Abstract
Isothiocyanates (ITCs) belong to a group of natural products that possess a highly reactive electrophilic −N=C=S functional group. They are stored in plants as precursor molecules, glucosinolates, which are processed by the tyrosinase enzyme upon plant tissue damage to release ITCs, along with other products. Isolated from broccoli, sulforaphane is by far the most studied antioxidant ITC, acting primarily through the induction of a transcription factor, the nuclear factor erythroid 2–related factor 2 (Nrf2), which upregulates downstream antioxidant genes/proteins. Paradoxically, sulforaphane, as a pro-oxidant compound, can also increase the levels of reactive oxygen species, a mechanism which is attributed to its anticancer effect. Beyond highlighting the common pro-oxidant and antioxidant effects of sulforaphane, the present paper was designed to assess the diverse anti-inflammatory mechanisms reported to date using a variety of in vitro and in vivo experimental models. Sulforaphane downregulates the expression of pro-inflammatory cytokines, chemokines, adhesion molecules, cycloxyhenase-2, and inducible nitric oxide synthase. The signalling pathways of nuclear factor κB, activator protein 1, sirtuins 1, silent information regulator sirtuin 1 and 3, and microRNAs are among those affected by sulforaphane. These anti-inflammatory actions are sometimes due to direct action via interaction with the sulfhydryl structural moiety of cysteine residues in enzymes/proteins. The following are among the topics discussed in this paper: paradoxical signalling pathways such as the immunosuppressant or immunostimulant mechanisms; crosstalk between the oxidative and inflammatory pathways; and effects dependent on health and disease states.
1. Overview of Chemistry and Biological Relevance
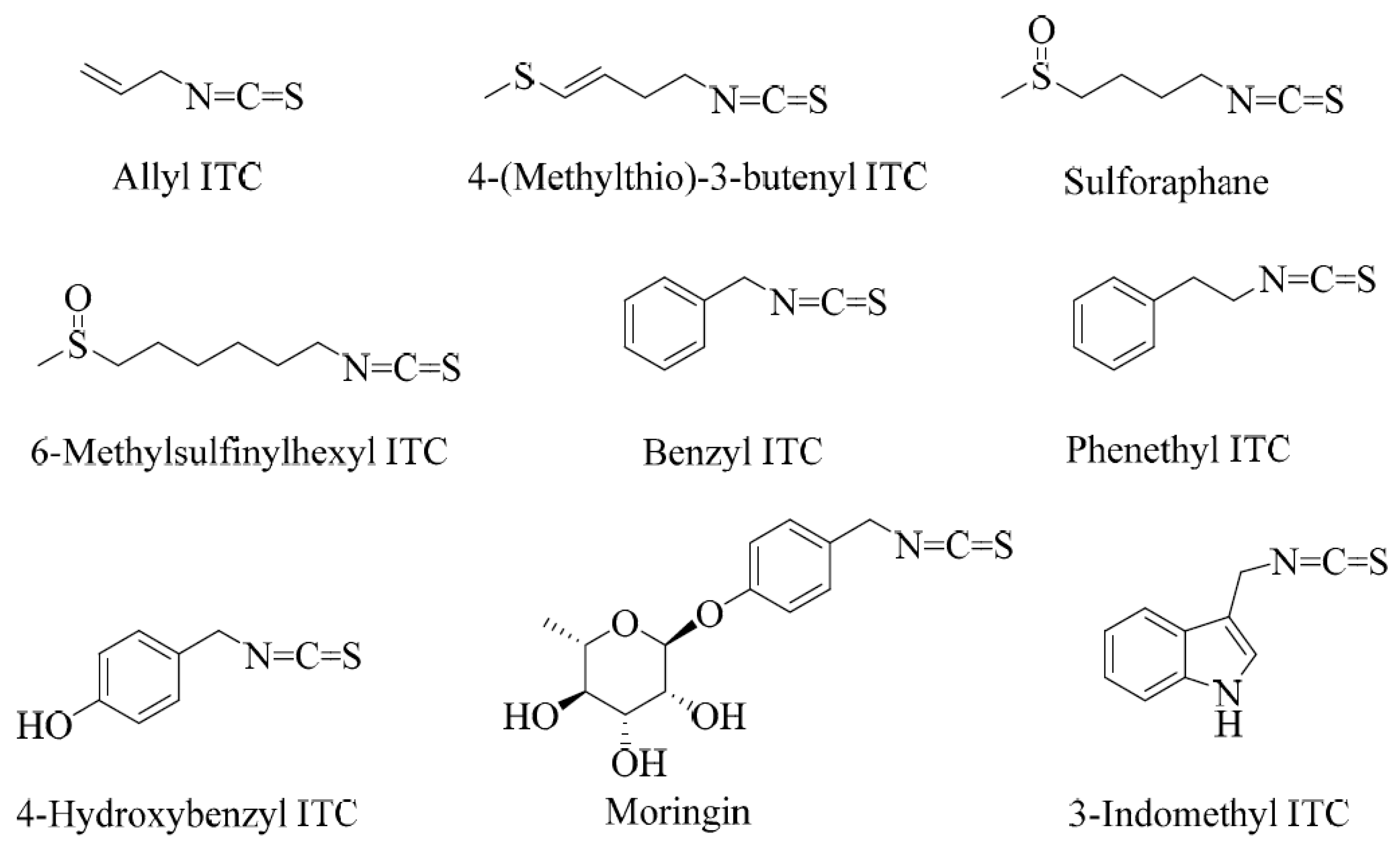
Sulforaphane is a small-molecular-weight sulphur-containing compound that belongs to a structural group of natural products called isothiocyanates (ITCs). Other examples of ITCs include benzyl, phenethyl, and allyl ITCs (Figure 1). The distinguishing feature of this class of compounds is the highly reactive electrophilic −N=C=S structural moiety, which undergoes several reactions in biological systems. The structural diversity of isothiocyanates in nature is represented by the side-chain R group (R-N=C=S), which can be made of aralkyls such as benzyl, 2-phenyl and 4-hydroxybenzyl, indoles such as indol-3-methyl or 4-hydroxyindol-3-ylmethy, or several aliphatic chain derivatives. Plants that produce ITCs store them in special cellular and subcellular sites in the form of precursor compounds called glucosinolates.
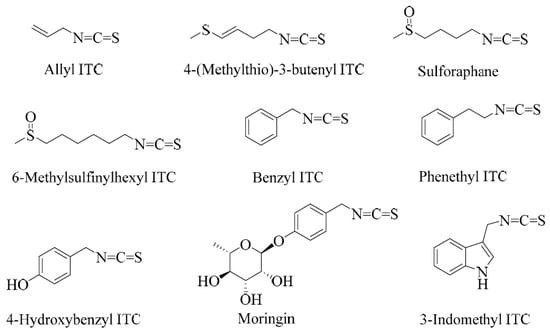
Figure 1.
Examples of some common isothiocyanates from plant sources.
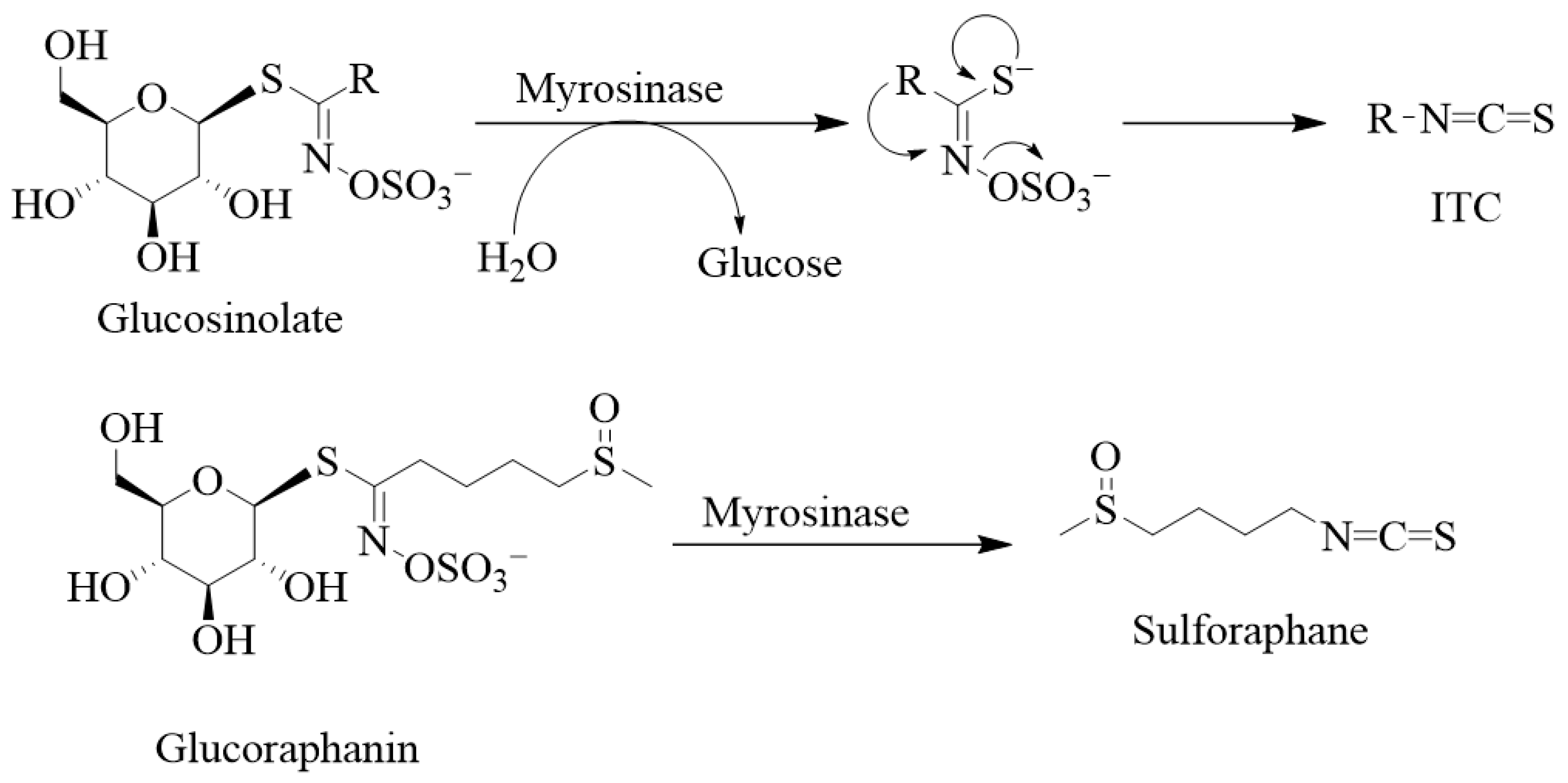
Structurally, the glucosinolates are thiohydroximates that contain an S-linked β-glucopyranosyl and O-linked sulphate residues (Figure 2). As it has been said above for ITCs, structural diversity is based on the nature of the R group of either the alkyl, aralkyl, or indolyl side chains that derive from amino acids (e.g., phenylalanine, tryptophan, and methionine). Agerbirk and Olsen [1] reported in the year 2012 the existence of around 132 glucosinolates isolated from plants, and this number has been further increased over the past decade. Glucosinolates and their ITC products are generally considered defensive or protective chemicals against herbivores, insects, and pathogens [2,3]. Upon damage of the plant tissues, say by herbivores, glucosinolates encounter the enzyme myrosinase (β-D-thioglucosidase; EC 3.2.1.147), leading to their breakdown into various products such as nitriles, thiocyanates, and isothiocyanates, with the latter being the most stable. Other products known to derive from hydrolysis by the enzyme include epithionitriles, hydroxynitriles, oxazolidine-2-thiones, and indoles. Thus, the key to ITC production is that the relevant enzyme (myrosinase) and substrates (glucosinolates) are stored in plants in separate cellular compartments [4], and the hydrolysis reaction is only initiated upon cellular damage. The enzyme cleaves the thio-linked glucose from glucosinolates to form an unstable intermediate, thiohydroximate-O-sulphonate. The final product depends on a variety of factors, including the immediate pH, temperature, specifier proteins, and ferrous ions [5,6,7]. For example, a neutral or alkaline pH can spontaneously lead to ITC formation. As shown by Williams et al. [8] on myrosinase activity in Lepidium sativum and Nasturtium officinale seeds, a pH of less than 4 along with ferrous ions (Fe2+) and the epithiospecifier protein favour nitrile formation. Further variations in the degradation products also depend on the nature of the side chain.
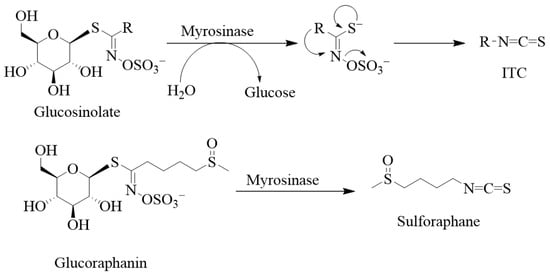
Figure 2.
Degradation of glucosinolates and some examples of isothiocyanates. Glucosinolates are composed of the thiohydroximate-O-sulphonate group linked to glucose. Removing the glucose unit from glucosinolates by myrosinase leads to the unstable aglucon, meaning that thiohydroximate-O-sulphate is released. Hence, myrosinase catalyses the hydrolysis of these S-glucosides to give D-glucose and an aglycone fragment, and the aglycon further rearranges itself to give sulphate and products, mostly isothiocyanate. In the case of sulforaphane, the glucosinolate precursor is glucoraphanin.
Glucosinolate- or ITC-producing plants mostly belong to plants of the order Brassicales, which include families such as the Brassicaceae, the Capparaceae, and the Caricaceae. The Brassicaceae family, which is also called the family of cruciferous vegetables, has around 346 accepted genera and over 3000 species, including cultivated crops such as broccoli, cabbages, Brussel sprouts, cauliflowers, mustard seeds, etc. Sulforaphane is among the best investigated ITCs, and, biogenically, it is derived from the glucosinolate glucoraphanin (Figure 2). Broccoli is the best-known source of sulforaphane and the C3-C6 aliphatic chain, as the R group is commonly found in Brassica species. Allyl ITC is the main component behind the pungency of wasabi (Japanese horseradish; Wasabia japonica) and black mustard (Brassica nigra) seeds and is formed from its corresponding glucosinolate, sinigrin [9,10,11]. In contrast, white mustard (Sinapis alba L.) seeds have p-hydroxybenzyl ITC as a main component for their flavour and aroma. This is generated via a myrosinase action on the corresponding glucosinolate, sinalbin [12]. 4-(Methylthio)-3-butenyl ITC is a component of Japanese white radish (Raphanus sativus), while 6-methylsufinylhexyl ITC has been isolated in a good amount from wasabi [13]. Benzyl ITC has been isolated from papaya (Carica papaya Linn.) peel, pulp, and seeds [14] and phenethyl ITC from watercress (Nasturtium officinale) [15]. The sugar derivatives of hydroxybenzyl ITC are represented by moringin, isolated from Moringa oleifera and M. stenopetala [16].
The bioactivity and electrophilic nature of ITCs are attributes of the R−N=C=S structural skeleton, which undergoes several reactions in biological systems. The electron-deficient central carbon atom in such structure is susceptible to nucleophilic attacks by the electron-rich centre. For example, thiol groups such as glutathione can be added to ITC molecules. The implication of this reaction is huge and includes the generation of reactive oxygen species (ROS) by depleting glutathione, through a direct covalent reaction with the glutathione S-transferase (GST) of other thiol-containing biological molecules such as proteins and enzymes. Hence, sulforaphane is among the best-known compounds generating the oxidative stress-based induction of apoptosis in cancer cells. In fact, by far the most studied biological activity of isothiocyanates is in the cancer field, where sulforaphane from broccoli, among others, has been shown to prevent carcinogenesis [17,18,19] and induce apoptosis in cancer cells [20,21,22]. For example, sulforaphane suppresses the incidence of tumours in mice exposed to UV light [23]. Sulforaphane also suppresses cancer cell migration and invasion [24], prostate carcinogenesis and pulmonary metastasis [25], and carcinogenicity induced by cadmium [26]. The immunomodulatory effect of sulforaphane in cancer is also well established and includes boosting the activity of immune cells such as natural killer cells against cancer [25]. It augments natural killer cell- and antibody-dependent cellular cytotoxicity by enhancing the production of cytokines IL-2 and IFN-γ [27]. In B16F-10 melanoma-induced metastasis-bearing C57BL/6 mice, sulforaphane has been shown to enhance natural killer cell activity while also enhancing antibody-dependent cellular cytotoxicity in metastatic tumour-bearing animals [28]. It is also worth noting that electrophiles and oxidants are detoxified in the body by phase II metabolic enzymes such as GST and NAD(P)H:quinone oxidoreductase 1 (NQO1). Interestingly, the antioxidant and antimutagenic effects of ITCs, including sulforaphane, are associated with the induction of GST and NQO1 [29,30]. Not surprisingly, isothiocyanates have numerous other biological activities, such as antifungal and bactericidal [31,32,33,34] as well as antiparasitic [35] activities. In this paper, the anti-inflammatory mechanisms of sulforaphane are scrutinised by assessing all the publications sourced from the literature (Web of Science, PubMed, and ScienceDirect) until February 2024.
2. Anti-Inflammatory Effects In Vivo
Readers should note that the present paper was designed to present a mechanistic overview of sulforaphane as an anti-inflammatory agent. This was achieved (see below) by assessing the in vitro studies in which sulforaphane had been shown to target inflammatory cells such as leukocytes, astrocytes, and endothelial, epithelial, and other cell types (Table 1, Table 2, Table 3, Table 4 and Table 5). With this in mind, it is worth highlighting that the anti-inflammatory effect of sulforaphane has further been confirmed through in vivo studies. These include experimental models in rats such as acetaminophen hepatitis [36] and hepatic ischaemia/reperfusion injury, where the expression of inflammatory mediators (cyclooxygenase-1 (COX-2), tumour necrosis factor- (TNF-, interleukin (IL)-6, and monocyte chemoattractant protein-1 (MCP-1)) and reactive oxygen species (ROS) production have been demonstrated to be inhibited. At the same time, the expression of the oxidative stress regulator transcription factor, the nuclear factor erythroid 2-related factor 2 (Nrf2), and its downstream gene/protein products (NQO1, haeme oxygenase-1 (HO-1), glutathione (GSH), catalase (CAT), and superoxide dismutase (SOD)) have been shown to be upregulated. Interestingly, the Nrf-2 inhibitor ML385 has been shown to reverse the observed anti-inflammatory and antioxidant effect [37]. Other rat models of inflammation in the liver where sulforaphane has shown positive results include non-alcoholic fatty liver disease [38] and sodium valproate-induced acute liver injury [39]. In a rat heart inflammation model, sulforaphane ameliorated doxorubicin-induced chronic heart failure [40] and reduced fibrosis and the scores of post-myocardial infarction associated with an increased HO-1 level [41], as well as the positive anti-inflammatory score in acrolein-induced cardiomyopathy [42], the cardiac ischaemia/reperfusion model [43], and cuprizone-induced cardiotoxicity [44]. Ischaemia/reperfusion injuries in rats have been further used to demonstrate the anti-inflammatory activity of sulforaphane in various organs, including the lungs [45] and retina [46,47]. Inflammation associated with diabetes in rats has been extensively used in sulforaphane studies and includes efficacy in diabetic neuropathy [48], a diabetic model of renal inflammation [49], experimental diabetic peripheral neuropathy in rats [50], and streptozotocin (STZ)-induced diabetic rats [51]. Further research on renal inflammation in rats has included folic acid-induced acute renal injury [52] and cisplatin-induced nephropathy where the expression of pro-inflammatory cytokine (TNF-α) and nuclear factor κB (NF-κB) were suppressed [53]. Other inflammation models in rats for sulforaphane research were cancer-induced bone pain [54], carrageenan-induced oedema [55], the traumatic haemorrhagic shock model [56], chromium-induced lung injury [57], arsenic-induced nephrotoxicity [58], ioversol-induced nephropathy [59], the neuroinflammation and spatial learning model [60], age-related renal injury in rats [61], anti-nociceptive and anti-inflammatory effects on a sciatic endometriosis rat model [62], and chronic renal allograft dysfunction [63].

Table 1.
Anti-inflammatory effect of sulforaphane via the modulation of leucocyte function.

Table 2.
Anti-inflammatory effect of sulforaphane via the modulation of astrocytes and glial cells.

Table 3.
Anti-inflammatory effect of sulforaphane via the modulation of endothelial cells.

Table 4.
Anti-inflammatory effect of sulforaphane via the modulation of epithelial cells.

Table 5.
Anti-inflammatory effect of sulforaphane via the modulation of other cell types.
Most research papers on the anti-inflammatory effect of sulforaphane in vivo are based on studies using mice experimental models. These include the classic anti-inflammatory in vivo model using carrageenan-induced oedema neuropathic pain [163], acrylamide-induced neuropathy [164], collagen-induced arthritis [79,165], osteoarthritis [166], adjuvant-induced chronic pain [167,168], chronic constriction injury-induced neuropathic pain [169], and acute gout [98] models. Other experimental models of inflammation employing mice are the demonstration of the anti-inflammatory potential of sulforaphane in lung diseases using cigarette smoke-induced alveolar damage [66], cigarette smoke-exposed asthmatic mice [170], bleomycin-induced pulmonary fibrosis [171], ovalbumin (OVA)-sensitised and cigarette smoke-induced airway inflammation [172], chlorine-induced airway hyper-responsiveness [173], haemorrhagic shock-induced lung injury [174], OVA-induced chronic allergic airways [175], lipopolysaccharide (LPS)-induced acute lung injury [176], respiratory syncytial virus (RSV)-induced bronchopulmonary inflammation [177], and the pulmonary arterial hypertension model [178].
Mice models of gut inflammation have been effectively used to show the anti-inflammatory effect of sulforaphane, such as those using dextran sodium sulphate (DSS)-induced gut inflammation [73,179], DSS-induced ulcerative colitis [118,180,181], high-fat high-cholesterol diet-induced gut inflammation [182], 5-fluorouracil-induced intestinal injury [183], necrotizing enterocolitis [184], gut inflammation associated with bladder cancer [185], the genetic model of intestinal polyps [186], and the genetic model of gastrointestinal dysfunction [187]. Anti-inflammatory effects in the liver of mice have been shown for sulforaphane using experimental models including the following: carbon tetrachloride-induced acute liver injury [188]; high-fat diet-induced non-alcoholic fatty hepatic steatosis and liver disease [189,190,191]; high-fat diet-induced [192,193]; LPS-induced acute liver injury [128,194,195,196]; hepatic ischaemia/reperfusion injury [197]; sickle cell disease model [198]; and cadmium-induced hepatotoxicity [199]. Inflammation associated with diabetes in mice has been effectively ameliorated by sulforaphane, as shown in high-fat diet- or STZ-induced diabetes and cardiomyopathy [200,201], db/db diabetic mice cardiomyopathy [202], diabetic cardiomyopathy in both type 1 and type 2 diabetes [203,204], high-fat diet-induced diabetes [205,206], ob/ob diabetic mice [90], obesity- and type 2 diabetes-associated pain [207], STZ-induced diabetes [208,209], STZ-induced diabetic nephropathy [210], type 1 diabetic OVE26 mice [211], and diabetes-induced vascular inflammation and pathogenesis [212]. Using a high-fat diet model, the anti-inflammatory effects of sulforaphane in mice have been further shown in [213,214] as well as in American diet-induced inflammation [215], TNF-α-induced vascular inflammation [121,122], and high-fat diet-induced obesity [196] models.
Infection models in mice have been used to show the anti-inflammatory effect of sulforaphane as demonstrated using helicobacter pylori infection [216], SARS-CoV-2 infection [217], LP-BM5 leukaemia retrovirus infection [218], and microcystin-LR (MC-LR)-induced inflammation [219]. These effects have been further validated using experimental autoimmune encephalomyelitis [220], autoimmune encephalomyelitis [74,221], LPS-induced acute inflammation [222], necrotizing enterocolitis [223], respiratory syncytial virus (RSV)-induced bronchopulmonary inflammation, epithelial injury [177], and LPS-induced endotoxemia [224] models.
Other anti-inflammatory effects of sulforaphane in mice have been based on ultraviolet B (UVB)-induced skin inflammation [225,226], the genetic model of muscular dystrophy [227,228], ischemia/reperfusion-induced muscular injury [229], ischaemia/reperfusion injury and cardiac allograft vasculopathy [127], vascular remodelling in hypoxic pulmonary hypertension [230], aged mice [231], angiotensin II-induced renal inflammation and injury [232], ischaemia/reperfusion injury [233], hypoxia-induced cardiomyopathy [234], the genetic model of kidney disease [235], retinitis Pigmentosa [236], atopic dermatitis [237], UV radiation-induced inflammation [238], acute exhaustive exercise-induced organ damage and inflammation [239], radiation-induced skin damage [240], oxazolone-induced chronic itch model [241], and acute pancreatitis in mouse [158] experimental models.
In the CNS domain, experimental models in mice for the anti-inflammatory effect of sulforaphane have included spinal cord injury [242,243], depression-like behaviour [244], LPS-induced depression-like behaviours [245,246,247], LPS-induced spatial learning and memory dysfunction [248], the transgenic model of Alzheimer’s disease [249], the genetic model of autism [250], contusion spinal cord injury [251], MG132-mediated spatial memory loss [252], LPS-induced depressive disorder [253], and the platelet aggregation and thrombus-associated cerebral microcirculation [254]. Some experimental models employing rabbits have also been used to demonstrate the anti-inflammatory effect of sulforaphane [255,256,257,258].
3. Anti-inflammatory Studies In Vitro
The effects of sulforaphane as an anti-inflammatory agent in vitro have been shown in cellular models using leucocytes (Table 1), astrocytes and glial (Table 2), endothelial (Table 3), epithelial (Table 4), and many other (Table 5) cell types. These effects are attributed to the suppression of the expression or activity of the various inflammatory mediators described below.
3.1. Anti-inflammatory Effect of Sulforaphane through the Suppression of Pro-Inflammatory Cytokines and Chemokine Production
Pro-inflammatory cytokines such as IL-1β, TNF-α, IL-6, and IL-8 play a pivotal role in the pathogenesis of various chronic inflammatory diseases. Numerous therapeutic approaches using antibodies target these cytokines or their receptors. Good examples of these agents are marketed drugs for chronic inflammatory diseases, including the following: adalimumab, infliximab, and certolizumab against TNF-α; rilonacept, anakinra (receptor antagonist), and canakinumab against IL-1; and tocilizumab and siltuximab against IL-6. On the other hand, small-molecular-weight inhibitors target the signalling pathways of these cytokines’ production, such as those induced by LPS, pro-inflammatory cytokines, ROS, or other inducers. The expression of such cytokines by a variety of agents has been shown to be suppressed by sulforaphane in alveolar macrophages [70], peritoneal macrophages [72], adipose tissue macrophages [75], bone marrow-derived macrophages [73], THP-1 or peripheral blood mononuclear cell (PBMC)-derived macrophages [75,77,80,81,82,86,99,100], murine RAW264.7 cells [89,90,91,93,95,97], dendritic cells [74,76], and T-cells [101]. Similarly, sulforaphane has been shown to suppress the expression of pro-inflammatory cytokines in microglial BV2 cells [103,110,113,115], N9 murine microglial cells [105], senescent astrocytes [108], primary co-cultures of rat microglial and astroglial cells [111], and Müller cells of the retina [51]. Other inflammation models in vitro where cytokine production has been suppressed by sulforaphane include mast cells [102], endothelial cells such as human umbilical vein endothelial cells (HUVECs) [121,122], saphenous vein endothelial cell [127], and transformed endothelial cells such as ECV304 [124]. Similarly, the upregulation of cytokines’ production in epithelial cells has been shown to be suppressed by sulforaphane, including in Caco-2 [135], human lung epithelial cells (BEAS-2B) [136,143,145], and primary mouse tracheal and human bronchial epithelial cells [139]. Several other cell types under inflammatory conditions have also responded to sulforaphane to downregulate the expression of pro-inflammatory cytokines [147,148,151,155]. On the other hand, the expression of anti-inflammatory cytokines such as IL-10 is promoted by sulforaphane, as shown in senescent astrocytes [108], the LPS-activated C6 astrocyte cell line [116], and human monocyte-derived dendritic cells [76].
