The Combinational Effect of Enhanced Infection Control Measures and Targeted Clinical Metagenomics Surveillance on the Burden of Endemic Carbapenem and Other β-Lactam Resistance Among Severely Ill Pediatric Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design, Patient Population, and Clinical Protocol
2.2. Targeted Molecular Analysis
2.3. Statistical Analysis
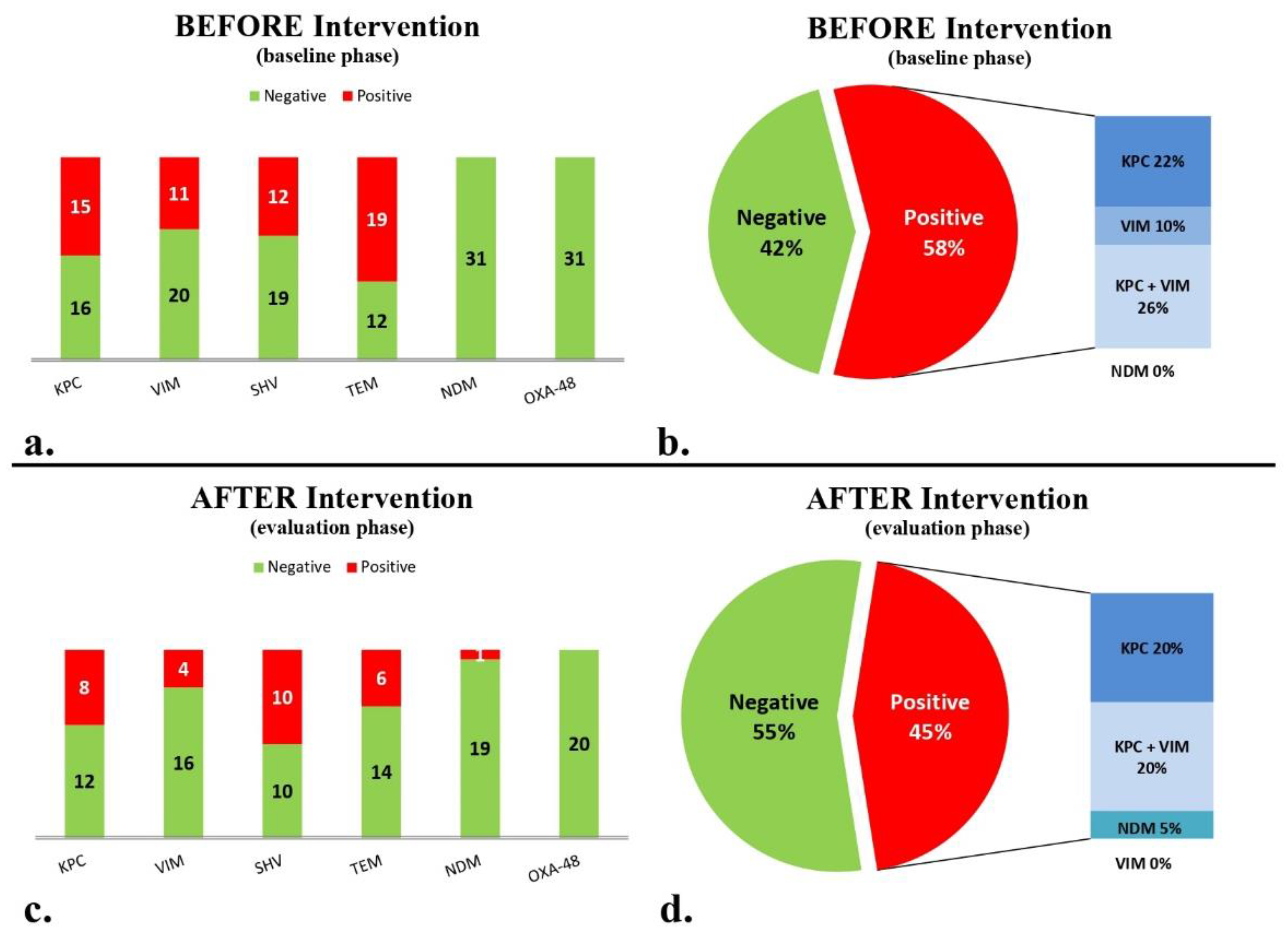
3. Results
3.1. Baseline, Intervention, and Maintenance Phase
3.2. AMR Gene Detection
3.3. Active Surveillance for Colonization: Comparing Targeted Molecular Analysis with Bacterial Cultures for Carbapenemase Genes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar] [PubMed] [PubMed Central]
- Barmpouni, M.; Gordon, J.P.; Miller, R.L.; Dennis, J.W.; Grammelis, V.; Rousakis, A.; Souliotis, K.; Poulakou, G.; Daikos, G.L.; Al-Taie, A. Clinical and Economic Value of Reducing Antimicrobial Resistance in the Management of Hospital-Acquired Infections with Limited Treatment Options in Greece. Infect. Dis. Ther. 2023, 12, 1891–1905. [Google Scholar] [CrossRef] [PubMed]
- Romandini, A.; Pani, A.; Schenardi, P.A.; Pattarino, G.A.C.; De Giacomo, C.; Scaglione, F. Antibiotic resistance in pediatric infections: Global emerging threats, Predicting the near future. Antibiotics 2021, 10, 393. [Google Scholar] [CrossRef]
- Naghavi, M.; Vollset, S.E.; Ikuta, K.S.; Swetschinski, L.R.; Gray, A.P.; Wool, E.E.; Aguilar, G.R.; Mestrovic, T.; Smith, G.; Han, C.; et al. Global burden of bacterial antimicrobial resistance 1990–2021: A systematic analysis with forecasts to 2050. Lancet 2024, 404, 1199–1226. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Burns, K.; Baño, J.R.; Borg, M.; Daikos, G.; Dumpis, U.; Lucet, J.C.; Moro, M.L.; Tacconelli, E.; Simonsen, G.S.; et al. Infection prevention and control measures and tools for the prevention of entry of carbapenem-resistant Enterobacteriaceae into healthcare settings: Guidance from the European Centre for Disease Prevention and Control. Antimicrob. Resist. Infect. Control 2017, 6, 113. [Google Scholar] [CrossRef]
- Hellenic Centre for Diseases Control and Prevention (KEELPNO/HCDCP). Action Plan for the Management of Infections by Multidrug-Resistance Gram-Negative Pathogens in Health Care Facilities: Procrustes, October 2010. Available online: https://eody.gov.gr/wp-content/uploads/2019/01/prokroustis-final-1.pdf (accessed on 20 July 2020).
- Karampatakis, T.; Antachopoulos, C.; Tsakris, A.; Roilides, E. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii in Greece: An extended review (2000–2015). Future Microbiol. 2017, 12, 801–815. [Google Scholar] [CrossRef] [PubMed]
