Functions of Differentially Regulated miRNAs in Breast Cancer Progression: Potential Markers for Early Detection and Candidates for Therapy
Abstract
1. Introduction
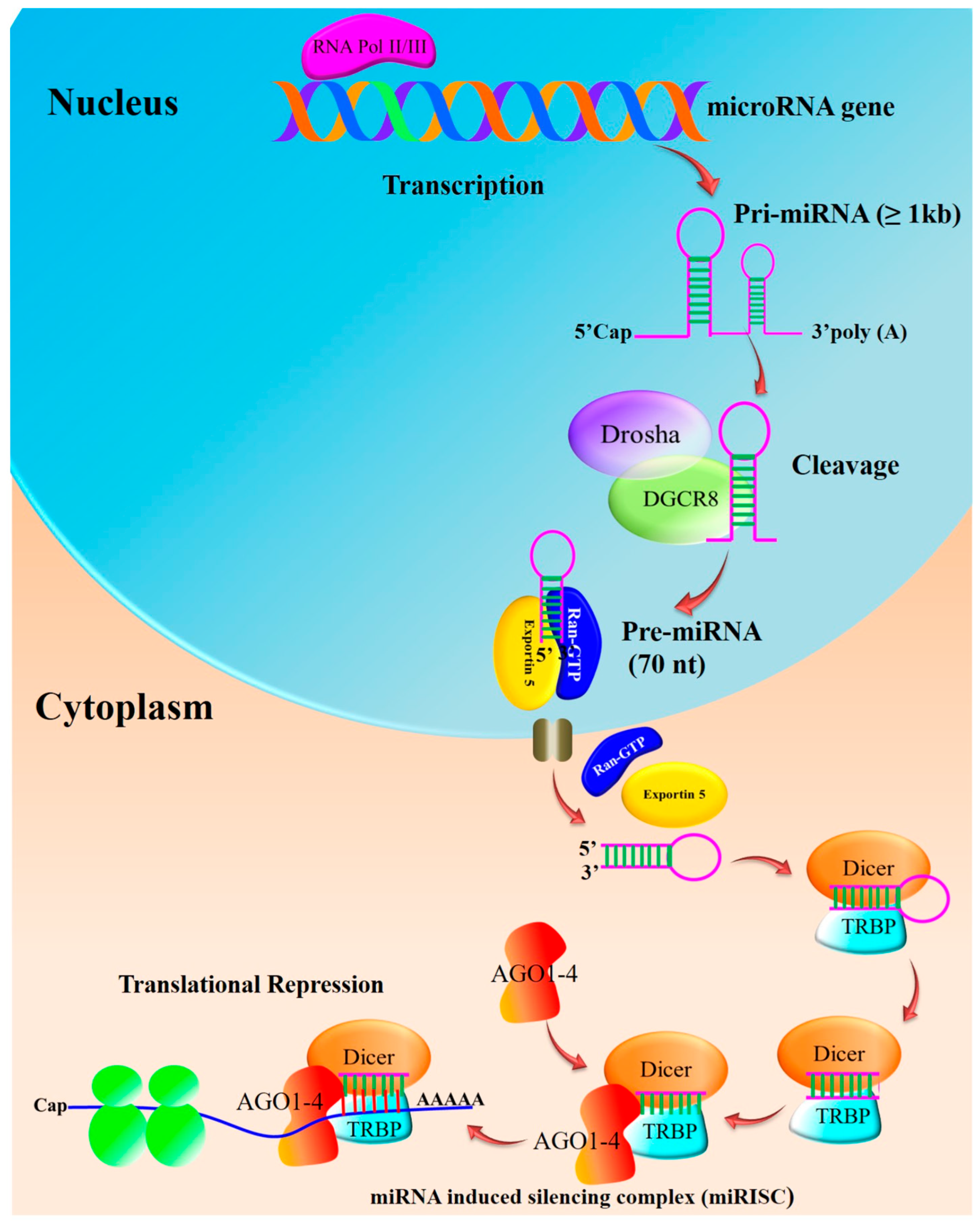
2. miRNA Biogenesis and Maturation
3. miRNA Targets and Their Role in Breast Cancer
- Oncogenic miRNAs (OncomiRs): these miRNAs are often upregulated in breast cancer and promote tumorigenesis by inhibiting tumor-suppressor genes or regulatory pathways.
- Tumor-Suppressive miRNAs: conversely, these miRNAs are downregulated in breast cancer and typically act to inhibit oncogenes or other pro-tumorigenic processes.
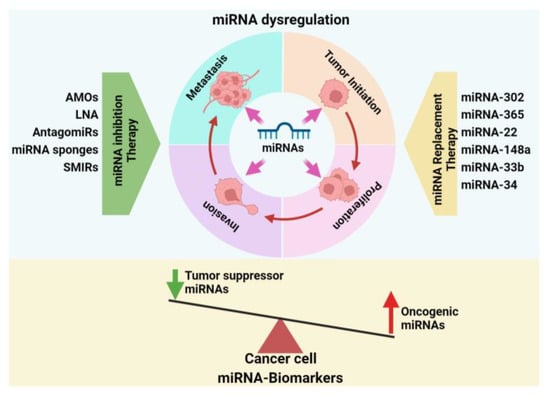
4. Significance of miRNAs in Breast Cancer Development
4.1. Breast Cancer Initiation and Progression
4.2. miRNA in Metastatic Breast Cancer
5. miRNA Expression in Breast Cancer Subtypes
6. miRNAs as Potential Diagnostic Biomarkers for Breast Cancer
7. miRNA-Based Therapeutic Approaches to Breast Cancer Treatment
7.1. miRNA Inhibition Therapy
7.1.1. Anti-miRNA Oligonucleotides (AMOs)
7.1.2. Locked Nucleic Acid (LNA)
7.1.3. AntagomiRs
7.1.4. miRNA Sponges
7.1.5. miRNA Small-Molecule Inhibitors (SMIRs)
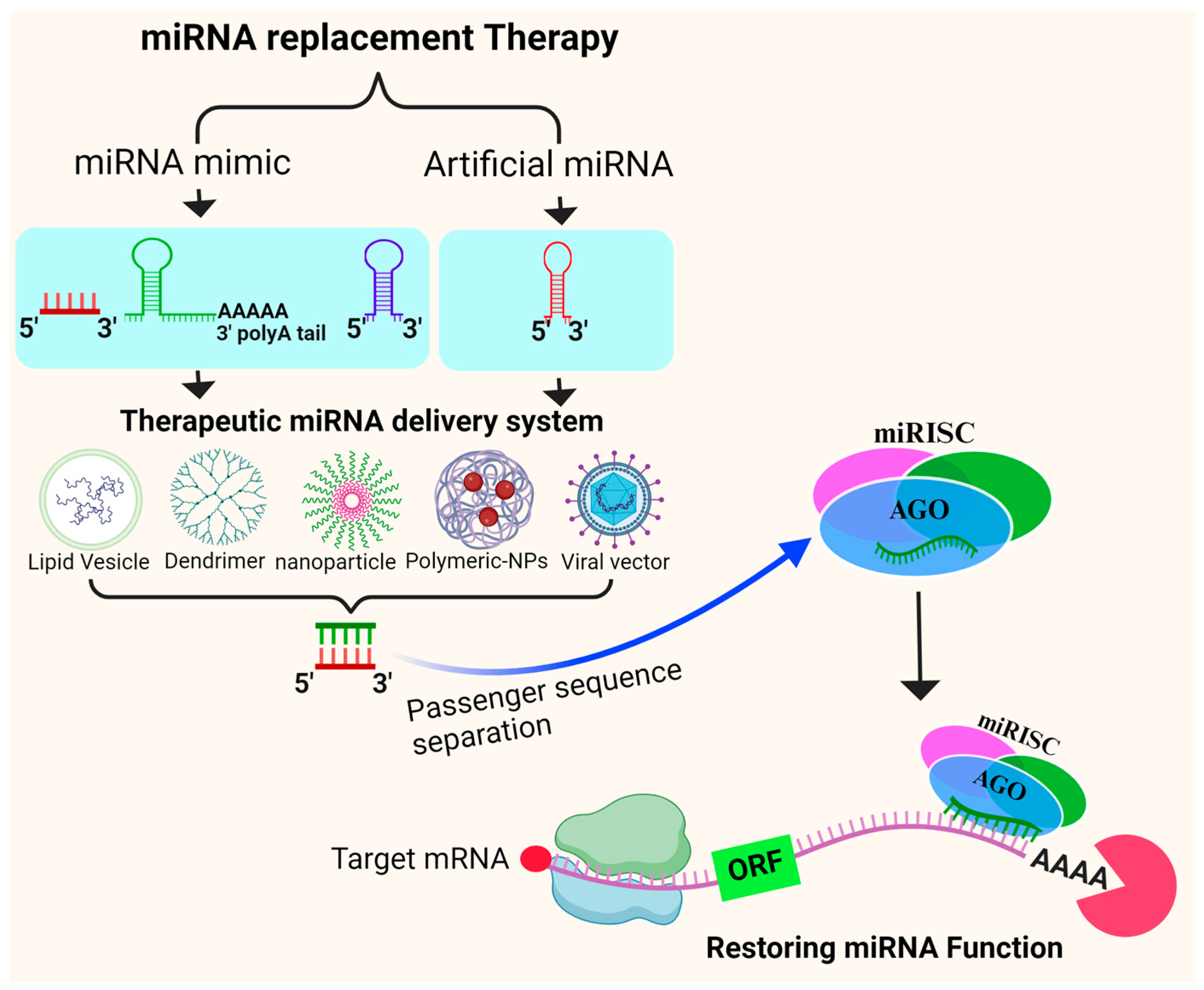
7.2. miRNA Replacement Therapy
8. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Lewandowska, A.; Rudzki, M.; Rudzki, S.; Lewandowski, T.; Laskowska, B. Environmental Risk Factors for Cancer—Review Paper. Ann. Agric. Environ. Med. 2019, 26, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Lakhani, S.R.; Ellis, I.O.; Schnitt, S.; Tan, P.H.; van de Vijver, M. (Eds.) WHO Classification of Tumours of the Breast; IARC: Lyon, France, 2012. [Google Scholar]
- Ferlay, J.; Ervik, M.; Lam, F.; Laversanne, M.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2024; Available online: https://gco.iarc.who.int/today (accessed on 12 December 2023).
