Time- and Region-Specific Selection of Reference Genes in the Rat Brain in the Lithium–Pilocarpine Model of Acquired Temporal Lobe Epilepsy
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and the Lithium–Pilocarpine Model of Temporal Lobe Epilepsy
2.2. Reverse Transcription Followed by Quantitative PCR (RT-qPCR)
2.3. Statistical Analysis
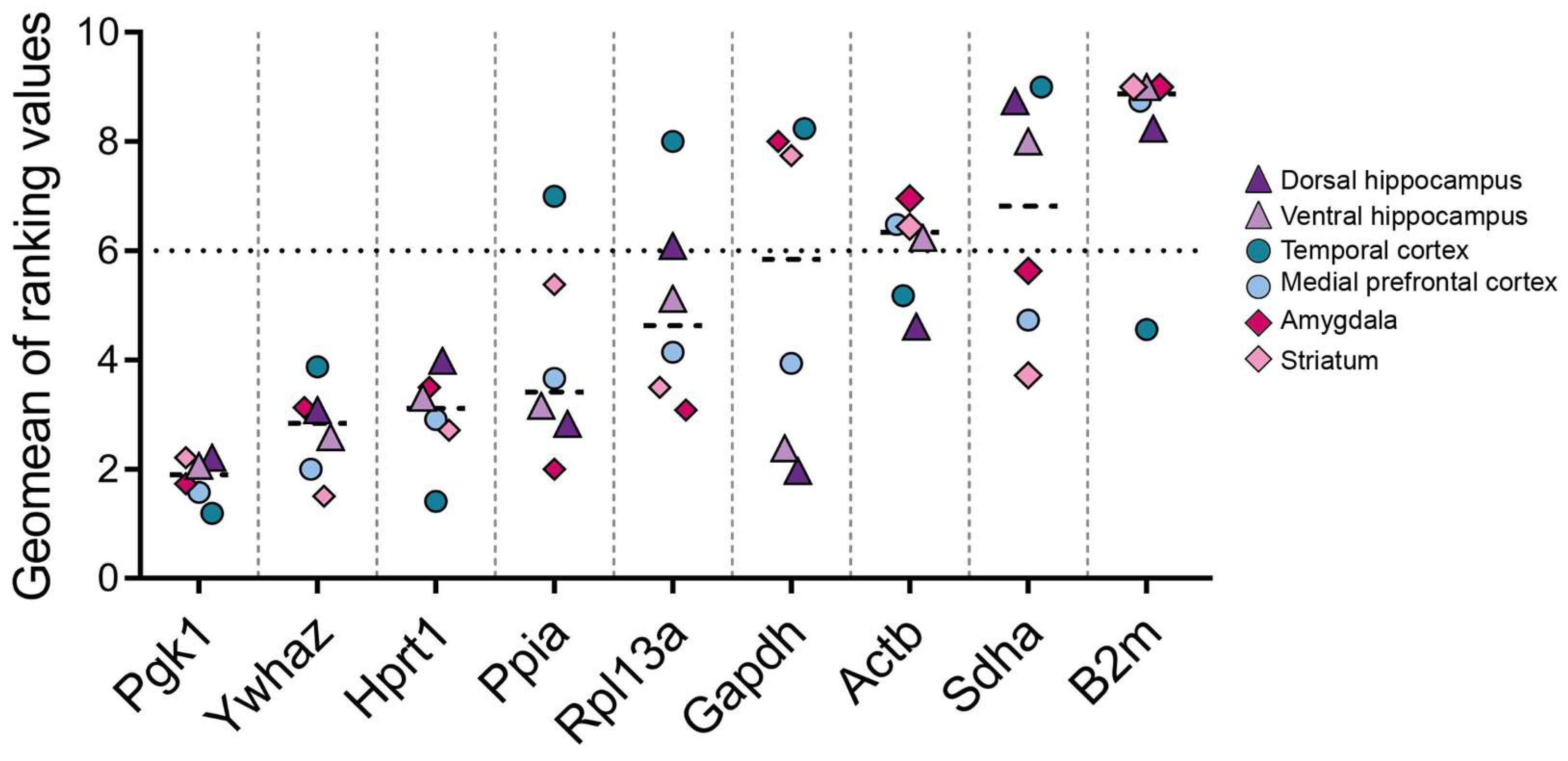
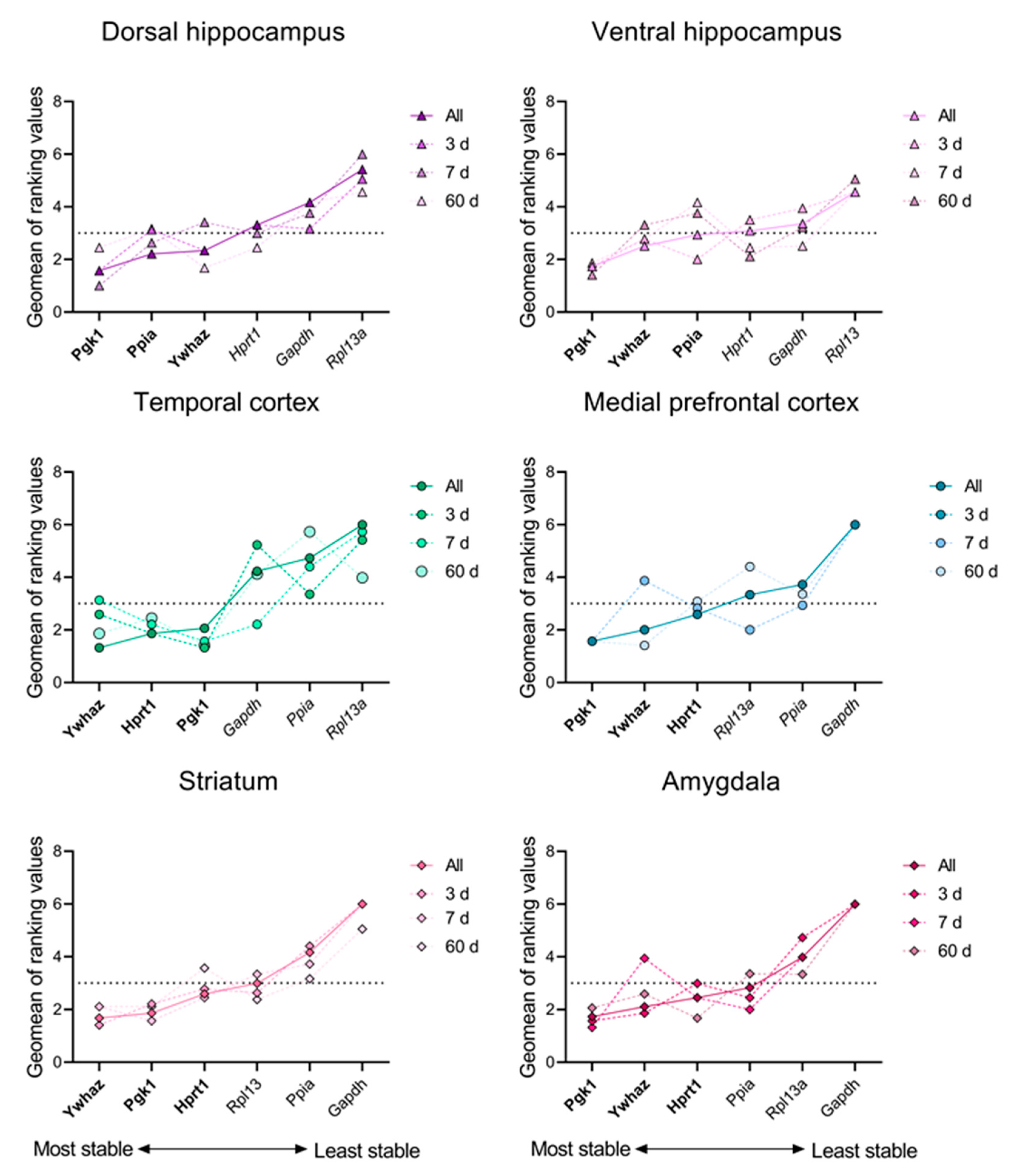
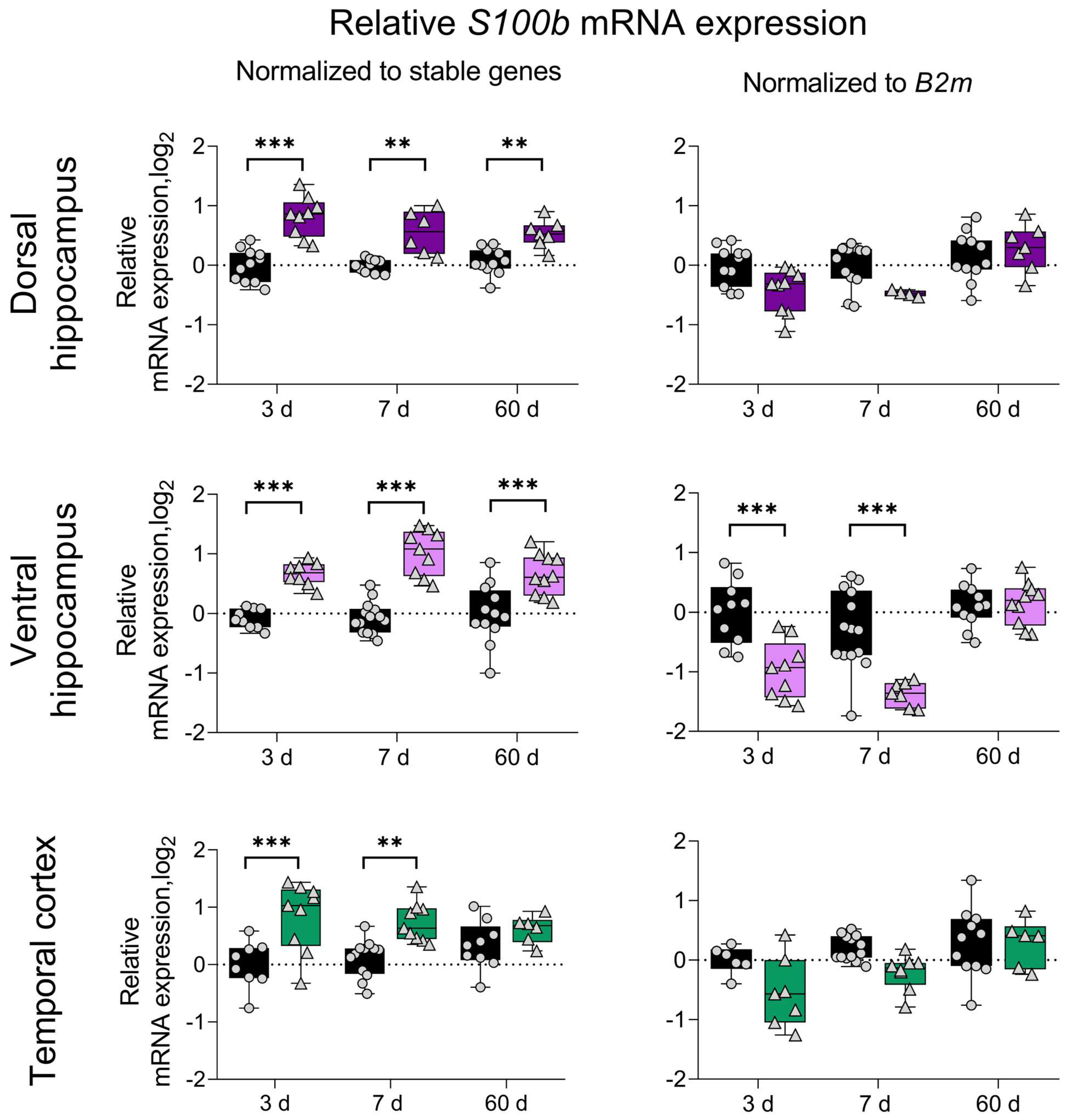
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Amygdala | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Delta CT | BestKeeper | NormFinder | GeNorm | Comprehensive Ranking | ||||||
| Rank | Gene | Average of STDV | Gene | Std Dev | Gene | Stability Value | Gene | Stability Value | Gene | Geomean of Ranking Values |
| 1 | Pgk1 | 0.32 | Ppia | 0.19 | Pgk1 | 0.089 | Hprt1 | Ywhaz | 0.215 | Pgk1 | 1.73 |
| 2 | Ywhaz | 0.35 | Rpl13a | 0.21 | Ywhaz | 0.203 | Ywhaz | 2.11 | ||
| 3 | Hprt1 | 0.35 | Pgk1 | 0.24 | Hprt1 | 0.216 | Pgk1 | 0.228 | Hprt1 | 2.45 |
| 4 | Ppia | 0.36 | Hprt1 | 0.31 | Ppia | 0.228 | Ppia | 0.268 | Ppia | 2.83 |
| 5 | Rpl13a | 0.42 | Ywhaz | 0.33 | Rpl13a | 0.328 | Rpl13a | 0.31 | Rpl13a | 3.98 |
| 6 | Gapdh | 0.56 | Gapdh | 0.44 | Gapdh | 0.512 | Gapdh | 0.392 | Gapdh | 6 |
| Medial Prefrontal Cortex | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Delta CT | BestKeeper | NormFinder | GeNorm | Comprehensive Ranking | ||||||
| Rank | Gene | Average of STDV | Gene | Std Dev | Gene | Stability Value | Gene | Stability Value | Gene | Geomean of Ranking Values |
| 1 | Pgk1 | 0.36 | Rpl13a | 0.23 | Pgk1 | 0.122 | Hprt1 | Ywhaz | 0.175 | Pgk1 | 1.57 |
