Hippo Signaling Dysregulation in Breast Cancer: Subtype-Independent Gene and miRNA Signatures
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Isolation of Total RNA
2.3. mRNA Profiling by Microarrays
2.4. Gene Expression Analysis by Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
2.6. miRNA Profiling and Target Prediction
2.7. Statistical Analysis
3. Results
3.1. mRNA Microarray-Based Gene Expression Profiling
3.2. RT-qPCR and ELISA Analysis of BIRC5, FGF1, RASSF6, SERPINE1 and STK4 Expression
3.3. Prediction of miRNA Targets
3.4. Overall Survival Outcomes Across Breast Cancer Subtypes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACTB | β-actin |
| BIRC5 | baculoviral IAP repeat containing 5 |
| ELISA | Enzyme-Linked Immunosorbent Assay |
| EMT | epithelial–mesenchymal transition |
| FGF1 | fibroblast growth factor 1 |
| LATS1 | large tumor suppressor kinase 1 |
| LATS2 | large tumor suppressor kinase 2 |
| OS | Overall survival |
| RASSF6 | Ras association domain family member 6 |
| RT-qPCR | Reverse Transcription Quantitative Polymerase Chain Reaction |
| SAV1 | Salvador |
| SERPINE1 | serpin family E member 1 |
| STK3 | serine/threonine-protein kinase 3 |
| STK4 | serine/threonine-protein kinase 4 |
| TAZ | transcriptional coactivator with PDZ-binding motif |
| TEAD | transcriptional enhanced associate domain |
| TNBC | triple-negative breast cancer |
| YAP | Yes-associated protein |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Gąska, I.; Czerw, A.; Pajewska, M.; Partyka, O.; Deptała, A.; Badowska-Kozakiewicz, A.; Budzik, M.; Sygit, K.; Wojtyła-Buciora, P.; Drobnik, J.; et al. The Cost of Breast Cancer: Economic and Social Perspective. Cancers 2025, 17, 3012. [Google Scholar] [CrossRef]
- Roy, M.; Fowler, A.M.; Ulaner, G.A.; Mahajan, A. Molecular Classification of Breast Cancer. PET Clin. 2023, 18, 441–458. [Google Scholar] [CrossRef]
- Fu, M.; Hu, Y.; Lan, T.; Guan, K.-L.; Luo, T.; Luo, M. The Hippo Signalling Pathway and Its Implications in Human Health and Diseases. Signal Transduct. Target. Ther. 2022, 7, 376. [Google Scholar] [CrossRef]
- Ma, S.; Meng, Z.; Chen, R.; Guan, K.-L. The Hippo Pathway: Biology and Pathophysiology. Annu. Rev. Biochem. 2019, 88, 577–604. [Google Scholar] [CrossRef]
- Sebio, A.; Lenz, H.-J. Molecular Pathways: Hippo Signaling, a Critical Tumor Suppressor. Clin. Cancer Res. 2015, 21, 5002–5007. [Google Scholar] [CrossRef]
- Ghaboura, N. Unraveling the Hippo Pathway: YAP/TAZ as Central Players in Cancer Metastasis and Drug Resistance. EXCLI J. 2025, 24, 612–637. [Google Scholar] [CrossRef] [PubMed]
- Misra, J.R.; Irvine, K.D. The Hippo Signaling Network and Its Biological Functions. Annu. Rev. Genet. 2018, 52, 65–87. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Pan, D. The Hippo Signaling Pathway in Development and Disease. Dev. Cell 2019, 50, 264–282. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Jiao, Z.; Yu, F.-X. The Hippo Signaling Pathway in Development and Regeneration. Cell Rep. 2024, 43, 113926. [Google Scholar] [CrossRef]