- Karampatakis, T.; Tsergouli, K.; Politi, L.; Diamantopoulou, G.; Iosifidis, E.; Antachopoulos, C.; Karyoti, A.; Sdougka, M.; Tsakris, A.; Roilides, E. Polyclonal predominance of concurrently producing OXA-23 and OXA-58 carbapenem-resistant Acinetobacter baumannii strains in a pediatric intensive care unit. Mol. Biol. Rep. 2019, 46, 3497–3500. [Google Scholar] [CrossRef] [PubMed]
- Schechner, V.; Straus-Robinson, K.; Schwartz, D.; Pfeffer, I.; Tarabeia, J.; Moskovich, R.; Chmelnitsky, I.; Schwaber, M.J.; Carmeli, Y.; Navon-Venezia, S. Evaluation of PCR-Based testing for surveillance of KPC-Producing Carbapenem-Resistant members of the Enterobacteriaceae family. J. Clin. Microbiol. 2009, 47, 3261–3265. [Google Scholar] [CrossRef] [PubMed]
- La, M.-V.; Lee, B.; Hong, B.Z.M.; Yah, J.Y.; Koo, S.-H.; Jiang, B.; Ng, L.S.Y.; Tan, T.-Y. Prevalence and antibiotic susceptibility of colistin-resistance gene (mcr-1) positive Enterobacteriaceae in stool specimens of patients attending a tertiary care hospital in Singapore. Int. J. Infect. Dis. 2019, 85, 124–126. [Google Scholar] [CrossRef] [PubMed]
- Crofts, T.S.; Gasparrini, A.J.; Dantas, G. Next-generation approaches to understand and combat the antibiotic resistome. Nat. Rev. Microbiol. 2017, 15, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Karampatakis, T.; Tsergouli, K.; Iosifidis, E.; Antachopoulos, C.; Volakli, E.; Karyoti, A.; Sdougka, M.; Tsakris, A.; Roilides, E. Effects of an active surveillance program and enhanced infection control measures on Carbapenem-Resistant Gram-Negative bacterial carriage and infections in pediatric intensive care. Microb. Drug Resist. 2019, 25, 1347–1356. [Google Scholar] [CrossRef] [PubMed]
- Herruzo, R.; Ruiz, G.; Perez-Blanco, V.; Gallego, S.; Mora, E.; Vizcaino, M.J.; Omeñaca, F. Bla-OXA48 gene microorganisms outbreak, in a tertiary Children’s Hospital, Over 3 years (2012–2014). Medicine 2017, 96, e7665. [Google Scholar] [CrossRef]
- Yang, P.; Chen, Y.; Jiang, S.; Shen, P.; Lu, X.; Xiao, Y. Association between antibiotic consumption and the rate of carbapenem-resistant Gram-negative bacteria from China based on 153 tertiary hospitals data in 2014. Antimicrob. Resist. Infect. Control 2018, 7, 137. [Google Scholar] [CrossRef]
- Renk, H.; Sarmisak, E.; Spott, C.; Kumpf, M.; Hofbeck, M.; Hölzl, F. Antibiotic stewardship in the PICU: Impact of ward rounds led by paediatric infectious diseases specialists on antibiotic consumption. Sci. Rep. 2020, 10, 8826. [Google Scholar] [CrossRef] [PubMed]
- WHO. Evidence of Hand Hygiene as the Building Block for Infection Prevention and Control. Available online: https://iris.who.int/bitstream/handle/10665/330079/WHO-HIS-SDS-2017.7-eng.pdf?sequence=1 (accessed on 20 July 2020).