| 2 | Ywhaz | 0.37 | Pgk1 | 0.24 | Ywhaz | 0.165 | Ywhaz | 2 | ||
| 3 | Hprt1 | 0.39 | Ppia | 0.25 | Hprt1 | 0.237 | Pgk1 | 0.193 | Hprt1 | 2.59 |
| 4 | Ppia | 0.43 | Ywhaz | 0.32 | Ppia | 0.289 | Ppia | 0.299 | Rpl13a | 3.34 |
| 5 | Rpl13a | 0.49 | Hprt1 | 0.37 | Rpl13a | 0.411 | Rpl13a | 0.351 | Ppia | 3.72 |
| 6 | Gapdh | 0.63 | Gapdh | 0.52 | Gapdh | 0.58 | Gapdh | 0.445 | Gapdh | 6 |
| Striatum | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Delta CT | BestKeeper | NormFinder | GeNorm | Comprehensive Ranking | ||||||
| Rank | Gene | Average of STDV | Gene | Std Dev | Gene | Stability Value | Gene | Stability Value | Gene | Geomean of Ranking Values |
| 1 | Ywhaz | 0.33 | Rpl13a | 0.27 | Pgk1 | 0.154 | Hprt1 | Ywhaz | 0.164 | Ywhaz | 1.68 |
| 2 | Pgk1 | 0.34 | Pgk1 | 0.29 | Ywhaz | 0.164 | Pgk1 | 1.86 | ||
| 3 | Hprt1 | 0.35 | Ppia | 0.36 | Hprt1 | 0.231 | Pgk1 | 0.232 | Hprt1 | 2.59 |
| 4 | Rpl13a | 0.39 | Ywhaz | 0.4 | Rpl13a | 0.289 | Ppia | 0.312 | Rpl13 | 2.99 |
| 5 | Ppia | 0.4 | Hprt1 | 0.44 | Ppia | 0.315 | Rpl13a | 0.327 | Ppia | 4.16 |
| 6 | Gapdh | 0.5 | Gapdh | 0.48 | Gapdh | 0.446 | Gapdh | 0.385 | Gapdh | 6 |
| Temporal Cortex | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Delta CT | BestKeeper | NormFinder | GeNorm | Comprehensive Ranking | ||||||
| Rank | Gene | Average of STDV | Gene | Std Dev | Gene | Stability Value | Gene | Stability Value | Gene | Geomean of Ranking Values |
| 1 | Ywhaz | 1.36 | Pgk1 | 0.45 | Ywhaz | 0.661 | Hprt1 | Ywhaz | 0.239 | Ywhaz | 1.32 |
| 2 | Pgk1 | 1.39 | Hprt1 | 0.5 | Hprt1 | 0.74 | Hprt1 | 1.86 | ||
| 3 | Hprt1 | 1.41 | Ywhaz | 0.53 | Pgk1 | 0.776 | Pgk1 | 0.318 | Pgk1 | 2.06 |