- Di, X.; Gao, X.; Peng, L.; Ai, J.; Jin, X.; Qi, S.; Li, H.; Wang, K.; Luo, D. Cellular Mechanotransduction in Health and Diseases: From Molecular Mechanism to Therapeutic Targets. Signal Transduct. Target. Ther. 2023, 8, 282. [Google Scholar] [CrossRef]
- Jiang, L.; Li, J.; Zhang, C.; Shang, Y.; Lin, J. YAP-mediated Crosstalk Between the Wnt and Hippo Signaling Pathways (Review). Mol. Med. Rep. 2020, 22, 4101–4106. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wei, W.; Tu, S.; Liang, B.; Li, C.; Li, Y.; Luo, W.; Wu, Y.; Dai, X.; Wang, Y.; et al. Upregulation of CYR61 by TGF-β and YAP Signaling Exerts a Counter-Suppression of Hepatocellular Carcinoma. J. Biol. Chem. 2024, 300, 107208. [Google Scholar] [CrossRef]
- Borreguero-Muñoz, N.; Fletcher, G.C.; Aguilar-Aragon, M.; Elbediwy, A.; Vincent-Mistiaen, Z.I.; Thompson, B.J. The Hippo Pathway Integrates PI3K–Akt Signals with Mechanical and Polarity Cues to Control Tissue Growth. PLoS Biol. 2019, 17, e3000509. [Google Scholar] [CrossRef]
- Paul, S.; Hagenbeek, T.J.; Tremblay, J.; Kameswaran, V.; Ong, C.; Liu, C.; Guarnaccia, A.D.; Mondo, J.A.; Hsu, P.L.; Kljavin, N.M.; et al. Cooperation Between the Hippo and MAPK Pathway Activation Drives Acquired Resistance to TEAD Inhibition. Nat. Commun. 2025, 16, 1743. [Google Scholar] [CrossRef]
- Kandettu, A.; Radhakrishnan, R.; Chakrabarty, S.; Sriharikrishnaa, S.; Kabekkodu, S.P. The Emerging Role of miRNA Clusters in Breast Cancer Progression. Biochim. Et Biophys. Acta (BBA)—Rev. Cancer 2020, 1874, 188413. [Google Scholar] [CrossRef]
- Sirek, T.; Sirek, A.; Zmarzły, N.; Opławski, M.; Król-Jatręga, K.; Boroń, D.; Chalcarz, M.; Ossowski, P.; Dziobek, K.; Strojny, D.; et al. Impact of MiRNAs on Wnt-Related Gene Activity in Breast Cancer. Sci. Rep. 2025, 15, 16211. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An Online Database for Prediction of Functional microRNA Targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Győrffy, B. Integrated Analysis of Public Datasets for the Discovery and Validation of Survival-Associated Genes in Solid Tumors. Innovation 2024, 5, 100625. [Google Scholar] [CrossRef]
- Győrffy, B. Transcriptome-Level Discovery of Survival-Associated Biomarkers and Therapy Targets in Non-Small-Cell Lung Cancer. Br. J. Pharmacol. 2024, 181, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G*Power 3: A Flexible Statistical Power Analysis Program for the Social, Behavioral, and Biomedical Sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]
- Meng, Z.; Moroishi, T.; Guan, K.-L. Mechanisms of Hippo Pathway Regulation. Genes. Dev. 2016, 30, 1–17. [Google Scholar] [CrossRef]