Moreover, in macrophages, the pro-inflammatory M1 marker’s morphology and genes (associated with pro-inflammatory cytokine production) are suppressed by sulforaphane, while it promotes the M2 marker genes associated with the anti-inflammatory mechanism [73,77,85,99]. In addition to IL-8, other chemokines’ expression, especially in the lung inflammation model, has been shown to be suppressed by sulforaphane. For example, the induced expression of MCP-1 [131,143] and MCP-1 and chemokine (C-X-C motif) ligand 1 (CXCL-1) [136] in airway epithelial cells is inhibited. Also, the expression of MCP-1 in HUVECs has been shown to be reduced by sulforaphane [119,121,122]. Hence, while multiple mechanisms may be implicated (see the following sections), the major anti-inflammatory mechanism of sulforaphane is attributed to the inhibition of the expression of pro-inflammatory cytokines and chemokines.
3.2. Anti-Inflammatory Effect of Sulforaphane through the Inhibition of the Expression of Adhesion Molecules
The major impact of pro-inflammatory cytokines such as IL-1 and TNF-α lies in their ability not only to induce the expression of other inflammatory cytokines or mediators but also the expression of key cell-surface adhesion molecules, primarily on endothelial and leucocyte cell surfaces. Over the last four decades, numerous research studies have shown that the expression levels of intracellular adhesion molecule 1 (ICAM-1), vascular cell adhesion molecule 1 (VCAM-1), and E-selectin on endothelial cell surfaces provide a good indication of the anti-inflammatory potential of therapeutic agents. In this connection, the endothelial cell surface expression of adhesion molecules such as ICAM-1 [124], VCAM-1 [121,122,129], ICAM-1 and VCAM-1 [119,120], and E-selectin and VCAM-1 [128] has been shown to be inhibited by sulforaphane. This is also evident in other cell types, including retinal pigment epithelial cells, where the suppression of the expression of ICAM-1 [134] and, in vascular smooth muscle cells (VSMCs), ICAM and VCAM [149] or VCAM-1 [150] has been observed. Similarly, the induced expression of ICAM-1 in epithelial cells by a variety of inflammatory mediators has been shown to be ameliorated [120,121,122,134]. Consequently, monocyte adhesion to activated endothelial cells has also been shown to be inhibited by sulforaphane [120,121,122]. Hence, the numerous in vivo anti-inflammatory effects of sulforaphane described in Section 2 are an attribute of both the suppression of the level of expression of pro-inflammatory cytokines as well as their effect on inflammatory cascades resulting from the reduction in adhesion molecules’ expression.
3.3. Anti-inflammatory Effect of Sulforaphane through the Suppression of COX-2 Expression
Classical anti-inflammatory compounds such as aspirin, indomethacin, ibuprofen, and diclofenac as well as the newer generation of selective COX-2 inhibitors (e.g., celecoxib) target the enzymatic activity of COX-2, while others, including steroidal anti-inflammatory agents, suppress the induced expression of COX-2. In the latter case, the expression of COX-2 has been shown to be suppressed by sulforaphane in activated peritoneal macrophages [72], PBMC [78], THP1 or PBMC [85], RAW264.7 cells [89,93,94,96,97], BV2 microglial cells [110,115], and mammary epithelial cells [130,140]. Other cells in which the expression of COX-2 has been shown to be suppressed by sulforaphane under inflammatory conditions include vascular smooth muscle cell lines [150], neuroblastoma SH-SY5Y cells [161], and rheumatoid arthritis synovial fibroblasts [157]. Hence, some of the anti-inflammatory effect of sulforaphane can be attributed to a reduction in expression of the key enzyme COX-2, thereby inhibiting the production of pro-inflammatory prostaglandins. Unlike COX-1, which is constitutively expressed and involved in normal physiological functions such as gastrointestinal tract (GIT) protection, targeting the inflammation-mediated or -induced expression of COX-2 by therapeutic agents avoids the general side effect of non-selective COX inhibitors.
3.4. Anti-inflammatory Effect of Sulforaphane through the Inhibition of iNOS Expression
Like COX-2, inducible nitric oxide synthase (iNOS) is an enzyme isoform associated with the inflammation-induced expression of inflammatory mediators, leading to nitric oxide (NO) overproduction. Inhibitors of iNOS have, thus, been sought after to treat inflammatory diseases [259,260]. The pro-inflammatory mediator (e.g., LPS and cytokines)-induced expression of iNOS has been shown to be suppressed by sulforaphane, such as in peritoneal macrophages [71], bone marrow-derived macrophages [72], stimulated RAW264.7 cells [89,90,91,92,93,96,97], PMBC-derived macrophages [78,99], BV2 microglial cells [110], astrocytes [116], mammary epithelial cells [130], neuroblastoma SH-SY5Y cells [161], C2C12 myoblasts [147], and vascular smooth muscle cells (VSMCs) [150]. With Macrophages being known to be factories of the NO which contributes to both oxidative stress and inflammation, some of the observed effects of sulforaphane are likely to be attributed to the inhibition of inflammation-associated iNOS expression.
3.5. Anti-Inflammatory Effect of Sulforaphane through the Inhibition of Inflammation-Associated Oxidative Stress
It is also worth noting that sulforaphane is widely known for its antioxidant effect primarily via the activation of the transcription factor Nrf2 (Table 1, Table 2, Table 3, Table 4 and Table 5). Induced ROS production in various cellular models has been shown to be suppressed by sulforaphane, including in neutrophiles [64], murine bone marrow-derived macrophages [72], PBMC [78], RAW264.7 cells [88,93], human lung epithelial cells (BEAS-2B) [136], N9 murine microglial cells [105], primary astroglial rat cultures or mouse cerebral cortices [106], primary cultures of cortical astrocytes from newborn pig brains [107], BV2 microglial cells [110], HUVECs [117], human brain endothelial cell [128], primary cultures of cerebral microvascular endothelial cells [107], mammary epithelial cells [130], airway epithelial [132,144], retinal pigment epithelial (ARPE-19) cells [133], Caco-2 cells [135], human lung epithelial cells (BEAS-2B) [136], primary mouse and tracheal and human bronchial epithelial cells [139], and human airway epithelial (NCI-H292) cells [144]. Readers should bear in mind that these are selective examples of sulforaphane’s effect on the amelioration of ROS production, specifically associated with inflammation experimental models. As described in Section 4, this antioxidant activity associated with inflammation is coupled with the induction of antioxidant defences through the Nrf2 signalling pathway (Table 1, Table 2, Table 3, Table 4 and Table 5).
4. Mechanistic Overview of the Anti-Inflammatory Effect of Sulforaphane
Mirroring the in vivo investigations, mechanistic studies on sulforaphane have been largely performed in vitro using various inflammatory models. The Nrf2-dependent and Nrf2-independent pathways of the key mechanisms are discussed below.
4.1. Induction of Nrf2
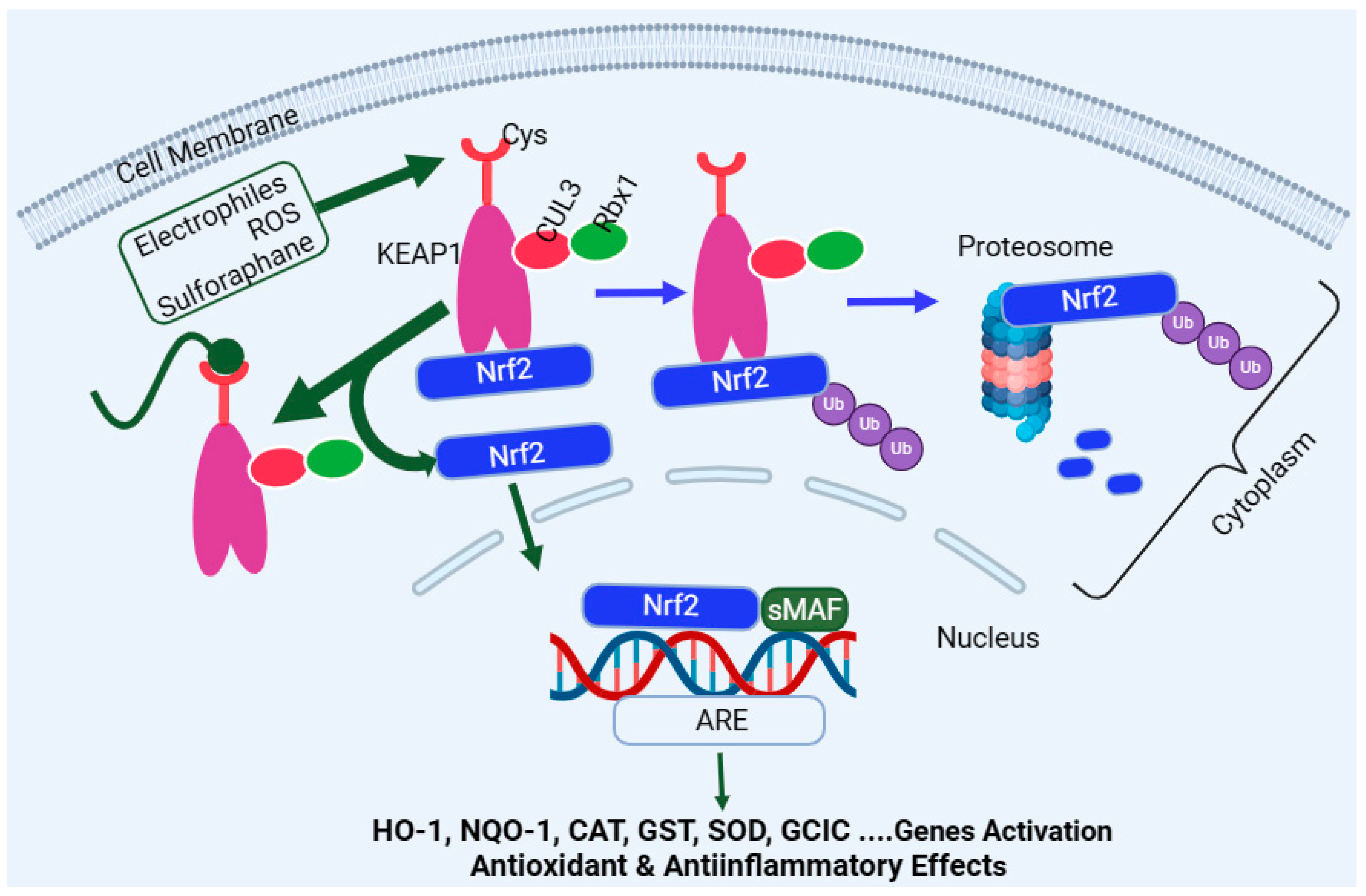
Encoded by the Nfe2l2 gene, Nrf2 is a master regulator of multiple antioxidant enzymes such as HO-1, NQO1, CAT, SOD, glutathione peroxidase (GPx), GST, therodoxins, and glutamate–cysteine ligase (GCL). In the latter case, GCL is a heterodimer of the GCL catalytic subunit (GCLC) and the GCL modifier subunit (GCLM). The target genes regulated by the Nrf2 pathway are far more than antioxidant enzymes and include detoxification enzymes, DNA repair enzymes, and molecular chaperone proteins. The activation or upregulation of the Nrf2 level leads to cytoprotection under stress conditions through various mechanisms, including the removal of ROS and oxidative stress, as well as anti-inflammatory and anti-apoptosis mechanisms. On this basis, Nrf2 is considered to be an evolutionarily conserved defence mechanism for a wide range of living organism to defend themselves against oxidative damage and xenobiotics [261]. Under normal physiological conditions, the Nrf2 in the cytoplasm binds to its negative regulator, the Kelch-like epichlorohydrin-associated protein (KEAP1), to form complexes. KEAP1 is the recognition site for redox-dependent CULLIN 3 (Cul3)-based (or CUL3–ring-box 1 (Rbx1)-containing) E3 ubiquitin ligase, which degrades Nrf2 (Figure 3)—i.e., KEAP1 acts as an adaptor protein for ubiquitin ligase, which is responsible for the ubiquitylation and subsequent degradation of Nrf2 through the proteasome (26s) system [262,263]. This system is collectively called the CULLIN-RING ubiquitin ligases complex, where CUL3 serves as a scaffold protein which forms a complex of E3 ligase with Rbx1 to recruit a cognate E2 enzyme. As one would expect, high levels of Nrf2 or Nrf2-target genes are observed in KEAP1 knockout mice and give these animals resistance to xenobiotics’ toxicity [264]. On the other hand, Nrf2-deficient mice have been shown to be susceptible to cigarette smoke-induced emphysema [265], hyperoxic lung injury [266], and pulmonary fibrosis [267]. One common activation pathway of Nrf2 is oxidative stress, ROS, or electrophiles, which directly interreacts with KEAP1 by oxidising or alkylating its cysteine residues, which are required for Nrf2 binding (Figure 3). This inactivates KEAP1 and removes the negative regulator of Nrf2 (removal of ubiquitination), leading to Nrf2 stabilisation and translocation into the nucleus to regulate target genes’ expression [268,269]. Upon entering the nucleus, Nrf2 heterodimerises with the small musculoaponeurotic fibrosarcoma (sMaf) protein family, which allows its binding to the antioxidant response element (ARE) sequence in the promoter regions of various target genes [270]. In addition to KEAP1, various protein kinases, such as mitogen-activated protein kinases (MAPKs) [271,272], protein kinase C (PKC) [271,273,274], AMP-activated protein kinase (AMPK) [275,276,277], PI3K [278,279], and glycogen synthase kinase-3 β (GSK3β) [280], induce the phosphorylation of Nrf2 and participate in Nrf2 transcription. The main regulatory mechanism of Nrf2 stability is, however, maintained through binding with KEAP1 (Figure 3).
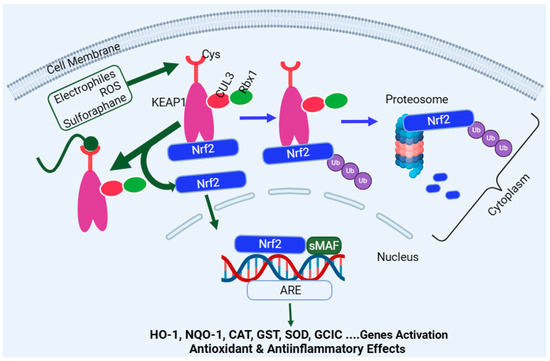
Figure 3.
Overview of the Nrf2 activation pathway. Nrf2 is inactivated under normal physiological condition through binding to KEAP1, leading to its proteasomal degradation. Reactive oxygen species, oxidative stress, or electrophiles such as sulforaphane can inactivate KEAP1 via interaction with redox-sensitive cysteine (Cys) residues. This allows the newly formed Nrf2 to be free to translocate to the nucleus and associate with sMAF to bind with the ARE domain of the DNA, leading to activation of target genes such as HO-1, NQO-1, CAT, GST, SOD, and GCLC. Abbreviations: sMAF, small musculoaponeurotic fibrosarcoma; ARE, antioxidant response element; CUL3, cullin-3; GCLC, glutamate–cysteine ligase catalytic; NQO1, NADPH quinone oxidoreductase enzyme; SOD, superoxide dismutase; HO-1, haeme oxygenase-1; CAT, catalase; and GST, glutathione S-transferase. This figure has been generated using Biorender.
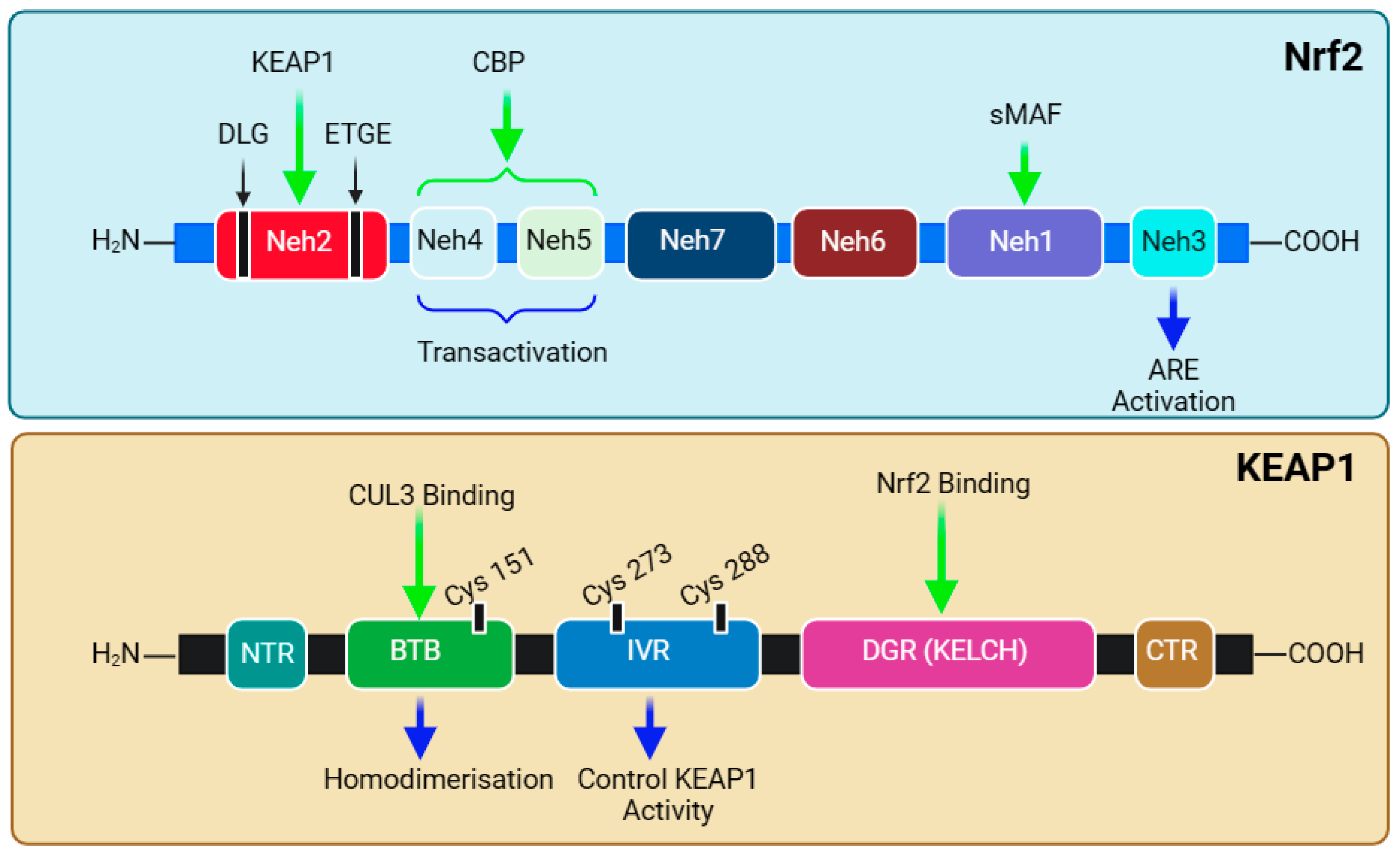
The functional adaptation of Nrf2 is based on its seven structural domains, Neh1–7, which attributes its stability and transcriptional activity to target genes (Figure 4). Notably, the N-terminal domain (Neh2) is responsible for binding with KEAP1 and, hence, governs the stability and ubiquitination of Nrf2. Studies have shown that high-affinity ETGE (Kd ~ 5 nM)- and low-affinity DLG (Kd ~ 1 μM)-binding motifs exist and coordinate Nrf2-KEAP1 binding [281,282,283,284]. The Neh2 domain contains seven lysine residues, which are critical substrates in the above-mentioned ubiquitination (binding with ubiquitin) and KEAP1-dependent Nrf2 degradation [269]. The mechanism of attachment and detachment of the Nrf2-KEAP1 in a model called Hinge and Latch mechanism has been described [285,286,287]. The next two domains, Neh4 and Neh5, are involved in interactions with nuclear co-factors and facilitate transcriptional activation. The synergistic cooperation of Neh4 and Neh5 to recruit a coactivator molecule called CBP (cAMP Responsive Element-Binding protein (CREB)-binding protein) has been suggested [288]. The activity of Nrf2 is enhanced by acetylation by CBP [289]. This is followed by the Neh7 domain, which is involved in repressing Nrf2 transcriptional activity via interaction with retinoic X receptor α [290]. Readers should note that Nrf2 was originally discovered in 1994 as a cap’n’collar (CNC) basic-region leucine zipper (bZIP) transcription factor [291]. Through a basic leucine zipper motif, the Neh1 domain is the site for binding Nrf2 to the ARE sequence and is involved in the interaction with the E2 ubiquitin-conjugating enzyme to govern Nrf2 stability and translocation [292]. Hence, the heterodimerisation of Nrf2 with Maf and DNA binding are functions of the Neh1 domain. KEAP1-independent Nrf2 degradation (e.g., by glycogen synthase kinase-3β (GSK-3β) is regulated by the Neh6 domain [293]. At the C-terminal, the Neh3 domain is involved in the interaction with a transcription coactivator called CHD6 (a chromo-ATPase/helicase DNA-binding protein) in the nucleus. Hence, three domains—Neh3, Neh4, and Neh5—are involved in the interaction with the coactivators for transactivation by Nrf2. The structural overview of both Nrf2 and KEAP1 is presented in Figure 4, and their relevance to sulforaphane bioactivity is discussed below.
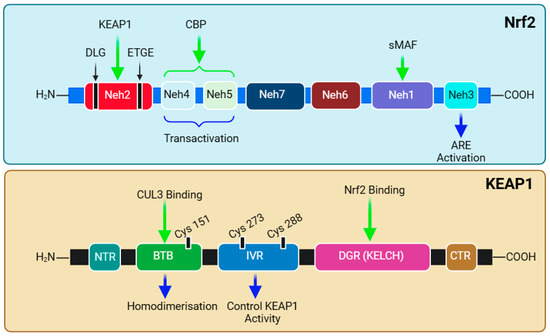
Figure 4.
Structural domains of Nrf2 and KEAP1. The protein structure of Nrf2 contains seven domains starting from the amino terminal, Neh2, through Neh4, Neh5, Neh7, Neh6, Neh1, and Neh3, at the carboxyl terminal’s end. The Neh2 domain contains DLG and ETGE as binding motifs for KEAP1. The Neh4 and Neh5 domains are known to be involved in NRF2 transactivation. The Neh1 domain has the DNA-binding motif through heterodimerisation with a small musculoaponeurotic fibrosarcoma (sMAF) protein. KEAP1 has three domains and a region on amino (NTR) and carboxyl (CTR) terminals. The Bric-à-Brac (BTB) domain is responsible for the homodimerisation of KEAP1 and binds to CUL3, and the intervening region (IVR) is involved in the control of KEAP1 activity, while the Kelch domain (KELCH) of DGR binds to the ETGE or DLG motif of the Neh2 domain of NRF2—i.e., one monomer of KEAP1 takes ETGE, while the other binds with DLG. This figure has been generated using Biorender.
The cysteine sulfhydryl groups of KEAP1 are the redox sensors regulating Nrf2’s transcriptional activity [294,295,296]. Under non-stressful or normal physiological conditions, the level of Nrf2 is kept low through constant degradation in the cytoplasm. The alteration in the sulfhydryl group of KEAP1 cysteines and the subsequent loss of E3 ligase activity are what that allows the newly synthesised and available Nrf2 to translocate to the nucleus (Figure 3). KEAP1 itself possesses several structural and functional domains (Figure 4), which are not detailed herein. Of note are the domains for the homodimerisation of the KEAP1 broad complex/tramtrack/bric-a-brac (BTB) domain, which binds CUL3 and is involved in KEAP1 homodimerisation [297]. The interaction of KEAP1 with the Neh2 domain of Nrf2 is a function of the so-called Kelch/DGR (double glycine repeat) domain, while the central intervening region (IVR) of KEAP1, which is rich in cysteine residues, controls KEAP1 activity. The DGR and carboxyl-terminal region (CTR) are also called the DC domain. From a functional point of view, forked-stem dimer structures with two large spheres enclosing the intervening double glycine repeat and C-terminal domains have been described by Ogura et al. [298].
The therapeutic implication of Nrf2 in the treatment of inflammatory diseases has been clinically proven. For example, dimethyl fumarate (DMF) has been effectively used in the treatment of relapsing-remitting multiple sclerosis [299,300,301]. The clinical application of DMF in the treatment of psoriasis has also been described [302]. Another potential drug at the development stage is bardoxolone (CDDO-Me), an oleanane-type triterpene acting as an Nrf2 inducer, which has shown promise in the treatment of diabetic nephropathy [303]. Overall, the activation of Nrf2 has been shown to reduce the pathological score of many diseases such as chronic obstructive pulmonary disease (COPD) [304] and sickle cell disease [305], among others. In this context, different therapeutic agents target the various cysteine residues located in KEAP1. Of these, sulforaphane is among the most extensively studied compounds acting as Nrf2 activators both in vitro and in vivo. It targets Cys151 (Figure 3) at the BTB domain of KEAP1 and blocks the KEAP1-CUL3 interaction, thereby reducing Nrf2 ubiquitination [306]. Diethyl maleate (DEM), DMF, and tert-butylhydroquinone are other examples of drugs acting in the same way as sulforaphane by preferentially targeting Cys151 [307]. The cyanoenone class of Nrf2 activators has also been shown to be targeting Cys151 in KEAP1, irrespective of their molecular size [308]. Andrographolide has also been shown to partially inhibit the interaction of KEAP1 with CUL3 by targeting Cys151 in KEAP1 [309], while 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2) is one exemplary drug preferentially targeting Cys288, and, in fact, the mutation of Cys151 does not affect Nrf2 induction by this compound [310]. Other agents such as 9-nitro-cotadec-9-enoic (OA-NO2) and 4-hydroxynonenal (4-HNE) target Cys151 but also, crucially, Cys273 and Cys288 [311]. In this regard, out of the twenty-seven cysteine residues known to occur in the human KEAP1 protein, the three that are most conserved across many living organisms and that are important targets for drugs are known to be Cys151, Cys273, and Cys288 [310]. A further group of compounds either targeting Cys288 or Cys226, Cys613, Cys622, and Cys624 is also known. Hence, drugs targeting cystine sensors of KEAP1 can be classified into several categories based on the cysteine residues they preferentially target. The readers should also note that not all therapeutic agents of Nrf2 induce work through the KEAP1 pathway. For example, curcumin has been shown to enhance the Nrf2 pathway by activating p38 MAPK to increase the Nrf2 level and the level of Nrf2-ARE interaction [312]. A further note on this subject is also that the upregulation of HO-1 activity and anti-inflammatory effects can be achieved by therapeutic agents independent of the Nrf2 pathway, as shown for some flavonoids [313].
Overall, numerous studies demonstrating the anti-inflammatory effect of sulforaphane have been shown to be associated with the induction of Nrf2 (Table 1, Table 2, Table 3, Table 4 and Table 5). In alveolar macrophages from patients with COPD, the use of siRNA to silence Nrf2 induction completely abolished the effect of sulforaphane upon recognition and the phagocytosis of clinical isolates of nontypeable Haemophilus influenza (NTHI) and Pseudomonas aeruginosa [65,66]. The induction of TNF-α, IL-1β, and IFN-β production in alveolar macrophages stimulated by LPS could also be suppressed by sulforaphane in the Nrf2-dependent pathway, as revealed by studies using Nrf2 (−/−) mice [71]. Along the same line of evidence, the deletion of KEAP1 in the lungs attenuates acute cigarette smoke-induced oxidative stress and inflammation [314]. The relevance of the induction of Nrf2 for the anti-inflammatory effect of sulforaphane in the various experimental models is clearly shown in Table 1, Table 2, Table 3, Table 4 and Table 5.
4.2. Inhibition of NF-κB
NF-κB is a generic name for a family of dimeric proteins as transcription factors induced by a variety of agents, including pro-inflammatory mediators. These proteins have subunits, five in mammals RelA (p65), RelB, p50 (from p105 precursor), p52 (p100 precursor), and c-Rel, which make dimers, but the most studied so far and the most widely expressed in cells is the p50/RelA dimer, NF-κB. All these subunits are known to have the Rel homology domain (RHD), which is critical for dimerisation, DNA binding, and the interaction with lκB inhibitors. In addition, RelA (also RelB) have transcription activation domains (TADs), which were all well characterised in the 1990s in [315] and the references therein. In unstimulated cells or under normal physiological conditions, the NF-κB dimers are bound to inhibitory molecules of the IκB family of proteins (inhibitors of NF-κB), and the activation of NF-κB requires cleavage from IκB by IκB kinase (IKK). The IKK is an enzyme complex that consists of two kinase subunits, IKKα (IKK1) and IKKβ (IKK2), and a regulatory subunit IKKγ (NEMO). The key to NF-κB activation is the phosphorylation of IκB proteins, inhibitory proteins, on specific serines in the N-terminal, leading to ubiquitination and the subsequent proteasomal degradation (Figure 5). Removing the inhibitory proteins from the complex releases NF-κB dimers to translocate to the nucleus and activate target inflammatory genes.