- Nguyen, D.X.; Bos, P.D.; Massagué, J. Metastasis: From Dissemination to Organ-Specific Colonization. Nat. Rev. Cancer 2009, 9, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.; Costa, A.; Norton, L.; Cameron, D.; Cufer, T.; Fallowfield, L.; Francis, P.; Gligorov, J.; Kyriakides, S.; Lin, N.; et al. 1st International Consensus Guidelines for Advanced Breast Cancer (ABC 1). Breast 2012, 21, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Yedjou, C.G.; Sims, J.N.; Miele, L.; Noubissi, F.; Lowe, L.; Fonseca, D.D.; Alo, R.A.; Payton, M.; Tchounwou, P.B. Health and Racial Disparity in Breast Cancer. Adv. Exp. Med. Biol. 2019, 1152, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015, 5, 1122–1143. [Google Scholar] [CrossRef]
- Rohan, T.; Ye, K.; Wang, Y.; Glass, A.G.; Ginsberg, M.; Loudig, O. MicroRNA expression in benign breast tissue and risk of subsequent invasive breast cancer. PLoS ONE 2018, 13, e0191814. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Khordadmehr, M.; Shahbazi, R.; Ezzati, H.; Jigari-Asl, F.; Sadreddini, S.; Baradaran, B. Key MicroRNAs in the Biology of Breast Cancer; Emerging Evidence in the Last Decade. J. Cell Physiol. 2019, 234, 8316–8326. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Kumar, S.; Keerthana, R.; Pazhanimuthu, A.; Perumal, P. Overexpression of Circulating MiRNA-21 and MiRNA-146a in Plasma Samples of Breast Cancer Patients. Indian J. Biochem. Biophys. 2013, 50, 210–214. [Google Scholar]
- Swellam, M.; El Magdoub, H.M.; Hassan, N.M.; Hefny, M.M.; Sobeih, M.E. Potential diagnostic role of circulating MiRNAs in breast cancer: Implications on clinicopathological characters. Clin. Biochem. 2018, 56, 47–54. [Google Scholar] [CrossRef]
- Savari, B.; Boozarpour, S.; Tahmasebi-Birgani, M.; Sabouri, H.; Hosseini, S.M. Overex-pression of MicroRNA-21 in the Serum of Breast Cancer Patients. MicorRNA 2020, 9, 58–63. [Google Scholar] [CrossRef]
- Clancy, J.W.; D’Souza-Schorey, C. Tumor-Derived Extracellular Vesicles: Multifunctional Entities in the Tumor Microenvironment. Annu. Rev. Pathol. Mech. Dis. 2023, 18, 205–229. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Ghamlouche, F.; Yehya, A.; Zeid, Y.; Fakhereddine, H.; Fawaz, J.; Liu, Y.-N.; Al-Sayegh, M.; Abou-Kheir, W. MicroRNAs as Clinical Tools for Diagnosis, Prognosis, and Therapy in Prostate Cancer. Transl. Oncol. 2023, 28, 101613. [Google Scholar] [CrossRef] [PubMed]
- Riolo, G.; Cantara, S.; Marzocchi, C.; Ricci, C. miRNA Targets: From Prediction Tools to Experimental Validation. Methods Protoc. 2020, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Saçar, M.D.; Hamzeiy, H.; Allmer, J. Can MiRBase provide positive data for machine learning for the detection of MiRNA hairpins? J. Integr. Bioinform. 2013, 10, 215. [Google Scholar] [CrossRef]
- Skoufos, G.; Kakoulidis, P.; Tastsoglou, S.; Zacharopoulou, E.; Kotsira, V.; Miliotis, M.; Mavromati, G.; Grigoriadis, D.; Zioga, M.; Velli, A.; et al. TarBase-v9.0 extends experimentally supported miRNA-gene interactions to cell-types and virally encoded miRNAs. Nucleic Acids Res. 2024, 52, D304–D310. [Google Scholar] [CrossRef]
- Loh, H.-Y.; Norman, B.P.; Lai, K.-S.; Rahman, N.M.A.N.A.; Alitheen, N.B.M.; Osman, M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 4940. [Google Scholar] [CrossRef]
- Richard, V.; Davey, M.G.; Annuk, H.; Miller, N.; Dwyer, R.M.; Lowery, A.; Kerin, M.J. MicroRNAs in Molecular Classification and Pathogenesis of Breast Tumors. Cancers 2021, 13, 5332. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The Role of MicroRNAs in Human Cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour Invasion and Metastasis Initiated by MicroRNA-10b in Breast Cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Lim, M.; Zhao, S.; Sano, Y.; Simone, B.A.; Savage, J.E.; Wickstrom, E.; Camphausen, K.; Pestell, R.G.; Simone, N.L. The Metastatic Potential of Triple-Negative Breast Cancer Is Decreased via Caloric Restriction-Mediated Reduction of the MiR-17~92 Cluster. Breast Cancer Res. Treat. 2014, 146, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.M.; Sultana, A.; Barua, D.; Islam, M.N.; Gupta, A.; Gupta, S. Differential Expression, Function and Prognostic Value of MiR-17–92 Cluster in ER-Positive and Triple-Negative Breast Cancer. Cancer Treat. Res. Commun. 2020, 25, 100224. [Google Scholar] [CrossRef]
- Frankel, L.B.; Christoffersen, N.R.; Jacobsen, A.; Lindow, M.; Krogh, A.; Lund, A.H. Programmed Cell Death 4 (PDCD4) Is an Important Functional Target of the MicroRNA MiR-21 in Breast Cancer Cells. J. Biol. Chem. 2008, 283, 1026–1033. [Google Scholar] [CrossRef]
- Zhao, D.; Tu, Y.; Wan, L.; Bu, L.; Huang, T.; Sun, X.; Wang, K.; Shen, B. In Vivo Monitoring of Angiogenesis Inhibition via Down-Regulation of Mir-21 in a VEGFR2-Luc Murine Breast Cancer Model Using Bioluminescent Imaging. PLoS ONE 2013, 8, e71472. [Google Scholar] [CrossRef]
- Zhu, S.; Wu, H.; Wu, F.; Nie, D.; Sheng, S.; Mo, Y.-Y. MicroRNA-21 Targets Tumor Suppressor Genes in Invasion and Metastasis. Cell Res. 2008, 18, 350–359. [Google Scholar] [CrossRef]
- Song, B.; Wang, C.; Liu, J.; Wang, X.; Lv, L.; Wei, L.; Xie, L.; Zheng, Y.; Song, X. MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J. Exp. Clin. Cancer Res. 2010, 29, 29. [Google Scholar] [CrossRef]
- Roscigno, G.; Puoti, I.; Giordano, I.; Donnarumma, E.; Russo, V.; Affinito, A.; Adamo, A.; Quintavalle, C.; Todaro, M.; Vivanco, M.D.; et al. MiR-24 Induces Chemotherapy Resistance and Hypoxic Advantage in Breast Cancer. Oncotarget 2017, 8, 19507–19521. [Google Scholar] [CrossRef]
- Lei, P.; Wang, W.; Sheldon, M.; Sun, Y.; Yao, F.; Ma, L. Role of Glucose Metabolic Reprogramming in Breast Cancer Progression and Drug Resistance. Cancers 2023, 15, 3390. [Google Scholar] [CrossRef]
- Hua, K.; Jin, J.; Zhao, J.; Song, J.; Song, H.; Li, D.; Maskey, N.; Zhao, B.; Wu, C.; Xu, H.; et al. MiR-135b, Upregulated in Breast Cancer, Promotes Cell Growth and Disrupts the Cell Cycle by Regulating LATS2. Int. J. Oncol. 2016, 48, 1997–2006. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, H.-W.; Lu, M.-H.; He, X.-H.; Li, Y.; Gu, H.; Liu, M.-F.; Wang, E.-D. MicroRNA-155 Functions as an OncomiR in Breast Cancer by Targeting the Suppressor of Cytokine Signaling 1 Gene. Cancer Res. 2010, 70, 3119–3127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-M.; Zhao, J.; Deng, H.-Y. MiR-155 Promotes Proliferation of Human Breast Cancer MCF-7 Cells through Targeting Tumor Protein 53-Induced Nuclear Protein 1. J. Biomed. Sci. 2013, 20, 79. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; He, L.; Coppola, M.; Guo, J.; Esposito, N.N.; Coppola, D.; Cheng, J.Q. MicroRNA-155 Regulates Cell Survival, Growth, and Chemosensitivity by Targeting FOXO3a in Breast Cancer. J. Biol. Chem. 2010, 285, 17869–17879. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.A.; Sossey-Alaoui, K.; Thompson, C.L.; Danielpour, D.; Schiemann, W.P. TGF-β Upregulates MiR-181a Expression to Promote Breast Cancer Metastasis. J. Clin. Investig. 2013, 123, 150–163. [Google Scholar] [CrossRef]