- Zhou, D.; Conrad, C.; Xia, F.; Park, J.-S.; Payer, B.; Yin, Y.; Lauwers, G.Y.; Thasler, W.; Lee, J.T.; Avruch, J.; et al. Mst1 and Mst2 Maintain Hepatocyte Quiescence and Suppress the Development of Hepatocellular Carcinoma Through Inactivation of the Yap1 Oncogene. Cancer Cell 2009, 16, 425–438. [Google Scholar] [CrossRef]
- Lin, X.; Cai, F.; Li, X.; Kong, X.; Xu, C.; Zuo, X.; Yang, Q. Prognostic Significance of Mammalian Sterile 20-like Kinase 1 in Breast Cancer. Tumour Biol. 2013, 34, 3239–3243. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.-Y.; Cai, F.-F.; Wang, M.-H.; Pan, X.; Wang, F.; Cai, L.; Cui, R.-R.; Chen, S.; Biskup, E. Mammalian Sterile 20-like Kinase 1 Expression and Its Prognostic Significance in Patients with Breast Cancer. Oncol. Lett. 2017, 14, 5457–5463. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Zhu, L.; Xiao, S.; Cui, Z.; Tang, J.; Yu, J.; Xie, M. MST1 Inhibits the Progression of Breast Cancer by Regulating the Hippo Signaling Pathway and May Serve as a Prognostic Biomarker. Mol. Med. Rep. 2021, 23, 383. [Google Scholar] [CrossRef]
- Li, L.; Fang, R.; Liu, B.; Shi, H.; Wang, Y.; Zhang, W.; Zhang, X.; Ye, L. Deacetylation of Tumor-Suppressor MST1 in Hippo Pathway Induces Its Degradation Through HBXIP-Elevated HDAC6 in Promotion of Breast Cancer Growth. Oncogene 2016, 35, 4048–4057. [Google Scholar] [CrossRef]
- Ma, G.; Xue, W.; Ni, J.; Tao, R. MiR-522-3p Targets Transcription Factor 4 to Overcome Cisplatin Resistance of Gastric Cells. J. Oncol. 2022, 2022, 6082373. [Google Scholar] [CrossRef]
- Miyamoto, M.; Sawada, K.; Nakamura, K.; Yoshimura, A.; Ishida, K.; Kobayashi, M.; Shimizu, A.; Yamamoto, M.; Kodama, M.; Hashimoto, K.; et al. Paclitaxel Exposure Downregulates miR-522 Expression and Its Downregulation Induces Paclitaxel Resistance in Ovarian Cancer Cells. Sci. Rep. 2020, 10, 16755. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, P.; Tan, Y.; Feng, Q.; Zhao, R. MicroRNA-522-3p Plays an Oncogenic Role in Glioblastoma Through Activating Wnt/β-Catenin Signaling Pathway via Targeting SFRP2. Neuroreport 2021, 32, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Bao, H.; Zhang, S.; Li, C.; Sun, G.; Sun, X.; Fu, T.; Wang, Y.; Liang, P. MicroRNA-522-3p Promotes Brain Metastasis in Non-Small Cell Lung Cancer by Targeting Tensin 1 and Modulating Blood-Brain Barrier Permeability. Exp. Cell Res. 2024, 442, 114199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Pan, Y.; Jin, L.; Yang, H.; Cao, P. Exosomal-miR-522-3p Derived from Cancer-Associated Fibroblasts Accelerates Tumor Metastasis and Angiogenesis via Repression Bone Morphogenetic Protein 5 in Colorectal Cancer. J. Gastroenterol. Hepatol. 2024, 39, 107–120. [Google Scholar] [CrossRef]
- Tan, S.M.; Kirchner, R.; Jin, J.; Hofmann, O.; McReynolds, L.; Hide, W.; Lieberman, J. Sequencing of Captive Target Transcripts Identifies the Network of Regulated Genes and Functions of Primate-Specific miR-522. Cell Rep. 2014, 8, 1225–1239. [Google Scholar] [CrossRef]
