Cell-Free Nuclear and Mitochondrial DNA as Potential Biomarkers for Assessing Sepsis Severity
Abstract
1. Introduction
2. Materials and Methods
2.1. Overview
2.2. Clinical Data
2.3. Analytical Assays
2.4. Statistical Analysis
3. Results
3.1. Patients’ Characteristics
3.2. cfDNA Concentration according to Sepsis Severity
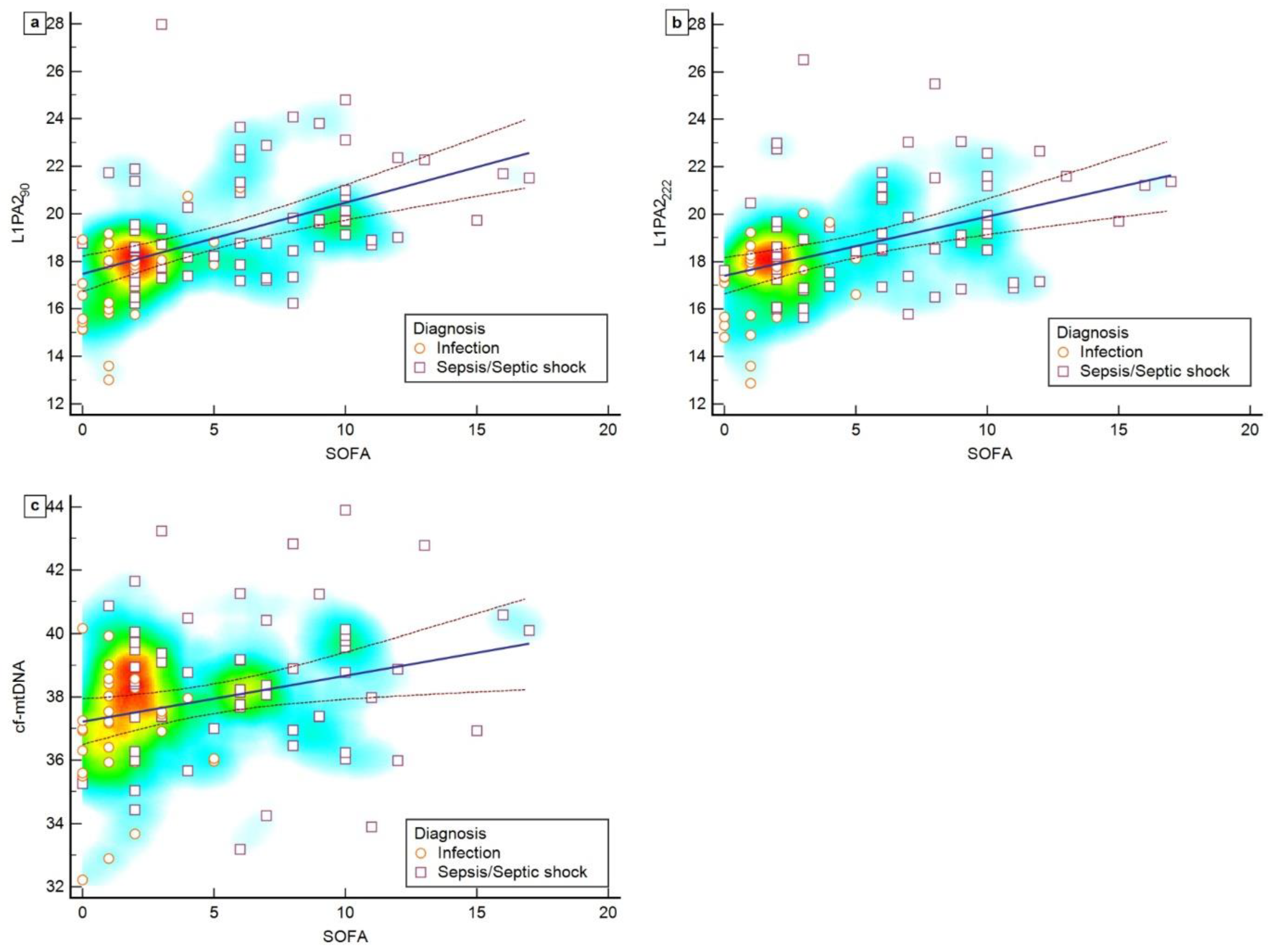
3.3. Correlation between cfDNA Levels and SOFA Scores
3.4. Potential Diagnostic Value of cfDNA
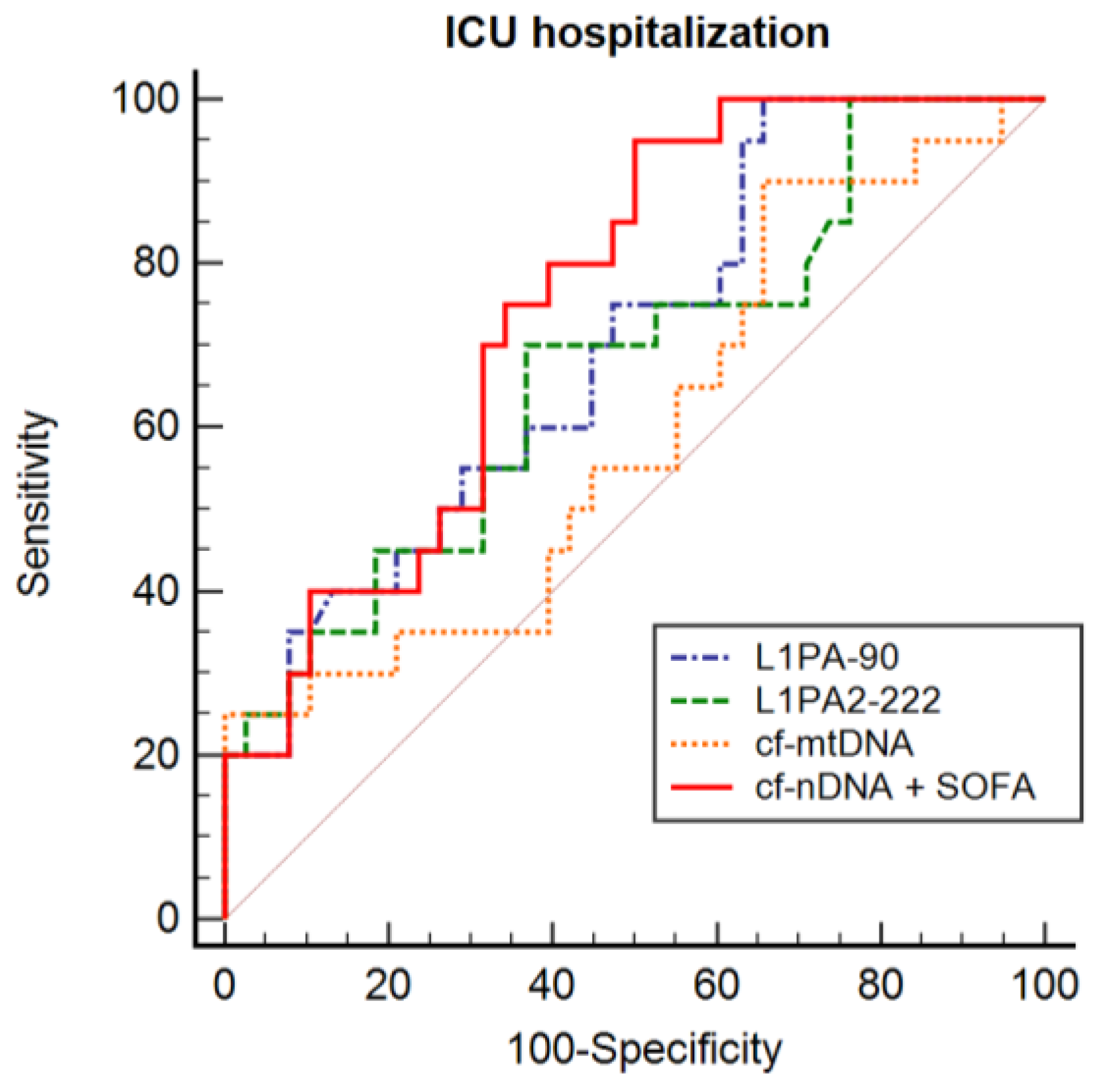
3.5. cfDNA and ICU Hospitalization
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singer, M.; Deutschman, C.S.; Seymour, C.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA J. Am. Med. Assoc. 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, Regional, and National Sepsis Incidence and Mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Machado, F.R.; Cavalcanti, A.B.; Bozza, F.A.; Ferreira, E.M.; Sousa, F.; Carrara, A.; Sousa, J.L. The Epidemiology of Sepsis in Brazilian Intensive Care Units (the Sepsis PREvalence Assessment Database, SPREAD): An Observational Study. Lancet Infect. Dis. 2017, 17, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Evans, L.; Rhodes, A.; Alhazzani, W.; Antonelli, M.; Coopersmith, C.M.; French, C.; Machado, F.R.; Mcintyre, L.; Ostermann, M.; Prescott, H.C.; et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock 2021. Intensive Care Med. 2021, 47, 1181–1247. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.B.; Jaehne, A.K.; Jayaprakash, N.; Semler, M.W.; Hegab, S.; Yataco, A.C.; Tatem, G.; Salem, D.; Moore, S.; Boka, K.; et al. Early Goal-Directed Therapy in Severe Sepsis and Septic Shock: Insights and Comparisons to ProCESS, ProMISe, and ARISE. Crit. Care 2016, 20, 160. [Google Scholar] [CrossRef] [PubMed]
- Lambden, S.; Laterre, P.F.; Levy, M.M.; Francois, B. The SOFA Score—Development, Utility and Challenges of Accurate Assessment in Clinical Trials. Crit Care 2019, 23, 374. [Google Scholar] [CrossRef] [PubMed]
- Shankar-Hari, M.; Phillips, G.S.; Levy, M.L.; Seymour, C.W.; Liu, V.X.; Deutschman, C.S.; Angus, D.C.; Rubenfeld, G.D.; Singer, M. Developing a Newdefinition and Assessing Newclinical Criteria for Septic Shock: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA J. Am. Med. Assoc. 2016, 315, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xu, R.; Zeng, Y.; Zhao, Y.; Hu, X. A Comparison of QSOFA, SIRS and NEWS in Predicting the Accuracy of Mortality in Patients with Suspected Sepsis: A Meta- Analysis. PLoS ONE 2022, 82, e0266755. [Google Scholar] [CrossRef] [PubMed]