| 4 | Gapdh | 1.73 | Ppia | 0.67 | Gapdh | 1.284 | Gapdh | 0.636 | Gapdh | 4.23 |
| 5 | Ppia | 2.29 | Gapdh | 0.72 | Ppia | 1.783 | Ppia | 1.332 | Ppia | 4.73 |
| 6 | Rpl13a | 2.86 | Rpl13a | 1 | Rpl13a | 2.674 | Rpl13a | 1.841 | Rpl13a | 6 |
| Dorsal Hippocampus | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Delta CT | BestKeeper | NormFinder | GeNorm | Comprehensive Ranking | ||||||
| Rank | Gene | Average of STDV | Gene | Std Dev | Gene | Stability Value | Gene | Stability Value | Gene | Geomean of Ranking Values |
| 1 | Pgk1 | 0.26 | Ppia | 0.2 | Pgk1 | 0.115 | Hprt1 | Ywhaz | 0.134 | Pgk1 | 1.57 |
| 2 | Ywhaz | 0.27 | Pgk1 | 0.2 | Ppia | 0.153 | Ppia | 2.21 | ||
| 3 | Ppia | 0.28 | Gapdh | 0.21 | Ywhaz | 0.18 | Pgk1 | 0.168 | Ywhaz | 2.34 |
| 4 | Hprt1 | 0.29 | Rpl13a | 0.25 | Gapdh | 0.22 | Ppia | 0.218 | Hprt1 | 3.31 |
| 5 | Gapdh | 0.32 | Ywhaz | 0.29 | Hprt1 | 0.225 | Gapdh | 0.252 | Gapdh | 4.16 |
| 6 | Rpl13a | 0.41 | Hprt1 | 0.3 | Rpl13a | 0.373 | Rpl13a | 0.305 | Rpl13a | 5.42 |
| Ventral Hippocampus | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Delta CT | BestKeeper | NormFinder | GeNorm | Comprehensive Ranking | ||||||
| Rank | Gene | Average of STDV | Gene | Std Dev | Gene | Stability Value | Gene | Stability Value | Gene | Geomean of Ranking Values |
| 1 | Pgk1 | 0.41 | Ppia | 0.32 | Pgk1 | 0.089 | Hprt1 | Ywhaz | 0.203 | Pgk1 | 1.73 |
| 2 | Ywhaz | 0.46 | Rpl13 | 0.33 | Gapdh | 0.297 | Ywhaz | 2.51 | ||
| 3 | Hprt1 | 0.48 | Gapdh | 0.34 | Ppia | 0.303 | Pgk1 | 0.302 | Ppia | 2.94 |
| 4 | Gapdh | 0.48 | Pgk1 | 0.34 | Ywhaz | 0.333 | Gapdh | 0.356 | Hprt1 | 3.08 |