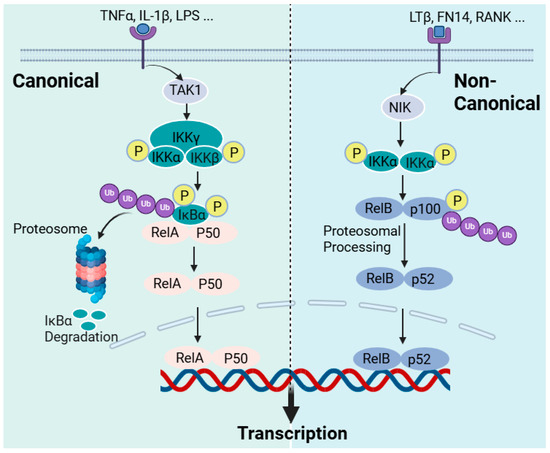
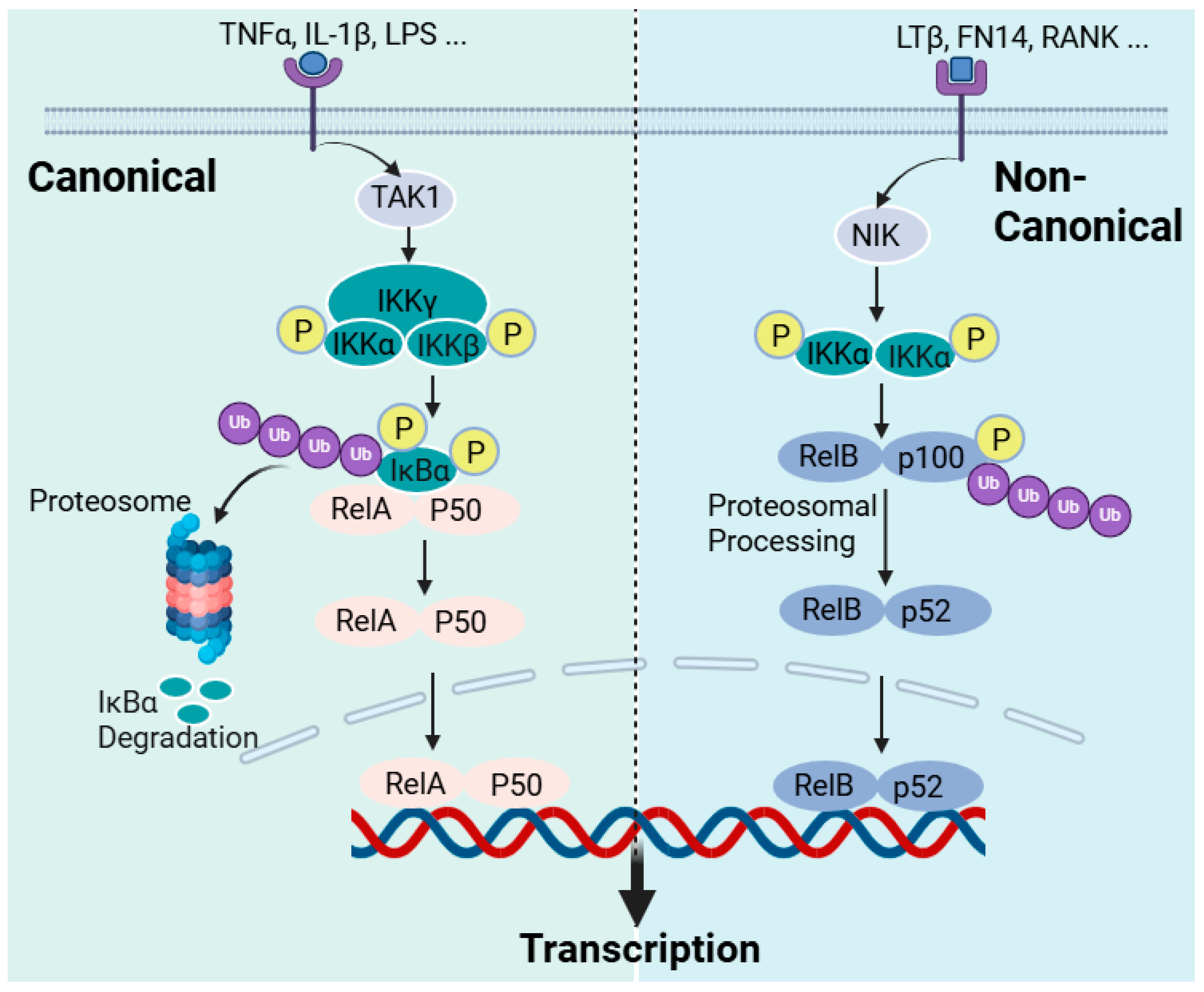
Figure 5.
The NF-κB activation pathways. The activation of cellular receptors to pro-inflammatory cytokines or bacterial and viral products leads to the activation of the NF-κB pathways through two principal routes: the canonical and non-canonical pathways. In the canonical NF-κB pathway, receptor activation (e.g., TNF-α, IL-1β, and LPS receptors) triggers the activation of the transforming growth factor beta-activated kinase 1 (TAK1), which activates the inhibitor of kappa B kinase (IKK) via phosphorylation at the IKKβ site. In turn, IKKβ then phosphorylates p105 bound to RelA to initiate proteasomal degradation or processing into p50. IKKβ also phosphorylates IκBα to initiate its proteasomal degradation. The free p50-RelA dimer then translocate to the nucleus to promote target gene activation. In the non-canonical pathway of NF-κB activation, receptor activation such as that of the lymphotoxin β receptor (LTβR), the B-cell-activating factor receptor (BAFF-R), Fn14, the Tweak receptor, and the receptor activator of NF-κB (RANK) leads to NF-κB-inducing kinase (NIK) activation, which phosphorylates the inhibitory kappa B kinase alpha (IKKα). IKKα, in turn, phosphorylates p100 for proteasomal processing into p52. The RelB/p52 dimer, as NF-κB, is then translocated to the nucleus. This figure has been generated using Biorender.
Readers who need to explore the detailed mechanism of the NF-κB activation pathways may refer to review articles on the subject [316,317]. Herein, a brief overview of NF-κB activation that is critically relevant to the discussion on the mechanism of action of sulforaphane is described. NF-κB has a diverse function in the cellular metabolism but is primarily involved in regulating the immune response following pathogenic or stress signals, leading to inflammation. In the innate immune response, immune cells such as macrophages and dendritic cells have receptors for pathogen-associated molecular patterns (PAMPs) that recognise bacteria and viruses. The toll-like receptor (TLR) family, out of which TLR4 is the best-known member, is among these receptors for bacterial products (LPS) that orchestrate the induction of NF-κB. These receptors are collectively called pattern recognition receptors (PRRs). Cell-mediated immunity consists of T lymphocytes-based antigen clearance (through the T-cell receptor (TCR)) and B lymphocyte (through the B-cell receptor) humoral immune responses, which also require NF-κB activation. In this classic example of immune cells’ activation by bacteria or their products, NF-κB is activated to initiate the inflammatory response, which includes the expression of pro-inflammatory cytokines (TNF-α, IL-1, and IL-6), chemokines, adhesion molecules, etc. This is what is called the “canonical” pathway of inflammation and can also be induced by pro-inflammatory cytokines such as TNF-α and IL-1, with the NF-κB involved being RelA- or cRel-containing complexes (Figure 5). Hence, the activation of TNF receptors, TLRs, T-cell receptors, and interleukin receptors typically leads to the activation of the canonical pathway of inflammation. Various inflammatory diseases, including rheumatoid arthritis, inflammatory bowel disease, asthma, and chronic obstructive pulmonary disease, are products of the exaggerated inflammatory response via the canonical NF-κB pathway. An “alternative” or “non-canonical” NF-κB activation pathway leading to the activation of RelB/p52 complexes can also occur by induction via lymphotoxin β, the B-cell-activating factor, and the receptor activator of the NF-κB ligand, among others. As indicated above, IKKα and IKKβ (catalytic units) and IKKγ (regulatory subunit) constitute the classical canonical NF-κB activation mechanism. IKKβ regulates the activation of the canonical pathway through the phosphorylation of IκBs and requires the IKKγ subunit, while IKKα is required for the activation of the alternative pathway (Figure 5). IKK itself is activated in the canonical pathway via receptor activation through phosphorylation by kinases and, most notably, the TGFβ-activated kinase 1 (TAK1; also known as MAP3K7), but there are many others, including RIP1 (receptor-interacting kinase 1) and TBK1 (TANK-binding kinase) [318]. The phosphorylation of IKKβ is also known to be involved in the processing of p105 to yield p50 [319]. Hence, the canonical activation of NF-κB is regulated through phosphorylation by IKKβ for the proteasomal degradation of IκBα, leading to RelA-p50 complexes’ translocation to the nucleus. On the other hand, the non-canonical pathway of NF-κB activation is the process of releasing RelB-p52 dimers through the phosphorylation of IκB, and, for this, IKK phosphorylation is a function of the NF-κB-inducing kinase (NIK; also known as MAP3K14). The link between NIK phosphorylation and the activation of TNF superfamily receptors (TNFSFRs) such as TNFSFR12A (Fn14, Tweak receptor), the lymphotoxin β receptor (LTβR), the B-cell-activating factor receptor (BAFF-R), the receptor activator of NF-κB (RANK), CD40, and CD27 has been established [320,321]. Further details are available from review articles on the subject [316,317]. The regulatory mechanism of NF-κB in the B-cell survival and maturation, dendritic cell activation, bone metabolism, and lymphoid organogenesis is known to involve the alternative or non-canonical pathway [322,323]. In terms of therapeutic intervention, the NF-κB pathway has been extensively studied in the last few decades as a potential target for anti-inflammatory compounds. By far, the most known specific target of this for natural products is IKK, predominantly targeting IKKβ. The diverse function of IKK, however, remains a challenge for the therapeutic application of these agents and, specifically, for targeting inflammatory diseases. Research on this field is extensively diverse, and the inhibitory compounds belong to diverse structural groups, including natural products such as terpenoids, flavonoids, or polyphenols, among others. Thiol-reactive compounds such as sulforaphane are included in this group [324].
On the above basis, sulforaphane has been shown to downregulate NF-κB activity in the following cell types under a variety of inflammatory conditions: bone marrow-derived dendritic cells [74]; T-cell lines [81]; PBMCs [82]; THP-1 [86,100]; RAW264.7 cells [94,95,97]; human mast cells [102]; mouse microglial BV2 cells [103,113,114]; N9 murine microglial cells [105]; primary rat microglia [114]; the C6 astrocyte cell line [116]; neuroblastoma SH-SY5Y cells [161]; N2a/APPswe cells [162]; HUVECs [121,122,126]; ECV304 [124,125]; human saphenous vein endothelial cells [127]; primary goat mammary epithelial cells [130]; human retinal pigment epithelial [134]; human mammary epithelial [140]; VSMCs [149,150]; primary human articular chondrocytes [153]; retinal pigment epithelial [154]; synoviocytes [155]; human embryonic kidney 293T (HEK293T) cells [156]; and mouse pancreatic acinar cells [158].
Mechanistically, sulforaphane, by virtue of forming dithiocarbamate through a reaction with thiol groups, can directly interact with DNA binding and the transactivation of NF-κB [325]. It has also been shown to suppress TNF-α-induced IκBα phosphorylation and IκBα degradation [326]. In TNF-α-stimulated HUVECs and human aortic endothelial cells, sulforaphane has inhibited the induced phosphorylation of IκB kinase (IKK) and IκB-α, as well as the binding of NF-κB to binding sites in the LIPG gene [126]. Through direct interaction with cysteines, sulforaphane also inhibits the IκBβ subunit and IκBα [95] and the phosphorylation of IκBα [102,126,130]. It is also reported to inhibit transcriptional activity as well as IκBα phosphorylation and degradation [121,122,124,140,150]. Evidence of the inhibition of the DNA-binding activity of NF-κB [153] as well as C/EBP and CREB binding [94] or the binding of NF-κB to binding sites in the LIPG gene, has also been shown [126]. Collectively, there is overwhelming evidence to suggest that the anti-inflammatory effect of sulforaphane is mediated through the downregulation of NF-κB at the various stages of its signalling cascade. The crosstalk between NF-κB activation and signalling through tyrosine kinases is discussed below (Section 4.4).
4.3. Nrf2-NF-κB Crosstalk
In the regulation of redox status and inflammation, there is a great deal of coordination between the Nrf2 and NF-κB pathways. Numerous reports have shown that stress conditions that activate the NF-κB pathway, such as that of TNF-α or pathologies such as cancer, also stimulate Nrf2’s protective mechanisms [327,328]. Generally, the activation of NF-κB can lead to the upregulation of Nrf2 or its target gene products, such as HO-1. In many respects, however, the inflammatory response mediated through the NF-κB pathway is counteracted by Nrf2 activation [329,330] (Figure 6). For example, the degradation of IKKβ by KEAP1, leading to decreased phosphorylation and the negative regulation of the NF-κB pathway, has been described [331]. The competition between Nrf2 and the NF-κB-p65 subunit to bind the transcriptional coactivator CBP has also been demonstrated [332]. The Nrf2 pathway has also been shown to suppress the transcription of several inflammatory cytokine genes [333]. In this regard, many anti-inflammatory agents that suppress NF-κB signalling have been shown to activate the Nrf2 pathway. These include α-tocopheryl [334].
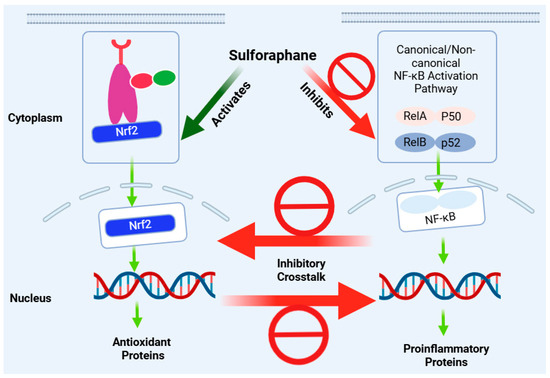
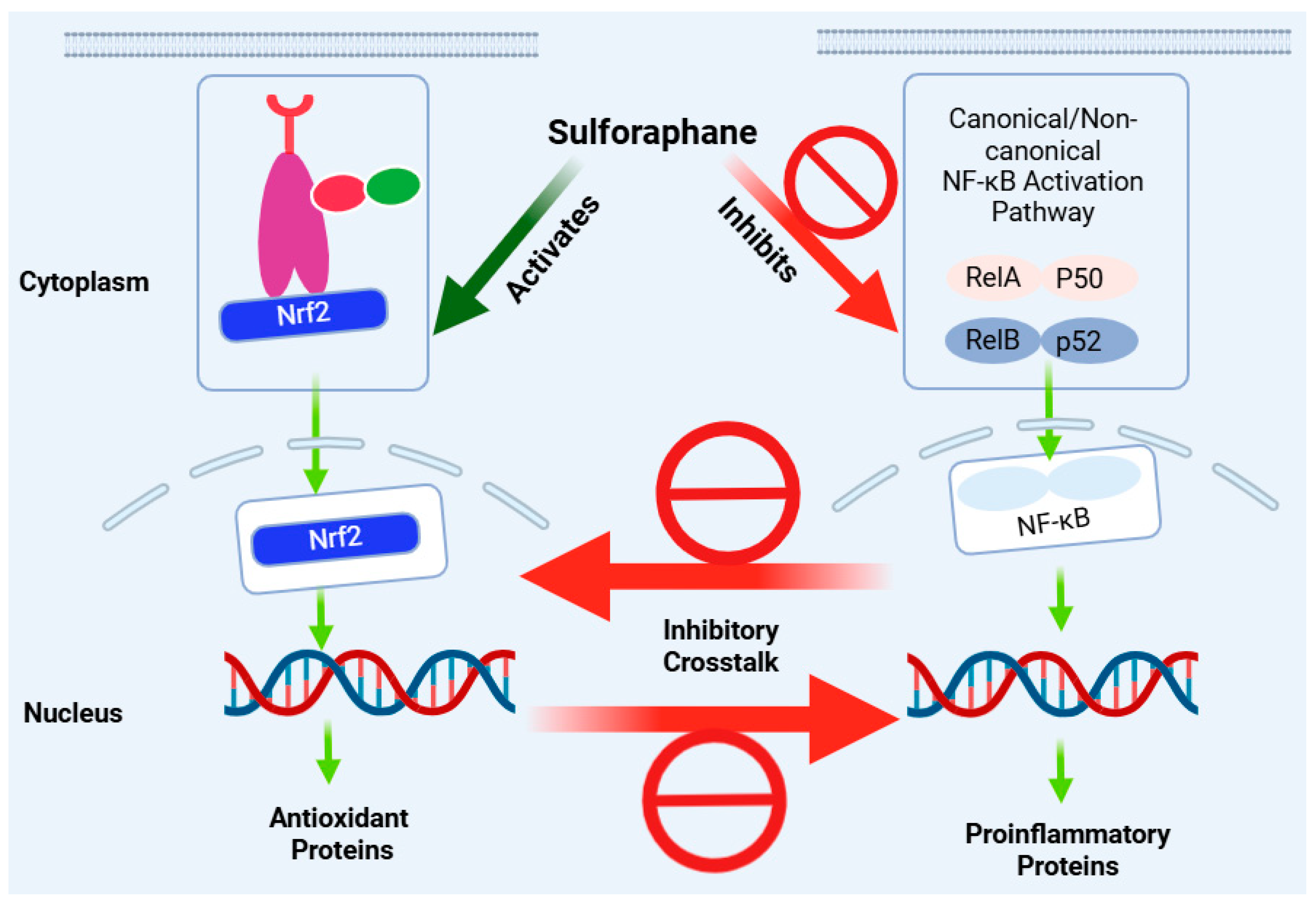
Figure 6.
Anti-inflammatory effect of sulforaphane via crosstalk of the Nrf2 and NF-κB pathways. Inhibition of the NF-κB pro-inflammatory pathway is mostly associated with the induction of the Nrf2 pathway as an antioxidant and anti-inflammatory mechanism. Both effects are simultaneously seen for sulforaphane in various experimental models, although the magnitude of modulation and interdependence of the two pathways may differ. This figure has been generated using Biorender.
In peritoneal macrophages, sulforaphane has been shown to inhibit the mRNA expression of TNF-α, IL-1β, COX-2, and iNOS induced by LPS, but this effect is evident in (+/+) but not in Nrf2 (−/−) peritoneal macrophages [71]. In almost all cases where both Nrf2 and NF-κB activation were studied, the effect of sulforaphane was associated with enhancing Nrf2 while, at the same time, downregulating NF-κB (Table 1, Table 2, Table 3, Table 4 and Table 5). Unequivocal evidence of crosstalk comes from knockdown studies where the effect of sulforaphane has been reported to be dependent on Nrf2 activation: for example, the inhibition of the NLRP3 inflammasome in an Nrf2-independent manner [98]. This could be a partial effect, not a complete one, as the knockdown of Nrf2 has been shown to partly (not completely) abolish the reduction in ROS, nitric oxide, and pro-inflammatory cytokines (TNF-α, IL-1β, and IL-6) [105]. Hence, multiple mechanisms of action, often paradoxical, may be at play in the anti-inflammatory effect of sulforaphane.
4.4. Signalling Paradox
In macrophages, the inhibitory effect of sulforaphane on cytokine expression and M1 markers’ C-C motif chemokine receptor 7, IL-23, and iNOS has been shown to be associated with the inhibition of mitogen-activated protein kinases (MAPK), p38, c-Jun N-terminal kinase (JNK), and phosphorylation [99]. Downregulating the phosphorylation of MAPK in PMA-activated human mast cells using sulforaphane has also been associated with the inhibition of NF-κB and cytokine expression [102]. PMA-activated endothelial cells (HUVECs) have also been shown to respond to sulforaphane treatment by reducing the level of pro-inflammatory cytokine (IL-1β and TNF-α) expression via the downregulation of the phosphorylation of p38, extracellular regulated kinases (ERK) 1/2, and JNK [123]. Another example of PMA-activated NF-κB activation can be found in the human mammary epithelial cells, where sulforaphane inhibits both IKK activation and ERK1/2—i.e., ERK1/2-IKKα-NF-κB signalling [140]. In ECV304 endothelial cells, the suppressive effect of sulforaphane on NF-κB translocation has been shown to be mediated via the inhibition of the phosphorylation of mainly p38 MAPK and JNK MAPK [125]. The expression of adhesion molecules on endothelial cell surfaces induced by TNF-α has also been shown to be inhibited by sulforaphane via the inhibition of the activation of p38 MAPK (not JNK), and, interestingly, this effect is not mediated via Nrf2 expression [129]. The phosphorylation levels of ERK and JNK MAPKs associated with cigarette smoke extract (CSE)- and particulate matter-induced inflammatory and chemokine gene (IL-1β, IL-6, IL-8, TNF-α, MCP-1, and CXCL-1) expression in human lung epithelial cells are further decreased by sulforaphane [136]. In BV2 microglial cells treated with advanced glycation end products, the expression of neuroinflammatory mediators and ROS is inhibited by sulforaphane in association with reduced levels of GSK3β activation and p38 phosphorylation (but not ERK and JNK phosphorylation) and the inhibition of NF-κB. All these data suggest that the anti-inflammatory effect of sulforaphane via suppressing the NF-κB activation pathway is associated with the suppression of the MAPK pathway and is not necessarily associated with Nrf2 activation. On the other hand, the induction of Nrf2 in microglial cells has been shown to be dependent on the phosphorylation of p38 and ERK1/2, and sulforaphane augments this activity [105]. It is also known that cigarette smoke generally activates MAPK signalling cascades in lung epithelial cells both in vitro and in vivo [335]. In this regard, the inhibition of p38 MAPK is proven to ameliorate allergen-induced pulmonary eosinophilia, mucus hypersecretion, and airway hyper-responsiveness or diseases such as asthma and COPD. While Nrf2 is experimentally proven to have a protective role in numerous airway diseases [336], the signalling pathway in collaboration or contradiction with NF-κB is still not clear. In a study using human lung epithelial cells, where inflammatory cytokines were inhibited by sulforaphane, MAPKs were inhibited while Nrf2 was activated [136]. Overall, sulforaphane seems to block the phosphorylation of MAPKs (p38, JNK, and ERK1/2), along with NF-κB p65 [113].
Studies have shown that the upregulation of phosphoinositide 3-kinase (PI3K) and Akt by therapeutic agents (e.g., ginkgolides and bilobalide) can activate Nrf2 [337]—i.e., Akt, as a downstream signal molecule of PI3K, can phosphorylate KEAP1 as well as have other effects, leading to Nrf2 activation. In this context the hypoxia- or cobalt chloride-induced upregulation of TLR4 mRNA and protein have been shown to be mediated by inhibiting PI3K/Akt activation, and sulforaphane reverses this effect to induce protection against oxidative stress in macrophages [88]. PI3K activation inhibits macrophage programming into M1, while Akt activation is a critical condition for M2 polarisation. Thus, sulforaphane suppresses the cobalt chloride-induced upregulation of TLR4 by inhibiting PI3K/Akt activation and the subsequent nuclear accumulation and transcriptional activation of HIF-1α [88].
4.5. Anti-Inflammatory Effect of Sulforaphane by Targeting Sirtuin 1 (SIRT1) Signalling
Sirtuin 1 (SIRT1) belongs to a family of proteins (SIRT1-7; silent information regulators or sirtuins) or deacetylase enzymes that function in collaboration with their essential co-factor, nicotinamide adenine dinucleotide (NAD+). They remove acetyl groups from the lysine residues of many proteins or histone substrates and function in the post-translational modification of proteins by mono-ADP ribosylation. As shown in the section below, SIRT1 also catalyses a range of substrates such as NF-κB, forkhead box class O family member proteins (FOXOs), peroxisome proliferator-activated receptor γ (PPARγ), peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α), and P53 [338,339,340]. In terms of inflammation, NF-κB and SIRT1 display antagonistic crosstalk, where NF-κB is acting in a pro-inflammatory manner while SIRT1 promotes anti-inflammatory responses. While SIRT1 overexpression reverses the inflammatory pathology, its underexpression promotes inflammation, as shown in experimental animals exposed to cigarette smoke or chronic obstructive pulmonary disease [341,342]. In macrophages, the knockdown of SIRT1 has been shown to increase the activation levels of NF-κB and pro-inflammatory cytokines [343]. These experiments have revealed that SIRT1 overexpression decreases the acetylation of RelA/p65 and NF-κB-dependent inflammation and/or NF-κB activity by inducing suppressor mechanisms. Hence, the upregulation of SIRT1 reduces COX-2 levels by inhibiting the activation of AP-1 and NF-κB [344]. Overall, SIRT1 activation could exert benefits in the treatment of inflammatory disorders, and its level/activity is suppressed by NF-κB through the modulation of downstream signalling. For example, through NF-κB induction, TNF-α inhibits the expression of peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1α), which plays critical role in SIRT1 signalling. Accordingly, SIRT1 activation suppresses the expression of TNF-α in macrophages [345]. Another important signalling molecule related to SIRT1 is the AMP-activated protein kinase (AMPK), which acts as an initial sensor to increase the level of NAD+ [346].
The inhibition of NF-κB p65 nuclear translocation by sulforaphane in human retinal pigment epithelial (ARPE-19) cells exposed to blue light is coupled with the Nrf2 pathway, which, in turn, is associated with increased protein expression of SIRT1 and PGC-1α gene expression [134]. Sulforaphane also upregulates the phosphorylated level of AMPK (p-AMPK), SIRT1, and PGC-1α to ameliorate LPS-induced changes in intestinal permeability, oxidative stress, inflammation, and apoptosis [135]. The upregulation of p-AMPK, SIRT1, and PGC-1α has also been observed in LPS-stimulated Caco-2 cells after treatment with sulforaphane, which is also coupled with the upregulation of antioxidant enzymes (e.g., SOD, GPx, and CAT) and the downregulation of inflammatory cytokines (IL-1β, IL-6, IL-8, and TNF-α) [135]. Oligomeric amyloid-β1-42 can trigger injury in the retinal pigment epithelium by repressing SIRT1, but treatment with sulforaphane can maintain cell viability and SIRT1 expression [347]. Sulforaphane prevents rat cardiomyocytes from hypoxia/reoxygenation injury in vitro by elevating the expression of SIRT1 in cardiomyocytes [348]. It also exerts an anti-apoptotic effect on chondrocytes and ameliorates the OA in vivo by activating SIRT1 [166]. In support of this mechanism, many natural products, including resveratrol as an allosteric activator of SIRT1 [349] and curcumin [350], have been shown to induce cardioprotective effects by activating SIRT1. Hence, SIRT1 activation is an emerging mechanism of action for the anti-inflammatory effect of sulforaphane, as depicted in Figure 7.