- Sharma, S.; Nagpal, N.; Ghosh, P.C.; Kulshreshtha, R. P53-MiR-191-SOX4 Regulatory Loop Affects Apoptosis in Breast Cancer. RNA 2017, 23, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Yu, H.; Yuan, J.; Guo, C.; Cao, H.; Li, W.; Xiao, C. MicroRNA-200b Impacts Breast Cancer Cell Migration and Invasion by Regulating Ezrin-Radixin-Moesin. Med. Sci. Monit. 2016, 22, 1946–1952. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, M.; Tong, Y.; Liu, X.; Zhang, L.; Dong, D.; Shao, J.; Zhou, Y. miR-206 Promotes Cancer Progression by Targeting Full-Length Neurokinin-1 Receptor in Breast Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819875168. [Google Scholar] [CrossRef]
- Harquail, J.; LeBlanc, N.; Ouellette, R.J.; Robichaud, G.A. MiRNAs 484 and 210 Regulate Pax-5 Expression and Function in Breast Cancer Cells. Carcinogenesis 2019, 40, 1010–1020. [Google Scholar] [CrossRef]
- McAnena, P.; Tanriverdi, K.; Curran, C.; Gilligan, K.; Freedman, J.E.; Brown, J.A.L.; Kerin, M.J. Circulating MicroRNAs MiR-331 and MiR-195 Differentiate Local Luminal a from Metastatic Breast Cancer. BMC Cancer 2019, 19, 436. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Gumireddy, K.; Schrier, M.; le Sage, C.; Nagel, R.; Nair, S.; Egan, D.A.; Li, A.; Huang, G.; Klein-Szanto, A.J.; et al. The MicroRNAs MiR-373 and MiR-520c Promote Tumour Invasion and Metastasis. Nat. Cell Biol. 2008, 10, 202–210. [Google Scholar] [CrossRef]
- Li, Z.; Meng, Q.; Pan, A.; Wu, X.; Cui, J.; Wang, Y.; Li, L. MicroRNA-455-3p Promotes Invasion and Migration in Triple Negative Breast Cancer by Targeting Tumor Suppressor EI24. Oncotarget 2017, 8, 19455–19466. [Google Scholar] [CrossRef]
- Matamala, N.; Vargas, M.T.; González-Cámpora, R.; Arias, J.I.; Menéndez, P.; Andrés-León, E.; Yanowsky, K.; Llaneza-Folgueras, A.; Miñambres, R.; Martínez-Delgado, B.; et al. MicroRNA Deregulation in Triple Negative Breast Cancer Reveals a Role of MiR-498 in Regulating BRCA1 Expression. Oncotarget 2016, 7, 20068–20079. [Google Scholar] [CrossRef]
- Jiang, H.; Wang, P.; Li, X.; Wang, Q.; Deng, Z.-B.; Zhuang, X.; Mu, J.; Zhang, L.; Wang, B.; Yan, J.; et al. Restoration of MiR17/20a in Solid Tumor Cells Enhances the Natural Killer Cell Antitumor Activity by Targeting Mekk2. Cancer Immunol. Res. 2014, 2, 789–799. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, C.; Wang, M.; Li, Z.; Casimiro, M.C.; Liu, M.; Wu, K.; Whittle, J.; Ju, X.; Hyslop, T.; et al. A Cyclin D1/MicroRNA 17/20 Regulatory Feedback Loop in Control of Breast Cancer Cell Proliferation. J. Cell Biol. 2008, 182, 509–517. [Google Scholar] [CrossRef]
- Zhang, H.; Cai, K.; Wang, J.; Wang, X.; Cheng, K.; Shi, F.; Jiang, L.; Zhang, Y.; Dou, J. MiR-7, Inhibited Indirectly by LincRNA HOTAIR, Directly Inhibits SETDB1 and Reverses the EMT of Breast Cancer Stem Cells by Downregulating the STAT3 Pathway. Stem Cells 2014, 32, 2858–2868. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Ding, C.; Zhang, H.; Gao, J. Let-7 MiRNAs Sensitize Breast Cancer Stem Cells to Radiation-Induced Repression through Inhibition of the Cyclin D1/Akt1/Wnt1 Signaling Pathway. Mol. Med. Rep. 2016, 14, 3285–3292. [Google Scholar] [CrossRef]
- Yu, F.; Deng, H.; Yao, H.; Liu, Q.; Su, F.; Song, E. Mir-30 Reduction Maintains Self-Renewal and Inhibits Apoptosis in Breast Tumor-Initiating Cells. Oncogene 2010, 29, 4194–4204. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Liu, A.Y.; Fan, C.; Zheng, H.; Li, Y.; Zhang, C.; Wu, S.; Yu, D.; Huang, Z.; Liu, F.; et al. MicroRNA-33b Inhibits Breast Cancer Metastasis by Targeting HMGA2, SALL4 and Twist1. Sci. Rep. 2015, 5, 9995. [Google Scholar] [CrossRef]
- Huang, Q.-D.; Zheng, S.-R.; Cai, Y.-J.; Chen, D.-L.; Shen, Y.-Y. IMP3 Promotes TNBC Stem Cell Property through MiRNA-34a Regulation. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2688–2696. [Google Scholar] [CrossRef]
- Ferracin, M.; Bassi, C.; Pedriali, M.; Pagotto, S.; D’Abundo, L.; Zagatti, B.; Corrà, F.; Musa, G.; Callegari, E.; Lupini, L.; et al. MiR-125b Targets Erythropoietin and Its Receptor and Their Expression Correlates with Metastatic Potential and ERBB2/HER2 Expression. Mol. Cancer 2013, 12, 130. [Google Scholar] [CrossRef]
- Feliciano, A.; Castellvi, J.; Artero-Castro, A.; Leal, J.A.; Romagosa, C.; Hernández-Losa, J.; Peg, V.; Fabra, A.; Vidal, F.; Kondoh, H.; et al. MiR-125b Acts as a Tumor Suppressor in Breast Tumorigenesis via Its Novel Direct Targets ENPEP, CK2-α, CCNJ, and MEGF9. PLoS ONE 2013, 8, e76247. [Google Scholar] [CrossRef]
- Chen, F.; Luo, N.; Hu, Y.; Li, X.; Zhang, K. MiR-137 Suppresses Triple-Negative Breast Cancer Stemness and Tumorigenesis by Perturbing BCL11A-DNMT1 Interaction. Cell. Phys. Biochem. 2018, 47, 2147–2158. [Google Scholar] [CrossRef]
- Zhou, L.L.; Dong, J.L.; Huang, G.; Sun, Z.L.; Wu, J. MicroRNA-143 Inhibits Cell Growth by Targeting ERK5 and MAP3K7 in Breast Cancer. Braz. J. Med. Biol. Res. 2017, 50, e5891. [Google Scholar] [CrossRef]
- Jiang, Q.; He, M.; Ma, M.-T.; Wu, H.-Z.; Yu, Z.-J.; Guan, S.; Jiang, L.-Y.; Wang, Y.; Zheng, D.-D.; Jin, F.; et al. MicroRNA-148a Inhibits Breast Cancer Migration and Invasion by Directly Targeting WNT-1. Oncol. Rep. 2016, 35, 1425–1432. [Google Scholar] [CrossRef]
- Xue, J.; Chen, Z.; Gu, X.; Zhang, Y.; Zhang, W. MicroRNA-148a inhibits migration of breast cancer cells by targeting MMP-13. Tumour Biol. 2016, 37, 1581–1590. [Google Scholar] [CrossRef]
- Yao, J.; Zhou, E.; Wang, Y.; Xu, F.; Zhang, D.; Zhong, D. microRNA-200a inhibits cell proliferation by targeting mitochondrial transcription factor A in breast cancer. DNA Cell Biol. 2014, 33, 291–300. [Google Scholar] [CrossRef]
- DeCastro, A.J.; Dunphy, K.A.; Hutchinson, J.; Balboni, A.L.; Cherukuri, P.; Jerry, D.J.; DiRenzo, J. MiR203 Mediates Subversion of Stem Cell Properties during Mammary Epithelial Differentiation via Repression of ΔNP63α and Promotes Mesenchymal-to-Epithelial Transition. Cell Death Dis. 2013, 4, e514. [Google Scholar] [CrossRef]
- Li, J.; Peng, S.; Zou, X.; Geng, X.; Wang, T.; Zhu, W.; Xia, T. Value of Negatively Correlated MiR-205-5p/HMGB3 and MiR-96-5p/FOXO1 on the Diagnosis of Breast Cancer and Benign Breast Diseases. Cancer Pathog. Ther. 2023, 1, 159–167. [Google Scholar] [CrossRef]
- Chao, C.-H.; Chang, C.-C.; Wu, M.-J.; Ko, H.-W.; Wang, D.; Hung, M.-C.; Yang, J.-Y.; Chang, C.-J. MicroRNA-205 Signaling Regulates Mammary Stem Cell Fate and Tumorigenesis. J. Clin. Investig. 2014, 124, 3093–3106. [Google Scholar] [CrossRef]
- Zhou, J.; Tian, Y.; Li, J.; Lu, B.; Sun, M.; Zou, Y.; Kong, R.; Luo, Y.; Shi, Y.; Wang, K.; et al. MiR-206 Is down-Regulated in Breast Cancer and Inhibits Cell Proliferation through the up-Regulation of CyclinD2. Biochem. Biophys. Res. Commun. 2013, 433, 207–212. [Google Scholar] [CrossRef]