| 5 | Ppia | 0.49 | Ywhaz | 0.49 | Hprt1 | 0.352 | Ppia | 0.399 | Gapdh | 3.36 |
| 6 | Rpl13a | 0.71 | Hprt1 | 0.5 | Rpl13 | 0.66 | Rpl13a | 0.504 | Rpl13a | 4.56 |
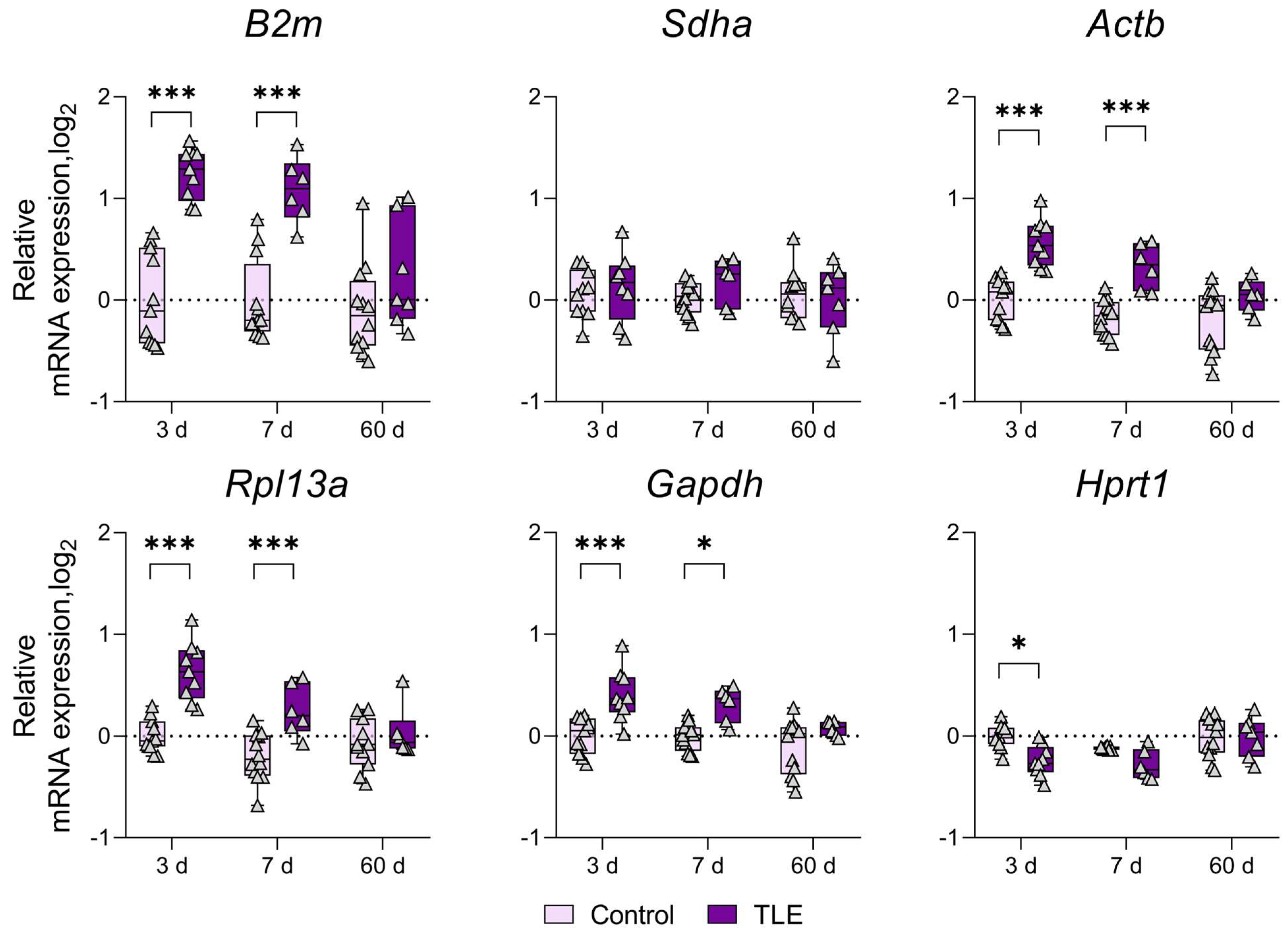
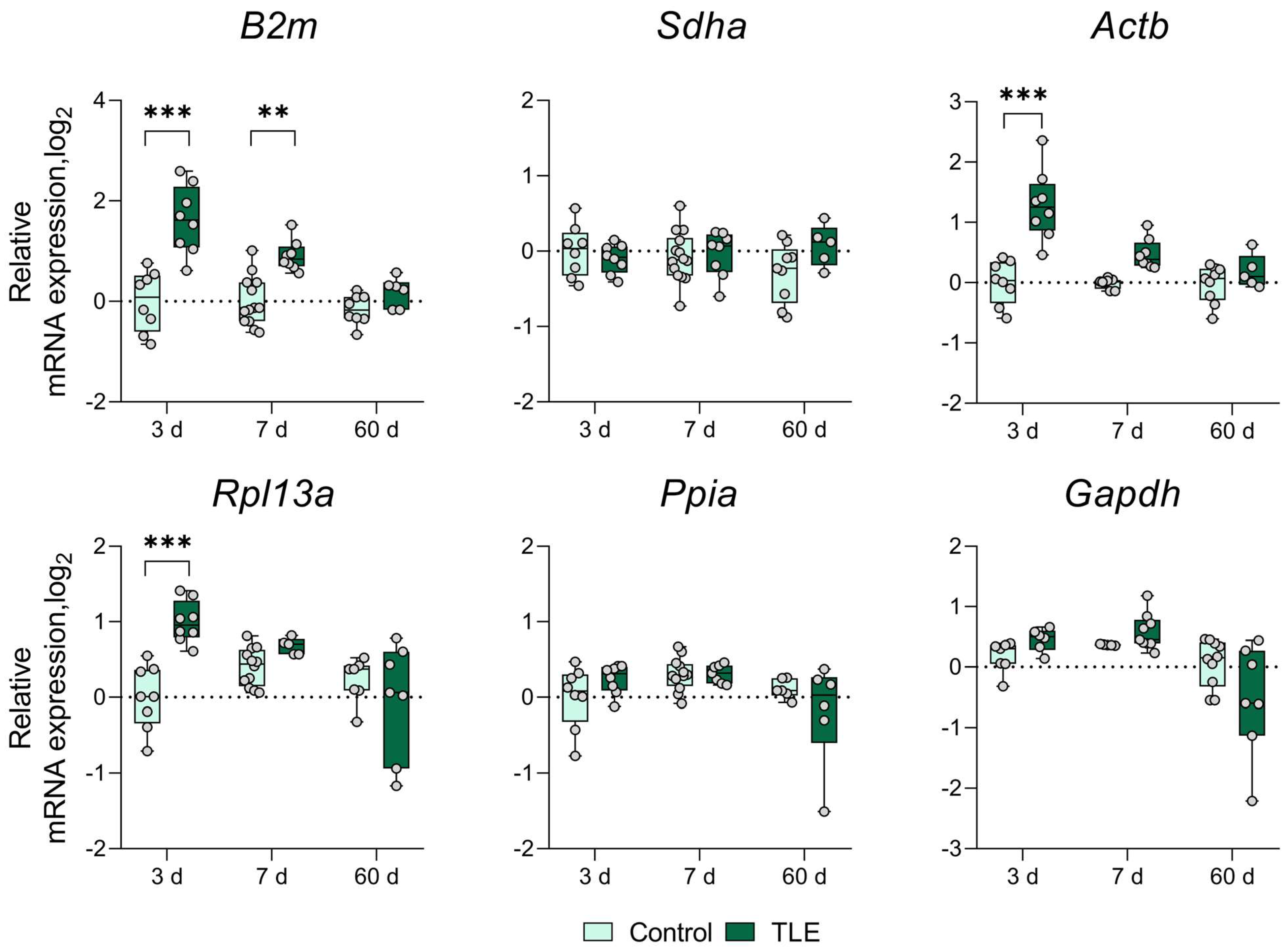
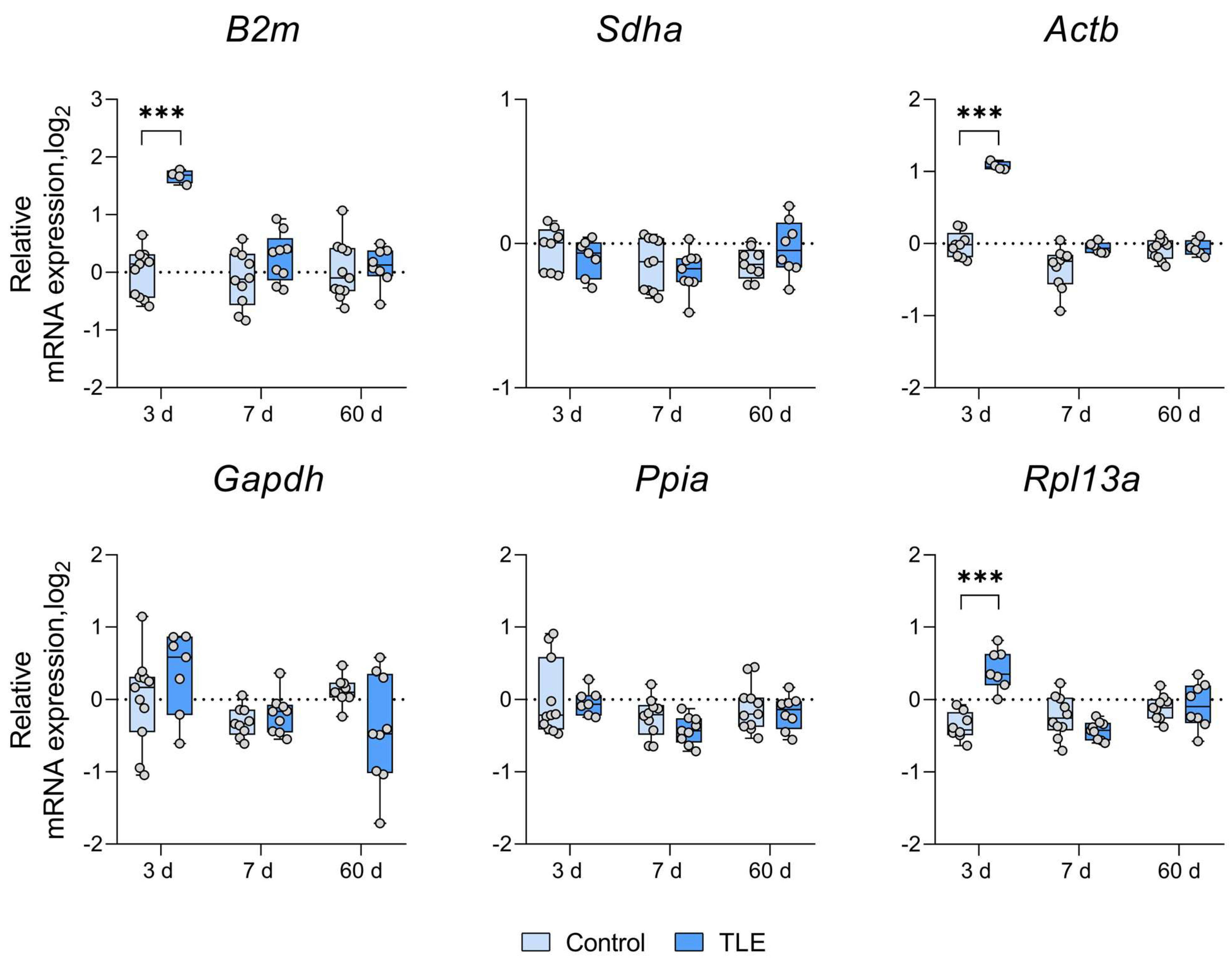

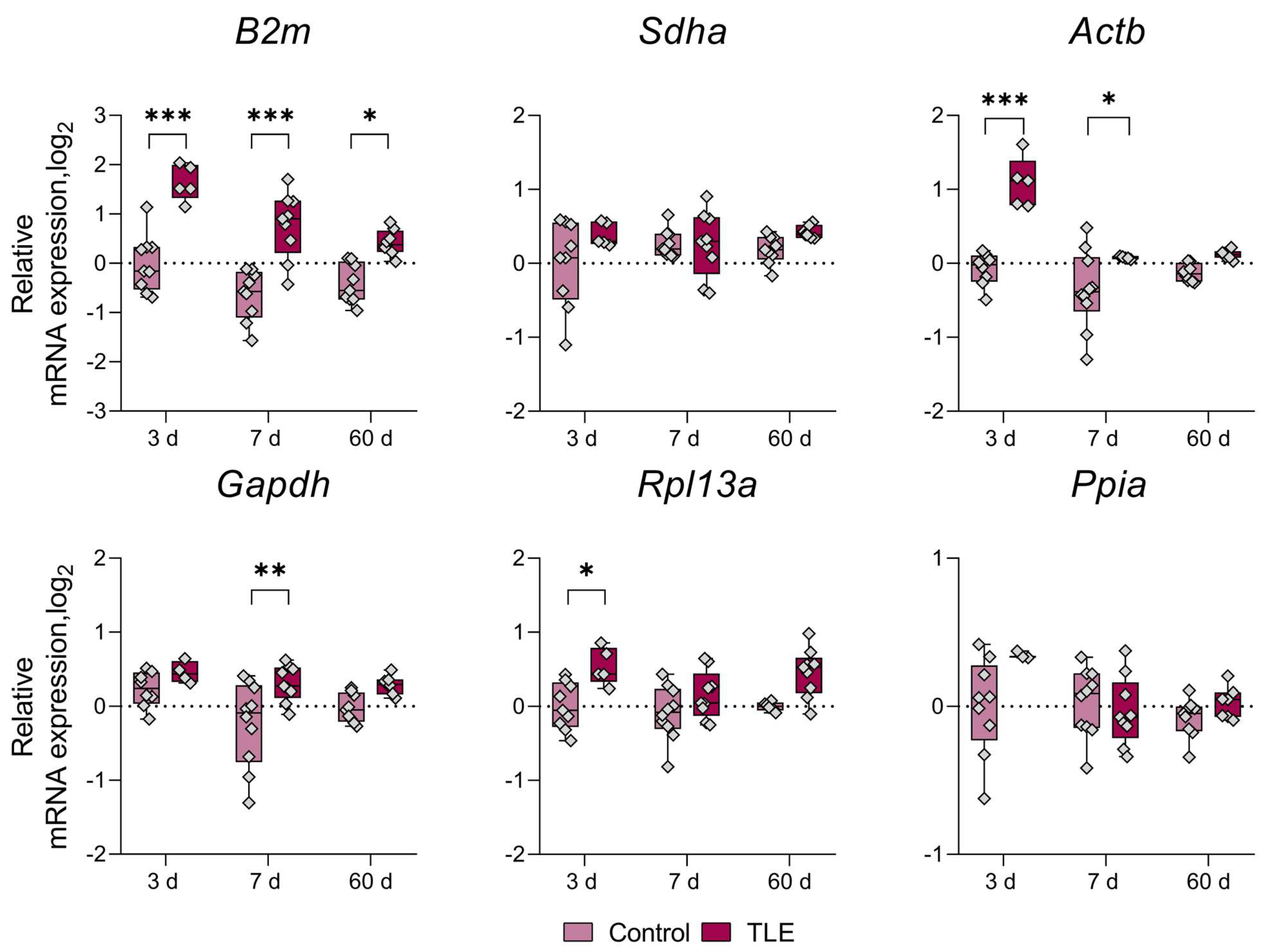
References
- Banerjee, P.N.; Filippi, D.; Allen Hauser, W. The descriptive epidemiology of epilepsy—A review. Epilepsy Res. 2009, 85, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Laxer, K.D.; Trinka, E.; Hirsch, L.J.; Cendes, F.; Langfitt, J.; Delanty, N.; Resnick, T.; Benbadis, S.R. The consequences of refractory epilepsy and its treatment. Epilepsy Behav. 2014, 37, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Löscher, W. Animal Models of Seizures and Epilepsy: Past, Present, and Future Role for the Discovery of Antiseizure Drugs. Neurochem. Res. 2017, 42, 1873–1888. [Google Scholar] [CrossRef] [PubMed]
- Lévesque, M.; Biagini, G.; de Curtis, M.; Gnatkovsky, V.; Pitsch, J.; Wang, S.; Avoli, M. The pilocarpine model of mesial temporal lobe epilepsy: Over one decade later, with more rodent species and new investigative approaches. Neurosci. Biobehav. Rev. 2021, 130, 274–291. [Google Scholar] [CrossRef] [PubMed]
- Kandratavicius, L.; Alves Balista, P.; Lopes-Aguiar, C.; Ruggiero, R.N.; Umeoka, E.H.; Garcia-Cairasco, N.; Bueno-Junior, L.S.; Leite, J.P. Animal models of epilepsy: Use and limitations. Neuropsychiatr. Dis. Treat. 2014, 10, 1693–1705. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Chapman, J.R.; Waldenström, J. With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef]