- Dong, Y.; Long, J.; Luo, X.; Xie, G.; Xiao, Z.J.; Tong, Y. Targeting of ΔNp63α by miR-522 Promotes the Migration of Breast Epithelial Cells. FEBS Open Bio 2021, 11, 468–481. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, W.; Wu, J.; Zhou, Z.; Ma, J. miR-522 Regulates Cell Proliferation, Migration, Invasion Capacities and Acts as a Potential Biomarker to Predict Prognosis in Triple-Negative Breast Cancer. Clin. Exp. Med. 2022, 22, 385–392. [Google Scholar] [CrossRef]
- Wen, Y.; Wang, Q.; Zhou, C.; Yan, D.; Qiu, G.; Yang, C.; Tang, H.; Peng, Z. Decreased Expression of RASSF6 Is a Novel Independent Prognostic Marker of a Worse Outcome in Gastric Cancer Patients after Curative Surgery. Ann. Surg. Oncol. 2011, 18, 3858–3867. [Google Scholar] [CrossRef]
- Ye, H.-L.; Li, D.-D.; Lin, Q.; Zhou, Y.; Zhou, Q.-B.; Zeng, B.; Fu, Z.-Q.; Gao, W.-C.; Liu, Y.-M.; Chen, R.-W.; et al. Low RASSF6 Expression in Pancreatic Ductal Adenocarcinoma Is Associated with Poor Survival. World J. Gastroenterol. 2015, 21, 6621–6630. [Google Scholar] [CrossRef]
- Tan, S.; Bian, X.; Wu, B.; Chen, X. RASSF6 Is Downregulated In Human Bladder Cancers And Regulates Doxorubicin Sensitivity And Mitochondrial Membrane Potential via the Hippo Signaling Pathway. Onco Targets Ther. 2019, 12, 9189–9200. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhao, T.-T.; Jin, F.; Li, J.-G.; Xu, Y.-Y.; Dong, H.-T.; Liu, Q.; Xing, P.; Zhu, G.-L.; Xu, H.; et al. Downregulation of RASSF6 Promotes Breast Cancer Growth and Chemoresistance Through Regulation of Hippo Signaling. Biochem. Biophys. Res. Commun. 2018, 503, 2340–2347. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Iwasa, H.; Hossain, S.; Xu, X.; Sawada, T.; Shimizu, T.; Maruyama, J.; Arimoto-Matsuzaki, K.; Hata, Y. Domain Analysis of Ras-Association Domain Family Member 6 upon Interaction with MDM2. FEBS Lett. 2017, 591, 260–272. [Google Scholar] [CrossRef]
- Hossain, S.; Iwasa, H.; Sarkar, A.; Maruyama, J.; Arimoto-Matsuzaki, K.; Hata, Y. The RASSF6 Tumor Suppressor Protein Regulates Apoptosis and Cell Cycle Progression via Retinoblastoma Protein. Mol. Cell Biol. 2018, 38, e00046-18. [Google Scholar] [CrossRef]
- Teven, C.M.; Farina, E.M.; Rivas, J.; Reid, R.R. Fibroblast Growth Factor (FGF) Signaling in Development and Skeletal Diseases. Genes. Dis. 2014, 1, 199–213. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewska, M.; Opalinski, L.; Haugsten, E.M.; Otlewski, J.; Wiedlocha, A. Crosstalk Between P38 and Erk 1/2 in Downregulation of FGF1-Induced Signaling. Int. J. Mol. Sci. 2019, 20, 1826. [Google Scholar] [CrossRef]
- Jeong, S.-H.; Kim, H.-B.; Kim, M.-C.; Lee, J.; Lee, J.H.; Kim, J.-H.; Kim, J.-W.; Park, W.-Y.; Kim, S.-Y.; Kim, J.B.; et al. Hippo-Mediated Suppression of IRS2/AKT Signaling Prevents Hepatic Steatosis and Liver Cancer. J. Clin. Investig. 2018, 128, 1010–1025. [Google Scholar] [CrossRef]
