.3390/biomedicines13102444
Chicago/Turabian StyleMarakhovskaya, Tatyana A., Dmitry V. Tabakov, Olga V. Glushkova, Zoya G. Antysheva, Yaroslava S. Kiseleva, Ekaterina S. Petriaikina, Nickolay A. Bugaev-Makarovskiy, Anna S. Tashchilova, Vasiliy E. Akimov, Julia A. Krupinova, and et al. 2025. "Preclinical Diagnosis of Type 1 Diabetes: Reality or Utopia" Biomedicines 13, no. 10: 2444. https://doi.org/10.3390/biomedicines13102444
APA StyleMarakhovskaya, T. A., Tabakov, D. V., Glushkova, O. V., Antysheva, Z. G., Kiseleva, Y. S., Petriaikina, E. S., Bugaev-Makarovskiy, N. A., Tashchilova, A. S., Akimov, V. E., Krupinova, J. A., Bogdanov, V. P., Frolova, T. M., Shchekina, V. S., Avsievich, E. S., Gorev, V. V., Rybkina, I. G., Osmanov, I. M., Kolomina, I. G., Khatkov, I. E., ... Skvortsova, V. I. (2025). Preclinical Diagnosis of Type 1 Diabetes: Reality or Utopia. Biomedicines, 13(10), 2444. https://doi.org/10.3390/biomedicines13102444