- Adegbite, B.R.; Edoa, J.R.; Ndoumba, W.F.N.; Mbadinga, L.B.D.; Mombo-Ngoma, G.; Jacob, S.T.; Rylance, J.; Hänscheid, T.; Adegnika, A.A.; Grobusch, M.P. A Comparison of Different Scores for Diagnosis and Mortality Prediction of Adults with Sepsis in Low-and-Middle-Income Countries: A Systematic Review and Meta-Analysis. EClinicalMedicine 2021, 42, 101184. [Google Scholar] [CrossRef]
- Charoensappakit, A.; Sae-khow, K.; Rattanaliam, P.; Vutthikraivit, N.; Pecheenbuvan, M.; Udomkarnjananun, S.; Leelahavanichkul, A. Cell-Free DNA as Diagnostic and Prognostic Biomarkers for Adult Sepsis: A Systematic Review and Meta-Analysis. Sci. Rep. 2023, 13, 19624. [Google Scholar] [CrossRef]
- de Miranda, F.S.; Barauna, V.G.; Dos Santos, L.; Costa, G.; Vassallo, P.F.; Campos, L.C.G. Properties and Application of Cell-Free DNA as a Clinical Biomarker. Int. J. Mol. Sci. 2021, 22, 9110. [Google Scholar] [CrossRef] [PubMed]
- Brazil. RESOLUTION No. 466, of 12 DECEMBER 2012. 2012, No. 466. Available online: https://conselho.saude.gov.br/resolucoes/2012/466_english.pdf (accessed on 5 March 2023).