- Ferguson, H.R.; Smith, M.P.; Francavilla, C. Fibroblast Growth Factor Receptors (FGFRs) and Noncanonical Partners in Cancer Signaling. Cells 2021, 10, 1201. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, Y.; Yu, J.; Guo, R. FGF Exhibits an Important Biological Role on Regulating Cell Proliferation of Breast Cancer When It Transports Into The Cell Nuclei. Cell Biochem. Biophys. 2022, 80, 311–320. [Google Scholar] [CrossRef]
- Szymczyk, J.; Czyrek, A.; Otlewski, J.; Zakrzewska, M. FGF1 Protects MCF-7 Cells against Taltobulin Through Both the MEKs/ERKs and PI3K/AKT Signaling Pathway. Biomedicines 2023, 11, 1856. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Castrejon, M.; Sankofi, B.M.; Murguia, S.J.; Udeme, A.-A.; Cen, H.H.; Xia, Y.H.; Thomas, N.S.; Berry, W.L.; Jones, K.L.; Richard, V.R.; et al. FGF1 Supports Glycolytic Metabolism Through the Estrogen Receptor in Endocrine-Resistant and Obesity-Associated Breast Cancer. Breast Cancer Res. 2023, 25, 99. [Google Scholar] [CrossRef]
- Lee, H.; Cho, S.W.; Cha, H.S.; Tae, K.; Choi, C.Y. Transient Activation of YAP/TAZ Confers Resistance to Morusin-Induced Apoptosis. BMC Mol. Cell Biol. 2025, 26, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Greene, M.I. Survivin as a Therapeutic Target for the Treatment of Human Cancer. Cancers 2024, 16, 1705. [Google Scholar] [CrossRef]
- Fäldt Beding, A.; Larsson, P.; Helou, K.; Einbeigi, Z.; Parris, T.Z. Pan-Cancer Analysis Identifies BIRC5 as a Prognostic Biomarker. BMC Cancer 2022, 22, 322. [Google Scholar] [CrossRef]
- Li, G.; Wang, Y.; Wang, W.; Lv, G.; Li, X.; Wang, J.; Liu, X.; Yuan, D.; Deng, S.; You, D. BIRC5 as a Prognostic and Diagnostic Biomarker in Pan-Cancer: An Integrated Analysis of Expression, Immune Subtypes, and Functional Networks. Front. Genet. 2024, 15, 1509342. [Google Scholar] [CrossRef] [PubMed]
- Oparina, N.; Erlandsson, M.C.; Fäldt Beding, A.; Parris, T.; Helou, K.; Karlsson, P.; Einbeigi, Z.; Bokarewa, M.I. Prognostic Significance of BIRC5/Survivin in Breast Cancer: Results from Three Independent Cohorts. Cancers 2021, 13, 2209. [Google Scholar] [CrossRef] [PubMed]
- Mehraj, U.; Aisha, S.; Sofi, S.; Mir, M.A. Expression Pattern and Prognostic Significance of Baculoviral Inhibitor of Apoptosis Repeat-Containing 5 (BIRC5) in Breast Cancer: A Comprehensive Analysis. Adv. Cancer Biol.—Metastasis 2022, 4, 100037. [Google Scholar] [CrossRef]
- Hamilton, A.M.; Walens, A.; Van Alsten, S.C.; Olsson, L.T.; Nsonwu-Farley, J.; Gao, X.; Kirk, E.L.; Perou, C.M.; Carey, L.A.; Troester, M.A.; et al. BIRC5 Expression by Race, Age and Clinical Factors in Breast Cancer Patients. Breast Cancer Res. 2024, 26, 50. [Google Scholar] [CrossRef] [PubMed]
- Al-Yahya, S.; Al-Saif, M.; Al-Ghamdi, M.; Moghrabi, W.; Khabar, K.S.A.; Al-Souhibani, N. Post-Transcriptional Regulation of BIRC5/Survivin Expression and Induction of Apoptosis in Breast Cancer Cells by Tristetraprolin. RNA Biol. 2023, 21, 1–15. [Google Scholar] [CrossRef]