- Schwarz, A.P.; Dyomina, A.V.; Zakharova, M.V.; Kovalenko, A.A.; Gryaznova, M.O.; Ischenko, A.M.; Zaitsev, A.V. The Reference Gene Validation in the Brain of Rats during Antioxidant and Anti-Inflammatory Treatment in the Lithium-Pilocarpine Model of Temporal Epilepsy. J. Evol. Biochem. Physiol. 2022, 58, 930–940. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, X.; Ren, J.; Wang, P.; Zhang, J.; Wei, Z.; Tian, Y. Validation of reference genes for quantitative real-time PCR in valproic acid rat models of autism. Mol. Biol. Rep. 2016, 43, 837–847. [Google Scholar] [CrossRef]
- Sadangi, C.; Rosenow, F.; Norwood, B.A. Validation of reference genes for quantitative gene expression analysis in experimental epilepsy. J. Neurosci. Res. 2017, 95, 2357–2366. [Google Scholar] [CrossRef]
- Schwarz, A.P.; Malygina, D.A.; Kovalenko, A.A.; Trofimov, A.N.; Zaitsev, A.V. Multiplex qPCR assay for assessment of reference gene expression stability in rat tissues/samples. Mol. Cell. Probes 2020, 53, 101611. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, A.P.; Nikitina, V.A.; Krytskaya, D.U.; Shcherbakova, K.P.; Trofimov, A.N. Reference gene expression stability within the rat brain under mild intermittent ketosis induced by supplementation with medium-chain triglycerides. PLoS ONE 2023, 18, e0273224. [Google Scholar] [CrossRef]
- Canto, A.M.; Godoi, A.B.; Matos, A.H.B.; Geraldis, J.C.; Rogerio, F.; Alvim, M.K.M.; Yasuda, C.L.; Ghizoni, E.; Tedeschi, H.; Veiga, D.F.T.; et al. Benchmarking the proteomic profile of animal models of mesial temporal epilepsy. Ann. Clin. Transl. Neurol. 