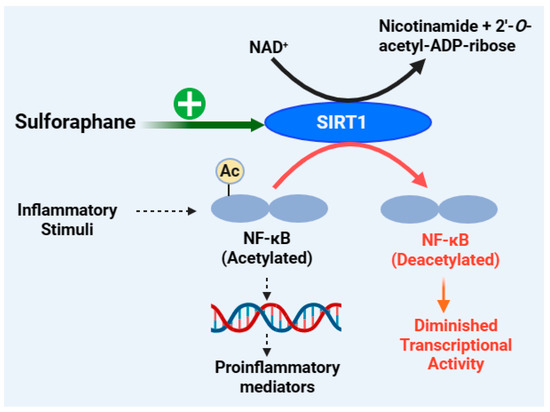
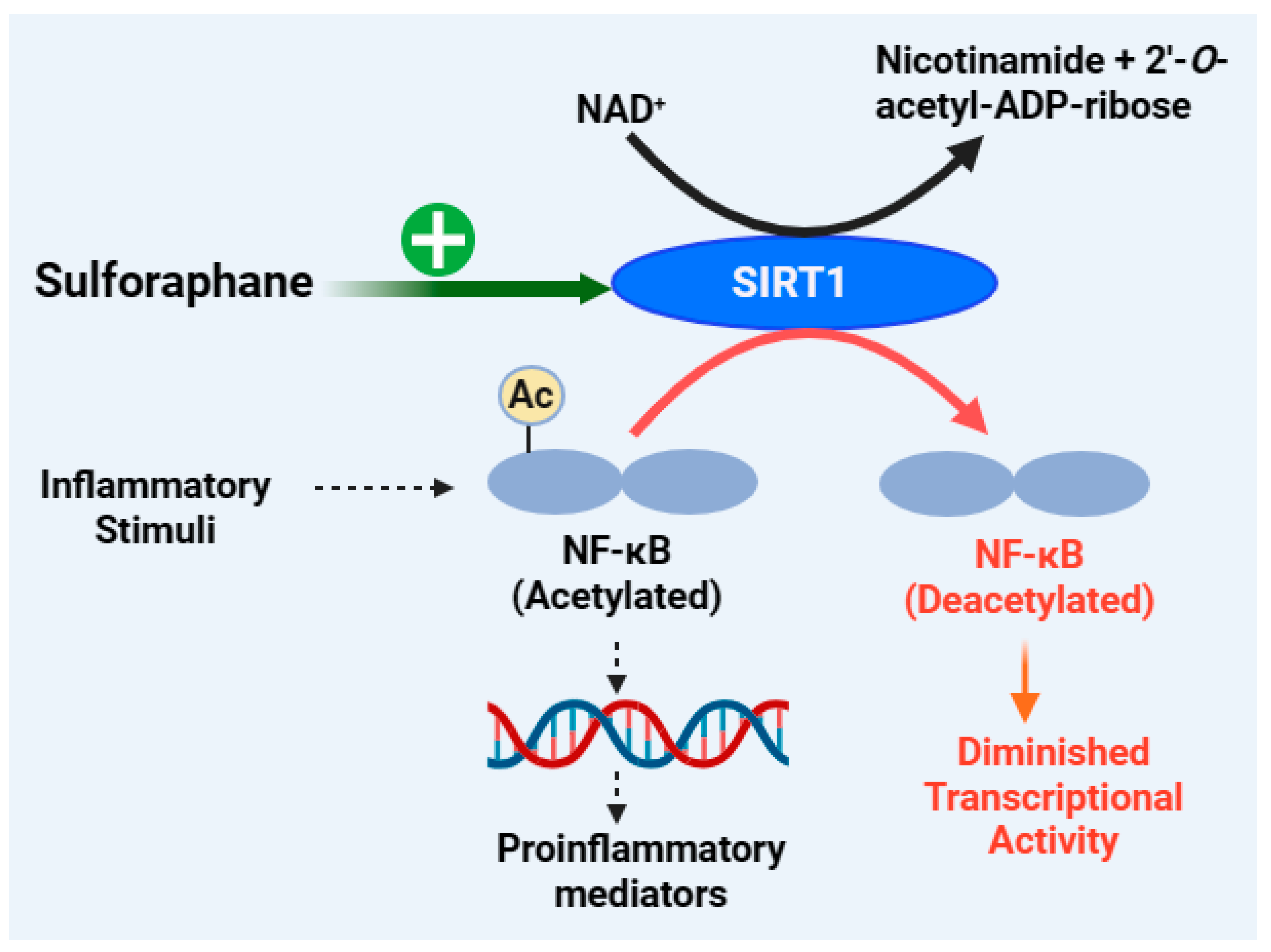
Figure 7.
Anti-inflammatory effect of sulforaphane through the SIRT1 mechanism. The normal function of NF-κB in the induction of inflammatory gene/protein (e.g., IL-1, IL-6, TNF, ICAM, VCAM, ELAM, iNOS, COX-2, etc.) expression requires the acetylation process. By deacetylating NF-κB, SIRT1 acts as a negative regulator of inflammation or NF-κB—i.e., SIRT1 denies NF-κB its DNA binding, transcriptional activity, and stability as well as interaction with protein modifiers. Boosting the expression of SIRT1 with sulforaphane thus reduces the NF-κB-dependent inflammatory response as well as other effects associated with the deacetylation activity of SIRT1 and several other inflammatory proteins. This figure has been generated using Biorender.
4.6. Anti-inflammatory Effect of Sulforaphane by Targeting STATs
The roles of the silent information regulator sirtuin 1 (STAT1) and STAT3, often as contradictory as pro- and anti-inflammatory mediators in macrophages, have been widely reported [351]. Thus, LPS and IL-6 can activate the M1 phenotype, and IL-10 acts as an anti-inflammatory cytokine through the modulation of STAT1 and STAT3 signalling [352]. STAT3 activation is generally considered to promote the anti-inflammatory M2 phenotype, in contrast to NF-κB activation, which promotes the M1 phenotype associated with the expression of pro-inflammatory cytokines. Thus, therapeutic agents that reduce the activation of STAT1 and/or suppress STAT3 may induce an anti-inflammatory effect by promoting the polarisation of M1 to the M2 phenotype [353]. For example, Sun et al. [73] have reported that sulforaphane enhances IL-10 production in macrophages while promoting STAT3 activation. On the other hand, the suppressive effect of sulforaphane on LPS-mediated increase in ICAM-1 and VCAM-1 expression on the endothelial (HUVEC) cell surface has been unequivocally proven (siMRA study) to be associated with the inhibition of STAT3 [120]. This finding agrees with that reported by Rakariyatham et al. [97] on macrophages stimulated by LPS, where the level of STAT3 phosphorylation was inhibited by sulforaphane. Furthermore, Jeong et al. [160] have shown that TNF-α-induced NF-κB as well as STAT1 activation in human keratinocytes is inhibited by sulforaphane. Hence, further research is needed to establish the cell- and/or inflammatory model-dependent role of STAT1/3 as a target for sulforaphane.
4.7. The Emerging Role of Activator Protein-1 (AP-1) as an Anti-Inflammatory Target for Sulforaphane
Just like NF-κB, activator protein-1 (AP-1) represent a family of transcription factors functioning as dimers to regulate immune function and oncogenic processes. Of note, it is linked to MAPK signalling and is involved in T-cell activation. AP-1 proteins play important roles in the development and maintenance of cancers, which are not described herein. Their role as a therapeutic target for inflammatory diseases has also been established [354] and includes chronic inflammatory diseases such as rheumatoid arthritis [355]. This is attributed to AP-1 being pro-inflammatory, as NF-κB, and able to regulate the expression of cytokines such as TNF-α and IL-1 through direct interaction with their promoter AP-1-binding motifs. As it has been shown for sulforaphane, the crosstalk between redox and inflammatory regulation for AP-1 follows the same line of evidence as NF-κB. Hence, the suppressive effect of sulforaphane on LPS-induced COX-2 expression has been shown to be associated with the inhibition of NF-κB, C/EBP, and CREB, as well as AP-1 [94]. By reducing the level of JNK phosphorylation, sulforaphane has been demonstrated to suppress NF-κB and AP-1 signalling in LPS-activated microglia [115]. The photoprotective effects of sulforaphane in human keratinocyte cells and BALB/c mice subjected to repetitive ultraviolet A (UVA) irradiation have been shown to be associated with the inhibition of MAPK/AP-1 signalling [356]. In a similar study, using UVB-induced AP-1 activation, sulforaphane directly inhibited the DNA-binding activity of AP-1 [357]. Further studies have shown that the inhibition of AP-1 by sulforaphane involves interaction with cysteine in the cFos DNA-binding domain [358]. Evidence is also rich on the anticancer effect of sulforaphane by targeting AP-1. In prostate cancer cells, for example, AP-1 activation is attenuated by the combinations of sulforaphane and epigallocatechin gallate [359]. Other known anti-inflammatory compounds of natural origin that have shown an effect through the AP-1-dependent pathway include quercetin [360], omega-3 docosahexaenoic fatty acid [361], ganglioside GM3 [362], and curcumol [363].
As with NF-κB, paradoxical evidence of AP-1’s role as a target for sulforaphane has been noted. For example, the treatment of breast cancer cells with sulforaphane inhibits the TPA-stimulated NF-κB-binding activity but not the AP-1-binding activity [364]. Hence, more research is needed, although, overall, AP-1 inhibition, just like NF-κB, appears to be a common mechanism for sulforaphane as an anti-inflammatory compound.
4.8. MicroRNAs as Potential Therapeutic Targets for Sulforaphane
MicroRNAs (miRNAs) are small-molecular-weight non-coding RNAs that suppress gene expression both by inhibiting protein translation and promoting mRNA cleavage. In principle, they can either enhance or inhibit inflammation, depending on the targeted mRNAs. As reviewed by Tahamtan et al. [365], several miRNAs are known to act as anti-inflammatory agents, and they include miR-10a, miR-21, miR-24, miR-106b, miR-124, miR-143, miR-145, miR-146, miR-155, and miR-375. They can act on several target transcription factors such as NF-κB and STATs, receptors such as TLR, TNF receptors, or even kinase enzymes. In the LPS-stimulated RAW264.7 cells, sulforaphane has been shown to suppress the miR-146a and miR-155 levels associated with inflammation [90,91]. The enhanced miR-423-5p levels associated with hepatic stellate cell activation in liver fibrosis have also been shown to be downregulated by sulforaphane [366]. In contradiction to such an effect, the level of miR-423-5p has been shown to be low in septic mice with a liver injury, and its overexpression alleviates acute liver injury and inflammatory responses [367]. Hence, more research in the miRNA field is needed to establish the contribution of miRNAs as targets for sulforaphane as an anti-inflammatory agent.
5. General Summary and Conclusions
The present analysis of sulforaphane as an anti-inflammatory agent clearly shows the crosstalk between inflammation and oxidative stress, specifically via NF-κB and Nrf2 signalling. Sulforaphane appears to suppress both inflammation and oxidative stress mostly by inhibiting NF-κB and enhancing Nrf2 signalling. The long list of publications based on sulforaphane suppressing inflammation in vivo and in vitro has been described in the previous sections through the effects attributed to the modulation of NF-κB and Nrf2. However, there have been paradoxical results against this conclusion, and more research is needed to clarify the degree of interdependence between the two pathways in mediating the anti-inflammatory effect of sulforaphane. The readers should also note that there are also some studies suggesting that sulforaphane could have pro- and anti-inflammatory effects, as revealed for placental cytokine production under some conditions of bacterial infections [368]. Furthermore, there are also studies that show that sulforaphane improves the oxidative status under ischaemia/reperfusion injury in rats without attenuating the inflammatory response [369]. Indeed, in some studies, the pro-oxidant nature of sulforaphane is well known and, with it, one of the mechanisms for its anticancer effect. As a pro-oxidant compound, sulforaphane synergises with anticancer agents such as cisplatin to induce apoptosis through ROS accumulation [370]. This paradoxical effect could be dose-dependent, as most of the anti-inflammatory effects in vitro are carried out at 5 or 10 µM, while higher doses are known to increase ROS and even inhibit ROS-scavenging enzymes such as GR and GPx activity in cancer cells [371]. By interfering with the glutathione recycling processes, sulforaphane can also induce oxidative stress and death through the p53-independent mechanism [371]. In the cancer domain, sulforaphane also augments the immune response instead of having an immunosuppressive effect, as shown in the WEHI-3-induced leukaemia mouse model, where it enhances phagocytosis by macrophages and natural killer cells [372]. Along the same line, Nrf2 activation by sulforaphane restores the age-related decrease in TH1 immunity by acting on dendritic cells, implying that it acts as an immunostimulant [373]. While it induces a cytoprotective effect via the Nrf2 mechanism in naïve cells, it actually promotes apoptosis in TNF-α-stimulated synoviocytes [155]. Hence, the effects of sulforaphane as an anti-inflammatory and cytoprotective agent depend on a variety of factors, including the health and disease states. Under hepatitis B virus (HBV) infection in vitro and in vivo, sulforaphane promotes macrophages to the pro-inflammatory M1 phenotype [374], while, under inflammatory conditions both in vitro and in vivo, it induces macrophages to change to the anti-inflammatory M2 phenotype [375], as explained in the various sections of this article.
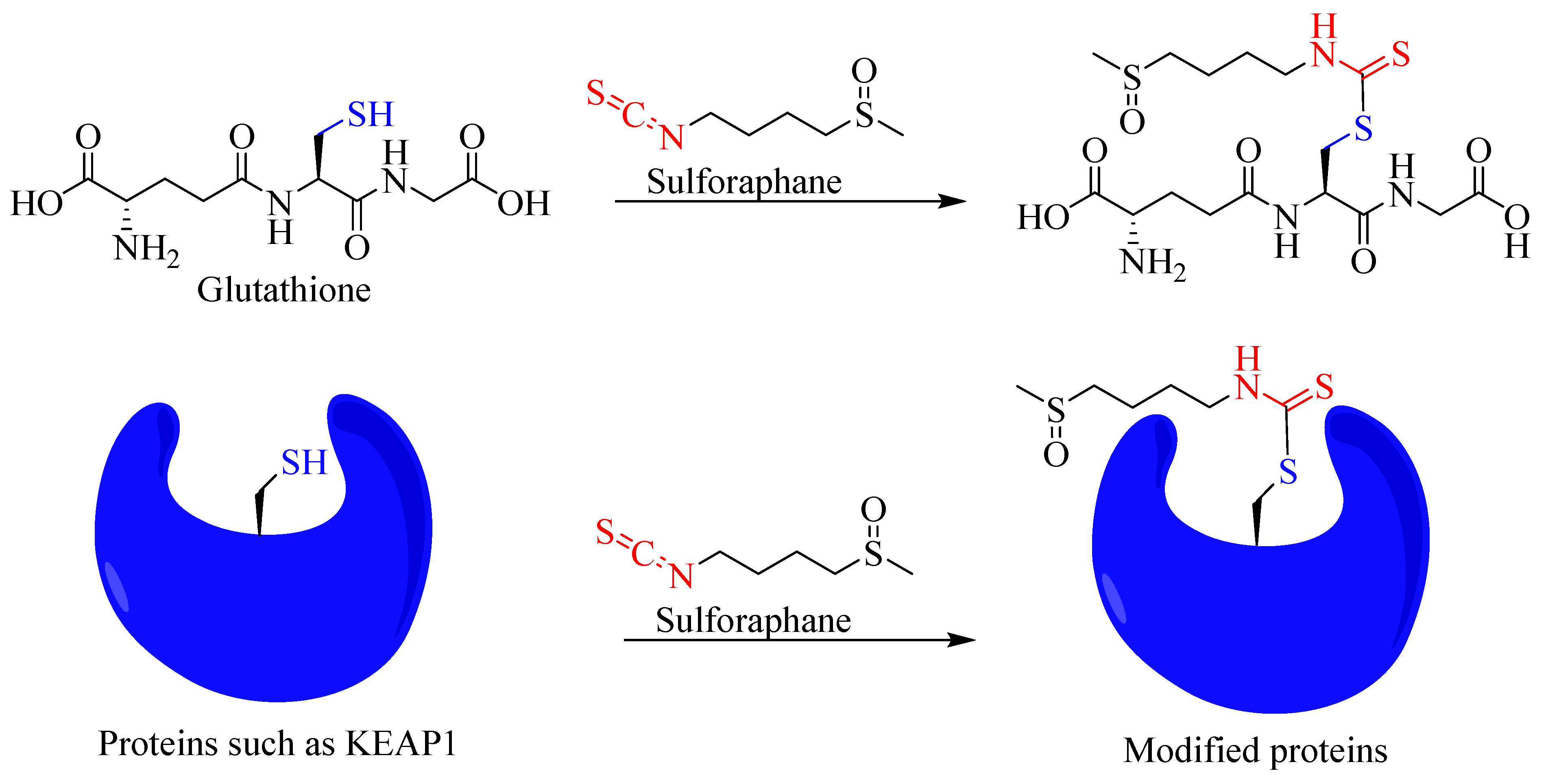
As readers can see from the extensive literature cited in this article in the context of inflammation as a disease target, sulforaphane is among the most widely studied natural products both in vitro and in vivo. Mechanistically, its paradoxical effects depending on health and pathophysiological states need further research. Most of the studies show that its effect via interactions with biological molecules such as the cysteine residue of proteins account to its diverse pharmacology. From small molecules such as glutathione to enzymes and transcriptional activities, sulforaphane has been shown to covalently interact with the -SH functional group to induce its diverse functions (Figure 8).
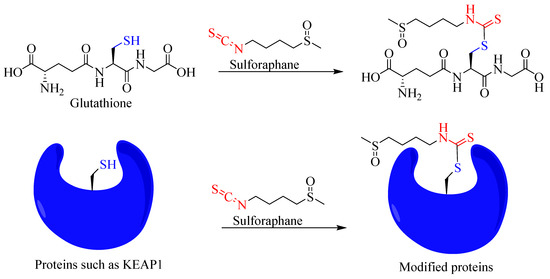
Figure 8.
Overview of covalent interaction of sulforaphane with the sulfhydryl group of biomolecules.
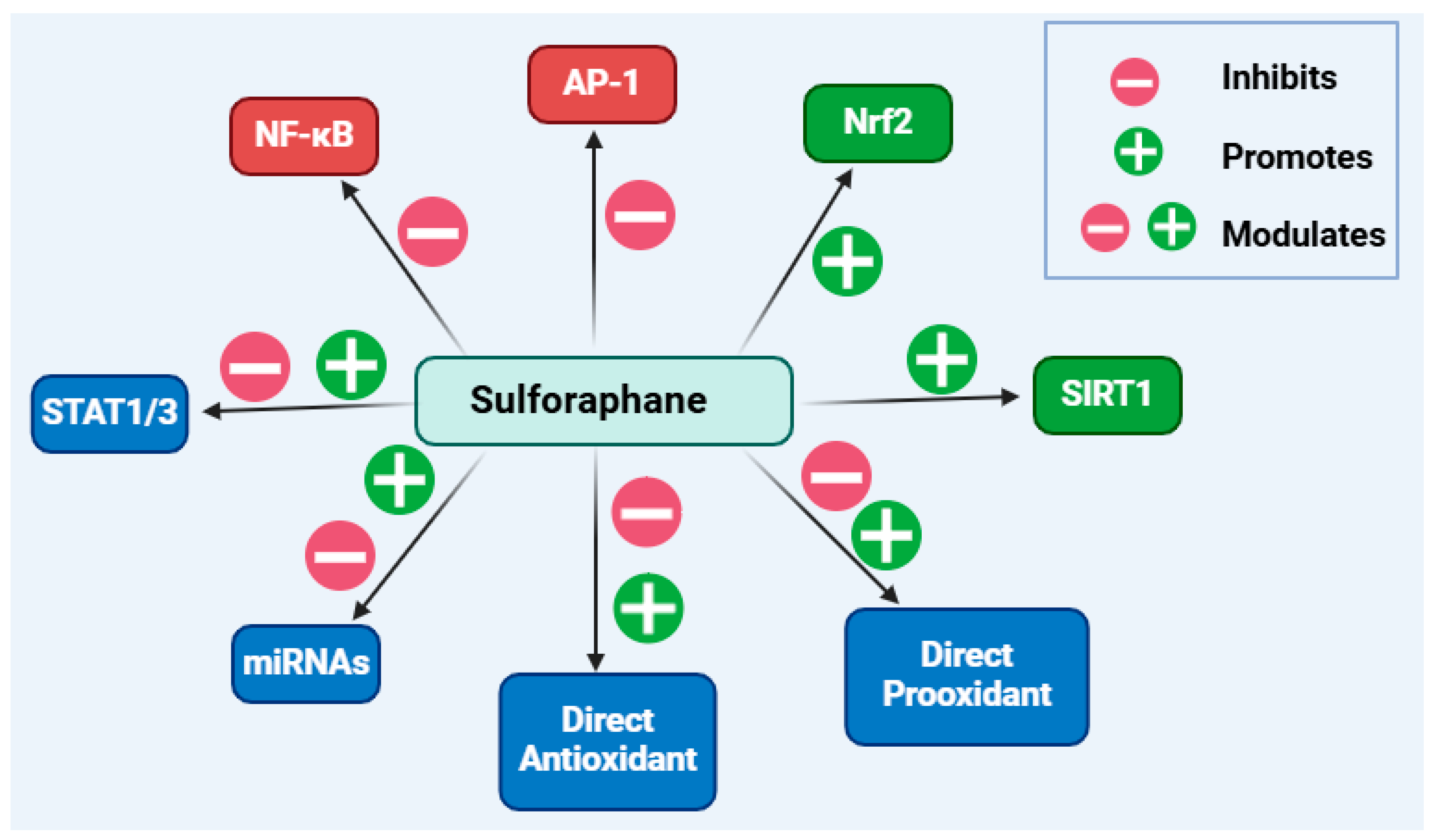
Overall, this article has shown the multiple targets for sulforaphane as an anti-inflammatory compound, which can be summarised as shown in Figure 9. On the basis of the available data, further research on leading optimisation and clinical studies on sulforaphane as an anti-inflammatory agent is well merited.
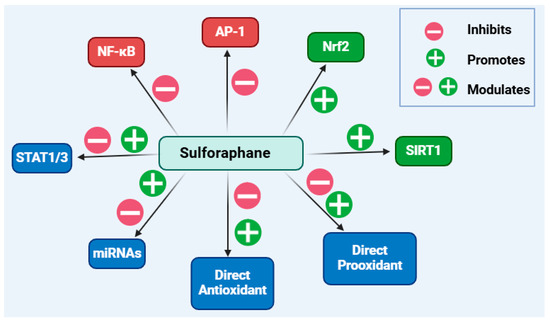
Figure 9.
Overview of the anti-inflammatory mechanisms of sulforaphane. Boosting the level and functional activity of Nrf2-dependent genes/proteins while inhibiting the pro-inflammatory mechanisms induced by the transcription factors NF-κB and AP-1 are its main anti-inflammatory mechanisms. Direct interaction of sulforaphane with biological molecules as a pro-oxidant or antioxidant compound may attribute to its biological actions. Evidence also suggests modulatory effects on STAT1 or STAT3 as well as some miRNAs, although further research is needed to clarify the detailed mechanisms of these effects. This figure has been generated using Biorender.
Funding
This work was not supported by internal or external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The author declares no conflicts of interest in this work.