- Lin, Z.-J.; Ming, J.; Yang, L.; Du, J.-Z.; Wang, N.; Luo, H.-J. Mechanism of Regulatory Effect of MicroRNA-206 on Connexin 43 in Distant Metastasis of Breast Cancer. Chin. Med. J. 2016, 129, 424–434. [Google Scholar] [CrossRef]
- Sun, X.; Li, Y.; Zheng, M.; Zuo, W.; Zheng, W. MicroRNA-223 Increases the Sensitivity of Triple-Negative Breast Cancer Stem Cells to TRAIL-Induced Apoptosis by Targeting HAX-1. PLoS ONE 2016, 11, e0162754. [Google Scholar] [CrossRef]
- Huang, X.; Lyu, J. Tumor Suppressor Function of MiR-483-3p on Breast Cancer via Targeting of the Cyclin E1 Gene. Exp. Ther. Med. 2018, 16, 2615–2620. [Google Scholar] [CrossRef]
- Luo, Q.; Li, X.; Gao, Y.; Long, Y.; Chen, L.; Huang, Y.; Fang, L. MiRNA-497 Regulates Cell Growth and Invasion by Targeting Cyclin E1 in Breast Cancer. Cancer Cell Int. 2013, 13, 95. [Google Scholar] [CrossRef]
- Li, D.; Song, H.; Wu, T.; Xie, D.; Hu, J.; Zhao, J.; Shen, Q.; Fang, L. MiR-519d-3p Suppresses Breast Cancer Cell Growth and Motility via Targeting LIM Domain Kinase 1. Mol. Cell Biochem. 2018, 444, 169–178. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Polyak, K. Breast Cancer: Origins and Evolution. J. Clin. Investig. 2007, 117, 3155–3163. [Google Scholar] [CrossRef]
- Sever, R.; Brugge, J.S. Signal Transduction in Cancer. Cold Spring Harb. Perspect. Med. 2015, 5, a006098. [Google Scholar] [CrossRef]
- Mai, Y.; Su, J.; Yang, C.; Xia, C.; Fu, L. The Strategies to Cure Cancer Patients by Eradicating Cancer Stem-like Cells. Mol. Cancer 2023, 22, 171. [Google Scholar] [CrossRef]
- Liu, H.; Patel, M.R.; Prescher, J.A.; Patsialou, A.; Qian, D.; Lin, J.; Wen, S.; Chang, Y.-F.; Bachmann, M.H.; Shimono, Y.; et al. Cancer Stem Cells from Human Breast Tumors Are Involved in Spontaneous Metastases in Orthotopic Mouse Models. Proc. Natl. Acad. Sci. USA 2010, 107, 18115–18120. [Google Scholar] [CrossRef]
- Chang, C.-J.; Yang, J.-Y.; Xia, W.; Chen, C.-T.; Xie, X.; Chao, C.-H.; Woodward, W.A.; Hsu, J.-M.; Hortobagyi, G.N.; Hung, M.-C. EZH2 Promotes Expansion of Breast Tumor Initiating Cells through Activation of RAF1-β-Catenin Signaling. Cancer Cell 2011, 19, 86–100. [Google Scholar] [CrossRef]
- Shimono, Y.; Zabala, M.; Cho, R.W.; Lobo, N.; Dalerba, P.; Qian, D.; Diehn, M.; Liu, H.; Panula, S.P.; Chiao, E.; et al. Downregulation of MiRNA-200c Links Breast Cancer Stem Cells with Normal Stem Cells. Cell 2009, 138, 592–603. [Google Scholar] [CrossRef]
- Zhu, Y.; Yu, F.; Jiao, Y.; Feng, J.; Tang, W.; Yao, H.; Gong, C.; Chen, J.; Su, F.; Zhang, Y.; et al. Reduced MiR-128 in Breast Tumor–Initiating Cells Induces Chemotherapeutic Resistance via Bmi-1 and ABCC5. Clin. Cancer Res. 2011, 17, 7105–7115. [Google Scholar] [CrossRef]
- Bockhorn, J.; Yee, K.; Chang, Y.-F.; Prat, A.; Huo, D.; Nwachukwu, C.; Dalton, R.; Huang, S.; Swanson, K.E.; Perou, C.M.; et al. MicroRNA-30c Targets Cytoskeleton Genes Involved in Breast Cancer Cell Invasion. Breast Cancer Res. Treat. 2013, 137, 373–382. [Google Scholar] [CrossRef]
- Yu, F.; Jiao, Y.; Zhu, Y.; Wang, Y.; Zhu, J.; Cui, X.; Liu, Y.; He, Y.; Park, E.-Y.; Zhang, H.; et al. MicroRNA 34c Gene Down-Regulation via DNA Methylation Promotes Self-Renewal and Epithelial-Mesenchymal Transition in Breast Tumor-Initiating Cells. J. Biol. Chem. 2012, 287, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Hwang-Verslues, W.W.; Chang, P.-H.; Wei, P.-C.; Yang, C.-Y.; Huang, C.-K.; Kuo, W.-H.; Shew, J.-Y.; Chang, K.-J.; Lee, E.Y.-H.P.; Lee, W.-H. MiR-495 Is Upregulated by E12/E47 in Breast Cancer Stem Cells, and Promotes Oncogenesis and Hypoxia Resistance via Downregulation of E-Cadherin and REDD1. Oncogene 2011, 30, 2463–2474. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, Y.; Tsuyada, A.; Ren, X.; Wu, X.; Stubblefield, K.; Rankin-Gee, E.K.; Wang, S.E. Transforming Growth Factor-β Regulates the Sphere-Initiating Stem Cell-like Feature in Breast Cancer through MiRNA-181 and ATM. Oncogene 2011, 30, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Riggio, A.I.; Varley, K.E.; Welm, A.L. The Lingering Mysteries of Metastatic Recurrence in Breast Cancer. Br. J. Cancer 2021, 124, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Baumann, Z.; Auf der Maur, P.; Bentires-Alj, M. Feed-forward Loops between Metastatic Cancer Cells and Their Microenvironment—The Stage of Escalation. EMBO Mol. Med. 2022, 14, e14283. [Google Scholar] [CrossRef]
- Huang, Y.; Hong, W.; Wei, X. The Molecular Mechanisms and Therapeutic Strategies of EMT in Tumor Progression and Metastasis. J. Hematol. Oncol. 2022, 15, 129. [Google Scholar] [CrossRef]
- Chua, H.L.; Bhat-Nakshatri, P.; Clare, S.E.; Morimiya, A.; Badve, S.; Nakshatri, H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: Potential involvement of ZEB-1 and ZEB-2. Oncogene 2007, 26, 711–724. [Google Scholar] [CrossRef]
- De, S.; Das, S.; Mukherjee, S.; Das, S.; Bandyopadhyay, S.S. Establishment of twist-1 and TGFBR2 as direct targets of microRNA-20a in mesenchymal to epithelial transition of breast cancer cell-line MDA-MB-231. Exp. Cell Res. 2017, 361, 85–92. [Google Scholar] [CrossRef]
- Kwak, S.Y.; Yoo, J.O.; An, H.J.; Bae, I.H.; Park, M.J.; Kim, J.; Han, Y.H. miR-5003-3p promotes epithelial-mesenchymal transition in breast cancer cells through Snail stabilization and direct targeting of E-cadherin. J. Mol. Cell Biol. 2016, 8, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Li, J.; Zhang, Y.; Wang, N.; Liang, H.; Liu, Y.; Zhang, C.Y.; Zen, K.; Gu, H. Slug-upregulated miR-221 promotes breast cancer progression through suppressing E-cadherin expression. Sci. Rep. 2016, 6, 25798. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The MiR-200 Family Determines the Epithelial Phenotype of Cancer Cells by Targeting the E-Cadherin Repressors ZEB1 and ZEB2. Genes Dev. 2008, 22, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Hosany, S.; Zhong, S.; Jiang, Y.; Zhang, F.; Lin, L.; Wang, X.; Gao, S.; Hu, X. MicroRNA-193a Inhibits Breast Cancer Proliferation and Metastasis by Downregulating WT1. PLoS ONE 2017, 12, e0185565. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nature Cell Biol. 2010, 12, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Martello, G.; Rosato, A.; Ferrari, F.; Manfrin, A.; Cordenonsi, M.; Dupont, S.; Enzo, E.; Guzzardo, V.; Rondina, M.; Spruce, T.; et al. A MicroRNA targeting dicer for metastasis control. Cell 2010, 141, 1195–1207. [Google Scholar] [CrossRef]
- Cochrane, D.R.; Cittelly, D.M.; Howe, E.N.; Spoelstra, N.S.; McKinsey, E.L.; LaPara, K.; Elias, A.; Yee, D.; Richer, J.K. MicroRNAs Link Estrogen Receptor Alpha Status and Dicer Levels in Breast Cancer. Horm. Cancer 2010, 1, 306–319. [Google Scholar] [CrossRef]
- Chen, P.-S.; Su, J.-L.; Cha, S.-T.; Tarn, W.-Y.; Wang, M.-Y.; Hsu, H.-C.; Lin, M.-T.; Chu, C.-Y.; Hua, K.-T.; Chen, C.-N.; et al. MiR-107 Promotes Tumor Progression by Targeting the Let-7 MicroRNA in Mice and Humans. J. Clin. Investig. 2011, 121, 3442–3455. [Google Scholar] [CrossRef]