2022, 9, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Pires, G.; Leitner, D.; Drummond, E.; Kanshin, E.; Nayak, S.; Askenazi, M.; Faustin, A.; Friedman, D.; Debure, L.; Ueberheide, B.; et al. Proteomic differences in the hippocampus and cortex of epilepsy brain tissue. Brain Commun. 2021, 3, fcab021. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, L.; Rizvanov, A.A.; Salafutdinov, I.I.; Dabirmanesh, B.; Sayyah, M.; Fathollahi, Y.; Khajeh, K. Hippocampal asymmetry: Differences in the left and right hippocampus proteome in the rat model of temporal lobe epilepsy. J. Proteomics 2017, 154, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, A.A.; Zakharova, M.V.; Schwarz, A.P.; Dyomina, A.V.; Zubareva, O.E.; Zaitsev, A.V. Changes in Metabotropic Glutamate Receptor Gene Expression in Rat Brain in a Lithium-Pilocarpine Model of Temporal Lobe Epilepsy. Int. J. Mol. Sci. 2022, 23, 2752. [Google Scholar] [CrossRef] [PubMed]
- Racine, R.J. Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalogr. Clin. Neurophysiol. 1972, 32, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Dyomina, A.V.; Zubareva, O.E.; Smolensky, I.V.; Vasilev, D.S.; Zakharova, M.V.; Kovalenko, A.A.; Schwarz, A.P.; Ischenko, A.M.; Zaitsev, A.V. Anakinra Reduces Epileptogenesis, Provides Neuroprotection, and Attenuates Behavioral Impairments in Rats in the Lithium-Pilocarpine Model of Epilepsy. Pharmaceuticals 2020, 13, 340. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genomics 2023, 23, 125. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Rektor, I.; Kuba, R.; Brázdil, M.; Chrastina, J. Do the basal ganglia inhibit seizure activity in temporal lobe epilepsy? Epilepsy Behav. 2012, 25, 56–59. [Google Scholar] [CrossRef]