References
- Agerbirk, N.; Olsen, C.E. Glucosinolate structures in evolution. Phytochemistry 2012, 77, 16–45. [Google Scholar] [CrossRef] [PubMed]
- Zasada, I.A.; Ferris, H. Nematode suppression with brassicaceous amendments: Application based upon glucosinolate profiles. Soil Biol. Biochem. 2004, 36, 1017–1024. [Google Scholar] [CrossRef]
- Pedras, M.S.; Chumala, P.B.; Suchy, M. Phytoalexins from Thlaspi arvense, a wild crucifer resistant to virulent Leptosphaeria maculans: Structures, syntheses and antifungal activity. Phytochemistry 2003, 64, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Kelly, P.J.; Bones, A.; Rossiter, J.T. Sub-cellular immunolocalization of the glucosinolate sinigrin in seedlings of Brassica juncea. Planta 1998, 206, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Backenköhler, A.; Eisenschmidt, D.; Schneegans, N.; Strieker, M.; Brandt, W.; Wittstock, U. Iron is a centrally bound cofactor of specifier proteins involved in glucosinolate breakdown. PLoS ONE 2018, 13, e0205755. [Google Scholar] [CrossRef] [PubMed]
- Eisenschmidt-Bönn, D.; Schneegans, N.; Backenkohler, A.; Wittstock, U.; Brandt, W. Structural diversification during glucosinolate breakdown: Mechanisms of thiocyanate, epithionitrile and simple nitrile formation. Plant J. 2019, 99, 329–343. [Google Scholar] [CrossRef] [PubMed]
- Kuchernig, J.C.; Burow, M.; Wittstock, U. Evolution of specifier proteins in glucosinolate-containing plants. BMC Evol. Biol. 2012, 12, 127. [Google Scholar] [CrossRef]
- Williams, D.J.; Critchley, C.; Pun, S.; Chaliha, M.; O’Hare, T.J. Differing mechanisms of simple nitrile formation on glucosinolate degradation in Lepidium sativum and Nasturtium officinale seeds. Phytochemistry 2009, 70, 1401–1409. [Google Scholar] [CrossRef]
- Li, L.; Lee, W.; Lee, W.J.; Auh, J.H.; Kim, S.S.; Yoon, J. Extraction of allyl isothiocyanate from wasabi (Wasabia japonica Matsum) using supercritical carbon dioxide. Food Sci. Biotechnol. 2010, 19, 405–410. [Google Scholar] [CrossRef]
- Yu, E.Y.; Pickering, I.J.; George, G.N.; Prince, R.C. In situ observation of the generation of isothiocyanates from sinigrin in horseradish and wasabi. Biochim. Biophys. Acta 2001, 1527, 156–160. [Google Scholar] [CrossRef]
- Dai, R.; Lim, L.T. Release of Allyl Isothiocyanate from Mustard Seed Meal Powder. J. Food Sci. 2014, 79, 1. [Google Scholar] [CrossRef] [PubMed]
- Popova, I.E.; Morra, M.J. Simultaneous Quantification of Sinigrin, Sinalbin, and Anionic Glucosinolate Hydrolysis Products in Brassica juncea and Sinapis alba Seed Extracts Using Ion Chromatography. J. Agric. Food Chem. 2014, 62, 10687–10693. [Google Scholar] [CrossRef] [PubMed]
- Morimitsu, Y.; Nakagawa, Y.; Hayashi, K.; Fujii, H.; Kumagai, T.; Nakamura, Y.; Osawa, T.; Horio, F.; Itoh, K.; Iida, K.; et al. A sulforaphane analogue that potently activates the Nrf2-dependent detoxification pathway. J. Biol. Chem. 2002, 277, 3456–3463. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wen, Y.; Chen, J.; Zhang, Y.; Zhang, H.; Sui, J.; Yi, G.; He, X. Rapid and sensitive analysis of benzyl isothiocyanate in peel, pulp, and seeds of Carica papaya Linn. by headspace gas chromatography-mass spectrometry. SN Appl. Sci. 2021, 3, 374. [Google Scholar] [CrossRef]
- Kyriakou, S.; Michailidou, K.; Amery, T.; Stewart, K.; Winyard, P.G.; Trafalis, D.T.; Franco, R.; Pappa, A.; Panayiotidis, M.I. Polyphenolics, Glucosinolates and Isothiocyanates Profiling of Aerial Parts of Nasturtium officinale (Watercress). Front. Plant Sci. 2022, 13, 998755. [Google Scholar] [CrossRef] [PubMed]
- Habtemariam, S. Methodology for rapid isolation of moringin: Potential anticancer compound from the seeds of Moringa stenopetala. Pharm. Anal. Acta 2017, 8, 558. [Google Scholar] [CrossRef]
- Conaway, C.C.; Yang, Y.M.; Chung, F.L. Isothiocyanates as cancer chemopreventive agents: Their biological activities and metabolism in rodents and humans. Curr. Drug Metab. 2002, 3, 233–255. [Google Scholar] [CrossRef] [PubMed]
- Manyes, L.; Luciano, F.B.; Manes, J.; Meca, G. In vitro antifungal activity of allyl isothiocyanate (AITC) against Aspergillus parasiticus and Penicillium expansum and evaluation of the AITC estimated daily intake. Food Chem. Toxicol. 2015, 83, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. The molecular basis that unifies the metabolism, cellular uptake and chemopreventive activities of dietary isothiocyanates. Carcinogenesis 2012, 33, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.R.; Hu, R.; Keum, Y.S.; Hebbar, B.; Shen, G.; Nair, S.S.; Kong, A.N.T. Effects of glutathione on antioxidant response element-mediated gene expression and apoptosis elicited by sulforaphane. Cancer Res. 2003, 63, 7520–7525. [Google Scholar]
- Singh, S.V.; Srivastava, S.K.; Choi, S.; Lew, K.L.; Antosiewicz, J.; Xiao, D.; Zeng, Y.; Watkins, S.C.; Johnson, C.S.; Trump, D.L.; et al. Sulforaphane-induced cell death in human prostate cancer cells is initiated by reactive oxygen species. J. Biol. Chem. 2005, 280, 19911–19924. [Google Scholar] [CrossRef] [PubMed]
- Rudolf, E.; Andělová, H.; Červinka, M. Activation of several concurrent proapoptic pathways by sulforaphane in human colon cancer cells SW620. Food Chem. Toxicol. 2009, 47, 2366–2373. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Jenkins, S.N.; Fahey, J.W.; Ye, L.; Wehage, S.L.; Liby, K.T.; Stephenson, K.K.; Wade, K.L.; Talalay, P. Protection against UV-light-induced skin carcinogenesis in SKH-1 high-risk mice by sulforaphane-containing broccoli sprout extracts. Cancer Lett. 2006, 240, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.L.; Adhikary, G.; Grun, D.; Kaetzel, D.M.; Eckert, R.L. The Ezh2 polycomb group protein drives an aggressive phenotype in melanoma cancer stem cells and is a target of diet derived sulforaphane. Mol. Carcinog. 2016, 55, 2024–2036. [Google Scholar] [CrossRef]
- Singh, S.V.; Warin, R.; Xiao, D.; Powolny, A.A.; Stan, S.D.; Arlotti, J.A.; Zeng, Y.; Hahm, E.R.; Marynowski, S.W.; Bommareddy, A.; et al. Sulforaphane Inhibits Prostate Carcinogenesis and Pulmonary Metastasis in TRAMP Mice in Association with Increased Cytotoxicity of Natural Killer Cells. Cancer Res. 2009, 69, 2117–2125. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mandal, A.K.; Son, Y.O.; Pratheeshkumar, P.; Wise, J.T.F.; Wang, L.; Zhang, Z.; Shi, X.; Chen, Z. Roles of ROS, Nrf2, and autophagy in cadmium-carcinogenesis and its prevention by sulforaphane. Toxicol. Appl. Pharmacol. 2018, 353, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Thejass, P.; Kuttan, G. Augmentation of natural killer cell and antibody-dependent cellular cytotoxicity in BALB/c mice by sulforaphane, a naturally occurring isothiocyanate from broccoli through enhanced production of cytokines IL-2 and IFNγ. Immunopharmacol. Immunotoxicol. 2006, 28, 443–457. [Google Scholar] [CrossRef]
- Thejass, P.; Kuttan, G. Modulation of cell-mediated immune response in B16F-10 melanoma-induced metastatic tumor-bearing C57BL/6 mice by sulforaphane. Immunopharmacol. Immunotoxicol. 2007, 29, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Boddupalli, S.; Mein, J.R.; James, D.R.; Lakkanna, S. Induction of phase 2 antioxidant enzymes by broccoli sulforaphane: Perspectives in maintaining the antioxidant activity of vitamins a, c, and e. Front. Genet. 2012, 3, 7. [Google Scholar] [CrossRef]
- Emmert, S.W.; Desai, D.; Amin, S.; Richie, J.P., Jr. Enhanced Nrf2-Dependant Induction of Glutathione in Mouse Embryonic Fibroblasts by Isoselenocyanate Analog of Sulforaphane. Bioorg. Med. Chem. Lett. 2010, 20, 2675–2679. [Google Scholar] [CrossRef]
- Dias, C.; Aires, A.; Saavedra, M.J. Antimicrobial activity of isothiocyanates from cruciferous plants against methicillin-resistant Staphylococcus aureus (MRSA). Int. J. Mol. Sci. 2014, 15, 19552–19561. [Google Scholar] [CrossRef] [PubMed]
- Sotelo, T.; Lema, M.; Soengas, P.; Cartea, M.E.; Velasco, P. In vitro activity of glucosinolates and their degradation products against brassica-pathogenic bacteria and fungi. Appl. Environ. Microbiol. 2015, 81, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Mayton, H.S.; Oliver, C.; Vaughn, S.F.; Loria, R. Correlation of fungicidal activity of Brassica species with allyl isothiocyanate production in macerated leaf tissue. Phytopathology 1996, 86, 267–271. [Google Scholar] [CrossRef]
- Smolinska, U.; Horbowicz, M. Fungicidal activity of volatiles from selected cruciferous plants against resting propagules of soil-borne fungal pathogens. J. Phytopathol. 1999, 147, 119–124. [Google Scholar] [CrossRef]
- Hashimoto, T.; Yoshioka, S.; Iwanaga, S.; Kanazawa, K. Anti-Malarial Activity of Allyl Isothiocyanate and N-acetyl-S-(N-allylthiocarbamoyl)-l-Cysteine. Mol. Nutr. Food Res. 2023, 67, e2300185. [Google Scholar] [CrossRef]
- Dokumacioglu, E.; Iskender, H.; Aktas, M.S.; Hanedan, B.; Dokumacioglu, A.; Sen, T.M.; Musmul, A. The effect of sulforaphane on oxidative stress and inflammation in rats with toxic hepatitis induced by acetaminophene. Bratisl. Med. J.-Bratisl. Lek. Listy 2017, 118, 453–459. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, W.L.; Xie, D.Q.; Jia, W. Sulforaphane alleviates hepatic ischemia-reperfusion injury through promoting the activation of Nrf-2/HO-1 signaling. Transpl. Immunol. 2021, 68, 101439. [Google Scholar] [CrossRef]
- Mansour, S.Z.; Moustafa, E.M.; Moawed, F.S.M. Modulation of endoplasmic reticulum stress via sulforaphane-mediated AMPK upregulation against nonalcoholic fatty liver disease in rats. Cell Stress Chaperones 2022, 27, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Nazmy, E.A.; El-Khouly, O.A.; Atef, H.; Said, E. Sulforaphane protects against sodium valproate-induced acute liver injury. Can. J. Physiol. Pharmacol. 2017, 95, 420–426. [Google Scholar] [CrossRef]
- Bai, Y.; Chen, Q.; Sun, Y.P.; Wang, X.; Lv, L.; Zhang, L.P. Sulforaphane protection against the development of doxorubicin-induced chronic heart failure is associated with Nrf2 Upregulation. Cardiovasc. Ther. 2017, 35, e12277. [Google Scholar] [CrossRef]
- Fernandes, R.O.; De Castro, A.L.; Bonetto, J.H.P.; Ortiz, V.D.; Müller, D.D.; Campos-Carraro, C.; Barbosa, S.; Neves, L.T.; Xavier, L.L.; Schenkel, P.C.; et al. Sulforaphane effects on postinfarction cardiac remodeling in rats: Modulation of redox-sensitive prosurvival and proapoptotic proteins. J. Nutr. Biochem. 2016, 34, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Emeka, P.M.; Ibrahim, H.I.M.; Morsy, M.A.; Alhaider, I.A.; Hussian, S.; Ahmed, E.A. Attenuation of Cardiomyopathy Induced in Sub-Chronic Exposure of Acrolein by Sulforaphane via Indirect PPARy Expression Promoter. Ind. J. Pharm. Educ. Res. 2021, 55, 1048–1059. [Google Scholar] [CrossRef]
- Silva-Palacios, A.; Ostolga-Chavarría, M.; Sánchez-Garibay, C.; Rojas-Morales, P.; Galván-Arzate, S.; Buelna-Chontal, M.; Pavón, N.; Pedraza-Chaverrí, J.; Königsberg, M.; Zazueta, C. Sulforaphane protects from myocardial ischemia-reperfusion damage through the balanced activation of Nrf2/AhR. Free Rad. Biol. Med. 2019, 143, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Fouad, G.I. Sulforaphane, an Nrf-2 Agonist, Modulates Oxidative Stress and Inflammation in a Rat Model of Cuprizone-Induced Cardiotoxicity and Hepatotoxicity. Cardiovasc. Toxicol. 2023, 23, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, S.X.; Zhang, Y.; Li, F.H.; Yu, C.X. Sulforaphane alleviates lung ischemia-reperfusion injury through activating Nrf-2/HO-1 signaling. Exp. Ther. Med. 2023, 25, 265. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; He, M.H.; Liu, R.X.; Brecha, N.C.; Yu, A.C.H.; Pu, M.L. Sulforaphane Protects Rodent Retinas against Ischemia-Reperfusion Injury through the Activation of the Nrf2/HO-1 Antioxidant Pathway. PLoS ONE 2014, 9, e114186. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.R.; Cao, X.N.; Gong, L.; Li, W.G. Sulforaphane alleviates retinal ganglion cell death and inflammation by suppressing NLRP3 inflammasome activation in a rat model of retinal ischemia/reperfusion injury. Inter. J. Immunopathol. Pharmacol. 2019, 33, 2058738419861777. [Google Scholar] [CrossRef] [PubMed]
- Negi, G.; Kumar, A.; Sharma, S.S. Nrf2 and NF-κB Modulation by Sulforaphane Counteracts Multiple Manifestations of Diabetic Neuropathy in Rats and High Glucose-Induced Changes. Curr. Neurovasc. Res. 2011, 8, 294–304. [Google Scholar] [CrossRef]
- Khaleel, S.A.; Raslan, N.A.; Alzokaky, A.A.; Ewees, M.G.; Ashour, A.A.; Abdel-Hamied, H.E.; Abd-Allah, A.R. Contrast media (meglumine diatrizoate) aggravates renal inflammation, oxidative DNA damage and apoptosis in diabetic rats which is restored by sulforaphane through Nrf2/HO-1 reactivation. Chem. Biol. Inter. 2019, 309, 108689. [Google Scholar] [CrossRef]
- Moustafa, P.E.; Abdelkader, N.F.; El Awdan, S.A.; El-Shabrawy, O.A.; Zaki, H.F. Extracellular Matrix Remodeling and Modulation of Inflammation and Oxidative Stress by Sulforaphane in Experimental Diabetic Peripheral Neuropathy. Inflammation 2018, 41, 1460–1476. [Google Scholar] [CrossRef]
- Li, S.; Yang, H.W.; Chen, X.L. Protective effects of sulforaphane on diabetic retinopathy: Activation of the Nrf2 pathway and inhibition of NLRP3 inflammasome formation. Exp. Anim. 2019, 68, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Zaghlool, S.S.; Abdelaal, N.; El-Shoura, E.A.M.; Mahmoud, N.I.; Ahmed, Y.M. Restoring glomerular filtration rate by sulforaphane modulates ERK1/2/ JNK/p38MAPK, IRF3/iNOS, Nrf2/HO-1 signaling pathways against folic acid-induced acute renal injury in rats. Int. Immunopharmacol. 2023, 123, 110777. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Beltrán, C.E.; Mukhopadhyay, P.; Horváth, B.; Rajesh, M.; Tapia, E.; García-Torres, I.; Pedraza-Chaverri, J.; Pacher, P. Sulforaphane, a natural constituent of broccoli, prevents cell death and inflammation in nephropathy. J. Nutr. Biochem. 2012, 23, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Xu, M.; Xu, L.S.; Ni, H.D.; Zhao, B.X.; Ni, C.B. Sulforaphane alleviates hyperalgesia and enhances analgesic potency of morphine in rats with cancer-induced bone pain. Eur. J. Pharmacol. 2021, 909, 174412. [Google Scholar] [CrossRef] [PubMed]
- Guadarrama-Enríquez, O.; Moreno-Pérez, G.F.; Gonáalez-Trujano, M.E.; Angeles-López, G.E.; Ventura-Martínez, R.; Díaz-Reval, I.; Cano-Martínez, A.; Pellicer, F.; Baenas, N.; Moreno, D.A.; et al. Antinociceptive and antiedema effects produced in rats by Brassica oleraceavar. sprouts involving sulforaphane. Inflammopharmacology 2023, 31, 3217–3226. [Google Scholar] [CrossRef] [PubMed]
- Guan, Z.H.; Zhou, L.M.; Zhang, Y.; Chen, H.P.; Shao, F.F. Sulforaphane Ameliorates the Liver Injury of Traumatic Hemorrhagic Shock Rats. J. Surg. Res. 2021, 267, 293–301. [Google Scholar] [CrossRef]
- Lv, Y.Y.; Jiang, H.J.; Li, S.Y.; Han, B.; Liu, Y.; Yang, D.Q.; Li, J.; Yang, Q.; Wu, P.; Zhang, Z. Sulforaphane prevents chromium-induced lung injury in rats via activation of the Akt/GSK-3β/Fyn pathway. Environ. Pollut. 2020, 259, 113812. [Google Scholar] [CrossRef] [PubMed]
- Thangapandiyan, S.; Ramesh, M.; Miltonprabu, S.; Hema, T.; Jothi, G.B.; Nandhini, V. Sulforaphane potentially attenuates arsenic-induced nephrotoxicity via the PI3K/Akt/Nrf2 pathway in albino Wistar rats. Environ. Sci. Pollut. Res. 2019, 26, 12247–12263. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.D.; Wen, L.T.; Dong, M.; Lu, X.J. Sulforaphane activates the cerebral vascular Nrf2 ARE pathway and suppresses inflammation to attenuate cerebral vasospasm in rat with subarachnoid hemorrhage. Brain Res. 2016, 1653, 1–7. [Google Scholar] [CrossRef]
- Hernández-Rabaza, V.; Cabrera-Pastor, A.; Taoro-González, L.; Malaguarnera, M.; Agustí, A.; Llansola, M.; Felipo, V. Hyperammonemia induces glial activation, neuroinflammation and alters neurotransmitter receptors in hippocampus, impairing spatial learning: Reversal by sulforaphane. J. Neuroinflamm. 2016, 13, 41. [Google Scholar] [CrossRef]
- Mohammad, R.S.; Lokhandwala, M.F.; Banday, A.A. Age-Related Mitochondrial Impairment and Renal Injury Is Ameliorated by Sulforaphane via Activation of Transcription Factor NRF2. Antioxidants 2022, 11, 156. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, Z.W.; Lu, X.F.; Meng, J.; Qin, X.Y.; Jiang, J. Anti-nociceptive and anti-inflammatory effects of sulforaphane on sciatic endometriosis in a rat model. Neurosci. Lett. 2020, 723, 134858. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.Y.; Zhou, Q.; Xia, Y.; You, X.; Zhao, Z.H.; Li, Y.Q.; Zou, H.Q. The Association Between Oxidative Stress Alleviation via Sulforaphane-Induced Nrf2-HO-1/NQO-1 Signaling Pathway Activation and Chronic Renal Allograft Dysfunction Improvement. Kidney Blood Press. Res. 2018, 43, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi-Onogi, S.; Ma, S.H.; Ruhee, R.T.; Tong, Y.S.; Seki, Y.; Suzuki, K. Sulforaphane Attenuates Neutrophil ROS Production, MPO Degranulation and Phagocytosis, but Does Not Affect NET Formation Ex Vivo and In Vitro. Int. J. Mol. Sci. 2023, 24, 8479. [Google Scholar] [CrossRef] [PubMed]
- Harvey, C.J.; Thimmulappa, R.K.; Sethi, S.; Kong, X.; Yarmus, L.; Brown, R.H.; Feller-Kopman, D.; Wise, R.; Biswal, S. Targeting Nrf2 Signaling Improves Bacterial Clearance by Alveolar Macrophages in Patients with COPD and in a Mouse Model. Sci. Trans. Med. 2011, 3, 78ra32. [Google Scholar] [CrossRef] [PubMed]
- Harvey, C.J.; Thimmulappa, R.K.; Sethi, S.; Yarmus, L.B.; Feller-Kopman, D.J.; Wise, R.A.; Biswal, S. Nrf2-dependent immunomodulation by sulforaphane improves bacterial phagocytosis in COPD macrophages and inhibits bacterial burden and inflammation in cigarette smoke-exposed mice. Am. J. Respir. Crit. Care Med. 2010, 181, A3828. [Google Scholar]
- Malhotra, D.; Thimmulappa, R.K.; Mercado, N.; Ito, K.; Kombairaju, P.; Kumar, S.; Ma, J.; Feller-Kopman, D.; Wise, R.; Barnes, P.; et al. Denitrosylation of HDAC2 by targeting Nrf2 restores glucocorticosteroid sensitivity in macrophages from COPD patients. J. Clin. Investig. 2011, 121, 4289–4302. [Google Scholar] [CrossRef]
- Staitieh, B.S.; Egea, E.E.; Fan, X.; Amah, A.; Guidot, D.M. Chronic Alcohol Ingestion Impairs Rat Alveolar Macrophage Phagocytosis via Disruption of RAGE Signaling. Am. J. Med. Sci. 2018, 355, 497–505. [Google Scholar] [CrossRef]
- Staitieh, B.S.; Ding, L.M.; Neveu, W.A.; Spearman, P.; Guidot, D.M.; Fan, X. HIV-1 decreases Nrf2/ARE activity and phagocytic function in alveolar macrophages. J. Leuk. Biol. 2017, 102, 517–525. [Google Scholar] [CrossRef]
- Yang, Q.; Pröll, M.J.; Salilew-Wondim, D.; Zhang, R.; Tesfaye, D.; Fan, H.T.; Cinar, M.U.; Große-Brinkhaus, C.; Tholen, E.; Islam, M.A.; et al. LPS-induced expression of CD14 in the TRIF pathway is epigenetically regulated by sulforaphane in porcine pulmonary alveolar macrophages. Innate Immunity 2016, 22, 682–695. [Google Scholar] [CrossRef]
- Lin, W.; Wu, R.T.; Wu, T.Y.; Khor, T.O.; Wang, H.; Kong, A.N. Sulforaphane suppressed LPS-induced inflammation in mouse peritoneal macrophages through Nrf2 dependent pathway. Biochem. Pharmacol. 2008, 76, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Bahiraii, S.; Brenner, M.; Yan, F.F.; Weckwerth, W.; Heiss, E.H. Sulforaphane diminishes moonlighting of pyruvate kinase M2 and interleukin 1β expression in M1 (LPS) macrophages. Front. Immunol. 2022, 13, 935692. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.Y.; Tang, J.Q.; Li, C.; Liu, J.; Liu, H.J. Sulforaphane attenuates dextran sodium sulphate induced intestinal inflammation viaIL-10/STAT3 signaling mediated macrophage phenotype switching. Food Sci. Hum. Wellness 2022, 11, 129–142. [Google Scholar] [CrossRef]
- Geisel, J.; Brück, J.; Glocova, I.; Dengler, K.; Sinnberg, T.; Rothfuss, O.; Walter, M.; Schulze-Osthoff, K.; Röcken, M.; Ghoreschi, K. Sulforaphane Protects from T Cell-Mediated Autoimmune Disease by Inhibition of IL-23 and IL-12 in Dendritic Cells. J. Immunol. 2014, 192, 3530–3539. [Google Scholar] [CrossRef] [PubMed]
- Williams, E.J.; Guilleminault, L.; Berthon, B.S.; Eslick, S.; Wright, T.; Karihaloo, C.; Gately, M.; Baines, K.J. Wood LG. Sulforaphane reduces pro-inflammatory response to palmitic acid in monocytes and adipose tissue macrophages. J. Nutr. Biochem. 2022, 104, 108978. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Prades, L.; Brasal-Prieto, M.; Alba, G.; Martin, V.; Montserrat-de la Paz, S.; Cejudo-Guillen, M.; Santa-Maria, C.; Dakhaoui, H.; Granados, B.; Sobrino, F.; et al. Sulforaphane Reduces the Chronic Inflammatory Immune Response of Human Dendritic Cells. Nutrients 2023, 15, 3405. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Bonay, M.; Vanhee, V.; Vinit, S.; Deramaudt, T.B. Comparative effectiveness of 4 natural and chemical activators of Nrf2 on inflammation, oxidative stress, macrophage polarization, and bactericidal activity in an in vitromacrophage infection model. PLoS ONE 2020, 15, e0234484. [Google Scholar] [CrossRef]
- Qin, W.S.; Deng, Y.H.; Cui, F.C. Sulforaphane protects against acrolein-induced oxidative stress and inflammatory responses: Modulation of Nrf-2 and COX-2 expression. Arch. Med. Sci. 2016, 12, 871–880. [Google Scholar] [CrossRef]
- Moon, S.J.; Jhun, J.; Ryu, J.; Kwon, J.Y.; Kim, S.Y.; Jung, K.; Cho, M.L.; Min, J.K. The anti-arthritis effect of sulforaphane, an activator of Nrf2, is associated with inhibition of both B cell differentiation and the production of inflammatory cytokines. PLoS ONE 2021, 16, e0245986. [Google Scholar]
- Mazarakis, N.; Anderson, J.; Toh, Z.Q.; Higgins, R.A.; Ha Do, L.A.; Luwor, R.B.; Licciardi, P.V. Examination of Novel Immunomodulatory Effects of L-Sulforaphane. Nutrients 2021, 13, 602. [Google Scholar] [CrossRef]
- Jamal, I.; Paudel, A.; Thompson, L.; Abdelmalek, M.; Khan, I.A.; Singh, V.B. Sulforaphane prevents the reactivation of HIV-1 by suppressing NFκB signaling. J. Virus Erad. 2023, 9, 100341. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, A.; Ahmad, S.F.; Al-Ayadhi, L.Y.; Attia, S.M.; Al-Harbi, N.O.; Alzahrani, K.S.; Bakheet, S.A. Differential regulation of Nrf2 is linked to elevated inflammation and nitrative stress in monocytes of children with autism. Psychoneuroendocrinology 2020, 113, 104554. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.L.; Liu, X.J.; Bao, H.R. Sulforaphane suppresses lipopolysaccharide- and Pam3CysSerLys4-mediated inflammation in chronic obstructive pulmonary disease via toll-like receptors. FEBS Open Bio 2021, 11, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.L.; Liu, X.J.; Bao, H.R.; Zhang, Y.; Tan, E.L. Sulforaphane Suppressed LPS and Pam3CSK4 Mediated Inflammation in COPD Through MyD88-Dependent Toll-Like Receptors Pathway. Chest 2016, 149, 351A. [Google Scholar] [CrossRef]
- Pal, S.; Konkimalla, V.B. Sulforaphane regulates phenotypic and functional switching of both induced and spontaneously differentiating human monocytes. Int. Immunopharmacol. 2016, 35, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Haodang, L.; Lianmei, Q.; Ranhui, L.; Liesong, C.; Jun, H.; Yihua, Z.; Cuiming, Z.; Yimou, W.; Xiaoxing, Y. HO-1 mediates the anti-inflammatory actions of Sulforaphane in monocytes stimulated with a mycoplasmal lipopeptide. Chem.-Biol. Inter. 2019, 306, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Harada, N.; Kanayama, M.; Maruyama, A.; Yoshida, A.; Tazumi, K.; Hosoya, T.; Mimura, J.; Toki, T.; Maher, J.M.; Yamamoto, M.; et al. Nrf2 regulates ferroportin 1-mediated iron efflux and counteracts lipopolysaccharide-induced ferroportin 1 mRNA suppression in macrophages. Arch. Biochem. Biophys. 2011, 508, 101–109. [Google Scholar] [CrossRef]
- Kim, S.Y.; Jeong, E.; Joung, S.M.; Lee, J.Y. PI3K/Akt contributes to increased expression of Toll-like receptor 4 in macrophages exposed to hypoxic stress. Biochem. Biophys. Res. Commun. 2012, 419, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Ranaweera, S.S.; Dissanayake, C.Y.; Natraj, P.; Lee, Y.J.; Han, C.H. Anti-inflammatory effect of sulforaphane on BPS-stimulated RAW 264.7 cells and ob/ob mice. J. Vet. Sci. 2020, 21, e91. [Google Scholar] [CrossRef]
- Ruhee, R.T.; Ma, S.H.; Suzuki, K. Sulforaphane Protects Cells against Lipopolysaccharide-Stimulated Inflammation in Murine Macrophages. Antioxidants 2019, 8, 577. [Google Scholar] [CrossRef]