| Genes | Primer Pairs (5′ to 3′) | Tm Optimal | Amplicon |
|---|---|---|---|
| blaKPC | AACCTCGTCGCGGAACCATT | 61 °C | 785 bp |
| AATCCCTCGAGCGCGAGTCTA | |||
| blaVIM | AGCGGTGAGTATCCGACA | 56.7 °C | 261 bp |
| ATGAAAGTGCGTGGAGAC | |||
| blaNDM | AAGCTGAGCACCGCATTAGCC | 60.2 °C | 217 bp |
| CGCCATCCCTGACGATCAAACC | |||
| blaOXA-48-like | ATAAGCAGCAAGGATTTACCAA | 55.5 °C | 516 bp |
| ACCAGCCAATCTTAGGTTCG | |||
| blaTEM | AACTGGATCTCAACAGCGGTA | 56.8 °C | 510 bp |
| CTGCAACTTTATCCGCCTCC | |||
| blaSHV | CGCTTTCCCATGATGAGCACCT | 61.6 °C | 324 bp |
| CGCCTCATTCAGTTCCGTTTCCC |
| Number of Patients | ||
|---|---|---|
| AMR Gene Detection | Baseline Phase (n = 31) | Maintenance Phase (n = 20) |
| Targeted Molecular Analysis | 18 (58%) | 9 (45%) |
| Initial culture-based analysis | 5 (16%) | 3 (15%) |
| Later-stage culture-based analysis | 2 (6.5%) | 4 (20%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giampani, A.; Simitsopoulou, M.; Sdougka, M.; Paschaloudis, C.; Roilides, E.; Iosifidis, E. The Combinational Effect of Enhanced Infection Control Measures and Targeted Clinical Metagenomics Surveillance on the Burden of Endemic Carbapenem and Other β-Lactam Resistance Among Severely Ill Pediatric Patients. Biomedicines 2025, 13, 31. https://doi.org/10.3390/biomedicines13010031
Giampani A, Simitsopoulou M, Sdougka M, Paschaloudis C, Roilides E, Iosifidis E. The Combinational Effect of Enhanced Infection Control Measures and Targeted Clinical Metagenomics Surveillance on the Burden of Endemic Carbapenem and Other β-Lactam Resistance Among Severely Ill Pediatric Patients. Biomedicines. 2025; 13(1):31. https://doi.org/10.3390/biomedicines13010031
Chicago/Turabian StyleGiampani, Athina, Maria Simitsopoulou, Maria Sdougka, Christos Paschaloudis, Emmanuel Roilides, and Elias Iosifidis. 2025. "The Combinational Effect of Enhanced Infection Control Measures and Targeted Clinical Metagenomics Surveillance on the Burden of Endemic Carbapenem and Other β-Lactam Resistance Among Severely Ill Pediatric Patients" Biomedicines 13, no. 1: 31. https://doi.org/10.3390/biomedicines13010031
APA StyleGiampani, A., Simitsopoulou, M., Sdougka, M., Paschaloudis, C., Roilides, E., & Iosifidis, E. (2025). The Combinational Effect of Enhanced Infection Control Measures and Targeted Clinical Metagenomics Surveillance on the Burden of Endemic Carbapenem and Other β-Lactam Resistance Among Severely Ill Pediatric Patients. Biomedicines, 13(1), 31. https://doi.org/10.3390/biomedicines13010031