- Gao, Y.; Hong, Y.; Huang, L.; Zheng, S.; Zhang, H.; Wang, S.; Yao, Y.; Zhao, Y.; Zhu, L.; Xu, Q.; et al. β2-microglobulin functions as an endogenous NMDAR antagonist to impair synaptic function. Cell 2023, 186, 1026–1038.e20. [Google Scholar] [CrossRef]
- Schwarz, A.P.; Kovalenko, A.A.; Malygina, D.A.; Postnikova, T.Y.; Zubareva, O.E.; Zaitsev, A.V. Reference Gene Validation in the Brain Regions of Young Rats after Pentylenetetrazole-Induced Seizures. Biomedicines 2020, 8, 239. [Google Scholar] [CrossRef]
- Stassen, Q.E.M.; Riemers, F.M.; Reijmerink, H.; Leegwater, P.A.J.; Penning, L.C. Reference genes for reverse transcription quantitative PCR in canine brain tissue. BMC Res. Notes 2015, 8, 761. [Google Scholar] [CrossRef][Green Version]
- Gan, Y.; Ye, F.; He, X.X. The role of YWHAZ in cancer: A maze of opportunities and challenges. J. Cancer 2020, 11, 2252–2264. [Google Scholar] [CrossRef]
- Aroniadou-Anderjaska, V.; Fritsch, B.; Qashu, F.; Braga, M.F.M. Pathology and pathophysiology of the amygdala in epileptogenesis and epilepsy. Epilepsy Res. 2008, 78, 102–116. [Google Scholar] [CrossRef] [PubMed]
- Fanselow, M.S.; Dong, H.-W. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron 2010, 65, 7–19. [Google Scholar] [CrossRef] [PubMed]
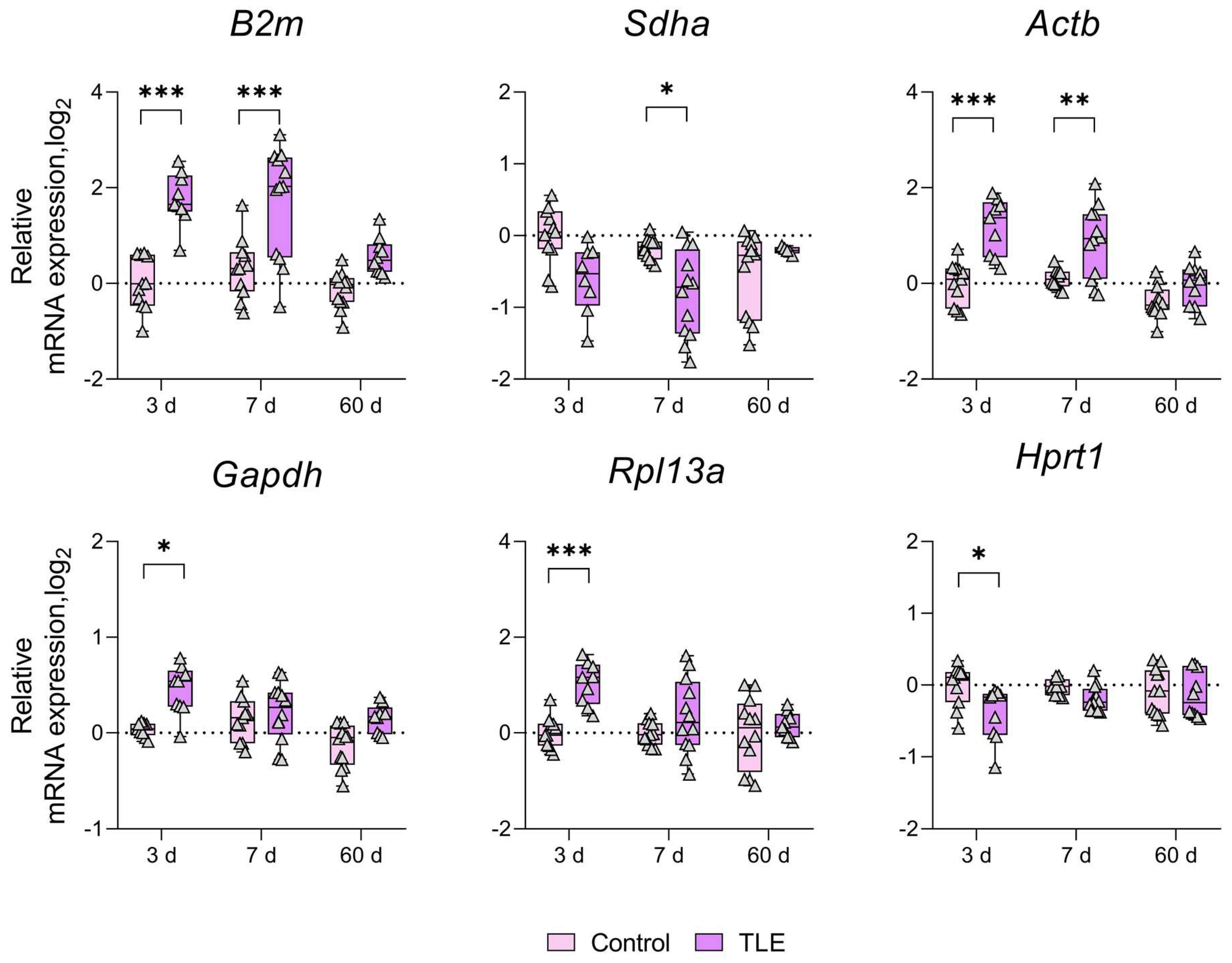
| Gene | B2m | Sdha | Actb | Gapdh | Rpl13a | Ppia | Hprt1 | |
|---|---|---|---|---|---|---|---|---|
| Brain Region | ||||||||
| Temporal Cortex | ↑ 3 d | ↑ 3 d | ↑ 3 d | * | ||||
| ↑ 7 d | ||||||||
| Ventral Hippocampus | ↑ 3 d | ↑ 3 d | ↑ 3 d | ↑ 3 d | * | ↓ 3 d | ||
| ↑ 7 d | ↓ 7 d | ↑ 7 d | ||||||