- Saleh, H.A.; Ramdan, E.; Elmazar, M.M.; Azzazy, H.M.E.; Abdelnaser, A. Comparing the protective effects of resveratrol, curcumin and sulforaphane against LPS/IFN-γ-mediated inflammation in doxorubicin-treated macrophages. Sci. Rep. 2021, 11, 545. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Tatsunami, R.; Wakame, K. Epalrestat suppresses inflammatory response in lipopolysaccharide-stimulated RAW264.7 cells. Allergol. Immunopathol. 2021, 49, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Vuong, L.D.; Nguyen, Q.N.; Truong, V.L. Anti-inflammatory and anti-oxidant effects of combination between sulforaphane and acetaminophen in LPS-stimulated RAW 264.7 macrophage cells. Immunopharmacol. Immunotoxicol. 2019, 41, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Woo, K.J.; Kwon, T.K. Sulforaphane suppresses lipopolysaccharide-induced cyclooxygenase-2 (COX-2) expression through the modulation of multiple targets in COX-2 gene promoter. Int. Immunopharmacol. 2007, 7, 1776–1783. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S.A.; Shelar, S.B.; Dang, T.M.; Lee, B.N.C.; Yang, H.; Ong, S.M.; Ng, H.L.; Chui, W.K.; Wong, S.C.; Chew, E.H. Sulforaphane and its methylcarbonyl analogs inhibit the LPS-stimulated inflammatory response in human monocytes through modulating cytokine production, suppressing chemotactic migration and phagocytosis in a NF-κB- and MAPK-dependent manner. Int. Immunopharmacol. 2015, 24, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.S.; Qiu, P.J.; Xu, G.; Wu, X.; Dong, P.; Yang, G.P.; Zheng, J.; McClements, D.J.; Xiao, H. Synergistic Anti-inflammatory Effects of Nobiletin and Sulforaphane in Lipopolysaccharide-Stimulated RAW 264.7 Cells. J. Agric. Food Chem. 2012, 60, 2157–2164. [Google Scholar] [CrossRef]
- Rakariyatham, K.; Wu, X.; Tang, Z.H.; Han, Y.H.; Wang, Q.; Xiao, H. Synergism between luteolin and sulforaphane in anti-inflammation. Food Funct. 2018, 9, 5115–5123. [Google Scholar] [CrossRef] [PubMed]
- Greaney, A.J.; Maier, N.K.; Leppla, S.H.; Moayeri, M. Sulforaphane inhibits multiple inflammasomes through an Nrf2-independent mechanism. J. Leukoc. Biol. 2016, 99, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Deramaudt, T.B.; Ali, M.; Vinit, S.; Bonay, M. Sulforaphane reduces intracellular survival of Staphylococcus aureus in macrophages through inhibition of JNK and p38 MAPK-induced inflammation. Int. J. Mol. Med. 2020, 45, 1927–1941. [Google Scholar] [CrossRef]
- Jhang, K.A.; Park, J.S.; Kim, H.S.; Chong, Y.H. Sulforaphane rescues amyloid-β peptide-mediated decrease in MerTK expression through its anti-inflammatory effect in human THP-1 macrophages. J. Neuroinflamm. 2018, 15, 75. [Google Scholar] [CrossRef]
- Liang, J.; Jahraus, B.; Balta, E.; Ziegler, J.A.; Hübner, K.; Blank, N.; Niesler, B.; Wabnitz, G.H.; Samstag, Y. Sulforaphane Inhibits Inflammatory Responses of Primary Human T-Cells by Increasing ROS and Depleting Glutathione. Front. Immunol. 2018, 9, 2584. [Google Scholar] [CrossRef]
- Jeon, M.; Lee, J.; Lee, H.K.; Cho, S.; Lim, J.H.; Choi, Y.; Pak, S.; Jeong, H.J. Sulforaphane mitigates mast cell-mediated allergic inflammatory reactions in in silicosimulation and in vitro models. Immunopharmacol. Immunotoxicol. 2020, 42, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Brasil, F.B.; de Almeida, F.J.S.; Luckachaki, M.D.; Dall’Oglio, E.L.; de Oliveira, M.R. The isothiocyanate sulforaphane prevents mitochondrial impairment and neuroinflammation in the human dopaminergic SH-SY5Y and in the mouse microglial BV2 cells: Role for heme oxygenase-1. Metab. Brain Dis. 2023, 38, 419–435. [Google Scholar] [CrossRef]
- Chilakala, R.R.; Manchikalapudi, A.L.; Kumar, A.; Sunkaria, A. Sulforaphane Attenuates Aβ Oligomers Mediated Decrease in Phagocytic Activity of Microglial Cells. Neuroscience 2020, 429, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Eren, E.; Tufekci, K.U.; Isci, K.B.; Tastan, B.; Genc, K.; Genc, S. Sulforaphane Inhibits Lipopolysaccharide-Induced Inflammation, Cytotoxicity, Oxidative Stress, and miR-155 Expression and Switches to Mox Phenotype through Activating Extracellular Signal-Regulated Kinase 1/2-Nuclear Factor Erythroid 2-Related Factor 2/Antioxidant Response Element Pathway in Murine Microglial Cells. Front. Immunol. 2018, 9, 36. [Google Scholar]
- Iizumi, T.; Takahashi, S.; Mashima, K.; Minami, K.; Izawa, Y.; Abe, T.; Hishiki, T.; Suematsu, M.; Kajimura, M.; Suzuki, N. A possible role of microglia-derived nitric oxide by lipopolysaccharide in activation of astroglial pentose-phosphate pathway via the Keap1/Nrf2 system. J. Neuroinflamm. 2016, 13, 99. [Google Scholar] [CrossRef]
- Liu, J.X.; Chandaka, G.K.; Zhang, R.; Parfenova, H. Acute antioxidant and cytoprotective effects of sulforaphane in brain endothelial cells and astrocytes during inflammation and excitotoxicity. Pharmacol. Res. Perspect. 2020, 8, e630. [Google Scholar] [CrossRef]
- Maciel-Barón, L.A.; Morales-Rosales, S.L.; Silva-Palacios, A.; Rodríguez-Barrera, R.H.; García-Alvarez, J.A.; Luna-López, A.; Pérez, V.I.; Torres, C.; Königsberg, M. The secretory phenotype of senescent astrocytes isolated from Wistar newborn rats changes with anti-inflammatory drugs, but does not have a short-term effect on neuronal mitochondrial potential. Biogerontology 2018, 19, 415–433. [Google Scholar] [CrossRef]
- Michalska, P.; Buendia, I.; Duarte, P.; FernandezMendivil, C.; Negredo, P.; Cuadrado, A.; López, M.G.; Leon, R. Melatonin-sulforaphane hybrid ITH12674 attenuates glial response in vivoby blocking LPS binding to MD2 and receptor oligomerization. Pharmacol. Res. 2020, 152, 104597. [Google Scholar] [CrossRef]
- Subedi, L.; Lee, J.H.; Gaire, B.P.; Kim, S.Y. Sulforaphane Inhibits MGO-AGE-Mediated Neuroinflammation by Suppressing NF-κB, MAPK, and AGE-RAGE Signaling Pathways in Microglial Cells. Antioxidants 2020, 9, 792. [Google Scholar] [CrossRef]
- Wierinckx, A.; Brevé, J.; Mercier, D.; Schultzberg, M.; Drukarch, B.; Van Dam, A.M. Detoxication enzyme inducers modify cytokine production in rat mixed glial cells. J. Neuroimmunol. 2005, 166, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Gao, M.H.; Wu, J.J.; Hu, P.L.; Xu, X.; Zhang, Y.R.; Wang, D.; Chen, Z.; Huang, C. Sulforaphane triggers a functional elongation of microglial process via the Akt signal. J. Nutr. Biochem. 2019, 67, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.S.; Yang, C.H.; Huang, W.H.; Du, S.H.; Mai, H.T.; Xiao, J.J.; Lu, T.M. Sulforaphane attenuates microglia-mediated neuronal necroptosis through down-regulation of MAPK/NF-κB signaling pathways in LPS-activated BV-2 microglia. Pharmacol. Res. 2018, 133, 218–235. [Google Scholar] [CrossRef] [PubMed]
- Brandenburg, L.O.; Kipp, M.; Lucius, R.; Pufe, T.; Wruck, C.J. Sulforaphane suppresses LPS-induced inflammation in primary rat microglia. Inflamm. Res. 2010, 59, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Subedi, L.; Lee, J.H.; Yumnam, S.; Ji, E.; Kim, S.Y. Anti-Inflammatory Effect of Sulforaphane on LPS-Activated Microglia Potentially through JNK/AP-1/NF-κB Inhibition and Nrf2/HO-1 Activation. Cells 2019, 8, 194. [Google Scholar] [CrossRef]
- Bobermin, L.D.; Weber, F.B.; dos Santos, T.M.; Bello-Klein, A.; Wyse, A.T.S.; Goncalves, C.A.; Quincozes-Santos, A. Sulforaphane Induces Glioprotection After LPS Challenge. Cell. Mol. Neurobiol. 2022, 42, 829–846. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.; Machado, M.R.; Alves, J.V.; Fraga-Silva, T.F.C.; Martins, R.B.; Campos, L.C.B.; Francisco, D.F.; Couto, A.E.S.; Bonato, V.L.D.; Arruda, E.; et al. Cytokine storm in individuals with severe COVID-19 decreases endothelial cell antioxidant defense via downregulation of the Nrf2 transcriptional factor. Am. J. Physiol. Heart Circ. Physiol. 2023, 325, H252–H263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tan, L.X.; Li, C.; Wu, H.; Ran, D.; Zhang, Z.Y. Sulforaphane alter the microbiota and mitigate colitis severity on mice ulcerative colitis induced by DSS. AMB Express 2020, 10, 119. [Google Scholar] [CrossRef]
- Matsui, T.; Nakamura, N.; Ojima, A.; Nishino, Y.; Yamagishi, S. Sulforaphane reduces advanced glycation end products (AGEs)-induced inflammation in endothelial cells and rat aorta. Nutr. Metab. Cardiovasc. Dis. 2016, 26, 797–807. [Google Scholar] [CrossRef]
- Cho, Y.S.; Kim, C.H.; Ha, T.S.; Ahn, H.Y. Inhibition of STAT3 phosphorylation by sulforaphane reduces adhesion molecule expression in vascular endothelial cell. Can. J. Physiol. Pharmacol. 2016, 94, 1220–1226. [Google Scholar] [CrossRef]
- Nallasamy, P.; Babu, P.V.A.; Shah, H.; Brooke, E.A.S.; Zhu, H.; Zhen, W.; Liu, D.; Li, Y.; Jia, Z. Sulforaphane at Physiological Concentrations Inhibits TNF-α-Induced Monocyte Adhesion to Human Vascular Endothelial Cells and Improves Vascular Inflammation in Mice Through a Nuclear Factor-κB-Mediated Mechanism. Arterioscl. Thromb. Vasc. Biol. 2014, 34, 454. [Google Scholar] [CrossRef]
- Nallasamy, P.; Si, H.W.; Babu, P.V.A.; Pan, D.K.; Fu, Y.; Brooke, E.A.S.; Shah, H.; Zhen, W.; Zhu, H.; Liu, D.; et al. Sulforaphane reduces vascular inflammation in mice and prevents TNF-α-induced monocyte adhesion to primary endothelial cells through interfering with the NF-κB pathway. J. Nutr. Biochem. 2014, 25, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.K.; Han, M.S.; Bae, J.S. Sulforaphane inhibits endothelial protein C receptor shedding in vitroand in vivo. Vasc. Pharmacol. 2014, 63, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.K.; Bae, J.S. Antithrombotic activities of sulforaphane via inhibiting platelet aggregation and FIIa/FXa. Arch. Pharm. Res. 2014, 37, 1454–1463. [Google Scholar] [CrossRef]
- Shan, Y.; Lin, N.; Yang, X.; Tan, J.; Zhao, R.; Dong, S.; Wang, S. Sulforaphane inhibited the expressions of intercellular adhesion molecule-1 and vascular cell adhesion molecule-1 through MyD88-dependent toll-like receptor-4 pathway in cultured endothelial cells. Nutr. Metab. Cardiovas. Dis. 2012, 22, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Kivelä, A.M.; Mäkinen, P.I.; Jyrkkänen, H.K.; Mella-Aho, E.; Xia, Y.F.; Kansanen, E.; Leinonen, H.; Verma, I.M.; Ylä-Herttuala, S.; Levonen, A.L. Sulforaphane inhibits endothelial lipase expression through NF-κB in endothelial cells. Atherosclerosis 2010, 213, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, N.; Kawajiri, H.; Badiwala, M.V.; Butany, J.; Li, R.K.; Billia, F.; Rao, V. Protective role of Nrf2 against ischemia reperfusion injury and cardiac allograft vasculopathy. Am. J. Transplant. 2020, 20, 1262–1271. [Google Scholar] [CrossRef] [PubMed]
- Holloway, P.M.; Gillespie, S.; Becker, F.; Vital, S.A.; Nguyen, V.; Alexander, J.S.; Evans, P.C.; Gavins, F.N.E. Sulforaphane induces neurovascular protection against a systemic inflammatory challenge via both Nrf2-dependent and independent pathways. Vasc. Pharmacol. 2016, 85, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Dodd, G.; Kunsch, C. Sulforaphane inhibits TNF-α-induced activation of p38 MAP kinase and VCAM-1 and MCP-1 expression in endothelial cells. Inflamm. Res. 2009, 58, 513–521. [Google Scholar] [CrossRef]
- Shao, D.; Shen, W.X.; Miao, Y.Y.; Gao, Z.; Pan, M.H.; Wei, Q.; Yan, Z.; Zhao, X.; Ma, B. Sulforaphane prevents LPS-induced inflammation by regulating the Nrf2-mediated autophagy pathway in goat mammary epithelial cells and a mouse model of mastitis. J. Anim. Sci. Biotechnol. 2023, 14, 61. [Google Scholar] [CrossRef]
- Gasparello, J.; D’Aversa, E.; Papi, C.; Gambari, L.; Grigolo, B.; Borgatti, M.; Finotti, A.; Gambari, R. Sulforaphane inhibits the expression of interleukin-6 and interleukin-8 induced in bronchial epithelial IB3-1 cells by exposure to the SARS-CoV-2 Spike protein. Phytomedicine 2021, 87, 153583. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Zhang, H.; Liu, Q.Y.; Jiang, B.; Chen, J.J.; Zhang, T. Sulforaphane attenuates oxidative stress and inflammation induced by fine particulate matter in human bronchial epithelial cells. J. Funct. Foods 2021, 81, 104460. [Google Scholar] [CrossRef]
- Sim, H.; Lee, W.; Choo, S.; Park, E.K.; Baek, M.C.; Lee, I.K.; Park, D.H.; Bae, J.S. Sulforaphane Alleviates Particulate Matter-Induced Oxidative Stress in Human Retinal Pigment Epithelial Cells. Front. Med. 2021, 8, 685032. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.M.; Cheng, K.C.; Huang, J.Y.; Wang, S.Y.; Lin, Y.N.; Tseng, Y.T.; Hsieh, C.W.; Wung, B.S. Sulforaphane inhibits blue light-induced inflammation and apoptosis by upregulating the SIRT1/PGC-1α/Nrf2 pathway and autophagy in retinal pigment epithelial cells. Toxicol. Appl. Pharmacol. 2021, 421, 115545. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Wu, Q. Sulforaphane protects intestinal epithelial cells against lipopolysaccharide-induced injury by activating the AMPK/SIRT1/PGC-1alpha pathway. Bioengineered 2021, 12, 4349–4360. [Google Scholar] [CrossRef]
- Son, E.S.; Park, J.W.; Kim, Y.J.; Jeong, S.H.; Hong, J.H.; Kim, S.H.; Kyung, S.Y. Effects of antioxidants on oxidative stress and inflammatory responses of human bronchial epithelial cells exposed to particulate matter and cigarette smoke extract. Toxicology 2020, 67, 104883. [Google Scholar] [CrossRef]
- Frias, D.P.; Gomes, R.L.N.; Yoshizaki, K.; Carvalho-Oliveira, R.; Matsuda, M.; Junqueira, M.D.; Teodoro, W.R.; Vasconcellos, P.C.; Pereira, D.C.A.; Conceição, P.R.D.; et al. Nrf2 positively regulates autophagy antioxidant response in human bronchial epithelial cells exposed to diesel exhaust particles. Sci. Rep. 2020, 10, 3704. [Google Scholar] [CrossRef] [PubMed]
- London, N.R.; Tharakan, A.; Lane, A.P.; Biswal, S.; Ramanathan, M. Nuclear erythroid 2-related factor 2 activation inhibits house dust mite-induced sinonasal epithelial cell barrier dysfunction. Int. Forum Allergy Rhinol. 2017, 7, 536–541. [Google Scholar] [CrossRef]
- Danyal, K.; de Jong, W.; O’Brien, E.; Bauer, R.A.; Heppner, D.E.; Little, A.C.; Hristova, M.; Habibovic, A.; van der Vliet, A. Acrolein and thiol-reactive electrophiles suppress allergen-induced innate airway epithelial responses by inhibition of DUOX1 and EGFR. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016, 311, L913–L923. [Google Scholar] [CrossRef]
- Kim, H.N.; Kim, D.H.; Kim, E.H.; Lee, M.H.; Kundu, J.K.; Na, H.K.; Cha, Y.N.; Surh, Y.J. Sulforaphane inhibits phorbol ester-stimulated IKK-NF-κB signaling and COX-2 expression in human mammary epithelial cells by targeting NF-κB activating kinase and ERK. Cancer Lett. 2014, 351, 41–49. [Google Scholar] [CrossRef]
- Ye, L.; Yu, T.; Li, Y.Q.; Chen, B.N.; Zhang, J.S.; Wen, Z.Y.; Zhang, B.; Zhou, X.; Li, X.; Li, F.; et al. Sulforaphane Enhances the Ability of Human Retinal Pigment Epithelial Cell against Oxidative Stress, and Its Effect on Gene Expression Profile Evaluated by Microarray Analysis. Oxid. Med. Cell. Longev. 2013, 2013, 413024. [Google Scholar] [CrossRef] [PubMed]
- Kesic, M.J.; Simmons, S.O.; Bauer, R.; Jaspers, I. Nrf2 expression modifies influenza A entry and replication in nasal epithelial cells. Free Rad. Biol. Med. 2011, 51, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Starrett, W.; Blake, D.J. Sulforaphane inhibits de novo synthesis of IL-8 and MCP-1 in human epithelial cells generated by cigarette smoke extract. J. Immunotoxicol. 2011, 8, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Xiangdong, Z.; Hongmei, Y.; Xiaohong, N.; Xiaoyan, X. Regulation of neutrophil elastase-induced MUC5AC expression by nuclear factor erythroid-2 related factor 2 in human airway epithelial cells. J. Investig. Med. 2010, 58, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Ritz, S.A.; Wan, J.X.; Diaz-Sanchez, D. Sulforaphane-stimulated phase II enzyme induction inhibits cytokine production by airway epithelial cells stimulated with diesel extract. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007, 292, L33–L39. [Google Scholar] [CrossRef] [PubMed]
- Al-Bakheit, A.; Abu-Qatouseh, L. Sulforaphane from broccoli attenuates inflammatory hepcidin by reducing IL-6 secretion in human HepG2 cells. J. Funct. Foods 2020, 75, 104210. [Google Scholar] [CrossRef]
- Wang, M.F.; Liu, M.L.; Xu, W.Z.; Teng, Z.Y.; Wu, X.W.; Gan, L.; Zhang, Y.A. Sulforaphane reduces lipopolysaccharide-induced inflammation and enhances myogenic differentiation of mouse embryonic myoblasts via the toll-like receptor 4 and NLRP3 pathways. Adv. Clin. Exp. Med. 2023, 32, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Faridvand, Y.; Haddadi, P.; Nejabati, H.R.; Ghaffari, S.; Zamani-Gharehchamani, E.; Nozari, S.; Nouri, M.; Jodati, A. Sulforaphane modulates CX3CL1/CX3CR1 axis and inflammation in palmitic acid-induced cell injury in C2C12 skeletal muscle cells. Mol. Biol. Rep. 2020, 47, 7971–7977. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.S.; Joung, H.; Kim, Y.S.; Shim, Y.S.; Ahn, Y.; Jeong, M.H.; Kee, H.J. Sulforaphane inhibits restenosis by suppressing inflammation and the proliferation of vascular smooth muscle cells. Atherosclerosis 2012, 225, 41–49. [Google Scholar] [CrossRef]
- Kim, H.A.; Yeo, Y.; Jung, H.A.; Jung, Y.O.; Park, S.J.; Kim, S.J. Phase 2 enzyme inducer sulforaphane blocks prostaglandin and nitric oxide synthesis in human articular chondrocytes and inhibits cartilage matrix degradation. Rheumatology 2012, 51, 1006–1016. [Google Scholar] [CrossRef]
- Zhao, X.D.; Zhou, Y.T.; Lu, X.J. Sulforaphane enhances the activity of the Nrf2-ARE pathway and attenuates inflammation in OxyHb-induced rat vascular smooth muscle cells. Inflamm. Res. 2013, 62, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, H.J.; Um, S.H.; Sohn, E.H.; Kim, B.O.; Moon, E.Y.; Rhee, D.K.; Pyo, S. Sulforaphane suppresses vascular adhesion molecule-1 expression in TNF-α-stimulated mouse vascular smooth muscle cells: Involvement of the MAPK, NF-κB and AP-1 signaling pathways. Vasc. Pharmacol. 2012, 56, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Davidson, R.K.; Jupp, O.; de Ferrars, R.; Kay, C.D.; Culley, K.L.; Norton, R.; Driscoll, C.; Vincent, T.L.; Donell, S.T.; Bao, Y.; et al. Sulforaphane Represses Matrix-Degrading Proteases and Protects Cartilage From Destruction In Vitro and In Vivo. Arthritis Rheum. 2013, 65, 3130–3140. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, Y.H.; Zhou, H.Y.; Cui, K.M. Sulforaphane alleviates LPS-induced inflammatory injury in ARPE-19 cells by repressing the PWRN2/NF-kB pathway. Immunopharmacol. Immunotoxicol. 2022, 44, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Fragoulis, A.; Laufs, J.; Müller, S.; Soppa, U.; Siegl, S.; Reiss, L.K.; Tohidnezhad, M.; Rosen, C.; Tenbrock, K.; Varoga, D.; et al. Sulforaphane has opposing effects on TNF-alpha stimulated and unstimulated synoviocytes. Arthritis Res. Ther. 2012, 14, R220. [Google Scholar] [CrossRef] [PubMed]
- Folkard, D.L.; Melchini, A.; Traka, M.H.; Al-Bakheit, A.; Saha, S.; Mulholland, F.; Watson, A.; Mithen, R.F. Suppression of LPS-induced transcription and cytokine secretion by the dietary isothiocyanate sulforaphane. Mol. Nutr. Food Res. 2014, 58, 2286–2296. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Lee, W.S.; Lee, E.G.; Sung, M.S.; Yoo, W.H. Sulforaphane Inhibits IL-1β-Induced Proliferation of Rheumatoid Arthritis Synovial Fibroblasts and the Production of MMPs, COX-2, and PGE2. Inflammation 2014, 37, 1496–1503. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.J.; Shang, H.X.; Chen, Y.Q.; Pan, L.L.; Bhatia, M.; Sun, J. Sulforaphane Protects Pancreatic Acinar Cell Injury by Modulating Nrf2-Mediated Oxidative Stress and NLRP3 Inflammatory Pathway. Oxid. Med. Cell. Longev. 2016, 2016, 7864150. [Google Scholar] [CrossRef]
- Ernst, I.M.; Kleszczynski, K.; Wagner, A.E.; Kruse, N.; Zillikens, D.; Rimbach, G.; Fischer, W. Sulforaphane and phenylethyl isothiocyanate protect human skin from UV-induced inflammation and apoptosis. Exp. Dermatol. 2013, 22, E19. [Google Scholar]
- Jeong, S.I.; Choi, B.M.; Jang, S. Sulforaphane Suppresses TARC/CCL17 and MDC/CCL22 Expression Through Heme Oxygenase-1 and NF-κB in Human Keratinocytes. Arch. Pharm. Res. 2010, 33, 1867–1876. [Google Scholar] [CrossRef]
- de Oliveira, M.R.; Brasil, F.B.; Fürstenau, C.R. Sulforaphane Attenuated the Pro-Inflammatory State Induced by Hydrogen Peroxide in SH-SY5Y Cells Through the Nrf2/HO-1 Signaling Pathway. Neurotox. Res. 2018, 34, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.F.; Zhang, J.L.; Chang, N. Epigenetic modification of Nrf2 by sulforaphane increases the antioxidative and anti-inflammatory capacity in a cellular model of Alzheimer’s disease. Eur. J. Pharmacol. 2018, 824, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Díaz, A.F.; Polo, S.; Gallardo, N.; Leánez, S.; Pol, O. Analgesic and Antidepressant Effects of Oltipraz on Neuropathic Pain in Mice by Modulating Microglial Activation. J. Clin. Med. 2019, 8, 890. [Google Scholar] [CrossRef]
- Davuljigari, C.B.; Ekuban, F.A.; Zong, C.; Fergany, A.A.M.; Morikawa, K.; Ichihara, G. Nrf2 Activation Attenuates Acrylamide-Induced Neuropathy in Mice. Int. J. Mol. Sci. 2021, 22, 5995. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.S.; Yoo, S.A.; Kim, H.S.; Kim, H.A.; Yea, K.; Ryu, S.H.; Chung, Y.J.; Cho, C.S.; Kim, W.U. Inhibition of Synovial Hyperplasia, Rheumatoid T Cell Activation, and Experimental Arthritis in Mice by Sulforaphane, a Naturally Occurring Isothiocyanate. Arthritis Rheum. 2010, 62, 159–170. [Google Scholar] [CrossRef]
- Chen, M.; Huang, L.; Lv, Y.; Li, L.; Dong, Q. Sulforaphane protects against oxidative stress-induced apoptosis via activating SIRT1 in mouse osteoarthritis. Mol. Med. Rep. 2021, 24, 612. [Google Scholar] [CrossRef]
- Redondo, A.; Chamorro, P.A.F.; Riego, G.; Leánez, S.; Pol, O. Treatment with Sulforaphane Produces Antinociception and Improves Morphine Effects during Inflammatory Pain in Mice. J. Pharmacol. Exp. Ther. 2017, 363, 293–302. [Google Scholar] [CrossRef]
- Silva Rodrigues, J.F.; Figueiredo, C.; Muniz, T.F.; de Aquino, A.F.S.; Nina, L.N.D.; Sousa, N.C.; Nascimento da Silva, L.C.; de Souza, B.G.G.F.; da Penha, T.A.; Abreu-Silva, A.L.; et al. Sulforaphane Modulates Joint Inflammation in a Murine Model of Complete Freund’s Adjuvant-Induced Mono-Arthritis. Molecules 2018, 23, 988. [Google Scholar] [CrossRef]
- Wang, C.; Wang, C.P. Anti-nociceptive and anti-inflammatory actions of sulforaphane in chronic constriction injury-induced neuropathic pain mice. Inflammopharmacology 2017, 25, 99–106. [Google Scholar] [CrossRef]
- Sakurai, H.; Morishima, Y.; Ishii, Y.; Yoshida, K.; Nakajima, M.; Tsunoda, Y.; Hayashi, S.Y.; Kiwamoto, T.; Matsuno, Y.; Kawaguchi, M.; et al. Sulforaphane ameliorates steroid insensitivity through an Nrf2-dependent pathway in cigarette smoke-exposed asthmatic mice. Free Rad. Biol. Med. 2018, 129, 473–485. [Google Scholar] [CrossRef]
- Yan, B.D.; Ma, Z.S.; Shi, S.M.; Hu, Y.X.; Ma, T.G.; Rong, G.; Yang, J.L. Sulforaphane prevents bleomycin-induced pulmonary fibrosis in mice by inhibiting oxidative stress via nuclear factor erythroid 2-related factor-2 activation. Mol. Med. Rep. 2017, 15, 4005–4014. [Google Scholar] [CrossRef]
- Sakurai, H.; Morishima, Y.; Tsunoda, Y.; Lin, S.Y.; Kiwamoto, T.; Matsuno, Y.; Ishii, Y.; Hizawa, N. Sulforaphane Abrogates Steroid Insensitivity In Cigarette Smoke-Induced Airway Inflammation Through Nrf2 Dependent Pathway. Am. J. Resp. Crit. Care Med. 2017, 195, A5295. [Google Scholar]
- Ano, S.; Panariti, M.; Allard, B.; O’Sullivan, M.; McGovern, T.K.; Hamamoto, Y.; Shii, Y.; Yamamoto, M.; Powell, W.S.; Martin, J.G. Inflammation and airway hyperresponsiveness after chlorine exposure are prolonged by Nrf2 deficiency in mice. Free Rad. Biol. Med. 2017, 102, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.Q.; Greven, J.; Fragoulis, A.; Horst, K.; Bläsius, F.; Wruck, C.; Pufe, T.; Kobbe, P.; Hildebrand, F.; Lichte, P. Sulforaphane-Dependent Up-Regulation of NRF2 Activity Alleviates Both Systemic Inflammatory Response and Lung Injury After Hemorrhagic Shock/Resuscitation in Mice. Shock 2022, 57, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Royce, S.G.; Licciardi, P.V.; Beh, R.C.; Bourke, J.E.; Donovan, C.; Hung, A.N.; Khurana, I.; Liang, J.J.; Maxwell, S.; Mazarakis, N.; et al. Sulforaphane prevents and reverses allergic airways disease in mice via anti-inflammatory, antioxidant, and epigenetic mechanisms. Cel. Mol. Life Sci. 2022, 79, 579. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.J.; Xu, F.; Yan, X.X.; Li, S.; Li, H.T. Sulforaphane exerts anti-inflammatory effects against lipopolysaccharide-induced acute lung injury in mice through the Nrf2/ARE pathway. Int. J. Mol. Med. 2016, 37, 182–188. [Google Scholar] [CrossRef]
- Cho, H.Y.; Imani, F.; Miller-DeGraff, L.; Walters, D.; Melendi, G.A.; Yamamoto, M.; Polack, F.P.; Kleeberger, S.R. Antiviral Activity of Nrf2 in a Murine Model of Respiratory Syncytial Virus Disease. Am. J. Resp. Crit. Care Med. 2009, 179, 138–150. [Google Scholar] [CrossRef]
- Kang, Y.; Zhang, G.Y.; Huang, E.C.; Huang, J.P.; Cai, J.; Cai, L.; Wang, S.; Keller, B.B. Sulforaphane prevents right ventricular injury and reduces pulmonary vascular remodeling in pulmonary arterial hypertension. Am. J. Physiol. Heart Circ. Physiol. 2020, 318, H853–H866. [Google Scholar] [CrossRef]
- Holman, J.; Hurd, M.; Moses, P.L.; Mawe, G.M.; Zhang, T.; Ishaq, S.L.; Li, Y.Y. Interplay of broccoli/broccoli sprout bioactives with gut microbiota in reducing inflammation in inflammatory bowel diseases. J. Nutr. Biochem. 2023, 113, 109238. [Google Scholar] [CrossRef]
- Wagner, A.E.; Will, O.; Sturm, C.; Lipinski, S.; Rosenstiel, P.; Rimbach, G. DSS-induced acute colitis in C57BL/6 mice is mitigated by sulforaphane pre-treatment. J. Nutr. Biochem. 2013, 24, 2085–2091. [Google Scholar] [CrossRef]