- Mathews, S.G.; Krishna, R.B.D.; M, L.; K, N.; Murali, S.; Agarwal, P.; Rani, E.; F, A.M. The Role of the Plasminogen Activator Inhibitor 1 (PAI1) in Ovarian Cancer: Mechanisms and Therapeutic Implications. Glob. Med. Genet. 2024, 11, 358–365. [Google Scholar] [CrossRef]
- Kong, H.-J.; Kwon, E.-J.; Kwon, O.-S.; Lee, H.; Choi, J.-Y.; Kim, Y.-J.; Kim, W.; Cha, H.-J. Crosstalk Between YAP and TGFβ Regulates SERPINE1 Expression in Mesenchymal Lung Cancer Cells. Int. J. Oncol. 2021, 58, 111–121. [Google Scholar] [CrossRef]
- Chen, S.; Li, Y.; Zhu, Y.; Fei, J.; Song, L.; Sun, G.; Guo, L.; Li, X. SERPINE1 Overexpression Promotes Malignant Progression and Poor Prognosis of Gastric Cancer. J. Oncol. 2022, 2022, 2647825. [Google Scholar] [CrossRef]
- Polo-Generelo, S.; Rodríguez-Mateo, C.; Torres, B.; Pintor-Tortolero, J.; Guerrero-Martínez, J.A.; König, J.; Vázquez, J.; Bonzón-Kulichenco, E.; Padillo-Ruiz, J.; de la Portilla, F.; et al. Serpine1 mRNA Confers Mesenchymal Characteristics to the Cell and Promotes CD8+ T Cells Exclusion from Colon Adenocarcinomas. Cell Death Discov. 2024, 10, 116. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Chen, S.; Zhu, C.; Shi, Y.; Dang, S.; Zhang, W.; Li, W. Pan-Cancer Analysis of SERPINE Family Genes as Biomarkers of Cancer Prognosis and Response to Therapy. Front. Mol. Biosci. 2023, 10, 1277508. [Google Scholar] [CrossRef]
- Ferroni, P.; Roselli, M.; Portarena, I.; Formica, V.; Riondino, S.; LA Farina, F.; Costarelli, L.; Melino, A.; Massimiani, G.; Cavaliere, F.; et al. Plasma Plasminogen Activator Inhibitor-1 (PAI-1) Levels in Breast Cancer—Relationship with Clinical Outcome. Anticancer. Res. 2014, 34, 1153–1161. [Google Scholar]
- Zhang, Q.; Lei, L.; Jing, D. Knockdown of SERPINE1 Reverses Resistance of Triple-negative Breast Cancer to Paclitaxel via Suppression of VEGFA. Oncol. Rep. 2020, 44, 1875–1884. [Google Scholar] [CrossRef]
- Su, Y.-H.; Wu, Y.-Z.; Ann, D.K.; Chen, J.L.-Y.; Kuo, C.-Y. Obesity Promotes Radioresistance Through SERPINE1-Mediated Aggressiveness and DNA Repair of Triple-Negative Breast Cancer. Cell Death Dis. 2023, 14, 53. [Google Scholar] [CrossRef]
- Fang, C.; Zhao, Y.; Guo, B. MiR-199b-5p Targets HER2 in Breast Cancer Cells. J. Cell Biochem. 2013, 114, 1457–1463. [Google Scholar] [CrossRef]
- Fang, C.; Wang, F.-B.; Li, Y.; Zeng, X.-T. Down-Regulation of miR-199b-5p Is Correlated with Poor Prognosis for Breast Cancer Patients. Biomed. Pharmacother. 2016, 84, 1189–1193. [Google Scholar] [CrossRef]
- Wu, A.; Chen, Y.; Liu, Y.; Lai, Y.; Liu, D. miR-199b-5p Inhibits Triple Negative Breast Cancer Cell Proliferation, Migration and Invasion by Targeting DDR1. Oncol. Lett. 2018, 16, 4889–4896. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Qiu, W.; Xiao, Y.; Ma, J.; Xu, F.; Zhang, K.; Gao, Y.; Chen, Q.; Li, Y.; Li, H.; et al. MiR-199b-5p Suppresses Tumor Angiogenesis Mediated by Vascular Endothelial Cells in Breast Cancer by Targeting ALK1. Front. Genet. 2019, 10, 1397. [Google Scholar] [CrossRef] [PubMed]