- Wu, Y.; Shi, W.; Tang, T.; Wang, Y.; Yin, X.; Chen, Y.; Zhang, Y.; Xing, Y.; Shen, Y.; Xia, T.; et al. MiR-29a Contributes to Breast Cancer Cells Epithelial–Mesenchymal Transition, Migration, and Invasion via down-Regulating Histone H4K20 Trimethylation through Directly Targeting SUV420H2. Cell Death Dis. 2019, 10, 176. [Google Scholar] [CrossRef]
- Ren, L.; Chen, H.; Song, J.; Chen, X.; Lin, C.; Zhang, X.; Hou, N.; Pan, J.; Zhou, Z.; Wang, L.; et al. MiR-454-3p-Mediated Wnt/β-Catenin Signaling Antagonists Suppression Promotes Breast Cancer Metastasis. Theranostics 2019, 9, 449–465. [Google Scholar] [CrossRef]
- Dobson, J.R.; Taipaleenmäki, H.; Hu, Y.-J.; Hong, D.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B.; Pratap, J. Hsa-Mir-30c Promotes the Invasive Phenotype of Metastatic Breast Cancer Cells by Targeting NOV/CCN3. Cancer Cell Int. 2014, 14, 73. [Google Scholar] [CrossRef]
- Huang, L.; Dai, T.; Lin, X.; Zhao, X.; Chen, X.; Wang, C.; Li, X.; Shen, H.; Wang, X. MicroRNA-224 Targets RKIP to Control Cell Invasion and Expression of Metastasis Genes in Human Breast Cancer Cells. Biochem. Biophys. Res. Commun. 2012, 425, 127–133. [Google Scholar] [CrossRef]
- Kong, W.; Yang, H.; He, L.; Zhao, J.; Coppola, D.; Dalton, W.S.; Cheng, J.Q. MicroRNA-155 Is Regulated by the Transforming Growth Factor β/Smad Pathway and Contributes to Epithelial Cell Plasticity by Targeting RhoA. Mol. Cell. Biol. 2008, 28, 6773–6784. [Google Scholar] [CrossRef]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Tang, P.; Tse, G.M. Immunohistochemical Surrogates for Molecular Classification of Breast Carcinoma: A 2015 Update. Arch. Pathol. Lab. Med. 2016, 140, 806–814. [Google Scholar] [CrossRef]
- Russnes, H.G.; Lingjærde, O.C.; Børresen-Dale, A.-L.; Caldas, C. Breast Cancer Molecular Stratification. Am. J. Pathol. 2017, 187, 2152–2162. [Google Scholar] [CrossRef]
- Parise, C.A.; Caggiano, V. Breast Cancer Survival Defined by the ER/PR/HER2 Subtypes and a Surrogate Classification according to Tumor Grade and Immunohistochemical Biomarkers. J. Cancer Epidemiol. 2014, 2014, 469251. [Google Scholar] [CrossRef]
- Davies, C.; Godwin, J.; Gray, R.; Clarke, M.; Cutter, D.; Darby, S.; McGale, P.; Pan, H.C.; Taylor, C.; Wang, Y.C.; et al. Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: Patient-level meta-analysis of randomised trials. Lancet 2011, 378, 771–784. [Google Scholar] [CrossRef]
- Normann, L.S.; Aure, M.R.; Leivonen, S.K.; Haugen, M.H.; Hongisto, V.; Kristensen, V.N.; Mælandsmo, G.M.; Sahlberg, K.K. MicroRNA in combination with HER2-targeting drugs reduces breast cancer cell viability in vitro. Sci. Rep. 2021, 11, 10893. [Google Scholar] [CrossRef]
- Søkilde, R.; Persson, H.; Ehinger, A.; Pirona, A.C.; Fernö, M.; Hegardt, C.; Larsson, C.; Loman, N.; Malmberg, M.; Rydén, L.; et al. Refinement of Breast Cancer Molecular Classification by MiRNA Expression Profiles. BMC Genom. 2019, 20, 503. [Google Scholar] [CrossRef]
- Amiruddin, A.; Massi, M.N.; Islam, A.A.; Patellongi, I.; Pratama, M.Y.; Sutandyo, N.; Natzir, R.; Hatta, M.; Md Latar, N.H.; Wahid, S. MicroRNA-221 and Tamoxifen Resistance in Luminal-Subtype Breast Cancer Patients: A Case-Control Study. Ann. Med. Surg. 2022, 73, 103092. [Google Scholar] [CrossRef]
- Fan, T.; Mao, Y.; Sun, Q.; Liu, F.; Lin, J.; Liu, Y.; Cui, J.; Jiang, Y. Branched Rolling Circle Amplification Method for Measuring Serum Circulating MicroRNA Levels for Early Breast Cancer Detection. Cancer Sci. 2018, 109, 2897–2906. [Google Scholar] [CrossRef]
- McDermott, A.M.; Miller, N.; Wall, D.; Martyn, L.M.; Ball, G.; Sweeney, K.J.; Kerin, M.J. Identification and Validation of Oncologic MiRNA Biomarkers for Luminal A-like Breast Cancer. PLoS ONE 2014, 9, e87032. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, N.; Liu, L.; Dong, H.; Wu, C. Correlation between MicroRNA-21, MicroRNA-206 and Estrogen Receptor, Progesterone Receptor, Human Epidermal Growth Factor Receptor 2 in Breast Cancer. Clin. Biochem. 2019, 71, 52–57. [Google Scholar] [CrossRef]
- Lowery, A.J.; Miller, N.; Devaney, A.; McNeill, R.E.; Davoren, P.A.; Lemetre, C.; Benes, V.; Schmidt, S.; Blake, J.; Ball, G.; et al. MicroRNA Signatures Predict Oestrogen Receptor, Progesterone Receptor and HER2/Neureceptor Status in Breast Cancer. Breast Cancer Res. 2009, 11, R27. [Google Scholar] [CrossRef]
- Amorim, M.; Lobo, J.; Fontes-Sousa, M.; Estevão-Pereira, H.; Salta, S.; Lopes, P.; Coimbra, N.; Antunes, L.; Palma de Sousa, S.; Henrique, R.; et al. Predictive and Prognostic Value of Selected MicroRNAs in Luminal Breast Cancer. Front. Genet. 2019, 10, 815. [Google Scholar] [CrossRef]
- Ulianova, E.P.; Tokmakov, V.V.; Shatova, I.S.; Sagakyants, A.B.; Goncharova, A.S.; Zaikina, E.V.; Chernikova, E.N.; Bakulina, S.M.; Pushkareva, T.F.; Kit, O.I.; et al. Evaluation of Prognostic Significance of MicroRNA in Tumors of Luminal, Primary Operable Breast Cancer without Her2 Neu Overexpression in Postmenopausal Women. J. Clin. Oncol. 2020, 38, e12558. [Google Scholar] [CrossRef]
- Ohzawa, H.; Miki, A.; Teratani, T.; Shiba, S.; Sakuma, Y.; Nishimura, W.; Noda, Y.; Fukushima, N.; Fujii, H.; Hozumi, Y.; et al. Usefulness of MiRNA Profiles for Predicting Pathological Responses to Neoadjuvant Chemotherapy in Patients with Human Epidermal Growth Factor Receptor 2-Positive Breast Cancer. Oncol. Lett. 2017, 13, 1731–1740. [Google Scholar] [CrossRef]
- Han, S.-H.; Kim, H.J.; Gwak, J.M.; Kim, M.; Chung, Y.R.; Park, S.Y. MicroRNA-222 Expression as a Predictive Marker for Tumor Progression in Hormone Receptor-Positive Breast Cancer. J. Breast Cancer 2017, 20, 35. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, X.; Niu, W.; Wang, H.; Wen, Q.; Fan, S.; Zhao, R.; Li, Z.; Xiong, W.; Peng, S.; et al. Elevated MicroRNA-125b Levels Predict a Worse Prognosis in HER2-Positive Breast Cancer Patients. Oncol. Lett. 2017, 13, 867–874. [Google Scholar] [CrossRef]
- Wu, X.; Somlo, G.; Yu, Y.; Palomares, M.R.; Li, A.X.; Zhou, W.; Chow, A.; Yen, Y.; Rossi, J.J.; Gao, H.; et al. De Novo Sequencing of Circulating MiRNAs Identifies Novel Markers Predicting Clinical Outcome of Locally Advanced Breast Cancer. J. Transl. Med. 2012, 10, 42. [Google Scholar] [CrossRef]
- Souza, K.C.B.; Evangelista, A.F.; Leal, L.F.; Souza, C.P.; Vieira, R.A.; Causin, R.L.; Neuber, A.C.; Pessoa, D.P.; Passos, G.A.S.; Reis, R.M.V.; et al. Identification of Cell-Free Circulating MicroRNAs for the Detection of Early Breast Cancer and Molecular Subtyping. J. Oncol. 2019, 2019, 8393769. [Google Scholar] [CrossRef]
- Mattie, M.D.; Benz, C.C.; Bowers, J.; Sensinger, K.; Wong, L.; Scott, G.K.; Fedele, V.; Ginzinger, D.; Getts, R.; Haqq, C. Optimized High-Throughput MicroRNA Expression Profiling Provides Novel Biomarker Assessment of Clinical Prostate and Breast Cancer Biopsies. Mol. Cancer 2006, 5, 24. [Google Scholar] [CrossRef]