- He, C.X.; Gao, M.F.; Zhang, X.H.; Lei, P.; Yang, H.T.; Qing, Y.P.; Zhang, L.A. The Protective Effect of Sulforaphane on Dextran Sulfate Sodium-Induced Colitis Depends on Gut Microbial and Nrf2-Related Mechanism. Front. Nutr. 2022, 9, 893344. [Google Scholar] [CrossRef] [PubMed]
- Zandani, G.; Anavi-Cohen, S.; Sela, N.; Nyska, A.; Madar, Z. Broccoli consumption attenuates inflammation and modulates gut microbiome composition and gut integrity-related factors in mice fed with a high-fat high-cholesterol diet. Food Nutr. Res. 2021, 65, 7631. [Google Scholar] [CrossRef]
- Wei, L.Y.; Wang, J.J.; Yan, L.; Shui, S.S.; Wang, L.; Zheng, W.; Liu, S.; Liu, C.; Zheng, L. Sulforaphane attenuates 5-fluorouracil induced intestinal injury in mice. J. Funct. Foods 2020, 69, 103965. [Google Scholar] [CrossRef]
- Wang, X.H.; Mi, Y.H.; Xiong, X.Y.; Bao, Z.K. The Protective Effect of Sulforaphane on ER-induced Apoptosis and Inflammation in Necrotizing Enterocolitis Mice. Comb. Chem. High Throughput Screen. 2023, 26, 1186–1195. [Google Scholar] [CrossRef] [PubMed]
- He, C.X.; Huang, L.; Lei, P.; Liu, X.D.; Li, B.L.; Shan, Y.J. Sulforaphane Normalizes Intestinal Flora and Enhances Gut Barrier in Mice with BBN-Induced Bladder Cancer. Mol. Nutr. Food Res. 2018, 62, 1800427. [Google Scholar] [CrossRef] [PubMed]
- Khor, T.O.; Hu, R.; Shen, G.X.; Jeong, W.S.; Hebbar, V.; Chen, C.; Xu, C.; Nair, S.; Reddy, B.; Chada, K.; et al. Pharmacogenomics of cancer chemopreventive isothiocyanate compound sulforaphane in the intestinal polyps of ApcMin/+ mice. Biopharm. Drug Dispos. 2006, 27, 407–420. [Google Scholar] [CrossRef]
- Swiderski, K.; Read, S.J.; Chan, A.D.S.; Chung, J.D.; Trieu, J.; Naim, T.; Koopman, R.; Lynch, G.S. Investigating the Potential for Sulforaphane to Attenuate Gastrointestinal Dysfunction in mdx Dystrophic Mice. Nutrients 2021, 13, 4559. [Google Scholar] [CrossRef] [PubMed]
- Ning, C.Q.; Gao, X.G.; Wang, C.Y.; Huo, X.K.; Liu, Z.H.; Sun, H.J.; Yang, X.; Sun, P.; Ma, X.; Meng, Q.; et al. Hepatoprotective effect of ginsenoside Rg1 from Panax ginseng on carbon tetrachloride-induced acute liver injury by activating Nrf2 signaling pathway in mice. Environ. Toxicol. 2018, 33, 1050–1060. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.K.; Ren, Z.N.; Zhu, S.L.; Wu, Y.Z.; Wang, G.; Zhang, H.; Chen, W.; He, Z.; Ye, X.L.; Zhai, Q.X. Sulforaphane ameliorates non-alcoholic fatty liver disease in mice by promoting FGF21/FGFR1 signaling pathway. Acta Pharmacol. Sin. 2022, 43, 1473–1483. [Google Scholar] [CrossRef]
- Yang, G.; Yeon, S.H.; Lee, H.E.; Kang, H.C.; Cho, Y.Y.; Lee, H.S.; Lee, J.Y. Suppression of NLRP3 inflammasome by oral treatment with sulforaphane alleviates acute gouty inflammation. Rheumatology 2018, 57, 727–736. [Google Scholar] [CrossRef]
- Xu, Y.; Huang, X.; Huangfu, B.; Hu, Y.; Xu, J.; Gao, R.; Huang, K.; He, X. Sulforaphane Ameliorates Nonalcoholic Fatty Liver Disease Induced by High-Fat and High-Fructose Diet via LPS/TLR4 in the Gut-Liver Axis. Nutrients 2023, 15, 743. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.X.; Sun, S.Y.; Liang, L.; Lou, C.X.; He, Q.J.; Ran, M.J.; Zhang, L.; Zhang, J.; Yan, C.; Yuan, H.; et al. Role of the Aryl Hydrocarbon Receptor and Gut Microbiota-Derived Metabolites Indole-3-Acetic Acid in Sulforaphane Alleviates Hepatic Steatosis in Mice. Front. Nutr. 2021, 8, 756565. [Google Scholar] [CrossRef] [PubMed]
- Promsuwan, S.; Sawamoto, K.; Xu, L.; Nagashimada, M.; Nagata, N.; Takiyama, Y. A natural Nrf2 activator glucoraphanin improves hepatic steatosis in high-fat diet-induced obese male mice associated with AMPK activation. Diabetol. Inter. 2023, 5, 86–98. [Google Scholar] [CrossRef]
- Mao, B.Y.; Ren, B.J.; Wu, J.Y.; Tang, X.; Zhang, Q.X.; Zhao, J.X.; Zhang, L.; Chen, W.; Cui, S. The Protective Effect of Broccoli Seed Extract against Lipopolysaccharide-Induced Acute Liver Injury via Gut Microbiota Modulation and Sulforaphane Production in Mice. Foods 2023, 12, 2786. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.C.; Bae, J.S. Suppressive Effects of Sulforaphane on TGFBIp-mediated Sepsis. Nat. Prod. Commun. 2017, 12, 1627–1630. [Google Scholar] [CrossRef]
- Lee, C.; Yang, S.; Lee, B.S.; Jeong, S.Y.; Kim, K.M.; Ku, S.K.; Bae, J.S. Hepatic protective effects of sulforaphane through the modulation of inflammatory pathways. J. Asian Nat. Prod. Res. 2020, 22, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.Q.; Greven, J.; Qin, K.; Fragoulis, A.; Horst, K.; Blaesius, F.; Wruck, C.; Pufe, T.; Kobbe, P.; Hildebrand, F.; et al. Sulforaphane Exerts Beneficial Immunomodulatory Effects on Liver Tissue via a Nrf2 Pathway-Related Mechanism in a Murine Model of Hemorrhagic Shock and Resuscitation. Front. Immunol. 2022, 13, 822895. [Google Scholar] [CrossRef] [PubMed]
- Panda, H.; Keleku-Lukwete, N.; Kuga, A.; Fuke, N.; Suganuma, H.; Suzuki, M.; Yamamoto, M. Dietary supplementation with sulforaphane attenuates liver damage and heme overload in a sickle cell disease murine model. Exp. Hematol. 2019, 77, 51–60. [Google Scholar] [CrossRef]
- He, Q.F.; Luo, Y.J.; Xie, Z.Q. Sulforaphane ameliorates cadmium induced hepatotoxicity through the up-regulation of /Nrf2/ARE pathway and the inactivation of NF-κB. J. Funct. Foods 2021, 77, 104297. [Google Scholar] [CrossRef]
- Zhang, Z.G.; Wang, S.D.; Zhou, S.S.; Yan, X.Q.; Wang, Y.G.; Chen, J.; Mellen, N.; Kong, M.; Gu, J.; Tan, Y.; et al. Sulforaphane prevents the development of cardiomyopathy in type 2 diabetic mice probably by reversing oxidative stress-induced inhibition of LKB1/AMPK pathway. J. Mol. Cell. Cardiol. 2014, 77, 42–52. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Zhang, D.D.; Dou, M.M.; Li, Z.B.; Zhang, J.; Zhao, X.Y. Dendrobium officinale Kimura et Migo attenuates diabetic cardiomyopathy through inhibiting oxidative stress, inflammation and fibrosis in streptozotocin-induced mice. Biomed. Pharmacother. 2016, 84, 1350–1358. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, S.D.; Ji, H.L.; Zhang, Z.G.; Chen, J.; Tan, Y.; Wintergerst, K.; Zheng, Y.; Sun, J.; Cai, L. Broccoli sprout extract prevents diabetic cardiomyopathy via Nrf2 activation in T2DM mice. Sci. Rep. 2016, 6, 30252. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.L.; Cheng, Y.L.; Wu, H.; Kong, L.L.; Wang, S.D.; Xu, Z.; Zhang, Z.; Tan, Y.; Keller, B.B.; Zhou, H.; et al. Metallothionein Is Downstream of Nrf2 and Partially Mediates Sulforaphane Prevention of Diabetic Cardiomyopathy. Diabetes 2017, 66, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, X.X.; Zhou, W.Q.; Men, H.B.; Bao, T.; Sun, Y.K.; Wang, Q.; Tan, Y.; Keller, B.B.; Tong, Q.; et al. Ferroptosis is essential for diabetic cardiomyopathy and is prevented by sulforaphane viaAMPK/NRF2 pathways. Acta Pharm. Sin. B 2022, 12, 708–722. [Google Scholar] [CrossRef]
- Sun, Y.K.; Zhou, S.S.; Guo, H.; Zhang, J.; Ma, T.J.; Zheng, Y.; Zhang, Z.; Cai, L. Protective effects of sulforaphane on type 2 diabetes-induced cardiomyopathy via AMPK-mediated activation of lipid metabolic pathways and NRF2 function. Metab.-Clin. Exp. 2020, 102, 154002. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Z.; Sun, W.; Tan, Y.; Liu, Y.; Zheng, Y.; Liu, Q.; Cai, L.; Sun, J. Sulforaphane Attenuation of Type 2 Diabetes-Induced Aortic Damage Was Associated with the Upregulation of Nrf2 Expression and Function. Oxid. Med. Cell. Longev. 2014, 2014, 123963. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, C.; Leánez, S.; Pol, O. The Inhibitory Effects of Cobalt Protoporphyrin IX and Cannabinoid 2 Receptor Agonists in Type 2 Diabetic Mice. Int. J. Mol. Sci. 2017, 18, 2268. [Google Scholar] [CrossRef]
- Bai, Y.; Cui, W.P.; Xin, Y.; Miao, X.; Barati, M.T.; Zhang, C.; Chen, Q.; Tan, Y.; Cui, T.; Zheng, Y.; et al. Prevention by sulforaphane of diabetic cardiomyopathy is associated with up-regulation of Nrf2 expression and transcription activation. J. Mol. Cell. Cardiol. 2013, 57, 82–95. [Google Scholar] [CrossRef]
- Lv, J.J.; Bao, S.Y.; Liu, T.H.; Wei, L.M.; Wang, D.M.; Ye, W.K.; Wang, N.; Song, S.; Li, J.; Chudhary, M.; et al. Sulforaphane delays diabetes-induced retinal photoreceptor cell degeneration. Cell Tissue Res. 2020, 382, 477–486. [Google Scholar] [CrossRef]
- Cui, W.P.; Bai, Y.; Miao, X.; Luo, P.; Chen, Q.; Tan, Y.; Rane, M.J.; Miao, L.; Cai, L. Prevention of Diabetic Nephropathy by Sulforaphane: Possible Role of Nrf2 Upregulation and Activation. Oxid. Med. Cell. Longev. 2012, 2012, 821936. [Google Scholar] [CrossRef]
- Wang, J.Q.; Wang, S.D.; Wang, W.N.; Chen, J.; Zhang, Z.G.; Zheng, Q.; Liu, Q.; Cai, L. Protection against diabetic cardiomyopathy is achieved using a combination of sulforaphane and zinc in type 1 diabetic OVE26 mice. J. Cell. Mol. Med. 2019, 23, 6319–6330. [Google Scholar] [CrossRef]
- Miao, X.; Bai, Y.; Sun, W.X.; Cui, W.P.; Xin, Y.; Wang, Y.H.; Tan, Y.; Miao, L.; Fu, Y.; Su, G.; et al. Sulforaphane prevention of diabetes-induced aortic damage was associated with the up-regulation of Nrf2 and its down-stream antioxidants. Nutr. Metab. 2012, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Nagata, N.; Xu, L.; Kohno, S.; Ushida, Y.; Aoki, Y.; Umeda, R.; Fuke, N.; Zhuge, F.; Ni, Y.; Nagashimada, M.; et al. Glucoraphanin Ameliorates Obesity and Insulin Resistance Through Adipose Tissue Browning and Reduction of Metabolic Endotoxemia in Mice. Diabetes 2017, 66, 1222–1236. [Google Scholar] [CrossRef]
- Tian, S.H.; Wang, Y.F.; Li, X.F.; Liu, J.; Wang, J.; Lu, Y.J. Sulforaphane Regulates Glucose and Lipid Metabolisms in Obese Mice by Restraining JNK and Activating Insulin and FGF21 Signal Pathways. J. Agri. Food Chem. 2021, 69, 13066–13079. [Google Scholar] [CrossRef] [PubMed]
- Totsch, S.K.; Meir, R.Y.; Quinn, T.L.; Lopez, S.A.; Gower, B.A.; Sorge, R.E. Effects of a Standard American Diet and an anti-inflammatory diet in male and female mice. Eur. J. Pain 2018, 22, 1203–1213. [Google Scholar] [CrossRef]
- Yanaka, A.; Fahey, J.W.; Fukumoto, A.; Nakayama, M.; Inoue, S.; Zhang, S.H.; Tauchi, M.; Suzuki, H.; Hyodo, I.; Yamamoto, M. Dietary Sulforaphane-Rich Broccoli Sprouts Reduce Colonization and Attenuate Gastritis in Helicobacter pylori-Infected Mice and Humans. Cancer Prev. Res. 2009, 2, 353–360. [Google Scholar] [CrossRef]
- Ordonez, A.A.; Bullen, C.K.; Villabona-Rueda, A.F.; Thompson, E.A.; Turner, M.L.; Merino, V.F.; Yan, Y.; Kim, J.; Davis, S.L.; Komm, O.; et al. Sulforaphane exhibits antiviral activity against pandemic SARS-CoV-2 and seasonal HCoV-OC43 coronaviruses in vitro and in mice. Commun. Biol. 2022, 5, 242. [Google Scholar] [CrossRef]
- Ho, J.N.; Kang, E.R.; Yoon, H.G.; Jeon, H.; Jun, W.; Watson, R.R.; Lee, J. Inhibition of Premature Death by Isothiocyanates through Immune Restoration in LP-BM5 Leukemia Retrovirus-Infected C57BL/6 Mice. Biosci. Biotechnol. Biochem. 2011, 75, 1234–1239. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.Y.; Mi, L.X.; Liu, J.; Song, L.R.; Chung, F.L.; Gan, N.Q. Sulforaphane prevents microcystin-LR-induced oxidative damage and apoptosis in BALB/c mice. Toxicol. Appl. Pharmacol. 2011, 255, 9–17. [Google Scholar] [CrossRef]
- Yoo, I.H.; Kim, M.J.; Kim, J.; Sung, J.J.; Park, S.T.; Ahn, S.W. The Anti-Inflammatory Effect of Sulforaphane in Mice with Experimental Autoimmune Encephalomyelitis. J. Korean Med. Sci. 2019, 34, e197. [Google Scholar] [CrossRef]
- Li, B.; Cui, W.; Liu, J.; Li, R.; Liu, Q.; Xie, X.H.; Ge, X.-L.; Zhang, J.; Song, X.-J.; Wang, Y.; et al. Sulforaphane ameliorates the development of experimental autoimmune encephalomyelitis by antagonizing oxidative stress and Th17-related inflammation in mice. Exp. Neurol. 2013, 250, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Stefanson, A.; Bakovic, M. Dietary polyacetylene falcarinol upregulated intestinal heme oxygenase-1 and modified plasma cytokine profile in late phase lipopolysaccharide-induced acute inflammation in CB57BL/6 mice. Nutr. Res. 2020, 80, 89–105. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.K.; Mi, Y.H.; Xiong, X.Y.; Wang, X.H. Sulforaphane Ameliorates the Intestinal Injury in Necrotizing Enterocolitis by Regulating the PI3K/Akt/GSK-3β Signaling Pathway. Can. J. Gastroenterol. Hepatol. 2022, 2022, 6529842. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Huang, Y.H.; Song, L.; Tong, X.H.; Li, J.F.; Lin, S.; Chen, X.; Zhang, J.C.; Zhang, Z.L.; Zeng, Q.Y. Lipopolysaccharide-induced endotoxaemia during adolescence promotes stress vulnerability in adult mice via deregulation of nuclear factor erythroid 2-related factor 2 in the medial prefrontal cortex. Psychopharmacology 2023, 240, 713–724. [Google Scholar] [CrossRef]
- Saw, C.L.; Huang, M.T.; Liu, Y.; Khor, T.O.; Conney, A.H.; Kong, A.N. Impact of Nrf2 on UVB-Induced Skin Inflammation/Photoprotection and Photoprotective Effect of Sulforaphane. Mol. Carcinog. 2011, 50, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Shibata, A.; Nakagawa, K.; Yamanoi, H.; Tsuduki, T.; Sookwong, P.; Higuchi, O.; Kimura, F.; Miyazawa, T. Sulforaphane suppresses ultraviolet B-induced inflammation in HaCaT keratinocytes and HR-1 hairless mice. J. Nutr. Biochem. 2010, 21, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.C.; Li, S.J.; Yang, C.L.; Xue, R.L.; Xi, Y.Y.; Wang, L.; Zhao, Q.L.; Li, D.J. Sulforaphane Attenuates Muscle Inflammation in Dystrophin-deficient mdx Mice via NF-E2-related Factor 2 (Nrf2)-mediated Inhibition of NF-κB Signaling Pathway. J. Biol. Chem. 2015, 290, 17784–17795. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.C.; Yang, C.L.; Xue, R.L.; Li, S.J.; Zhang, T.; Pan, L.; Ma, X.; Wang, L.; Li, D. Sulforaphane alleviates muscular dystrophy in mdx mice by activation of Nrf2. J. Appl. Physiol. 2015, 118, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.H.; Wang, J.Q.; Bi, W.; Zhang, F.; Chi, K.; Shi, L.; Yuan, T.; Ma, K.; Gao, X. Sulforaphane Ameliorates Limb Ischemia/Reperfusion-Induced Muscular Injury in Mice by Inhibiting Pyroptosis and Autophagy via the Nrf2-ARE Pathway. Evid. Based Complement. Alteren. Med. 2022, 2022, 4653864. [Google Scholar] [CrossRef]
- Pan, J.J.; Wang, R.; Pei, Y.D.; Wang, D.Y.; Wu, N.; Ji, Y.K.; Tang, Q.; Liu, L.; Cheng, K.; Liu, Q.; et al. Sulforaphane alleviated vascular remodeling in hypoxic pulmonary hypertension via inhibiting inflammation and oxidative stress. J. Nutr. Biochem. 2023, 111, 109182. [Google Scholar] [CrossRef]
- Townsend, B.E.; Chen, Y.J.; Jeffery, E.H.; Johnson, R.W. Dietary broccoli mildly improves neuroinflammation in aged mice but does not reduce lipopolysaccharide-induced sickness behavior. Nutr. Res. 2014, 34, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Pourafshar, S.; Cechova, S.; Sung, S.S.J.; Ruiz, P.; Yang, G.; Le, T. Sulforaphane Rich Broccoli Powder Attenuates the Augmented Angiotensin II Induced Renal Inflammation and Injury in GSTM1 Deficient Mice. Hypertension 2018, 72, AP158. [Google Scholar] [CrossRef]
- Franke, M.; Bieber, M.; Kraft, P.; Weber, A.N.R.; Stoll, G.; Schuhmann, M.K. The NLRP3 inflammasome drives inflammation in ischemia/reperfusion injury after transient middle cerebral artery occlusion in mice. Brain Behav. Immun. 2021, 92, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Q.; Zhang, J.; Chen, L.P.; Cai, J.; Li, Z.J.; Zhang, Z.G.; Zheng, Q.; Wang, Y.; Zhou, S.; Liu, Q.; et al. Combination of Broccoli Sprout Extract and Zinc Provides Better Protection against Intermittent Hypoxia-Induced Cardiomyopathy Than Monotherapy in Mice. Oxid. Med. Cell. Longev. 2019, 2019, 2985901. [Google Scholar] [CrossRef] [PubMed]
- Gigliotti, J.C.; Tin, A.; Pourafshar, S.; Cechova, S.; Wang, Y.T.; Sung, S.J.; Bodonyi-Kovacs, G.; Cross, J.V.; Yang, G.; Nguyen, N.; et al. GSTM1 Deletion Exaggerates Kidney Injury in Experimental Mouse Models and Confers the Protective Effect of Cruciferous Vegetables in Mice and Humans. J. Am. Soc. Nephrol. 2020, 31, 102–116. [Google Scholar] [CrossRef] [PubMed]
- Canto, A.; Martínez-González, J.; Miranda, M.; Olivar, T.; Almansa, I.; Hernández-Rabaza, V. Sulforaphane Modulates the Inflammation and Delays Neurodegeneration on a Retinitis Pigmentosa Mice Model. Front. Pharmacol. 2022, 13, 811257. [Google Scholar] [CrossRef]
- Alyoussef, A. Attenuation of experimentally induced atopic dermatitis in mice by sulforaphane: Effect on inflammation and apoptosis. Toxicol. Mech. Meth. 2022, 32, 224–232. [Google Scholar] [CrossRef]
- Knatko, E.V.; Ibbotson, S.H.; Zhang, Y.; Higgins, M.; Fahey, J.W.; Talalay, P.; Dawe, R.S.; Ferguson, J.; Huang, J.T.; Clarke, R.; et al. Nrf2 Activation Protects against Solar-Simulated Ultraviolet Radiation in Mice and Humans. Cancer Prev. Res. 2015, 8, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Ruhee, R.T.; Ma, S.H.; Suzuki, K. Protective Effects of Sulforaphane on Exercise-Induced Organ Damage via Inducing Antioxidant Defense Responses. Antioxidants 2020, 9, 136. [Google Scholar] [CrossRef]
- Jiang, X.; Wei, J.; Zhao, Q.; Wang, B.; Wang, H.; Dong, L. Sulforaphane Attenuates Radiation-Induced Skin Damage By Regulating the Effects of Anti-Inflammation and Anti-Oxidation. Int. J. Rad. Oncol. Biol. Phys. 2019, 105, E659–E660. [Google Scholar] [CrossRef]
- Moriyama, M.; Konno, M.; Serizawa, K.; Yuzawa, N.; Majima, Y.; Hayashi, I.; Suzuki, T.; Kainoh, M. Anti-pruritic effect of isothiocyanates: Potential involvement of toll-like receptor 3 signaling. Pharmacol. Res. Perspect. 2022, 10, e01038. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Wang, H.D.; Wang, X.L.; Liao, H.; Zhao, X.Z. Transcription Factor Nrf2 Protects the Spinal Cord from Inflammation Produced by Spinal Cord Injury. J. Surg. Res. 2011, 170, E105–E115. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Wang, H.D.; Wang, X.L.; Qiao, L.A.; Yin, H.X. Sulforaphane Attenuates Matrix Metalloproteinase-9 Expression Following Spinal Cord Injury in Mice. Ann. Clin. Lab. Sci. 2010, 40, 354–360. [Google Scholar] [PubMed]
- He, L.J.; Zheng, Y.; Huang, L.X.; Ye, J.Y.; Ye, Y.Y.; Luo, H.Y. Nrf2 regulates the arginase 1+ microglia phenotype through the initiation of TREM2 transcription, ameliorating depression-like behavior in mice. Transl. Psychiatry 2022, 12, 459. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.C.; Yao, W.; Dong, C.; Yang, C.; Ren, Q.; Ma, M. Prophylactic effects of sulforaphane on depression-like behavior and dendritic changes in mice after inflammation. J. Nutr. Biochem. 2017, 39, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Gao, Q.; Zhao, P.; Gao, Y.; Xi, Y.; Wang, X.; Liang, Y.; Shi, H.; Ma, Y. Sulforaphane produces antidepressant- and anxiolytic-like effects in adult mice. Behav. Brain Res. 2016, 301, 55–62. [Google Scholar] [CrossRef]
- Ferreira-Chamorro, P.; Redondo, A.; Riego, G.; Leánez, S.; Pol, O. Sulforaphane Inhibited the Nociceptive Responses, Anxiety- and Depressive-Like Behaviors Associated with Neuropathic Pain and Improved the Anti-allodynic Effects of Morphine in Mice. Front Pharmacol. 2018, 9, 1332. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Xiong, B.R.; Zhan, B.; Li, S.; Huang, N.N.; Zhan, G.F. Sulforaphane Alleviates Lipopolysaccharide-induced Spatial Learning and Memory Dysfunction in Mice: The Role of BDNF-mTOR Signaling Pathway. Neuroscience 2018, 388, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.T.; Yang, H.Y.; Wang, W.; Wu, Q.Q.; Tian, Y.R.H.; Jia, J.P. Sulforaphane Inhibits the Generation of Amyloid-β Oligomer and Promotes Spatial Learning and Memory in Alzheimer’s Disease (PS1V97L) Transgenic Mice. J. Alz. Dis. 2018, 62, 1803–1813. [Google Scholar] [CrossRef]
- Nadeem, A.; Ahmad, S.F.; Al-Harbi, N.O.; Attia, S.M.; Bakheet, S.A.; Ibrahim, K. Nrf2 activator, sulforaphane ameliorates autism-like symptoms through suppression of Th17 related signaling and rectification of oxidant antioxidant imbalance in periphery and brain of BTBR T plus tf/J mice. Behav. Brain Res. 2019, 364, 213–224. [Google Scholar] [CrossRef]
- Benedict, A.L.; Mountney, A.; Hurtado, A.; Bryan, K.E.; Schnaar, R.L.; Dinkova-Kostova, A.T.; Talalay, P. Neuroprotective Effects of Sulforaphane after Contusive Spinal Cord Injury. J. Neurotrauma 2012, 29, 2576–2586. [Google Scholar] [CrossRef] [PubMed]
- Sunkaria, A.; Bhardwaj, S.; Yadav, A.; Halder, A.; Sandhir, R. Sulforaphane attenuates postnatal proteasome inhibition and improves spatial learning in adult mice. J. Nutr. Biochem. 2018, 51, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Martin-de-Saavedra, M.D.; Budni, J.; Cunha, M.P.; Gómez-Rangel, V.; Lorrio, S.; del Barrio, L.; Lastres-Becker, I.; Parada, E.; Tor, R.M. Nrf2 participates in depressive disorders through an anti-inflammatory mechanism. Psychoneuroendocrinology 2013, 38, 2010–2022. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, S.; Holloway, P.M.; Becker, F.; Rauzi, F.; Vital, S.A.; Taylor, K.A.; Stokes, K.Y.; Emerson, M.; Gavins, F.N.E. The isothiocyanate sulforaphane modulates platelet function and protects against cerebral thrombotic dysfunction. Br. J. Pharmacol. 2018, 175, 3333–3346. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.; Luong, L.; Naase, H.; Vives, M.; Jakaj, G.; Finch, J.; Boyle, J.; Mulholland, J.W.; Kwak, J.H.; Pyo, S.; et al. Sulforaphane pretreatment prevents systemic inflammation and renal injury in response to cardiopulmonary bypass. J. Thor. Cardiovasc. Surg. 2014, 148, 690–697. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shehatou, G.S.G.; Suddek, G.M. Sulforaphane attenuates the development of atherosclerosis and improves endothelial dysfunction in hypercholesterolemic rabbits. Exp. Biol. Med. 2016, 241, 426–436. [Google Scholar] [CrossRef]
- Sun, Z.J.; Niu, Z.Q.; Wu, S.S.; Shan, S.Q. Protective mechanism of sulforaphane in Nrf2 and anti-lung injury in ARDS rabbits. Exp. Ther. Med. 2018, 15, 4911–4915. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Zhu, D.; Chen, D.; Zhang, Q.; Dong, H.; Wu, W.; Lu, H.; Wu, G. Sulforaphane, a Natural Isothiocyanate Compound, Improves Cardiac Function and Remodeling by Inhibiting Oxidative Stress and Inflammation in a Rabbit Model of Chronic Heart Failure. Med. Sci. Monit. 2018, 24, 1473–1483. [Google Scholar] [CrossRef] [PubMed]
- Grottelli, S.; Amoroso, R.; Macchioni, L.; D’Onofrio, F.; Fettucciari, K.; Bellezza, I.; Maccallini, C. Acetamidine-based iNOS inhibitors as molecular tools to counteract inflammation in BV2 microglial cells. Molecules 2020, 25, 2646. [Google Scholar] [CrossRef]
- Gallorini, M.; Rapino, M.; Schweikl, H.; Cataldi, A.; Amoroso, R.; Maccallini, C. Selective inhibitors of the inducible nitric oxide synthase as modulators of cell responses in LPS-stimulated human monocytes. Molecules 2021, 26, 4419. [Google Scholar] [CrossRef]
- Fuse, Y.; Kobayashi, M. Conservation of the Keap1-Nrf2 system: An evolutionary journey through stressful space and time. Molecules 2017, 22, 436. [Google Scholar] [CrossRef] [PubMed]
- Cullinan, S.B.; Gordan, J.D.; Jin, J.; Harper, J.W.; Diehl, J.A. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 2004, 24, 8477–8486. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Kang, M.I.; Okawa, H.; Ohtsuji, M.; Zenke, Y.; Chiba, T.; Igarashi, K.; Yamamoto, M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004, 24, 7130–7139. [Google Scholar] [CrossRef] [PubMed]
- Okawa, H.; Motohashi, H.; Kobayashi, A.; Aburatani, H.; Kensler, T.W.; Yamamoto, M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem. Biophys. Res. Commun. 2006, 339, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Rangasamy, T.; Cho, C.Y.; Thimmulappa, R.K.; Zhen, L.; Srisuma, S.S.; Kensler, T.W.; Yamamoto, M.; Petrache, I.; Tuder, R.M.; Biswal, S. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J. Clin. Investig. 2004, 114, 1248–1259. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.Y.; Jedlicka, A.E.; Reddy, S.P.; Kensler, T.W.; Yamamoto, M.; Zhang, L.Y.; Kleeberger, S.R. Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell. Mol. Biol. 2002, 26, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.Y.; Reddy, S.P.; Yamamoto, M.; Kleeberger, S.R. The transcription factor NRF2 protects against pulmonary fibrosis. FASEB J. 2004, 18, 1258–1260. [Google Scholar] [CrossRef]
- Furukawa, M.; Xiong, Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell. Biol. 2005, 25, 162–171. [Google Scholar] [CrossRef]
- Zhang, D.D.; Lo, S.C.; Cross, J.V.; Templeton, D.J.; Hannink, M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 2004, 24, 10941–10953. [Google Scholar] [CrossRef]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional regulation by Nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef]
- Lee, K.M.; Kang, K.; Lee, S.B.; Nho, C.W. Nuclear factor-E2 (Nrf2) is regulated through the differential activation of ERK1/2 and PKC alpha/beta II by Gymnasterkoreayne B. Cancer Lett. 2013, 330, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Chen, C.; Mo, Y.Y.; Hebbar, V.; Owuor, E.D.; Tann, T.H.; Kong, A.N.T. Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. J. Biol. Chem. 2000, 275, 39907–39913. [Google Scholar] [CrossRef] [PubMed]