- Ruan, X.; Yan, W.; Cao, M.; Daza, R.A.M.; Fong, M.Y.; Yang, K.; Wu, J.; Liu, X.; Palomares, M.; Wu, X.; et al. Breast Cancer Cell-Secreted miR-199b-5p Hijacks Neurometabolic Coupling to Promote Brain Metastasis. Nat. Commun. 2024, 15, 4549. [Google Scholar] [CrossRef]
- Chang, C.-W.; Yu, J.-C.; Hsieh, Y.-H.; Yao, C.-C.; Chao, J.-I.; Chen, P.-M.; Hsieh, H.-Y.; Hsiung, C.-N.; Chu, H.-W.; Shen, C.-Y.; et al. MicroRNA-30a Increases Tight Junction Protein Expression to Suppress the Epithelial-Mesenchymal Transition and Metastasis by Targeting Slug in Breast Cancer. Oncotarget 2016, 7, 16462–16478. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-D.; Jiang, L.-H.; Sun, D.-W.; Li, J.; Tang, J.-H. miR-30a Inhibits the Biological Function of Breast Cancer Cells by Targeting Notch1. Int. J. Mol. Med. 2017, 40, 1235–1242. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xiao, B.; Shi, X.; Bai, J. miR-30a Regulates the Proliferation and Invasion of Breast Cancer Cells by Targeting Snail. Oncol. Lett. 2019, 17, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Mitsueda, R.; Nagata, A.; Toda, H.; Tomioka, Y.; Yasudome, R.; Kato, M.; Shinden, Y.; Nakajo, A.; Seki, N. Identification of Tumor-Suppressive miR-30a-3p Controlled Genes: ANLN as a Therapeutic Target in Breast Cancer. Noncoding RNA 2024, 10, 60. [Google Scholar] [CrossRef] [PubMed]
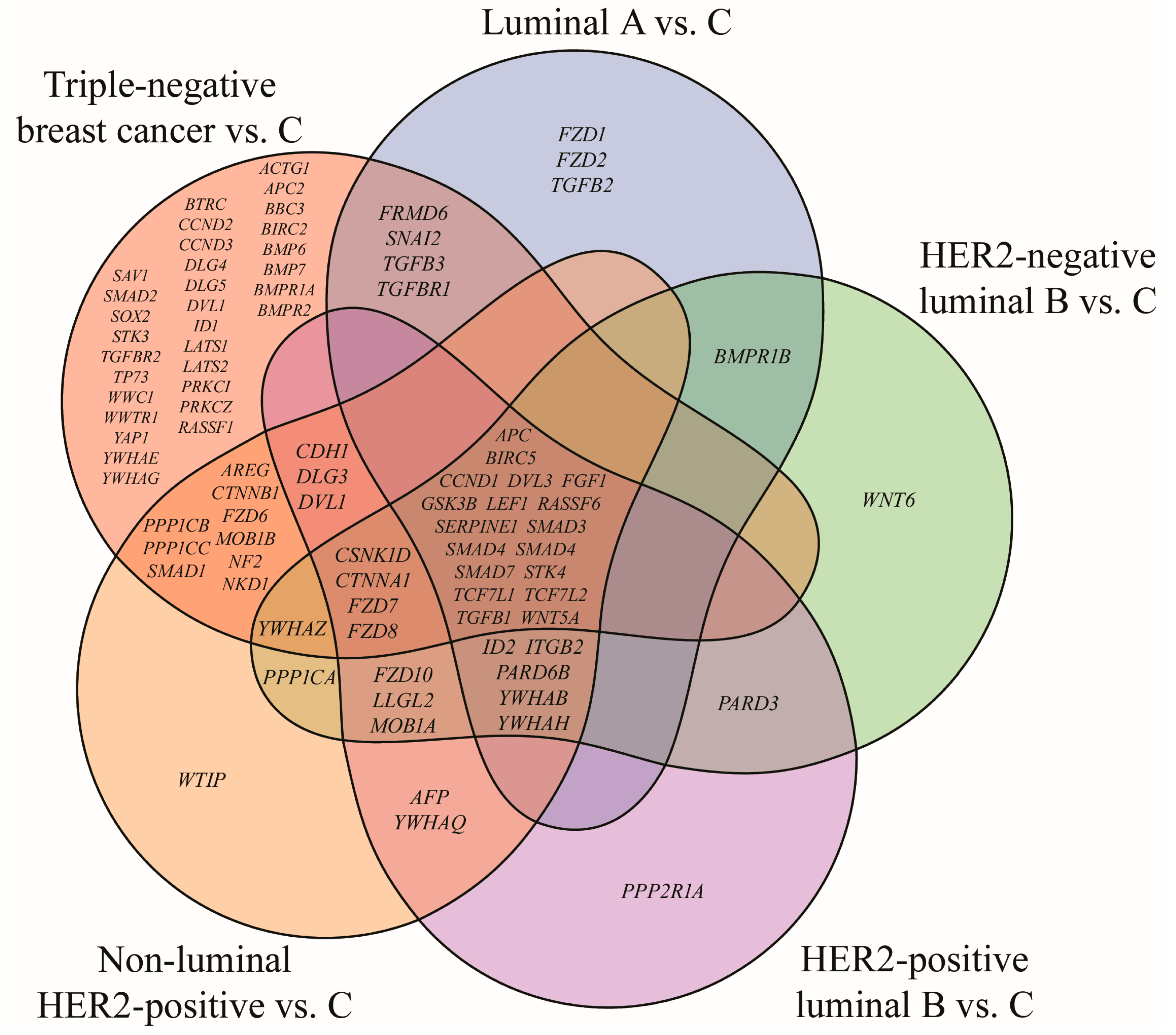

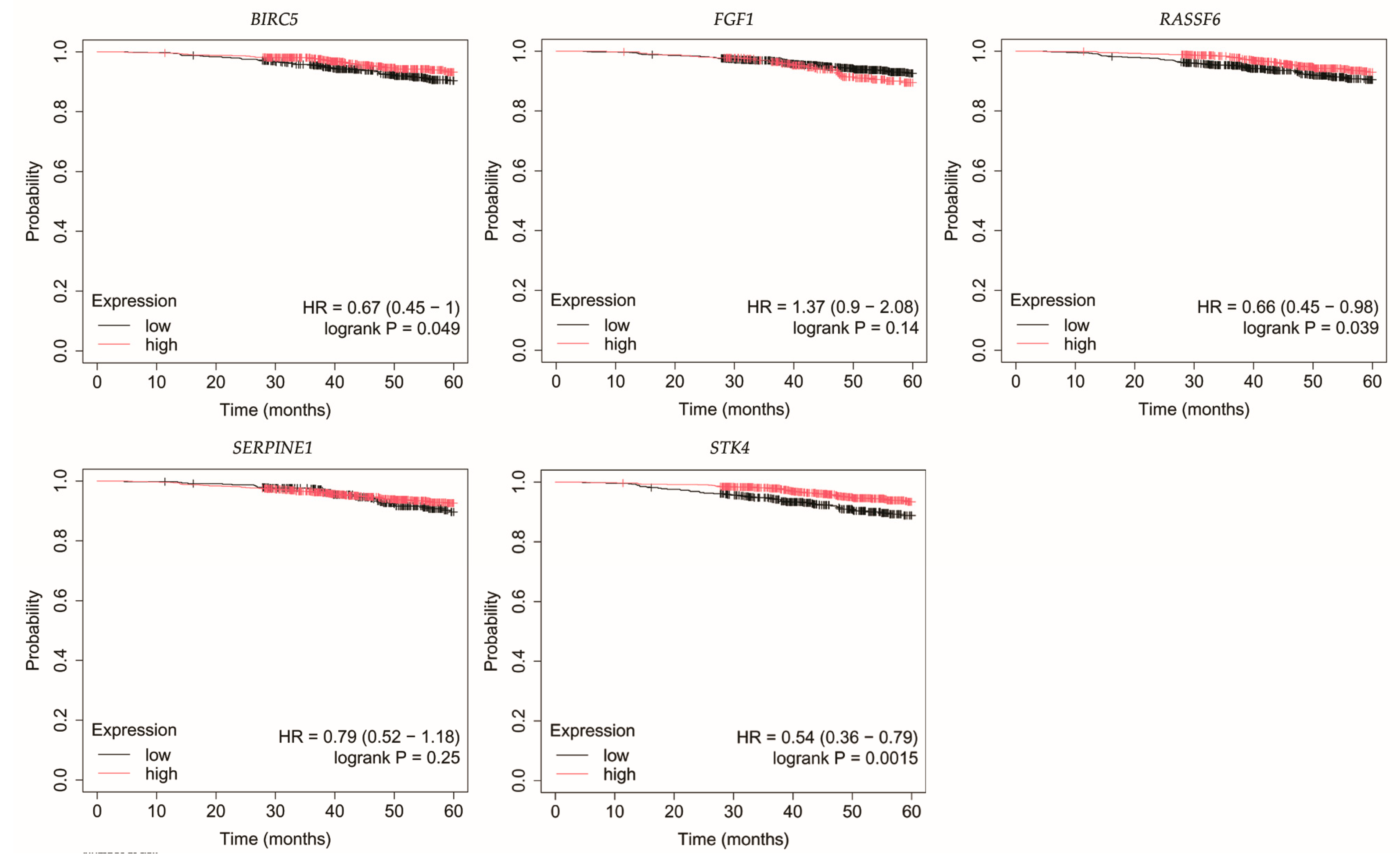


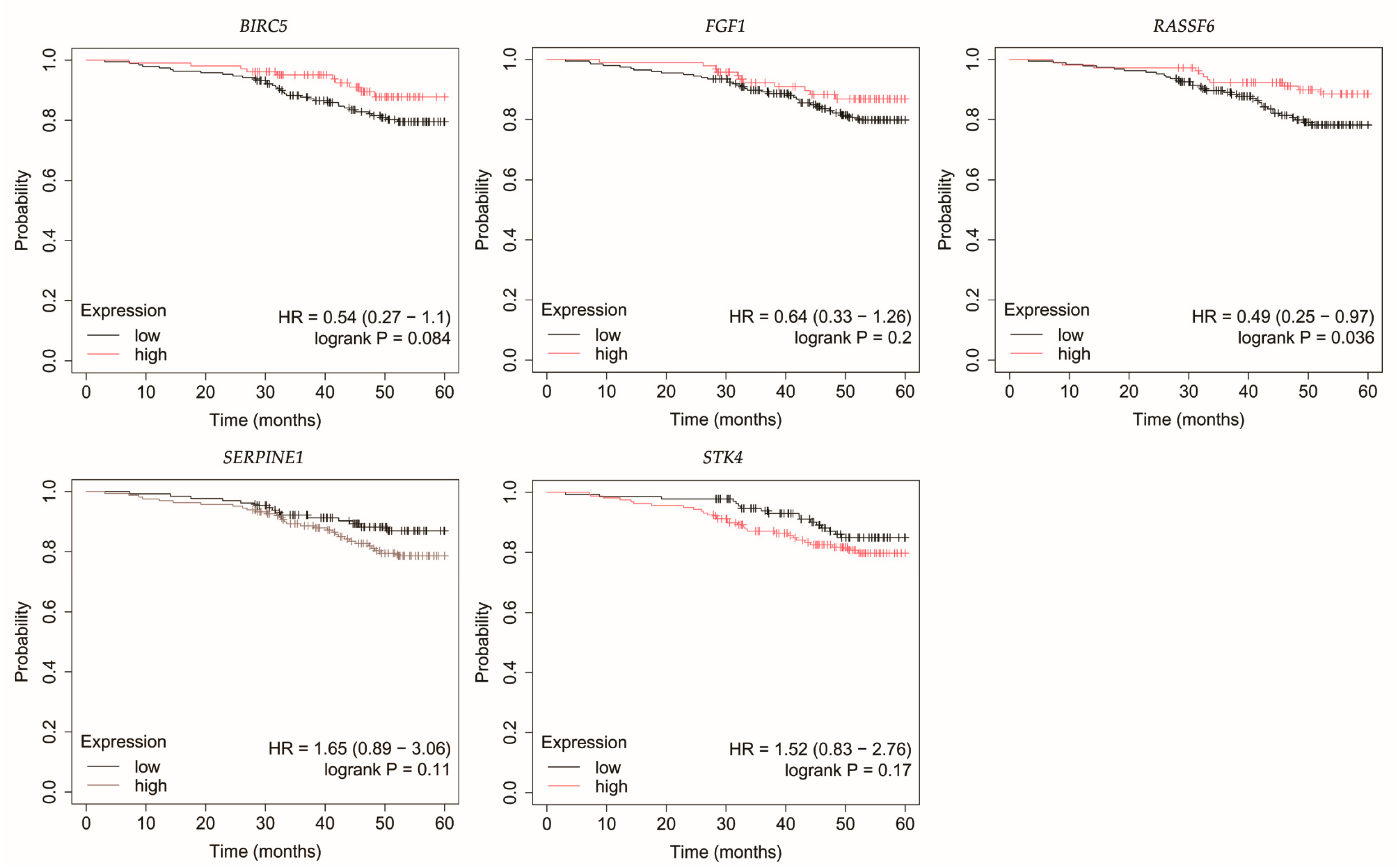
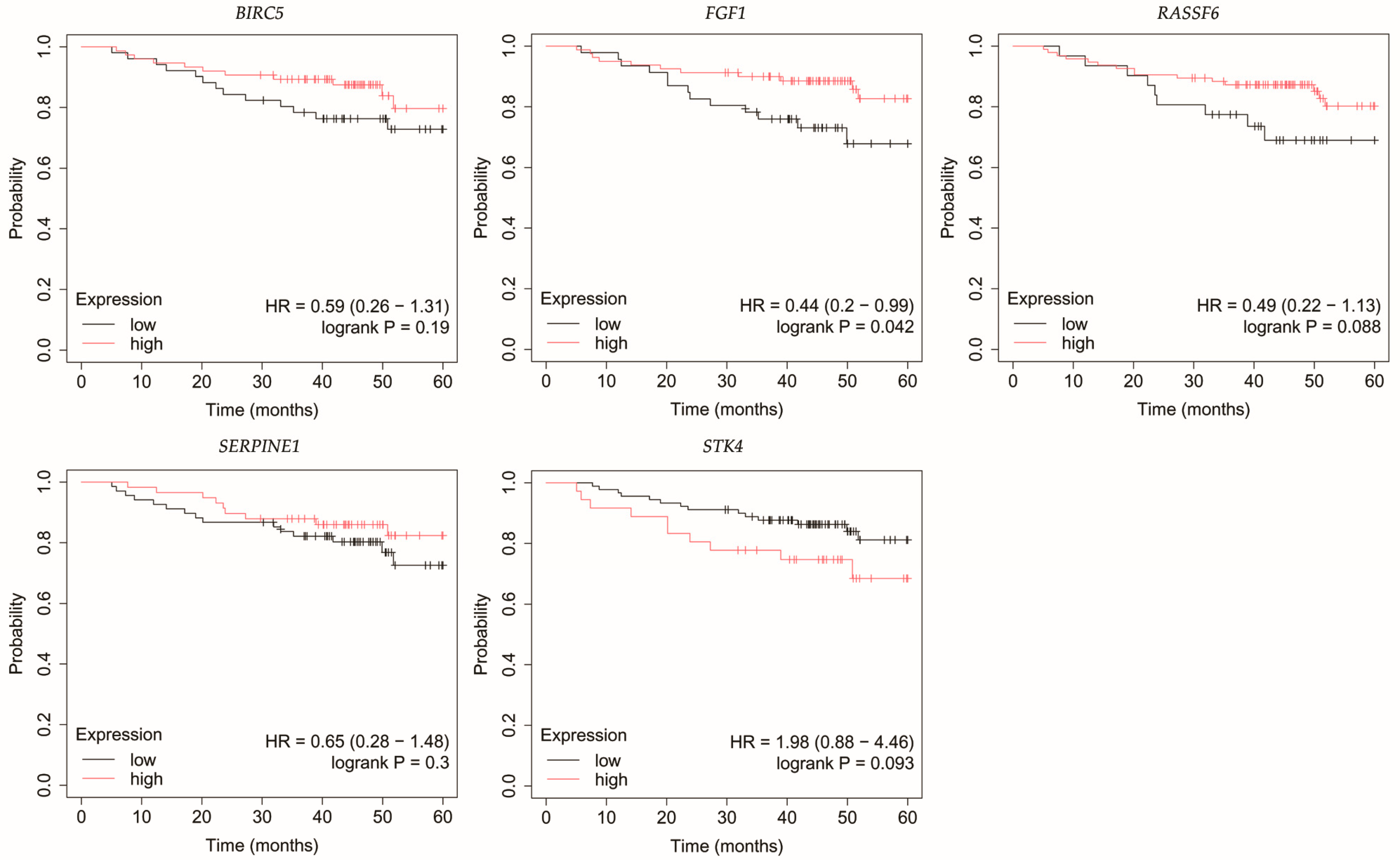
| Cancer Subtype | Cancer Grade | Age | BMI [kg/m2] | |||
|---|---|---|---|---|---|---|
| G1 | G2 | G3 | <50 years | >50 years | ||
| Luminal A | 23 | 48 | 59 | 43 | 87 | 30.8 ± 2.8 |
| HER2-negative luminal B | 31 | 57 | 12 | 32 | 68 | 30.2 ± 4.6 |
| HER2-positive luminal B | 23 | 57 | 16 | 19 | 77 | 32.1 ± 6.2 |
| Non-luminal HER2-positive | 9 | 12 | 15 | 9 | 27 | 33.2 ± 5.7 |
| TNBC | 14 | 21 | 8 | 10 | 33 | 34.7 ± 3 |
| mRNA | RT-qPCR Primers (5′–3′) |
|---|---|
| BIRC5 | Forward: CCACTGAGAACGAGCCAGACTT Reverse: GTATTACAGGCGTAAGCCACCG |
| FGF1 | Forward: ATGGCACAGTGGATGGGACAAG Reverse: TAAAAGCCCGTCGGTGTCCATG |
| RASSF6 | Forward: CGTATTAGTGAGCTGGACAGGAC Reverse: CTGGTTCATCCTTTGCATGTGGC |
| SERPINE1 | Forward: CTCATCAGCCACTGGAAAGGCA Reverse: GACTCGTGAAGTCAGCCTGAAAC |
| STK4 | Forward: CTGTGTAGCAGACATCTGGTCC Reverse: CTGGTTTTCGGAATGTGGGAGG |
| ACTB | Forward: TCACCCACACTGTGCCCATCTACGA Reverse: CAGCGGAACCGCTCATTGCCAATGG |
| ID | mRNA | Fold Change | ||||
|---|---|---|---|---|---|---|
| LumA vs. C | HER2- Negative LumB vs. C | HER2- Positive LumB vs. C | Non-Luminal HER2-Positive vs. C | TNBC vs. C | ||
| 202095_s_at | BIRC5 | 2.11 | 2.99 | 3.49 | 3.77 | 4.33 |
| 205117_at | FGF1 | −2.46 | −3.21 | −3.2 | −3.91 | −4.52 |
| 233463_at | RASSF6 | −2.47 | −2.97 | −3.13 | −3.27 | −3.54 |
| 235638_at | −3.02 | −3.33 | −3.51 | −3.4 | −3.95 | |