- UN. Declaration of Helsinki. Recommendations Guiding Doctors in Clinical Research. WHO Chron. 1965, 19, 31–32. [Google Scholar] [CrossRef]
- Tug, S.; Helmig, S.; Deichmann, E.R.; Schmeier-Jürchott, A.; Wagner, E.; Zimmermann, T.; Radsak, M.; Giacca, M.; Simon, P. Exercise-Induced Increases in Cell Free DNA in Human Plasma Originate Predominantly from Cells of the Haematopoietic Lineage. Exerc. Immunol. Rev. 2015, 21, 164–173. [Google Scholar]
- Tug, S.; Tross, A.; Hegen, P.; Wanja, E.; Neuberger, I.; Simon, P.; Helmig, S.; Scho, W. Acute Effects of Strength Exercises and Effects of Regular Strength Training on Cell Free DNA Concentrations in Blood Plasma. PLoS ONE 2017, 12, e0184668. [Google Scholar] [CrossRef]
- Beiter, T.; Fragasso, A.; Hudemann, J.; Nieß, A.M.; Simon, P. Short-Term Treadmill Running as a Model for Studying Cell-Free DNA Kinetics in Vivo. Clin. Chem. 2011, 57, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Meddeb, R.; Dache, Z.A.A.; Thezenas, S.; Otandault, A.; Tanos, R.; Pastor, B.; Sanchez, C.; Azzi, J.; Tousch, G.; Azan, S.; et al. Quantifying Circulating Cell-Free DNA in Humans. Sci. Rep. 2019, 9, 5220. [Google Scholar] [CrossRef] [PubMed]
- Barichello, T.; Generoso, J.S.; Singer, M.; Dal-Pizzol, F. Biomarkers for Sepsis: More than Just Fever and Leukocytosis—A Narrative Review. Crit. Care 2022, 26, 14. [Google Scholar] [CrossRef]
- Pierrakos, C.; Velissaris, D.; Bisdorff, M.; Marshall, J.C.; Vincent, J. Biomarkers of Sepsis: Time for a Reappraisal. Crit. Care 2020, 24, 287. [Google Scholar] [CrossRef]
- Pierrakos, C.; Vincent, J.L. Sepsis Biomarkers: A Review. Crit. Care 2010, 14, R15. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Ungerer, V.; Diehl, F.; Anker, P.; Dor, Y.; Fleischhacker, M.; Gahan, P.B.; Hui, L.; Holdenrieder, S.; Thierry, A.R. Towards Systematic Nomenclature for Cell-Free DNA. Hum. Genet. 2021, 140, 565–578. [Google Scholar] [CrossRef]
- Zeng, X.; Li, X.; Zhang, Y.; Cao, C.; Zhou, Q. IL6 Induces MtDNA Leakage to Affect the Immune Escape of Endometrial Carcinoma via CGAS-STING. J. Immunol. Res. 2022, 2022, 3815853. [Google Scholar] [CrossRef]
- Harrington, J.S.; Choi, A.M.K.; Nakahira, K. Mitochondrial DNA in Sepsis. Curr. Opin. Crit. Care 2017, 23, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Clementi, A.; Virzì, G.M.; Brocca, A.; Pastori, S.; De Cal, M.; Marcante, S.; Granata, A.; Ronco, C. The Role of Cell-Free Plasma DNA in Critically Ill Patients with Sepsis. Blood Purif. 2016, 41, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Duplessis, C.; Gregory, M.; Frey, K.; Bell, M.; Truong, L.; Schully, K.; Lawler, J.; Langley, R.J.; Kingsmore, S.F.; Woods, C.W.; et al. Evaluating the Discriminating Capacity of Cell Death (Apoptotic) Biomarkers in Sepsis. J. Intensive Care 2018, 5, 72. [Google Scholar] [CrossRef] [PubMed]
- Rannikko, J.; Seiskari, T.; Huttunen, R.; Tarkiainen, I.; Jylhävä, J.; Hurme, M.; Syrjänen, J.; Aittoniemi, J. Plasma Cell-Free DNA and QSOFA Score Predict 7-Day Mortality in 481 Emergency Department Bacteraemia Patients. J. Intern. Med. 2018, 284, 418–426. [Google Scholar] [CrossRef]
- Timmermans, K.; Kox, M.; Scheffer, G.J.; Pickkers, P. Plasma Nuclear and Mitochondrial Dna Levels, and Markers of Inflammation, Shock, and Organ Damage in Patients with Septic Shock. Shock 2016, 45, 607–612. [Google Scholar] [CrossRef]