- Toyama, T.; Kondo, N.; Endo, Y.; Sugiura, H.; Yoshimoto, N.; Iwasa, M.; Takahashi, S.; Fujii, Y.; Yamashita, H. High Expression of MicroRNA-210 Is an Independent Factor Indicating a Poor Prognosis in Japanese Triple-Negative Breast Cancer Patients. Jpn. J. Clin. Oncol. 2012, 42, 256–263. [Google Scholar] [CrossRef]
- Kalniete, D.; Nakazawa-Miklaševiča, M.; Štrumfa, I.; Āboliņš, A.; Irmejs, A.; Gardovskis, J.; Miklaševičs, E. High Expression of MiR-214 Is Associated with a Worse Disease-Specific Survival of the Triple-Negative Breast Cancer Patients. Hered. Cancer Clin. Pract. 2015, 13, 7. [Google Scholar] [CrossRef][Green Version]
- Yao, L.; Liu, Y.; Cao, Z.; Li, J.; Huang, Y.; Hu, X.; Shao, Z. MicroRNA-493 Is a Prognostic Factor in Triple-negative Breast Cancer. Cancer Sci. 2018, 109, 2294–2301. [Google Scholar] [CrossRef]
- Uva, P.; Cossu-Rocca, P.; Loi, F.; Pira, G.; Murgia, L.; Orrù, S.; Floris, M.; Muroni, M.R.; Sanges, F.; Carru, C.; et al. MiRNA-135b Contributes to Triple Negative Breast Cancer Molecular Heterogeneity: Different Expression Profile in Basal-like Versus Non-Basal-like Phenotypes. Int. J. Med. Sci. 2018, 15, 536–548. [Google Scholar] [CrossRef]
- Kolesnikov, N.N.; Veryaskina, Y.A.; Titov, S.E.; Rodionov, V.V.; Gening, T.P.; Abakumova, T.V.; Kometova, V.V.; Torosyan, M.K.; Zhimulev, I.F. Expression of Micrornas in Molecular Genetic Breast Cancer Subtypes. Cancer Treat. Res. Commun. 2019, 20, 100026. [Google Scholar] [CrossRef]
- Moi, L.; Braaten, T.; Al-Shibli, K.; Lund, E.; Busund, L.-T.R. Differential Expression of the MiR-17-92 Cluster and MiR-17 Family in Breast Cancer According to Tumor Type; Results from the Norwegian Women and Cancer (NOWAC) Study. J. Transl. Med. 2019, 17, 334. [Google Scholar] [CrossRef] [PubMed]
- Harris, E.E.R. Precision Medicine for Breast Cancer: The Paths to Truly Individualized Diagnosis and Treatment. Int. J. Breast Cancer. 2018, 2018, 4809183. [Google Scholar] [CrossRef]
- Hamam, R.; Hamam, D.; Alsaleh, K.A.; Kassem, M.; Zaher, W.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. Circulating microRNAs in breast cancer: Novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017, 8, e3045. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Zhao, H.; Shen, J.; Medico, L.; Wang, D.; Ambrosone, C.B.; Liu, S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS ONE 2010, 5, e13735. [Google Scholar] [CrossRef]
- Bahrami, A.; Aledavood, A.; Anvari, K.; Hassanian, S.M.; Maftouh, M.; Yaghobzade, A.; Salarzaee, O.; ShahidSales, S.; Avan, A. The prognostic and therapeutic application of microRNAs in breast cancer: Tissue and circulating microRNAs. J. Cell. Physiol. 2018, 233, 774–786. [Google Scholar] [CrossRef]
- Terkelsen, T.; Russo, F.; Gromov, P.; Haakensen, V.D.; Brunak, S.; Gromova, I.; Krogh, A.; Papaleo, E. Secreted Breast Tumor Interstitial Fluid MicroRNAs and Their Target Genes Are Associated with Triple-Negative Breast Cancer, Tumor Grade, and Immune Infiltration. Breast Cancer Res. 2020, 22, 73. [Google Scholar] [CrossRef]
- Li, J.; He, D.; Bi, Y.; Liu, S. The Emerging Roles of Exosomal MiRNAs in Breast Cancer Progression and Potential Clinical Applications. Breast Cancer Targets Ther. 2023, 15, 825–840. [Google Scholar] [CrossRef] [PubMed]
- Cardinali, B.; Tasso, R.; Piccioli, P.; Ciferri, M.C.; Quarto, R.; Del Mastro, L. Circulating miRNAs in Breast Cancer Diagnosis and Prognosis. Cancers 2022, 14, 2317. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhang, Q.; Xu, J.; Guo, L.; Li, X. Clinical Significance of Serum MiR-21 in Breast Cancer Compared with CA153 and CEA. Chin. J. Cancer Res. 2013, 25, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Canatan, D.; Sönmez, Y.; Yılmaz, Ö.; Çim, A.; Coşkun, H.Ş.; Sezgin Göksu, S.; Ucar, S.; Aktekin, M.R. MicroRNAs as biomarkers for breast cancer. Acta Biomed. 2021, 92, e2021028. [Google Scholar] [CrossRef]
- Shimomura, A.; Shiino, S.; Kawauchi, J.; Takizawa, S.; Sakamoto, H.; Matsuzaki, J.; Ono, M.; Takeshita, F.; Niida, S.; Shimizu, C.; et al. Novel Combination of Serum MicroRNA for Detecting Breast Cancer in the Early Stage. Cancer Sci. 2016, 107, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Heneghan, H.M.; Miller, N.; Lowery, A.J.; Sweeney, K.J.; Newell, J.; Kerin, M.J. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann. Surg. 2010, 251, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Milde-Langosch, K.; Steinbach, B.; Müller, V.; Pantel, K. Diagnostic Potential of PTEN-Targeting MiR-214 in the Blood of Breast Cancer Patients. Breast Cancer Res. Treat. 2012, 134, 933–941. [Google Scholar] [CrossRef]
- Chen, W.; Cai, F.; Zhang, B.; Barekati, Z.; Zhong, X.Y. The Level of Circulating MiRNA-10b and MiRNA-373 in Detecting Lymph Node Metastasis of Breast Cancer: Potential Biomarkers. Tumor Biol. 2013, 34, 455–462. [Google Scholar] [CrossRef]
- van Schooneveld, E.; Wouters, M.C.; Van der Auwera, I.; Peeters, D.J.; Wildiers, H.; Van Dam, P.A.; Vergote, I.; Vermeulen, P.B.; Dirix, L.Y.; Van Laere, S.J. Expression profiling of cancerous and normal breast tissues identifies microRNAs that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers. Breast Cancer Res. 2012, 14, R34. [Google Scholar] [CrossRef]
- Jung, E.J.; Santarpia, L.; Kim, J.; Esteva, F.J.; Moretti, E.; Buzdar, A.U.; Di Leo, A.; Le, X.F.; Bast, R.C., Jr.; Park, S.T.; et al. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer 2012, 118, 2603–2614. [Google Scholar] [CrossRef]
- Roth, C.; Rack, B.; Müller, V.; Janni, W.; Pantel, K.; Schwarzenbach, H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010, 12, R90. [Google Scholar] [CrossRef]
- Wu, Q.; Lu, Z.; Li, H.; Lu, J.; Guo, L.; Ge, Q. Next-Generation Sequencing of MicroRNAs for Breast Cancer Detection. J. Biomed. Biotechnol. 2011, 2011, 597145. [Google Scholar] [CrossRef]
- Wang, Y.; Yin, W.; Lin, Y.; Yin, K.; Zhou, L.; Du, Y.; Yan, T.; Lu, J. Downregulated Circulating MicroRNAs after Surgery: Potential Noninvasive Biomarkers for Diagnosis and Prognosis of Early Breast Cancer. Cell Death Discov. 2018, 4, 87. [Google Scholar] [CrossRef] [PubMed]
- Cuk, K.; Zucknick, M.; Heil, J.; Madhavan, D.; Schott, S.; Turchinovich, A.; Arlt, D.; Rath, M.; Sohn, C.; Benner, A.; et al. Circulating MicroRNAs in Plasma as Early Detection Markers for Breast Cancer. Int. J. Cancer 2013, 132, 1602–1612. [Google Scholar] [CrossRef]
- Ng, E.K.O.; Li, R.; Shin, V.Y.; Jin, H.C.; Leung, C.P.H.; Ma, E.S.K.; Pang, R.; Chua, D.; Chu, K.-M.; Law, W.L.; et al. Circulating MicroRNAs as Specific Biomarkers for Breast Cancer Detection. PLoS ONE 2013, 8, e53141. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. MiRNA-Based Biomarkers, Therapies, and Resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Wang, L.; Li, S.; Chen, F.; Au-Yeung, K.K.-W.; Shi, C. MicroRNA as an Important Target for Anticancer Drug Development. Front. Pharmacol. 2021, 12, 736323. [Google Scholar] [CrossRef] [PubMed]