| 202627_s_at | SERPINE1 | 3.17 | 2.68 | 2.77 | 3.36 | 3.97 |
| 202628_s_at | 3.21 | 2.43 | 2.87 | 3.5 | 3.52 | |
| 1568765_at | 2.88 | 3.1 | 3.07 | 3.49 | 3.57 | |
| 211085_s_at | STK4 | −2.02 | −2.22 | −2.25 | −2.63 | −3.75 |
| Protein [ng/mL] | Control | LumA | HER2-Negative LumB | HER2-Positive LumB | HER2-Positive | TNBC |
|---|---|---|---|---|---|---|
| BIRC5 | 1.13 ± 0.2 | 2.94 ± 0.2 * | 3.93 ± 0.3 * | 3.98 ± 0.3 * | 4.08 ± 0.3 * | 6.47 ± 0.3 * |
| FGF1 | 1.27 ± 0.1 | below detection threshold * | below detection threshold * | below detection threshold * | below detection threshold * | below detection threshold * |
| RASSF6 | 0.32 ± 0.1 | below detection threshold * | below detection threshold * | below detection threshold * | below detection threshold * | below detection threshold * |
| SERPINE1 | 6.23 ± 0.2 | 7.3 ± 0.1 * | 8.43 ± 0.2 * | 8.79 ± 0.2 * | 11.48 ± 0.2 * | 13.05 ± 0.2 * |
| STK4 | 2.34 ± 0.4 | below detection threshold * | below detection threshold * | below detection threshold * | below detection threshold * | below detection threshold * |
| mRNA | miRNA | Target Score | Fold Change | ||||
|---|---|---|---|---|---|---|---|
| LumA vs. C | HER2-Negative LumB vs. C | HER2-Positive LumB vs. C | HER2-Positive vs. C | TNBC vs. C | |||
| SERPINE1 | miR-199b-5p | 86 | −2.17 | −2.36 | −3.03 | −3.64 | −3.41 |
| miR-30a-3p | 80 | −2.69 | −3.04 | −4.19 | −4.44 | −5.28 | |
| STK4 | miR-522-3p | 89 | 2.05 | 2.17 | 2.38 | 2.33 | 2.56 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Król-Jatręga, K.; Mitka-Krysiak, E.; Boroń, K.; Zmarzły, N.; Ossowski, P.; Plata-Babula, A.; Ordon, P.; Kulej, W.; Sirek, T.; Gajdeczka, J.; et al. Hippo Signaling Dysregulation in Breast Cancer: Subtype-Independent Gene and miRNA Signatures. Biomedicines 2025, 13, 2342. https://doi.org/10.3390/biomedicines13102342
Król-Jatręga K, Mitka-Krysiak E, Boroń K, Zmarzły N, Ossowski P, Plata-Babula A, Ordon P, Kulej W, Sirek T, Gajdeczka J, et al. Hippo Signaling Dysregulation in Breast Cancer: Subtype-Independent Gene and miRNA Signatures. Biomedicines. 2025; 13(10):2342. https://doi.org/10.3390/biomedicines13102342
Chicago/Turabian StyleKról-Jatręga, Katarzyna, Elżbieta Mitka-Krysiak, Kacper Boroń, Nikola Zmarzły, Piotr Ossowski, Aleksandra Plata-Babula, Paweł Ordon, Wojciech Kulej, Tomasz Sirek, Julia Gajdeczka, and et al. 2025. "Hippo Signaling Dysregulation in Breast Cancer: Subtype-Independent Gene and miRNA Signatures" Biomedicines 13, no. 10: 2342. https://doi.org/10.3390/biomedicines13102342
APA StyleKról-Jatręga, K., Mitka-Krysiak, E., Boroń, K., Zmarzły, N., Ossowski, P., Plata-Babula, A., Ordon, P., Kulej, W., Sirek, T., Gajdeczka, J., Prudnikov, Y., Bereza, K., Nowotny-Czupryna, O., Boroń, D., & Grabarek, B. O. (2025). Hippo Signaling Dysregulation in Breast Cancer: Subtype-Independent Gene and miRNA Signatures. Biomedicines, 13(10), 2342. https://doi.org/10.3390/biomedicines13102342