- Schäfer, S.T.; Adamzik, M.; Schumak, B.; Scherag, A.; Engler, A.; Schönborn, N.; Walden, J.; Koch, S.; Baba, H.A.; Steinmann, J. Mitochondrial DNA. Anesthesiology 2016, 124, 923–933. [Google Scholar] [CrossRef]
- Bhagirath, V.C.; Dwivedi, D.J.; Liaw, P.C. Comparison of the Proinflammatory and Procoagulant Properties of Nuclear, Mitochondrial, and Bacterial DNA. Shock 2015, 44, 265–271. [Google Scholar] [CrossRef]
- Devonshire, A.S.; Whale, A.S.; Gutteridge, A.; Jones, G.; Cowen, S.; Foy, C.A.; Huggett, J.F. Towards Standardisation of Cell-Free DNA Measurement in Plasma: Controls for Extraction Efficiency, Fragment Size Bias and Quantification. Anal. Bioanal. Chem. 2014, 406, 6499–6512. [Google Scholar] [CrossRef]
- Moss, J.; Magenheim, J.; Neiman, D.; Zemmour, H.; Loyfer, N.; Korach, A.; Samet, Y.; Maoz, M.; Druid, H.; Arner, P.; et al. Comprehensive Human Cell-Type Methylation Atlas Reveals Origins of Circulating Cell-Free DNA in Health and Disease. Nat. Commun. 2018, 9, 5068. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Bakhrebah, M.A.; Alotaibi, J.; Natto, Z.S.; Alkhaibari, R.S.; Alawad, E.; Alshammari, H.M.; Alwarthan, S.; Alhajri, M.; Almogbel, M.S.; et al. Unleashing the Power of Artificial Intelligence for Diagnosing and Treating Infectious Diseases: A Comprehensive Review. J. Infect. Public Health 2023, 16, 1837–1847. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, D.; McGrath, J.; Martin-Loeches, I. Optimizing Artificial Intelligence in Sepsis Management: Opportunities in the Present and Looking Closely to the Future. J. Intensive Med. 2023, 4, 34–45. [Google Scholar] [CrossRef] [PubMed]




| All Cases (N = 94) | Infection (n = 32) | Sepsis (n = 30) | Septic Shock (n = 32) | p-Value † | |
|---|---|---|---|---|---|
| Age at diagnosis, median (IQR) | 65 (48.7–76) | 54.5 (35–68) | 61.5 (49.5–75.2) | 75 (68.2–79) | <0.0001 * |
| Male gender (%) | 53.8 | 35.5 | 53.3 | 71.9 | 0.015 * |
| All Cases (N = 94) | Infection (n = 32) | Sepsis (n = 30) | Septic Shock (n = 32) | p-Value † | |
|---|---|---|---|---|---|
| Source of diagnosis, (%) | |||||
| Emergency | 33.7 | 24.0 | 36.7 | 38.7 | |
| Internal medicine | 46.5 | 72.0 | 50.0 | 22.6 | 0.397 |
| ICU | 19.8 | 4.0 | 13.3 | 38.7 | |
| ICU hospitalization (%) | 46.9 | 14.8 | 54.2 | 70.0 | <0.0001 * |
| Length of stay, median (IQR) | 22 (10–35.5) | 11 (7–23.5) | 23.5 (10.2–36.0) | 26 (13.5–41) | 0.003 * |
| SOFA, median (IQR) | 3 (2–8) | 1 (0–2.2) | 4 (2–7.2) | 8 (5.2–10) | <0.0001 * |
| Positive qSOFA, (%) | 54.3 | 20.0 | 53.3 | 87.5 | <0.0001 * |
| Lethality, (%) | 28.0 | 9.7 | 20.0 | 53.1 | <0.0001 * |
| Comorbidities, (%) | |||||
| Hypertension | 45.7 | 31.3 | 33.3 | 71.9 | 0.001 * |
| Diabetes Mellitus | 24.5 | 12.5 | 26.7 | 34.4 | 0.115 |
| Cardiopathy | 28.7 | 18.8 | 16.7 | 50.0 | 0.007 * |
| Nephropathy | 12.8 | 18.8 | 3.3 | 15.6 | 0.164 |
| Hepatopathy | 7.4 | 3.1 | 13.3 | 6.3 | 0.293 |
| Neoplasia | 7.4 | 3.1 | 13.3 | 6.3 | 0.293 |
| Others | 21.3 | 18.8 | 16.7 | 28.1 | 0.542 |
| None | 24.5 | 37.5 | 30.0 | 6.3 | 0.006 * |
| Nosocomial infection, (%) | 60.5 | 57.7 | 48.0 | 73.3 | 0.156 |
| Focus of infection, (%) | |||||