- Hussen, B.M.; Rasul, M.F.; Abdullah, S.R.; Hidayat, H.J.; Faraj, G.S.H.; Ali, F.A.; Salihi, A.; Baniahmad, A.; Ghafouri-Fard, S.; Rahman, M.; et al. Targeting MiRNA by CRISPR/Cas in Cancer: Advantages and Challenges. Mil. Med. Res. 2023, 10, 32. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Saini, H.; Sharma, A.; Gupta, S.; Huddar, V.G.; Tripathi, R. Breast Cancer: MiRNAs Monitoring Chemoresistance and Systemic Therapy. Front. Oncol. 2023, 13, 1155254. [Google Scholar] [CrossRef] [PubMed]
- Sell, M.C.; Ramlogan-Steel, C.A.; Steel, J.C.; Dhungel, B.P. MicroRNAs in Cancer Metastasis: Biological and Therapeutic Implications. Expert. Rev. Mol. Med. 2023, 25, e14. [Google Scholar] [CrossRef]
- Shah, M.Y.; Ferrajoli, A.; Sood, A.K.; Lopez-Berestein, G.; Calin, G.A. MicroRNA Therapeutics in Cancer—An Emerging Concept. eBioMedicine 2016, 12, 34–42. [Google Scholar] [CrossRef]
- Munoz, J.P.; Perez-Moreno, P.; Perez, Y.; Calaf, G.M. The Role of MicroRNAs in Breast Cancer and the Challenges of Their Clinical Application. Diagnostics 2023, 13, 3072. [Google Scholar] [CrossRef]
- Balacescu, O.; Visan, S.; Baldasici, O.; Balacescu, L.; Vlad, C.; Achimas-Cadariu, P. miRNA-Based Therapeutics in Oncology, Realities, and Challenges. In Antisense Therapy; IntechOpen: London, UK, 2019. [Google Scholar]
- Okumura, S.; Hirano, Y.; Komatsu, Y. Stable Duplex-Linked Antisense Targeting MiR-148a Inhibits Breast Cancer Cell Proliferation. Sci. Rep. 2021, 11, 11467. [Google Scholar] [CrossRef]
- Van Der Ree, M.H.; Van Der Meer, A.J.; De Bruijne, J.; Maan, R.; Van Vliet, A.; Welzel, T.M.; Zeuzem, S.; Lawitz, E.J. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res. 2014, 111, 53–59. [Google Scholar] [CrossRef]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in Oligonucleotide Drug Delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef]
- Querfeld, C.; Pacheco, T.; Foss, F.M.; Halwani, A.S.; Porcu, P.; Seto, A.G.; Ruckman, J.; Landry, M.L.; Jackson, A.L.; Pestano, L.A.; et al. Preliminary results of a phase 1 trial evaluating MRG-106, a synthetic microRNA antagonist (LNA antimiR) of microRNA-155, in patients with CTCL. Blood 2016, 128, 1829. [Google Scholar] [CrossRef]
- De Cola, A.; Lamolinara, A.; Lanuti, P.; Rossi, C.; Iezzi, M.; Marchisio, M.; Todaro, M.; De Laurenzi, V. MiR-205-5p Inhibition by Locked Nucleic Acids Impairs Metastatic Potential of Breast Cancer Cells. Cell Death Dis. 2018, 9, 821. [Google Scholar] [CrossRef]
- Haftmann, C.; Riedel, R.; Porstner, M.; Wittmann, J.; Chang, H.-D.; Radbruch, A.; Mashreghi, M.-F. Direct Uptake of Antagomirs and Efficient Knockdown of MiRNA in Primary B and T Lymphocytes. J. Immunol. Methods 2015, 426, 128–133. [Google Scholar] [CrossRef]
- Ebert, M.S.; Sharp, P.A. Emerging Roles for Natural MicroRNA Sponges. Curr. Biol. 2010, 20, R858–R861. [Google Scholar] [CrossRef]
- Barta, T.; Peskova, L.; Hampl, A. MiRNAsong: A Web-Based Tool for Generation and Testing of MiRNA Sponge Constructs in Silico. Sci. Rep. 2016, 6, 36625. [Google Scholar] [CrossRef]
- Liang, A.-L.; Zhang, T.-T.; Zhou, N.; Wu, C.Y.; Lin, M.-H.; Liu, Y.-J. MiRNA-10b Sponge: An Anti-Breast Cancer Study in Vitro. Oncol. Rep. 2016, 35, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Mandal, C.C.; Ghosh-Choudhury, T.; Dey, N.; Choudhury, G.G.; Ghosh-Choudhury, N. miR-21 is targeted by omega-3 polyunsaturated fatty acid to regulate breast tumor CSF-1 expression. Carcinogenesis 2012, 33, 1897–1908. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Xiao, C.; Wan, X.; Cha, W.; Miao, Y.; Zhou, Y.; Qin, C.; Cui, T.; Su, F.; Shan, X. Small Molecules with Big Roles in MicroRNA Chemical Biology and MicroRNA-Targeted Therapeutics. RNA Biol. 2019, 16, 707–718. [Google Scholar] [CrossRef]
- Sun, J.; Xu, M.; Ru, J.; James-Bott, A.; Xiong, D.; Wang, X.; Cribbs, A.P. Small Molecule-Mediated Targeting of MicroRNAs for Drug Discovery: Experiments, Computational Techniques, and Disease Implications. Eur. J. Med. Chem. 2023, 257, 115500. [Google Scholar] [CrossRef]
- Melo, S.; Villanueva, A.; Moutinho, C.; Davalos, V.; Spizzo, R.; Ivan, C.; Rossi, S.; Setien, F.; Casanovas, O.; Simo-Riudalbas, L.; et al. Small Molecule Enoxacin Is a Cancer-Specific Growth Inhibitor That Acts by Enhancing TAR RNA-Binding Protein 2-Mediated MicroRNA Processing. Proc. Natl. Acad. Sci. USA 2011, 108, 4394–4399. [Google Scholar] [CrossRef]
- Monroig-Bosque, P.d.C.; Shah, M.Y.; Fu, X.; Fuentes-Mattei, E.; Ling, H.; Ivan, C.; Nouraee, N.; Huang, B.; Chen, L.; Pileczki, V.; et al. OncomiR-10b Hijacks the Small Molecule Inhibitor Linifanib in Human Cancers. Sci. Rep. 2018, 8, 13106. [Google Scholar] [CrossRef] [PubMed]
- Hosseinahli, N.; Aghapour, M.; Duijf, P.H.G.; Baradaran, B. Treating Cancer with MicroRNA Replacement Therapy: A Literature Review. J. Cell Physiol. 2018, 233, 5574–5588. [Google Scholar] [CrossRef]
- Liang, Z.; Ahn, J.; Guo, D.; Votaw, J.R.; Shim, H. MicroRNA-302 Replacement Therapy Sensitizes Breast Cancer Cells to Ionizing Radiation. Pharm. Res. 2013, 30, 1008–1016. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Z.; Wang, Q.; Xing, X.-J.; Zhao, Y. Overexpression of MicroRNA-365 Inhibits Breast Cancer Cell Growth and Chemo-Resistance through GALNT4. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4710–4718. [Google Scholar]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]

| MicroRNA | Target | Function | References |
|---|---|---|---|
| Oncogenic miRNAs in breast cancer | |||
| miRNA-10b | HOXD10 | Promotes cell migration, invasion, and metastasis | [25] |
| miRNA-17/92 cluster | COL4A3, LAMA3, TIMP2/3, ADORA1 | Promotes lymph node metastasis, enhanced cell proliferation, colony formation, migration, and invasion in Triple Negative Breast Cancer (TNBC) | [26,27] |
| miRNA-21 | PDCD4, PTEN, TPM1, TIMP3 | Promotes invasion, metastasis, and migration | [28,29,30,31] |
| miRNA-24 | Nanog, Oct-3/4, BimL, F1H1, HIF-1, Snail, VEGFA | Hypoxia-inducible miRNA | [32] |
| miRNA-122 | Pyruvate kinase and citrate synthase | Promotes metastasis by reprogrammed glucose metabolism | [33] |
| miRNA-135b | LATS2, CDK2, p-YAP | Promotes cell proliferation and S-G2/M cell cycle progression | [34] |
| miRNA-155 | SOCS1, TP53INP1, FOXO3 | Promotes cell growth, proliferation, and survival | [35,36,37] |
| miRNA-181a | Bim | Promotes epithelial-to-mesenchymal transition (EMT), migration, and invasion | [38] |
| miRNA-191-5p | SOX4, caspase-3, caspase-7, p53 | Promotes apoptosis resistance and doxorubicin resistance | [39] |
| miRNA-200b | Ezrin/Radixin/Moesin (ERM) | Promotes metastasis and invasion | [40] |
| miRNA-206 | NK-1 | Promotes breast cancer cell invasion, migration, proliferation, and colony formation in vitro | [41] |
| miRNA-210 | Pax-5 | Modulating EMT and hypoxia | [42] |
| miRNA-331 | HER2, HOTAIR, E2F1, DOHH | Promotes metastasis and invasion by elevation in plasma of metastatic breast cancer patients | [43] |
| miRNA-373 | CD44 | Promotes cell migration, invasion, and metastasis | [44] |