| Dorsal Hippocampus | ↑ 3 d | ↑ 3 d | ↑ 3 d | ↑ 3 d | * | ↓ 3 d | ||
| ↑ 7 d | ↑ 7 d | ↑ 7 d | ↑ 7 d | |||||
| Striatum | ↑ 3 d | ↑ 3 d | * | |||||
| Amygdala | ↑ 3 d | ↑ 3 d | ↑ 3 d | * | ||||
| ↑ 7 d | ↑ 7 d | ↑ 7 d | ||||||
| ↑ 60 d | ||||||||
| Medial Prefrontal Cortex | ↑ 3 d | ↑ 3 d | ↑ 3 d | * | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schwarz, A.P.; Zakharova, M.V.; Kovalenko, A.A.; Dyomina, A.V.; Zubareva, O.E.; Zaitsev, A.V. Time- and Region-Specific Selection of Reference Genes in the Rat Brain in the Lithium–Pilocarpine Model of Acquired Temporal Lobe Epilepsy. Biomedicines 2024, 12, 1100. https://doi.org/10.3390/biomedicines12051100
Schwarz AP, Zakharova MV, Kovalenko AA, Dyomina AV, Zubareva OE, Zaitsev AV. Time- and Region-Specific Selection of Reference Genes in the Rat Brain in the Lithium–Pilocarpine Model of Acquired Temporal Lobe Epilepsy. Biomedicines. 2024; 12(5):1100. https://doi.org/10.3390/biomedicines12051100
Chicago/Turabian StyleSchwarz, Alexander P., Maria V. Zakharova, Anna A. Kovalenko, Alexandra V. Dyomina, Olga E. Zubareva, and Aleksey V. Zaitsev. 2024. "Time- and Region-Specific Selection of Reference Genes in the Rat Brain in the Lithium–Pilocarpine Model of Acquired Temporal Lobe Epilepsy" Biomedicines 12, no. 5: 1100. https://doi.org/10.3390/biomedicines12051100
APA StyleSchwarz, A. P., Zakharova, M. V., Kovalenko, A. A., Dyomina, A. V., Zubareva, O. E., & Zaitsev, A. V. (2024). Time- and Region-Specific Selection of Reference Genes in the Rat Brain in the Lithium–Pilocarpine Model of Acquired Temporal Lobe Epilepsy. Biomedicines, 12(5), 1100. https://doi.org/10.3390/biomedicines12051100