| Unknown | 31.9 | 31.3 | 26.7 | 37.5 | 0.673 |
| Abdominal | 14.9 | 12.5 | 16.7 | 15.6 | 0.936 |
| Respiratory | 26.6 | 15.6 | 36.7 | 28.1 | 0.168 |
| Genitourinary tract | 17.0 | 28.1 | 10.0 | 12.5 | 0.158 |
| Skin-soft tissue/bone-joint | 6.4 | 9.4 | 6.7 | 3.1 | 0.687 |
| Catheter | 3.2 | 3.1 | 3.3 | 3.1 | 1.000 |
| All Cases (N = 94) | Infection (n = 32) | Sepsis (n = 30) | Septic Shock (n = 32) | p-Value † | |
|---|---|---|---|---|---|
| median (IQR) | |||||
| Leukocyte (WBCs/mm3) | 12,780 (8520–17,830) | 11,270 (9040–16,010) | 14,235 (6647–19,995) | 13,060 (7464–21,060) | 0.912 |
| Platelet (×103/mm3) | 224 (130–300) | 244 (154–312) | 248 (131–409) | 182 (108.5–259) | 0.184 |
| INR | 1.18 (1.03–1.35) | 1.08 (1.00–1.21) | 1.18 (1.03–1.40) | 1.25 (1.13–1.53) | 0.014 * |
| Total bilirubin (mg/dL) | 0.90 (0.52–1.79) | 0.58 (0.36–1.05) | 0.97 (0.49–5.02) | 1.11 (0.69–1.82) | 0.023 * |
| C-reactive protein (mg/L) | 112.5 (64.4–128.8) | 118.1 (50.1–132.0) | 111.6 (55.2–127.3) | 110.3 (66.1–129.1) | 0.972 |
| Creatinine (mg/dL) | 1.49 (0.74–2.32) | 0.78 (0.63–1.39) | 1.5 (0.78–2.45) | 1.83 (1.07–3.11) | 0.012 * |
| Lactate (mmol/L) | 1.9 (1.2–2.9) | 1.6 (1.1–2.5) | 1.7 (1.1–3.6) | 2.45 (1.8–3.5) | 0.030 * |
| cfDNA | Cut-Off Value ** | AUC (95% CI) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| L1PA290 | 18.07 | 0.817 (0.725–0.909) * | 77.0 | 79.3 |
| L1PA2222 | 18.27 | 0.741 (0.634–0.849) * | 66.6 | 81.4 |
| cf-mtDNA | 38.58 | 0.703 (0.596–0.810) * | 48.2 | 90.6 |
| cf-nDNA + SOFA | - | 0.916 (0.853–0.979) * | 88.1 | 80.0 |
| cfDNA | Cut-Off Value ** | AUC (95% CI) | Sensitivity z (%) | Specificity (%) |
|---|---|---|---|---|
| L1PA290 | 17.17 | 0.692 (0.559–0.806) * | 100.0 | 34.2 |
| L1PA2222 | 18.11 | 0.653 (0.520–0.770) * | 66.6 | 62.5 |
| cf-mtDNA | 40.17 | 0.596 (0.463–0.720) | 25.0 | 100.0 |
| cf-nDNA + SOFA | - | 0.752 (0.622–0.855) * | 95.2 | 50.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Miranda, F.S.; Claudio, L.M.A.M.; de Almeida, D.S.M.; Nunes, J.B.; Barauna, V.G.; Luiz, W.B.; Vassallo, P.F.; Campos, L.C.G. Cell-Free Nuclear and Mitochondrial DNA as Potential Biomarkers for Assessing Sepsis Severity. Biomedicines 2024, 12, 933. https://doi.org/10.3390/biomedicines12050933
de Miranda FS, Claudio LMAM, de Almeida DSM, Nunes JB, Barauna VG, Luiz WB, Vassallo PF, Campos LCG. Cell-Free Nuclear and Mitochondrial DNA as Potential Biomarkers for Assessing Sepsis Severity. Biomedicines. 2024; 12(5):933. https://doi.org/10.3390/biomedicines12050933
Chicago/Turabian Stylede Miranda, Felipe Silva, Livia Maria A. M. Claudio, Dayanne Silva M. de Almeida, Juliana Braga Nunes, Valério Garrone Barauna, Wilson Barros Luiz, Paula Frizzera Vassallo, and Luciene Cristina Gastalho Campos. 2024. "Cell-Free Nuclear and Mitochondrial DNA as Potential Biomarkers for Assessing Sepsis Severity" Biomedicines 12, no. 5: 933. https://doi.org/10.3390/biomedicines12050933
APA Stylede Miranda, F. S., Claudio, L. M. A. M., de Almeida, D. S. M., Nunes, J. B., Barauna, V. G., Luiz, W. B., Vassallo, P. F., & Campos, L. C. G. (2024). Cell-Free Nuclear and Mitochondrial DNA as Potential Biomarkers for Assessing Sepsis Severity. Biomedicines, 12(5), 933. https://doi.org/10.3390/biomedicines12050933