| miRNA-455-3p | EI24 | Promotes proliferation, invasion, and migration | [45] |
| miRNA-498 | BRCA1 | Promotes TNBC cell proliferation | [46] |
| miRNA-520c | CD44 | Promotes cell migration, invasion, and metastasis | [44] |
| Tumor-suppressor miRNAs in breast cancer | |||
| miRNA-17-92 | Mekk2, cyclin D1 | Promotes NK cell antitumoral activity and reduces metastasis, regulates G1 to S phase transition | [47,48] |
| miRNA-7 | SETDB1 | Inhibits cell invasion and metastasis, decreases the BCSC population, and partially reverses EMT | [49] |
| let-7d | Cyclin D1 | Induces stem cells radiation sensitization | [50] |
| miRNA-30 | Ubc9, ITGB3 | Inhibits self-renewal of breast tumor-initiating cells (BT-ICs), trigger apoptosis | [51] |
| miRNA-33b | HMGA2, SALL4, Twist1 | Regulates cell stemness and metastasis | [52] |
| miRNA-34a | IMP3 | Regulates TNBC stem cell property | [53] |
| miRNA-125b | ERBB2, EPO, EPOR, ENPEP, CK2-α, CCNJ, MEGF9 | Inhibits cell proliferation and differentiation, migration and invasion | [54,55] |
| miRNA-137 | FSTL1 | Suppresses TNBC stemness | [56] |
| miRNA-143 | ERK5, MAP3K7, Cyclin D1 | Anti-proliferative | [57] |
| miRNA-148a | WNT1, MMP13 | Inhibits cell proliferation, migration and invasion | [58,59] |
| miRNA-200a | TFAM | Regulates breast cancer cell growth and mtDNA copy number | [60] |
| miRNA-203 | ΔNp63α | Forfeiture of self-renewing capacity associated with epithelial stem cells, suppresses proliferation and colony formation | [61] |
| miRNA-205 | HMGB3, Notch-2 | Suppresses proliferation and invasion and inhibits EMT and stem cell properties | [62,63] |
| miRNA-206 | Cyclin D2, Cx43 | Reduces migration, invasion, and metastasis | [64,65] |
| miRNA-223 | HAX-1 | Re-sensitizes TNBC stem cells to tumor necrosis factor-related apoptosis | [66] |
| miRNA-483-3p | Cyclin E1, p-NPAT, NPAT, CDK2 | Anti-proliferative and G1-S cell cycle arrest | [67] |
| miRNA-497 | Cyclin E1 | Anti-proliferative and reduces migration | [68] |
| miRNA-519d-3p | LIMK1 | Suppresses growth and motility | [69] |
| Subtype | miRNA Signature | Findings | Analysis Type | References |
|---|---|---|---|---|
| Luminal-A | miRNA-99a/let-7c/ miRNA-125b | High expression in Luminal A compared to Luminal B tissue | Breast cancer tissues-cluster analysis of small RNAseq | [107] |
| Luminal-A | miRNA-221 | High expression | Plasma samples—analyzed using qRT-PCR | [108] |
| Luminal-A | miRNA-16, miRNA-21, miRNA-155, miRNA-195 | Higher expression than healthy controls | Serum circulating miRNAs analyzed using qRT-PCR | [109] |
| Luminal-A | miRNA-29a, miRNA-181a and miRNA-652 | Reliably differentiate between cancers and controls | Microarray analysis and confirmed using qRT-PCR | [110] |
| Luminal-A | miRNA-206 | Higher expression | Serum samples and analyzed using qRT-PCR | [111] |
| Luminal-B | miRNA-342 | Higher expression | Early-stage breast cancer specimens— microarray and qRT-PCR analysis | [112] |
| Luminal-B | miRNA-182-5p and miRNA-200b-3p | Significantly higher than in normal breast tissues | Breast cancer tissue: global focus miRNA PCR Panel and analyzed using qRT-PCR | [113] |
| Luminal-B | miRNA-21, miRNA-221, miRNA-200a and miRNA-196a | Overexpression in Luminal B breast cancer without overexpression of HER2/neu | Tumor tissues of patients with Luminal B breast cancer and analyzed using qRT-PCR | [114] |
| Luminal-B | miRNA-210 | Upregulated in the analyses of all 40 patients’ samples | FFPE blocks: miRNA microarray and confirmed using qRT-PCR | [115] |
| Luminal-B | miRNA-222 | Higher expression in Luminal-B/(HER2+) subtypes than in Luminal A and TNBC subtypes | Breast cancer tissues and analyzed using qRT-PCR | [116] |
| HER2-enriched | miRNA-125b | Significantly increased expression in breast cancer tissues compared with that in the non-cancerous tissues | FFPE breast cancer tissues, luciferase activity and qRT-PCR | [117] |
| HER2- enriched | miRNA-375 and miRNA-122 | High levels of circulating miRNA-375 and low levels of miRNA-122 were associated with HER2 status | Serum specimens: Solexa deep sequencing and confirmed using qRT-PCR | [118] |
| HER2- enriched | miRNA-548ar-5p, miRNA-584-3p, miRNA-615-3p, and miRNA-1283 | Significantly differential expression in the HER2-enriched subtype | Serum samples: Multiplexed gene expression analysis | [119] |
| HER2- enriched | let-7f, let-7g, miRNA-107, miRNA-10b, miRNA-126, miRNA-154 and miRNA-195 | Inversely correlated with HER2 overexpression | Primary breast cancer biopsies: microarray and confirmed using qRT-PCR | [120] |
| HER2- enriched | miRNA-520d, miRNA-376b | Highly expressed and accurately predicted HER2 status in early-stage breast tumors | Early-stage breast cancer specimens- microarray and qPCR analysis | [112] |
| TNBC or basal-like | miRNA-210 | Significantly higher expression compared to ER+/HER2− breast cancers | Breast cancer tissue and analyzed using qRT-PCR | [121] |
| TNBC or basal-like | miRNA-214 | Higher expression | FFPE tissues and confirmed using qRT-PCR | [122] |
| TNBC or basal-like | miRNA-493 | High expression | Tissue microarrays | [123] |
| TNBC or basal-like | miRNA-135b | High expression | TaqMan low-density array | [124] |
| TNBC or basal-like | miRNA-20a, miRNA-221 | Higher expression compared to Luminal A and Luminal B/HER2− breast cancer subtypes | Biopsies of tumor tissue; and confirmed using qRT-PCR | [125] |
| TNBC or basal-like | miRNA-17 family | Overexpressed in high grade and TNBC associated with aggressive behavior | FFPE- tissue: miRCURY LNA microarray and confirmed using qRT-PCR | [126] |
| TNBC or basal-like | miRNA-17-92 | Elevated in TNBC but reduced in ER-positive breast cancer and associated with poor outcome. The miRNA-17–92 expression enhanced cell growth and invasion of TNBC cells | Breast cancer cell lines: high-throughput mRNA sequencing and confirmed using qRT-PCR | [27] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Subramanian, K.; Sinha, R. Functions of Differentially Regulated miRNAs in Breast Cancer Progression: Potential Markers for Early Detection and Candidates for Therapy. Biomedicines 2024, 12, 691. https://doi.org/10.3390/biomedicines12030691
Subramanian K, Sinha R. Functions of Differentially Regulated miRNAs in Breast Cancer Progression: Potential Markers for Early Detection and Candidates for Therapy. Biomedicines. 2024; 12(3):691. https://doi.org/10.3390/biomedicines12030691
Chicago/Turabian StyleSubramanian, Kumar, and Raghu Sinha. 2024. "Functions of Differentially Regulated miRNAs in Breast Cancer Progression: Potential Markers for Early Detection and Candidates for Therapy" Biomedicines 12, no. 3: 691. https://doi.org/10.3390/biomedicines12030691
APA StyleSubramanian, K., & Sinha, R. (2024). Functions of Differentially Regulated miRNAs in Breast Cancer Progression: Potential Markers for Early Detection and Candidates for Therapy. Biomedicines, 12(3), 691. https://doi.org/10.3390/biomedicines12030691