- Buelna-Chontal, M.; Guevara-Chavez, J.G.; Silva-Palacios, A.; Medina-Campos, O.N.; Pedraza-Chaverri, J.; Zazueta, C. Nrf2-regulated antioxidant response is activated by protein kinase C in postconditioned rat hearts. Free Radic. Biol. Med. 2014, 74, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.C.; Nguyen, T.; Pickett, C.B. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 2002, 277, 42769–42774. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Jin, M.L.; Ko, M.J.; Park, G.; Choi, Y.W. Anti-neuroinflammatory effect of emodin in LPS-Stimulated microglia: Involvement of AMPK/Nrf2 activation. Neurochem. Res. 2016, 41, 2981–2992. [Google Scholar] [CrossRef] [PubMed]
- Sid, B.; Glorieux, C.; Valenzuela, M.; Rommelaere, G.; Najimi, M.; Dejeans, N.; Renard, P.; Verrax, J.; Calderon, P.B. AICAR induces Nrf2 activation by an AMPK-independent mechanism in hepatocarcinoma cells. Biochem. Pharmacol. 2014, 91, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Joo, M.S.; Kim, W.D.; Lee, K.Y.; Kim, J.H.; Koo, J.H.; Kim, S.G. AMPK facilitates nuclear accumulation of Nrf2 by phosphorylating at Serine 550. Mol. Cell. Biol. 2016, 36, 1931–1942. [Google Scholar] [CrossRef]
- Lee, J.; Hanson, J.M.; Chu, W.A.; Johnson, J.A. Phosphatidylinositol 3-kinase, not extracellular signal-regulated kinase, regulates activation of the antioxidant-responsive element in IMR-32 human neuroblastoma cells. J. Biol. Chem. 2001, 276, 20011–20016. [Google Scholar] [CrossRef] [PubMed]
- Nakaso, K.; Yano, H.; Fukuhara, Y.; Takeshima, T.; Wada-Isoe, K.; Nakashima, K. PI3K is a key molecule in the Nrf2-mediated regulation of antioxidative proteins by hemin in human neuroblastoma cells. FEBS Lett. 2003, 546, 181–184. [Google Scholar] [CrossRef]
- Cuadrado, A. Structural and functional characterization of Nrf2 degradation by glycogen synthase kinase 3/beta-TrCP. Free Radic. Biol. Med. 2015, 88, 147–157. [Google Scholar] [CrossRef]
- Fukutomi, T.; Takagi, K.; Mizushima, T.; Ohuchi, N.; Yamamoto, M. Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1. Mol. Cell. Biol. 2014, 34, 832–846. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.C.; Li, X.; Henzl, M.T.; Beamer, L.J.; Hannink, M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006, 25, 3605–3617. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: A two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 2006, 281, 24756–24768. [Google Scholar] [CrossRef] [PubMed]
- Tong, K.I.; Katoh, Y.; Kusunoki, H.; Itoh, K.; Tanaka, T.; Yamamoto, M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: Characterization of the two-site molecular recognition model. Mol. Cell. Biol. 2006, 26, 2887–2900. [Google Scholar] [CrossRef] [PubMed]
- Horie, Y.; Suzuki, T.; Inoue, J.; Iso, T.; Wells, G.; Moore, T.W.; Mizushima, T.; Dinkova-Kostova, A.T.; Kasai, T.; Kamei, T.; et al. Molecular basis for the disruption of Keap1-Nrf2 interaction via Hinge & Latch mechanism. Commun. Biol. 2021, 4, 576. [Google Scholar]
- Li, W.; Kong, A.N. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog. 2009, 48, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Tong, K.I.; Padmanabhan, B.; Kobayashi, A.; Shang, C.; Hirotsu, Y.; Yokoyama, S.; Yamamoto, M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol. Cell. Biol. 2007, 27, 7511–7521. [Google Scholar] [CrossRef] [PubMed]
- Katoh, Y.; Itoh, K.; Yoshida, E.; Miyagishi, M.; Fukamizu, A.; Yamamoto, M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 2001, 6, 857–868. [Google Scholar] [CrossRef]
- Sun, Z.; Chin, Y.E.; Zhang, D.D. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol. Cell. Biol. 2009, 29, 2658–2672. [Google Scholar] [CrossRef]
- Wang, H.; Liu, K.; Geng, M.; Gao, P.; Wu, X.; Hai, Y.; Li, Y.; Li, Y.; Luo, L.; Hayes, J.D.; et al. RXRalpha inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res. 2013, 73, 3097–3108. [Google Scholar] [CrossRef]
- Moi, P.; Chan, K.; Asunis, I.; Cao, A.; Kan, Y.W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 1994, 91, 9926–9930. [Google Scholar] [CrossRef] [PubMed]
- Plafker, K.S.; Nguyen, L.; Barneche, M.; Mirza, S.; Crawford, D.; Plafker, S.M. The ubiquitin-conjugating enzyme UbcM2 can regulate the stability and activity of the antioxidant transcription factor Nrf2. J. Biol. Chem. 2010, 285, 23064–23074. [Google Scholar] [CrossRef] [PubMed]
- Rada, P.; Rojo, A.I.; Chowdhry, S.; McMahon, M.; Hayes, J.D.; Cuadrado, A. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell. Biol. 2011, 31, 1121–1133. [Google Scholar] [CrossRef] [PubMed]
- Baird, L.; Yamamoto, M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell Biol. 2020, 40, e00099-20. [Google Scholar] [CrossRef] [PubMed]
- Sihvola, V.; Levonen, A.L. Keap1 as the redox sensor of the antioxidant response. Arch. Biochem. Biophys. 2017, 617, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Yamamoto, M. Stress-sensing mechanisms and the physiological roles of the Keap1-Nrf2 system during cellular stress. J. Biol. Chem. 2017, 292, 16817–16824. [Google Scholar] [CrossRef] [PubMed]
- Zipper, L.M.; Mulcahy, R.T. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J. Biol. Chem. 2002, 277, 36544–36552. [Google Scholar] [CrossRef] [PubMed]
- Ogura, T.; Tong, K.I.; Mio, K.; Maruyama, Y.; Kurokawa, H.; Sato, C.; Yamamoto, M. Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc. Natl. Acad. Sci. USA 2010, 107, 2842–2847. [Google Scholar] [CrossRef] [PubMed]
- Carlström, K.E.; Ewing, E.; Granqvist, M.; Gyllenberg, A.; Aeinehband, S.; Enoksson, S.L.; Checa, A.; Badam, T.V.S.; Huang, J.; Gomez-Cabrero, D.; et al. Therapeutic efficacy of dimethyl fumarate in relapsing-remitting multiple sclerosis associates with ROS pathway in monocytes. Nat. Commun. 2019, 10, 3081. [Google Scholar] [CrossRef]
- Fox, R.J.; Miller, D.H.; Phillips, J.T.; Hutchinson, M.; Havrdova, E.; Kita, M.; Yang, M.; Raghupathi, K.; Novas, M.; Sweetser, M.T.; et al. Placebo-controlled phase 3 study of oral BG-12 or glatiramer in multiple sclerosis. N. Engl. J. Med. 2012, 367, 1087–1097. [Google Scholar] [CrossRef]
- Gold, R.; Arnold, D.L.; Bar-Or, A.; Hutchinson, M.; Kappos, L.; Havrdova, E.; MacManus, D.G.; Yousry, T.A.; Pozzilli, C.; Selmaj, K.; et al. Long-term effects of delayed-release dimethyl fumarate in multiple sclerosis: Interim analysis of ENDORSE, a randomized extension study. Mult. Scler. 2017, 23, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Esposito, M.; Campanati, A.; Giunta, A.; Calianno, G.; Bianchi, L.; Diotallevi, F.; Offidani, A.M.; Fargnoli, M.C. Dimethyl Fumarate’s Effectiveness and Safety in Psoriasis: A Real-Life Experience During the COVID-19 Pandemic. Dermatol. Ther. 2022, 12, 671–681. [Google Scholar] [CrossRef] [PubMed]
- De Zeeuw, D.; Akizawa, T.; Audhya, P.; Bakris, G.L.; Chin, M.; Christ-Schmidt, H.; Goldsberry, A.; Houser, M.; Krauth, M.; Lambers Heerspink, H.J.; et al. Bardoxolone methyl in type 2 diabetes and stage 4 chronic kidney disease. N. Engl. J. Med. 2013, 369, 2492–2503. [Google Scholar] [CrossRef] [PubMed]
- Ishii, Y.; Itoh, K.; Morishima, Y.; Kimura, T.; Kiwamoto, T.; Iizuka, T.; Hegab, A.E.; Hosoya, T.; Nomura, A.; Sakamoto, T.; et al. Transcription factor Nrf2 plays a pivotal role in protection against elastase-induced pulmonary inflammation and emphysema. J. Immunol. 2005, 175, 6968–6975. [Google Scholar] [CrossRef]
- Keleku-Lukwete, N.; Suzuki, M.; Otsuki, A.; Tsuchida, K.; Katayama, S.; Hayashi, M.; Naganuma, E.; Moriguchi, T.; Tanabe, O.; Engel, J.D.; et al. Amelioration of inflammation and tissue damage in sickle cell model mice by Nrf2 activation. Proc. Natl. Acad. Sci. USA 2015, 112, 12169–12174. [Google Scholar] [CrossRef] [PubMed]
- Rachakonda, G.; Xiong, Y.; Sekhar, K.R.; Stamer, S.L.; Liebler, D.C. Freeman ML. Covalent modification at Cys151 dissociates the electrophile sensor Keap1 from the ubiquitin ligase CUL3. Chem. Res. Toxicol. 2008, 21, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Takaya, K.; Suzuki, T.; Motohashi, H.; Onodera, K.; Satomi, S.; Kensler, T.W.; Yamamoto, M. Validation of the multiple sensor mechanism of the Keap1-Nrf2 system. Free Radic. Biol. Med. 2012, 53, 817–827. [Google Scholar] [CrossRef]
- Naidu, S.D.; Muramatsu, A.; Saito, R.; Asami, S.; Honda, T.; Hosoya, T.; Itoh, K.; Yamamoto, M.; Suzuki, T.; Dinkova-Kostova, A.T. C151 in KEAP1 is the main cysteine sensor for the cyanoenone class of NRF2 activators, irrespective of molecular size or shape. Sci. Rep. 2018, 8, 8037. [Google Scholar] [CrossRef]
- Wong, D.P.W.; Ng, M.Y.; Leung, J.Y.; Boh, B.K.; Lim, E.C.; Tan, S.H.; Lim, S.; Seah, W.H.; Hu, C.Z.; Ho, B.C.; et al. Regulation of the NRF2 transcription factor by andrographolide and organic extracts from plant endophytes. PLoS ONE 2018, 13, e0204853. [Google Scholar] [CrossRef]
- Saito, R.; Suzuki, T.; Hiramoto, K.; Asami, S.; Naganuma, E.; Suda, H.; Iso, T.; Yamamoto, H.; Morita, M.; Baird, L.; et al. Characterizations of Three Major Cysteine Sensors of Keap1 in Stress Response. Mol. Cell. Biol. 2015, 36, 271–284. [Google Scholar] [CrossRef]
- Kansanen, E.; Bonacci, G.; Schopfer, F.J.; Kuosmanen, S.M.; Tong, K.I.; Leinonen, H.; Woodcock, S.R.; Yamamoto, M.; Carlberg, C.; Yla-Herttuala, S.; et al. Electrophilic nitro-fatty acids activate NRF2 by a KEAP1 cysteine 151-independent mechanism. J. Biol. Chem. 2011, 286, 14019–14027. [Google Scholar] [CrossRef]
- Balogun, E.; Hoque, M.; Gong, P.; Killeen, E.; Green, C.J.; Foresti, R.; Alam, J.; Motterlini, R. Curcumin activates the haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochem. J. 2003, 371, 887–895. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, C.Y.; Bai, L.P.; Pan, H.D.; Shu, L.M.; Kong, A.N.; Leung, E.L.; Liu, L.; Li, T. Flavonoids derived from liquorice suppress murine macrophage activation by up-regulating heme oxygenase-1 independent of Nrf2 activation. Int. Immunopharmacol. 2015, 28, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Blake, D.J.; Singh, A.; Kombairaju, P.; Malhotra, D.; Mariani, T.J.; Tuder, R.M.; Gabrielson, E.; Biswal, S. Deletion of Keap1 in the Lung Attenuates Acute Cigarette Smoke-Induced Oxidative Stress and Inflammation. Am. J. Resp. Cell Mol. Biol. 2010, 42, 524–536. [Google Scholar] [CrossRef] [PubMed]
- Oeckinghaus, A.; Ghosh, S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.; Vargas, J.; Hoffmann, A. Signaling via the NFκB system. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016, 8, 227–241. [Google Scholar] [CrossRef]
- Sun, S.C. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017, 17, 545–558. [Google Scholar] [CrossRef]
- Israël, A. The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harb. Perspect. Biol. 2010, 2, a000158. [Google Scholar] [CrossRef] [PubMed]
- Orian, A.; Gonen, H.; Bercovich, B.; Fajerman, I.; Eytan, E.; Iwai, K.; Schwartz, A.L.; Ciechanover, A. SCFb-TrCP ubiquitin ligase-mediated processing of NF-kB p105 requires phosphorylation of its C-terminus by IkB kinase. EMBO J. 2020, 19, 2580–2591. [Google Scholar] [CrossRef]
- Sun, S.C. The noncanonical NF-κB pathway. Immunol. Rev. 2012, 246, 125–140. [Google Scholar] [CrossRef]
- Yin, L.; Wu, L.; Wesche, H.; Arthur, C.D.; White, J.M.; Goeddel, D.V.; Schreiber, R.D. Defective lymphotoxin-beta receptor-induced NF-kappaB transcriptional activity in NIK-deficient mice. Sci. N. Y. 2001, 291, 2162–2165. [Google Scholar] [CrossRef] [PubMed]
- Dejardin, E.; Droin, N.M.; Delhase, M.; Haas, E.; Cao, Y.; Makris, C.; Li, Z.-W.; Karin, M.; Ware, C.F.; Green, D.R. The lymphotoxin-β receptor induces different patterns of gene expression via two NF-κB pathways. Immunity 2002, 17, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Dejardin, E. The alternative NF-κB pathway from biochemistry to biology: Pitfalls and promises for future drug development. Biochem. Pharmacol. 2006, 72, 1161–1179. [Google Scholar] [CrossRef] [PubMed]
- Gamble, C.; McIntosh, K.; Scott, R.; Ho, K.H.; Plevin, R.; Paul, A. Inhibitory Kappa B Kinases as Targets for Pharmacological Regulation. Br. J. Pharmacol. 2012, 165, 802–819. [Google Scholar] [CrossRef] [PubMed]
- Heiss, E.; Herhaus, C.; Klimo, K.; Bartsch, H.; Gerhäuser, C. Nuclear factor kappa B is a molecular target for sulforaphane-mediated anti-inflammatory mechanisms. J. Biol. Chem. 2001, 276, 32008–32015. [Google Scholar] [CrossRef] [PubMed]
- Moon, D.O.; Kim, M.O.; Kang, S.H.; Choi, Y.H.; Kim, G.Y. Sulforaphane suppresses TNF-alpha-mediated activation of NF-kappaB and induces apoptosis through activation of reactive oxygen species-dependent caspase-3. Cancer Lett. 2009, 274, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, A.; Martin-Moldes, Z.; Ye, J.; Lastres-Becker, I. Transcription factors NRF2 and NF-kappaB are coordinated effectors of the Rho family, GTP-binding protein RAC1 during inflammation. J. Biol. Chem. 2014, 289, 15244–15258. [Google Scholar] [CrossRef] [PubMed]
- Rushworth, S.A.; Zaitseva, L.; Murray, M.Y.; Shah, N.M.; Bowles, K.M.; MacEwa, D.J. The high Nrf2 expression in human acute myeloid leukemia is driven by NF-kappaB and underlies its chemo-resistance. Blood 2012, 120, 5188–5198. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Khor, T.O.; Xu, C.; Shen, G.; Jeong, W.S.; Yu, S.; Kong, A.N. Activation of Nrf2-antioxidant signaling attenuates NFkappaB-inflammatory response and elicits apoptosis. Biochem. Pharmacol. 2008, 76, 1485–1489. [Google Scholar] [CrossRef]
- Kim, S.W.; Lee, H.K.; Shin, J.H.; Lee, J.K. Up-down regulation of HO-1 and iNOS gene expressions by ethyl pyruvate via recruiting p300 to Nrf2 and depriving It from p65. Free Radic. Biol. Med. 2013, 65, 468–476. [Google Scholar] [CrossRef]
- Kim, J.E.; You, D.J.; Lee, C.; Ahn, C.; Seong, J.Y.; Hwang, J.I. Suppression of NF-kappaB signaling by KEAP1 regulation of IKKbeta activity through autophagic degradation and inhibition of phosphorylation. Cell. Signal. 2010, 22, 1645–1654. [Google Scholar] [CrossRef]
- Liu, G.H.; Qu, J.; Shen, X. NF-kappaB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochem. Biophys. Acta 2008, 1783, 713–727. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, E.H.; Suzuki, T.; Funayama, R.; Nagashima, T.; Hayashi, M.; Sekine, H.; Tanaka, N.; Moriguchi, T.; Motohashi, H.; Nakayama, K.; et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016, 7, 11624. [Google Scholar] [CrossRef] [PubMed]
- Bellezza, I.; Tucci, A.; Galli, F.; Grottelli, S.; Mierla, A.L.; Pilolli, F.; Minelli, A. Inhibition of NF-κB nuclear translocation via HO-1 activation underlies α-tocopheryl succinate toxicity. J. Nutr. Biochem. 2012, 23, 1583–1591. [Google Scholar] [CrossRef]
- Mossman, B.T.; Lounsbury, K.M.; Reddy, S.P. Oxidants and signaling by mitogen-activated protein kinases in lung epithelium. Am. J. Respir. Cell Mol. Biol. 2006, 34, 666–669. [Google Scholar] [CrossRef]
- Cho, H.Y.; Kleeberger, S.R. Nrf2 protects against airway disorders. Toxicol. Appl. Pharmacol. 2010, 244, 43–56. [Google Scholar] [CrossRef]
- Liu, Q.; Jin, Z.; Xu, Z.; Yang, H.; Li, L.; Li, G.; Li, F.; Gu, S.; Zong, S.; Zhou, J.; et al. Antioxidant effects of ginkgolides and bilobalide against cerebral ischemia injury by activating the Akt/Nrf2 pathway in vitro and in vivo. Cell Stress Chaperones 2019, 24, 441–452. [Google Scholar] [CrossRef]
- Vaziri, H.; Dessain, S.K.; Eaton, E.N.; Imai, S.I.; Frye, R.A.; Pandita, T.K.; Guarente, L.; Weinberg, R.A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 2001, 107, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Yeung, F.; Hoberg, J.E.; Ramsey, C.S.; Keller, M.D.; Jones, D.R.; Frye, R.A.; Mayo, M.W. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004, 23, 2369–2380. [Google Scholar] [CrossRef]
- Tikhanovich, I.; Cox, J.; Weinman, S.A. Forkhead box class O transcription factors in liver function and disease. J. Gastroenterol. Hepatol. 2013, 28, 125–131. [Google Scholar] [CrossRef]
- Rajendrasozhan, S.; Yang, S.R.; Kinnula, V.L.; Rahman, I. SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2008, 177, 861–870. [Google Scholar] [CrossRef]
- Yang, S.R.; Wright, J.; Bauter, M.; Seweryniak, K.; Kode, A.; Rahman, I. Sirtuin regulates cigarette smoke-induced proinflammatory mediator release via RelA/p65 NF-kappaB in macrophages in vitro and in rat lungs in vivo: Implications for chronic inflammation and aging. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007, 292, L567–L576. [Google Scholar] [CrossRef]
- Yoshizaki, T.; Schenk, S.; Imamura, T.; Babendure, J.L.; Sonoda, N.; Bae, E.J.; Oh, D.Y.; Lu, M.; Milne, J.C.; Westphal, C.; et al. SIRT1 Inhibits Inflammatory Pathways in Macrophages and Modulates Insulin Sensitivity. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E419–E428. [Google Scholar] [CrossRef]
- Yang, C.M.; Chen, Y.W.; Chi, P.L.; Lin, C.C.; Hsiao, L.D. Resveratrol Inhibits BK-Induced COX-2 Transcription by Suppressing Acetylation of AP-1 and NF-κB in Human Rheumatoid Arthritis Synovial Fibroblasts. Biochem. Pharmacol. 2017, 132, 77–91. [Google Scholar] [CrossRef]
- Chen, G.; Yu, W.; Chen, X. Sirt1 activator represses the transcription of TNF-α in THP-1 cells of a sepsis model via deacetylation of H4K16. Mol. Med. Rep. 2016, 14, 5544–5550. [Google Scholar] [CrossRef] [PubMed]
- Canto, C.; Gerhart-Hines, Z.; Feige, J.N.; Lagouge, M.; Noriega, L.; Milne, J.C.; Elliott, P.J.; Puigserver, P.; Auwerx, J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 2009, 458, 1056–1060. [Google Scholar] [CrossRef]
- Wang, S.; Tang, Y.J. Sulforaphane ameliorates amyloid-β-induced inflammatory injury by suppressing the PARP1/SIRT1 pathway in retinal pigment epithelial cells. Bioengineered 2021, 12, 7079–7089. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.P.; Wang, S.L.; Liu, B.; Tang, L.; Kuang, R.R.; Wang, X.B.; Zhao, C.; Song, X.D.; Cao, X.M.; Wu, X.; et al. Sulforaphane prevents rat cardiomyocytes from hypoxia/reoxygenation injury in vitro via activating SIRT1 and subsequently inhibiting ER stress. Acta Pharmacol. Sin. 2016, 37, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, B.P. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science 2013, 339, 1216–1219. [Google Scholar] [CrossRef]
- Xiao, J.; Sheng, C.; Zhang, X.; Guo, M.; Ji, X. Curcumin protects against myocardial infarction-induced cardiac fibrosis via SIRT1 activation in vivo and in vitro. Drug Des. Dev. Ther. 2016, 10, 1267–1277. [Google Scholar] [CrossRef]
- Butturini, E.; Carcereri de Prati, A.; Mariotto, S. Redox regulation of STAT1 and STAT3 signaling. Int. J. Mol. Sci. 2020, 21, 7034. [Google Scholar] [CrossRef] [PubMed]
- Okugawa, S.; Ota, Y.; Kitazawa, T.; Nakayama, K.; Yanagimoto, S.; Tsukada, K.; Kawada, M.; Kimura, S. Janus kinase 2 is involved in lipopolysaccharide-induced activation of macrophages. Am. J. Physiol. Cell Physiol. 2003, 285, C399–C408. [Google Scholar] [CrossRef] [PubMed]
- Yi, W.J.; Kim, T.S. Melatonin protects mice against stress-induced inflammation through enhancement of M2 macrophage polarization. Int. Immunopharmacol. 2017, 48, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Schonthaler, H.B.; Guinea-Viniegra, J.; Wagner, E.F. Targeting inflammation by modulating the Jun/AP-1 pathway. Ann. Rheum. Dis. 2011, 70 (Suppl. 1), 109–112. [Google Scholar] [CrossRef] [PubMed]
- Shiozawa, S.; Tsumiyama, K. Pathogenesis of rheumatoid arthritis and c-Fos/AP-1. Cell Cycle 2009, 8, 1539–1543. [Google Scholar] [CrossRef] [PubMed]
- Chaiprasongsuk, A.; Lohakul, J.; Soontrapa, K.; Sampattavanich, S.; Akarasereenont, P.; Panich, U. Activation of Nrf2 Reduces UVA-Mediated MMP-1 Upregulation via MAPK/AP-1 Signaling Cascades: The Photoprotective Effects of Sulforaphane and Hispidulin. J. Pharmacol. Exp. Ther. 2017, 360, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhang, Y.; Cooper, S.; Sikorski, E.; Rohwer, J.; Bowden, G.T. Phase II enzyme inducer, sulforaphane, inhibits UVB-induced AP-1 activation in human keratinocytes by a novel mechanism. Mol. Carcinog. 2004, 41, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, S.E.; Melton, T.F.; Olson, E.R.; Zhang, J.; Saboda, K.; Bowden, G.T. Inhibition of activator protein-1 by sulforaphane involves interaction with cysteine in the cFos DNA-binding domain: Implications for chemoprevention of UVB-induced skin cancer. Cancer Res. 2009, 69, 7103–7110. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Barve, A.; Khor, T.O.; Shen, G.X.; Lin, W.; Chan, J.Y.; Cai, L.; Kong, A.N. Regulation of Nrf2- and AP-1-mediated gene expression by epigallocatechin-3-gallate and sulforaphane in prostate of Nrf2-knockout or C57BL/6J mice and PC-3 AP-1 human prostate cancer cells. Acta Pharmacol. Sin. 2010, 31, 1223–1240. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, X.; Zhu, G.; Liu, H.; Chen, J.; Wang, Y.; He, X. Quercetin inhibits TNF-α induced HUVECs apoptosis and inflammation via downregulating NF-kB and AP-1 signaling pathway in vitro. Medicine 2020, 99, e22241. [Google Scholar] [CrossRef]
- Zgórzyńska, E.; Stulczewski, D.; Dziedzic, B.; Su, K.P.; Walczewska, A. Docosahexaenoic fatty acid reduces the pro-inflammatory response induced by IL-1β in astrocytes through inhibition of NF-κB and AP-1 transcription factor activation. BMC Neurosci. 2021, 22, 4. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kwak, C.H.; Ha, S.H.; Kwon, K.M.; Abekura, F.; Cho, S.H.; Chang, Y.C.; Lee, Y.C.; Ha, K.T.; Chung, T.W.; et al. Ganglioside GM3 suppresses lipopolysaccharide-induced inflammatory responses in rAW 264.7 macrophage cells through NF-κB, AP-1, and MAPKs signaling. J. Cell Biochem. 2018, 119, 1173–1182. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zong, C.; Gao, Y.; Cai, R.; Fang, L.; Lu, J.; Liu, F.; Qi, Y. Curcumol exhibits anti-inflammatory properties by interfering with the JNK-mediated AP-1 pathway in lipopolysaccharide-activated RAW264.7 cells. Eur. J. Pharmacol. 2014, 723, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.R.; Noh, E.M.; Han, J.H.; Kim, J.M.; Hwang, B.M.; Kim, B.S.; Lee, S.H.; Jung, S.H.; Youn, H.J.; Chung, E.Y.; et al. Sulforaphane controls TPA-induced MMP-9 expression through the NF-κB signaling pathway, but not AP-1, in MCF-7 breast cancer cells. BMB Rep. 2013, 46, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Tahamtan, A.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. Anti-inflammatory microRNAs and their potential for inflammatory diseases treatment. Front. Immunol. 2018, 9, 1377. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.H.; Li, J.W.; Sun, H.T.; He, S.Q.; Pang, J. Sulforaphane inhibits the activation of hepatic stellate cell by miRNA-423-5p targeting suppressor of fused. Hum. Cell 2019, 32, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xing, N.; Dong, L. Effect of MiR-423-5p expression on the severity of lipopolysaccharide-induced acute liver injury, inflammatory response and immune function in mice. Trop. J. Pharm. Res. 2022, 21, 761–767. [Google Scholar] [CrossRef]
- Arita, Y.; Park, H.J.; Cantillon, A.; Verma, K.; Menon, R.; Getahun, D.; Peltier, M.R. Pro- and anti-inflammatory effects of sulforaphane on placental cytokine production. J. Reprod. Immunol. 2019, 131, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Bonetto, J.H.; Fernandes, R.O.; Seolin, B.G.; Müller, D.D.; Teixeira, R.B.; Araujo, A.S.; Vassallo, D.; Schenkel, P.C.; Belló-Klein, A. Sulforaphane improves oxidative status without attenuating the inflammatory response or cardiac impairment induced by ischemia-reperfusion in rats. Can. J. Physiol. Pharmacol. 2016, 94, 508–516. [Google Scholar] [CrossRef]
- Lee, Y.J.; Lee, S.H. Pro-oxidant activity of sulforaphane and cisplatin potentiates apoptosis and simultaneously promotes autophagy in malignant mesothelioma cells. Mol. Med. Rep. 2017, 16, 2133–2141. [Google Scholar] [CrossRef]
- Ferreira de Oliveira, J.M.; Costa, M.; Pedrosa, T.; Pinto, P.; Remédios, C.; Oliveira, H.; Pimentel, F.; Almeida, L.; Santos, C. Sulforaphane induces oxidative stress and death by p53-independent mechanism: Implication of impaired glutathione recycling. PLoS ONE 2014, 9, e92980. [Google Scholar] [CrossRef] [PubMed]
- Shih, Y.L.; Wu, L.Y.; Lee, C.H.; Chen, Y.L.; Hsueh, S.C.; Lu, H.F.; Liao, N.C.; Chung, J.G. Sulforaphane promotes immune responses in a WEHI-3-induced leukemia mouse model through enhanced phagocytosis of macrophages and natural killer cell activities in vivo. Mol. Med. Rep. 2016, 13, 4023–4029. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Barajas, B.; Wang, M.; Nel, A.E. Nrf2 activation by sulforaphane restores the age-related decrease of TH1 immunity: Role of dendritic cells. J. Allergy Clin. Immunol. 2008, 121, 1255–1261. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wu, Y.; Xiang, X.; Lv, X.; He, M.; Xu, C.; Lai, G.; Xiang, T. Sulforaphane effectively inhibits HBV by altering Treg/Th17 immune balance and the MIF-macrophages polarizing axis in vitro and in vivo. Virus Res. 2024, 341, 199316. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, H.; Pan, C.; Li, R.; Zhao, W.; Song, T. Sulforaphane reduces adipose tissue fibrosis via promoting M2 macrophages polarization in HFD fed-mice. Biochim. Biophys. Acta Mol. Cell Res. 2024, 1871, 119626. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).